Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is primarily transmitted through aerosolized droplets.1 Patients with coronavirus disease 2019 (COVID-19) exhale SARS-CoV-2 RNA copies at a rate of 103 to 105 per minute.2 In the United States, in-center hemodialysis patients and staff are mandated to wear face masks while staying in the dialysis clinic. Previously, face mask sampling was applied to patients with common cold symptoms.3 Recently, a novel method was described to detect exhaled SARS-CoV-2 by using special sampling strips attached to the inside of a duck-billed mask.4 We tested the hypothesis that assaying masks worn by in-center hemodialysis patients for SARS-CoV-2 RNA could provide an opportunity to identify infected subjects.
Methods
In this institutional review board–approved exempt study (Western IRB ES-21–003), we collected deidentified masks in a dialysis clinic where hemodialysis patients with COVID-19 are dialyzed in a designated COVID-19 shift. In those patients, SARS-CoV-2 RNA has previously been diagnosed by RT-PCR in nasopharyngeal (NP) swabs. Per our policy, patients stay in the COVID-19 shift until tested negative in two consecutive NP swabs. We also collected masks in patients dialyzed in regular (i.e., non-COVID-19) shifts. Mask wearing is mandatory for everyone, including patients, while staying in the dialysis facility. Three-layer disposable masks were provided to each patient at the time of entry. Upon completion of the session (mostly 3–4 hours), patients’ masks were collected in individual Ziplock bags and then stored at room temperature for 13–24 hours until analysis.
The inner mask surface was swabbed for 30 seconds using a cotton-tipped applicator that was pre-soaked with 100 μl PBS. The swab head was then placed in a microcentrifuge tube containing 600 μl PBS and vortexed for 10 min at 1000 rpm.
RNA extraction and RT-PCR reaction were performed using TaqPath RT-PCR COVID-19 Combo Kit (Thermo Fisher A47814) as described previously.5 Bacteria MS2 Phage was added to samples prior to RNA extraction. We increased the PCR cycles to 45 for maximum detectability. The multiplex probes anneal to one MS2 sequence and three SARS-CoV-2 target sequences (S gene, N gene, ORF1ab). Test result interpretation is shown in Figure 1A.
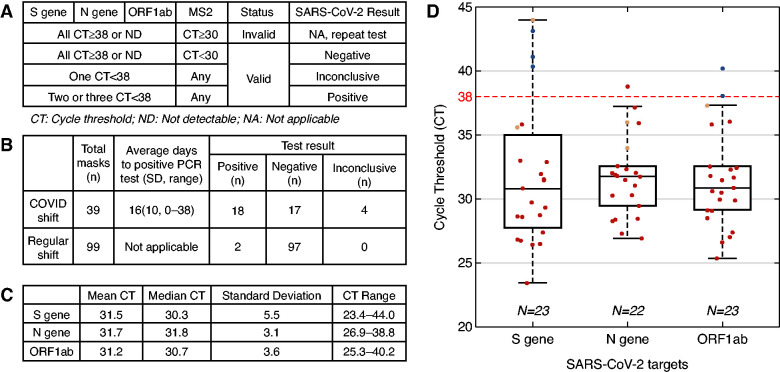
Figure 1.
Face masks worn by in-center hemodialysis patients can harbor SARS-CoV-2. (A) Interpretation of test results. (B) Test results for a total of 138 face masks collected from patients in COVID-19 and regular shifts. (C) Results for three SARS-CoV-2 RNA targets, S gene, N gene and ORF1ab, isolated from face masks worn by patients who tested positive for SARS-CoV-2 by nasopharyngeal swab. Note that two positive samples from regular shift are included in the calculation. (D) CT distribution for three SARS-CoV-2 targets for all 138 masks (undetectable CTs are excluded). Positive (red), negative (blue), and inconclusive (orange) results per definition shown in panel A. The horizontal dashed line indicates the CT cut-off for SARS-CoV-2 PCR validation.
Results
We collected a total of 138 face masks, 39 from 14 patients with confirmed COVID-19 dialyzed in a designated COVID-19 shift (7 females; median age, 59 years [range, 32–81]) and 99 masks from patients dialyzed in regular shifts. COVID-19 patients provided one to five masks on average 16±10 days (median, 15; range, 0–38) after COVID-19 diagnosis. Eighteen of the 39 masks from COVID-19 patients tested positive for SARS-CoV-2 (Figure 1B). Ninety-seven of 99 masks from regular shift patients tested negative (Figure 1B); the two positive masks came from an asymptomatic patient who was subsequently diagnosed with COVID-19 by NP swab. Overall, mean cycle thresholds (CT) were around 31.5 for S gene, N gene, and ORF1ab (Figure 1C). CT distributions per target are shown in Figure 1D.
Discussion
Although NP swab RT-PCR is standard for COVID-19 diagnosis, sample collection is uncomfortable to most subjects, neither efficient nor economic for screening purposes, and requires health care professionals. Our proof-of-principle study shows that face masks worn by in-center hemodialysis patients can harbor SARS-CoV-2. Therefore, we think that assaying of face masks should be explored further, as it could be useful especially when NP swabbing is not feasible or desirable. Although the minimum duration of mask use to retrieve SARS-CoV-2 RNA is unknown, it is interesting to note that Ma et al.2 detected exhaled virus after 5 min of breathing into a collection device. Besides RT-PCR testing, viral particles collected from masks can be used for SARS-CoV-2 sequencing.
There are limitations to our study: first, its exempt nature only allowed us to collect deidentified masks, preventing us from correlating mask results with the time elapsed since onset of COVID-19 symptoms and diagnosis. Second, no NP swabs could be collected concurrently with the masks, precluding quantitation of sensitivity and specificity of mask SARS-CoV-2 testing. However, since the only positive masks in the non-COVID-19 shift came from a patient who was subsequently diagnosed with COVID-19, test specificity is most likely high. Determining test sensitivity will require concurrent NP swab sampling in COVID-19 patients. The low positive rate in our study may be due to the long lag between COVID-19 diagnosis and mask testing, as SARS-CoV-2 shedding declines 4 days after symptom onset.6
It is currently unknown if fomite transmission of SARS-CoV-2 via used masks can occur. We are aware of two publications indicating that SARS-CoV-2 virus can remain viable for up to 7 days on surgical masks.7,8 Of note, in both reports, mask pieces were soaked in virus transportation medium. We used PBS-soaked cotton-tipped applicators to forcefully swab the masks. These treatments are very different from unintentional, brief touches. Disposable medical masks are usually made of non-woven polypropylene fabric with electrostatic properties that enhance virus retention. Therefore, we think that typical handling of a virus-laden mask is unlikely to result in significant viral transfer.
As incidence and prevalence of COVID-19 is declining, mask testing may lend itself to novel strategies, such as pool testing.9 Pool testing is particularly efficient in settings with a low disease prevalence.10 This approach will require the development of setting-specific workflows to optimize mask collection and processing.
Disclosures
The Renal Research Institute is a wholly owned subsidiary of Fresenius Medical Care. P. Kotanko and D. Maddux hold stock in Fresenius Medical Care. P. Kotanko reports research funding from Fresenius Medical Care, KidneyX, National Institutes of Health; honoraria from HSTalks; multiple patents in the kidney space; scientific advisor or Membership via Editorial Board of Blood Purification, Editorial Board of Kidney and Blood Pressure Research; and other interests/relationships with Fresenius Medical Care. N. Grobe reports scientific advisor or membership with American Chemical Society; and other interests/relationships as a member of Oligo Nation. D. Maddux reports current employment with Fresenius Medical Care North America and spouse employment with Fresenius Medical Care; scientific advisor or membership via spouse on Fresenius Medical Care Board and Goldfinch Bio Board; and other interests/relationships with Fresenius Medical Care Foundation. All remaining authors have nothing to disclose.
Funding
This study was supported in part by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK130067 and an award by the KidneyX COVID-19 Kidney Care Challenge, a joint effort of the US Department of Health and Human Services and the American Society of Nephrology.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Tang S, Mao Y, Jones RM, Tan Q, Ji JS, Li N, et al. : Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ Int 144: 106039, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma J, Qi X, Chen H, Li X, Zhang Z, Wang H, et al. : Coronavirus disease 2019 patients in earlier stages exhaled millions of severe acute respiratory syndrome coronavirus 2 per hour. Clin Infect Dis 72: 652–654, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huynh KN, Oliver BG, Stelzer S, Rawlinson WD, Tovey ER: A new method for sampling and detection of exhaled respiratory virus aerosols. Clin Infect Dis 46: 93–95, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Williams CM, Pan D, Decker J, Wisniewska A, Fletcher E, Sze S, et al. : Exhaled SARS-CoV-2 quantified by face-mask sampling in hospitalised patients with COVID-19. J Infect 82: 253–259, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Patel A, Tisdale L, Haq Z, Ye X, Lasky R, et al. : SARS-CoV-2 in spent dialysate from chronic peritoneal dialysis patients with COVID-19. Kidney360 2: 86–89, 2021. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J: Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 173: 262–267, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen H-L, Chan MCW, et al. : Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 1: e10, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Li T, Deng Y, Liu S, Zhang D, Li H, et al. : Stability of SARS-CoV-2 on environmental surfaces and in human excreta. J Hosp Infect 107: 105–107, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grobe N, Cherif A, Wang X, Dong Z, Kotanko P: Sample pooling: burden or solution? Clinical Microbiology and Infection 2021. Available at: https://www.sciencedirect.com/science/article/pii/S1198743X21001907. Accessed June 28, 2021 [DOI] [PMC free article] [PubMed]
- 10.Cherif A, Grobe N, Wang X, Kotanko P: Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw Open 3: e2013075, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]