Abstract
A line probe assay (INNO-LiPA HBV DR) detecting drug-resistant hepatitis B virus (HBV) strains was evaluated. Results concordant with sequence analysis were obtained with 48 of 56 serum samples from HBV-infected patients undergoing lamivudine therapy. In eight cases, additional minor subpopulations could be identified by the line probe assay.
Lamivudine, a nucleoside analogue, has become a potent tool for the therapy of chronic hepatitis B virus (HBV) infection (11). Studies performed with patients suffering from chronic hepatitis have shown that treatment with lamivudine leads to a decrease in the patient's HBV load in serum (5). This effect was also observed in patients after liver or renal transplantation (9, 10) and in patients with simultaneous human immunodeficiency virus (HIV) infection (14). It has been observed, however, that drug-resistant HBV variants may emerge during lamivudine treatment. Such lamivudine-resistant strains exhibit specific mutations, especially in the YMDD region of the HBV polymerase gene, including either an M552V mutation associated with an L528M mutation or an M552I mutation alone (8).
The identification of such mutations is of increasing importance, also in view of the use of alternative antiviral substances such as penciclovir or famciclovir, for which promising data for the treatment of chronic HBV infections have been presented (4, 7). The identification of resistant HBV strains is usually performed by sequence analysis. In addition to other alternative test systems, such as restriction fragment length polymorphism assays (2), a new test system using the line probe assay (LiPA) technology (Innogenetics, Ghent, Belgium) (13) has recently been presented for the detection of resistance mutations in the HBV polymerase gene.
This INNO-LiPA HBV DR has now been evaluated by testing serum samples from patients with chronic HBV infections and undergoing lamivudine treatment. The results have been compared to those obtained by a standard sequencing protocol that is routinely used for detection of drug-resistant HBV strains.
In the present study 32 serum samples from 10 renal transplant patients with chronic HBV infection were investigated at the onset and during the follow-up of therapy with 100 mg of lamivudine per day in six cases, 50 mg per day in two cases, and 25 mg per day in another two cases. In addition, 22 serum samples from 13 HIV patients were investigated during or after the completion of lamivudine therapy with a 300-mg dose of lamivudine per day. Also, two serum samples from two other patients with chronic HBV infection who were undergoing lamivudine therapy were tested.
The sequence analysis was performed as previously described (10), using the primers published by Bartholomew et al. (3).
The INNO-LiPA HBV DR was performed as recommended by Innogenetics. A part of HBV domains B and C of the pol gene is amplified. The biotinylated PCR fragments are reverse hybridized using typing-membrane-based INNO-LiPA HBV DR strips. After hybridization, streptavidin labeled with alkaline phosphatase was added and bound to the previously formed biotinylated hybrids. Incubation with the substrate BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium chromogen results in a purple-brown color development.
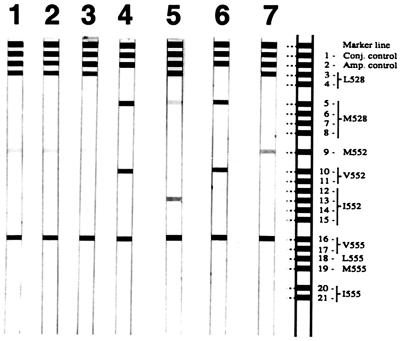
The serum samples tested contained virus levels between 103 and 109 copies of HBV DNA as determined by a previously described quantitative HBV DNA PCR assay (10). All samples were easily amplified by the PCR steps of both genotyping assays and yielded unambiguous lines on the LiPA strip and clear sequencing results. In addition, we have further investigated the sensitivity of the LiPA by applying it to samples from a proficiency panel for quantitative HBV DNA testing, which had been distributed by the European Union Quality Control Concerted Action (15). All of the samples of this panel that contained HBV DNA down to a concentration of 103 copies per ml were detected by the LiPA (Fig. 1), indicating a high sensitivity of the assay. This is of importance for the early detection of resistant strains, especially in view of previously published data that show that the appearance of resistance mutations may precede an increase in viral load by several months (13).
FIG. 1.
LiPA strips 1 to 4 show the development of resistant HBV in a renal transplant patient during lamivudine therapy. Samples were taken at the beginning of therapy (strip 1), after 3 and 6 months (strips 2 and 3), and after 9 months, when a resistant strain was detectable (strip 4). LiPA strips 5 and 6 show the results of patients D and E, respectively. LiPA strip 7 shows the results found by testing an HBV DNA standard with a low copy number (103 copies/ml). Conj., conjugate; amp., amplification.
A comparison of the results obtained by the two methods is presented in Table 1. Identical results were found with 48 of the 56 samples (86%). With eight serum samples, however, there were observed discrepancies between the results of LiPA and those of sequence analysis. In all of these cases, the LiPA indicated the presence of additional virus species. This finding is statistically significant as shown by the McNemar chi-square test (P = 0.004) and is probably due to the presence of minor viral subspecies that were missed by sequencing, which usually allows the detection of minor virus variants only if they constitute about 20% of the viral population (12). It is unlikely that the detection of minor viral subspecies was due to nonspecific hybridization because the selected probes in the LiPA have been shown to be very specific (13) and no unusual mutations in the polymerase gene were detected in any of the samples by sequence analysis.
TABLE 1.
Comparison of the results obtained by sequence analysis and INNO-LiPA HBV DR from 56 serum samplesa
| Results | Patient(s) | Sequence analysis
|
INNO-LiPA HBV DR
|
No. of samples | ||
|---|---|---|---|---|---|---|
| Strain type(s) detected | Codons | Strain type(s) detected | Codons | |||
| Concordant | 48 | |||||
| wt | L528, M552 | wt | L528, M552 | 30 | ||
| mt | M528, V552 | mt | M528, V552 | 14 | ||
| wt + mt | L + M528, M + V552 | wt + mt | L + M528, M + V552 | 4 | ||
| Discordant | 8 | |||||
| A, F | mt | M528, V552 | wt + mt | L + M528, M + V552 | 2 | |
| B | mt | M528, V552 | wt + mt | L + M528, V552 | 1 | |
| C | wt | L528, M552 | wt + mt | L + M528, M + V552 | 1 | |
| D | wt + mt | L528, I552 | wt + mt | L + M528, I552 | 1 | |
| E | mt | M528, V552 | mt | M528, V + I552 | 1 | |
| G | wt + mt | M528, M + V552 | wt + mt | L + M528, M + V552 | 1 | |
| H | wt + mt | L + M528, M552 | wt + mt | L + M528, M + V552 | 1 | |
wt, wild type; mt, mutant type. Underlining indicates differing results, and boldface indicates mutated residues.
The 8 discordant samples were taken from 3 of 32 renal transplant patient samples, from 4 of 22 sera taken from HIV patients, and from 1 of 2 samples taken from other patients with chronic hepatitis. This distribution between the patient groups was not statistically significant (chi-square test, P = 0.22).
The discrepant results are probably caused by the fact that, in patients during long-term therapy as well as after cessation of lamivudine treatment, various mixtures of wild-type and mutated virus strains may develop and are differentially detected by the two assays.
In two samples, the LiPA indicated the presence of additional mutations, which led in both cases to a combination of the M552I mutation with either the L528M mutation alone (patient D) or L528M together with the M552V mutation (patient E) (Fig. 1). In previous studies as well as in the present one, sequence analysis allowed in vivo the detection of either the M552I mutation only or the absolutely linked M552V and L528M mutations (1, 6, 8). In one published study it was possible only by cloning to show that both resistant variants were present in the same sample (8). The LiPA data shown here confirm that both mutational patterns can be present at the same time.
In conclusion, the INNO-LiPA HBV DR proved to be a sensitive tool for resistance testing and appears to be a technique which may be useful in the detection of HBV strains mutated at codons 528 and 552.
Acknowledgments
We thank Sylvia Malik, Thomas Urbanek, and Michaela Binder for excellent technical assistance and Steve Allison for critical reading of the manuscript.
REFERENCES
- 1.Allen M I, DesLauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Allen M I, Gauthier J, DesLauriers M, Bourne E J, Carrick K M, Baldanti F, Ross L L, Lutz M W, Condreay L D. Two sensitive PCR-based methods for detection of hepatitis B virus variants associated with reduced susceptibility to lamivudine. J Clin Microbiol. 1999;37:3338–3347. doi: 10.1128/jcm.37.10.3338-3347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzendahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 4.Colledge D, Locarnini S, Shaw T. Synergistic inhibition of hepadnaviral replication by lamivudine in combination with penciclovir in vitro. Hepatology. 1997;26:216–225. doi: 10.1053/jhep.1997.v26.pm0009214473. [DOI] [PubMed] [Google Scholar]
- 5.Lai C L, Chien R N, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 6.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 7.Main J, Brown J L, Howells C, Galassini R, Crossey M, Karayiannis P, Georgiou P, Atkinson G, Thomas H C. A double blind, placebo-controlled study to assess the effect of famciclovir on virus replication in patients with chronic hepatitis B virus infection. J Viral Hepat. 1996;3:211–215. doi: 10.1111/j.1365-2893.1996.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 8.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 9.Perrillo R, Rakela J, Dienstag J, Levy G, Martin P, Wright T, Caldwell S, Schiff E, Gish R, Villeneuve J P, Farr G, Anschuetz G, Crowther L, Brown N. Multicenter study of lamivudine therapy for hepatitis B after liver transplantation. Lamivudine Transplant Group. Hepatology. 1999;29:1581–1586. doi: 10.1002/hep.510290507. [DOI] [PubMed] [Google Scholar]
- 10.Puchhammer-Stockl E, Mandl C W, Kletzmayr J, Holzmann H, Hofmann A, Aberle S W, Heinz F X, Watschinger B, Hofmann H. Monitoring the virus load can predict the emergence of drug-resistant hepatitis B virus strains in renal transplantation patients during lamivudine therapy. J Infect Dis. 2000;181:2063–2066. doi: 10.1086/315519. [DOI] [PubMed] [Google Scholar]
- 11.Schalm S W, de Man R A, Heijtink R A, Niesters H G. New nucleoside analogues for chronic hepatitis B. J Hepatol. 1995;22:52–56. [PubMed] [Google Scholar]
- 12.Schuurman R. State of the art of genotypic HIV-1 drug resistance. Curr Opin Infect Dis. 1997;10:480–484. [Google Scholar]
- 13.Stuyver L, Van Geyt C, De Gendt S, Van Reybroeck G, Zoulim F, Leroux-Roels G, Rossau R. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000;38:702–707. doi: 10.1128/jcm.38.2.702-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thibault V, Benhamou Y, Seguret C, Bochet M, Katlama C, Bricaire F, Opolon P, Poynard T, Agut H. Hepatitis B virus (HBV) mutations associated with resistance to lamivudine in patients coinfected with HBV and human immunodeficiency virus. J Clin Microbiol. 1999;37:3013–3016. doi: 10.1128/jcm.37.9.3013-3016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thon E V, Schirm J, Van Loon A, Reid J, Klapper P E, Cleator G M. First EU-QCCA Hepatitis B Virus Proficiency Panel: summary of results. Manchester, United Kingdom: European Union Quality Control Concerted Action for Nucleic Acid Amplification in Diagnostic Virology; 2000. [Google Scholar]