Abstract
INTRODUCTION:
Conventional white light imaging (WLI) endoscopy is the most common screening technique used for detecting early esophageal squamous cell carcinoma (ESCC). Nevertheless, it is difficult to detect and delineate margins of early ESCC using WLI endoscopy. This study aimed to develop an artificial intelligence (AI) model to detect and delineate margins of early ESCC under WLI endoscopy.
METHODS:
A total of 13,083 WLI images from 1,239 patients were used to train and test the AI model. To evaluate the detection performance of the model, 1,479 images and 563 images were used as internal and external validation data sets, respectively. For assessing the delineation performance of the model, 1,114 images and 211 images were used as internal and external validation data sets, respectively. In addition, 216 images were used to compare the delineation performance between the model and endoscopists.
RESULTS:
The model showed an accuracy of 85.7% and 84.5% in detecting lesions in internal and external validation, respectively. For delineating margins, the model achieved an accuracy of 93.4% and 95.7% in the internal and external validation, respectively, under an overlap ratio of 0.60. The accuracy of the model, senior endoscopists, and expert endoscopists in delineating margins were 98.1%, 78.6%, and 95.3%, respectively. The proposed model achieved similar delineating performance compared with that of expert endoscopists but superior to senior endoscopists.
DISCUSSION:
We successfully developed an AI model, which can be used to accurately detect early ESCC and delineate the margins of the lesions under WLI endoscopy.
INTRODUCTION
Esophageal cancer is one of the major malignant tumors affecting public health. It ranks seventh in terms of incidence worldwide, and it was the sixth leading cause of cancer mortality in 2018 (1). In some parts of Asia and sub-Saharan Africa, esophageal squamous cell carcinoma (ESCC) accounts for most esophageal cancer cases (2). As most of the patients with ESCC are diagnosed at advanced stages, the overall 5-year survival rate remains lower than 20% (3). However, over recent years, remarkable progress has been achieved in the prognosis of ESCC because of the contribution of endoscopy in the diagnosis and treatment of early ESCC (4,5).
Although various endoscopic diagnostic techniques, such as conventional white light imaging (WLI), narrow-band imaging (NBI), magnifying endoscopy (ME), and iodine staining, are used to detect early ESCC (6), screening for early ESCC is generally performed by standard endoscopic observation with WLI, especially in some resource-limited regions where NBI and other advanced diagnostic techniques are unavailable (7). Yet, detecting early ESCC using WLI endoscopy is challenging even for experienced endoscopists. The reported sensitivity and specificity of WLI endoscopy for the diagnosis of early ESCC were only 62% and 79%, respectively (8,9). Meanwhile, as 1 of the main factors that influence endoscopic resection, the lateral extent of cancer has been reported to be significantly related to the rate of postoperative esophageal stricture after endoscopic treatment (10). In addition, a previous study suggested that a postoperative ulcer, which occupies two-thirds or more of the circumference of the esophagus, may result in the formation of a significant stricture (11). Thus, it is strongly recommended to evaluate the circumferential extent of the lesion before attempting treatment (12).
Over the recent years, there has been a significant improvement in the area of artificial intelligence (AI) in various medical fields (13,14). Deep convolutional neural networks (DCNNs) have been successfully used for real-time detection of early ESCC under WLI or NBI endoscopy (15–17). However, the application of DCNNs in detecting early ESCC under WLI is limited, and the detection accuracy can reach only 80% with the help of AI-aided diagnosis (18). Up till now, there are no reports on the development of DCNNs in delineating margins of early ESCC under WLI. Thus, a more advanced measurement approach to help endoscopists to achieve diagnosis and delineate early ESCC under WLI endoscopy is required.
Consequently, in this study, we trained an AI model to assist endoscopists in the diagnosis of early ESCC under WLI endoscopy. Furthermore, we evaluated the performance of our model in delineating margins of lesions using expert endoscopists' marked margins as the gold standard.
METHODS
Training data set collection
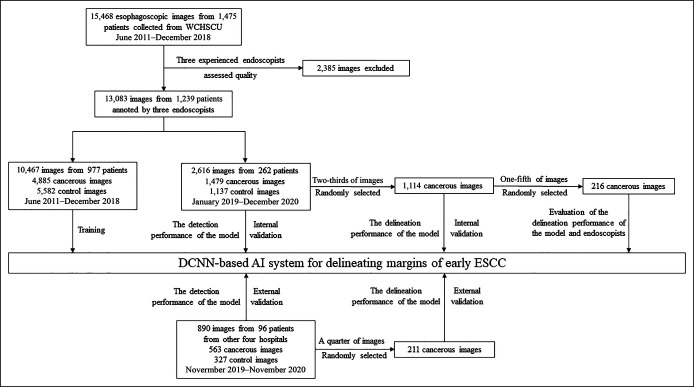
We gathered 13,083 endoscopic still images from 1,239 consecutive patients undergoing endoscopic resection in West China Hospital (WCH) in Chengdu between June 2011 and December 2018, which were used as an internal data set for training the model (Figure 1). All lesions were histologically proven to exhibit early ESCC with a negative resection margin. Early ESCC was defined as low-grade and high-grade intraepithelial neoplasia and cancer limited to the mucosa or submucosa. Images with poor quality (bleeding, halation, blur, and defocus) were excluded. Among the 13,083 nonmagnified WLI images saved in JPEG format, 4,885 images with early ESCCs and 5,582 WLI control images were used to train the model, while the other 2,616 images were used as an internal validation to evaluate the detection performance of the model. All the endoscopic images were captured using Olympus endoscopes (GIF-H290, GIF-H260Z, and GIF-H260; Olympus Medical Systems, Tokyo, Japan).
Figure 1.
Flowcharts of the data set for preprocessing, training, and validation of the model. DCNNs, deep convolutional neural networks; ESCC, esophageal squamous cell carcinoma; WCHSCU, West China Hospital Sichuan University.
Participants, annotation, and model development
First, we defined the manual delineation of early ESCC under WLI marked by expert endoscopists as the gold standard for evaluation of detection and delineation performance. For model development and testing, the margins of the lesions under WLI were manually delineated using polygonal frames by an endoscopy expert (C.C.W., with at least a 10-year experience of endoscopy and more than 10,000 endoscopy procedures from WCH), who took the margins of the lesions under NBI, iodine staining, and resection specimen as reference. Two-thirds of the images were manually annotated by taking iodine staining as a reference, and the remaining images were marked by taking NBI and resection specimen as a reference. Then, the marking was reconfirmed by another expert (B.H., with an experience of at least 1,000 endoscopic submucosal dissection procedures at WCH) until the 2 expert endoscopists reached a consensus on the delineation of each image (kappa score, 0.725). At last, all the manual markers of early ESCC under WLI confirmed by C.C.W. and B.H. were histologically demonstrated with negative resection margin and, therefore, were used as gold standards. A full description of the development of the model can be found in the Supplementary Material (see Supplementary Figure 1, Supplementary Digital Content 2, http://links.lww.com/CTG/A730).
Evaluating the detection and delineation performance of the model
Next, we used 2 other independent nonmagnified endoscopic data sets for testing the model. First, we used 1,479 images pertaining to 262 consecutive patients undergoing endoscopy resection in WCH between January 2019 and December 2020 as the internal test data set. In the external test data set, we collected 563 WLI images from 96 consecutive cases of early ESCC from other hospitals (Cangxi People's Hospital contributed 23 cases; Zigong Fourth People's Hospital contributed 22 cases; Nanchong Central Hospital contributed 27 cases; and Affiliated Hospital of North Sichuan Medical College contributed 24 cases) between November 2019 and November 2020. All cases were histologically confirmed with a negative resection margin. Suboptimal quality images were excluded from the study.
Second, the delineation performance evaluation was further conducted in another internal data set (two-thirds of the early ESCC images from the internal data set) and another external data set (a quarter of the early ESCC images from the external data set). To evaluate the model's delineating performance, we first calculated the intersection ratio between the gold standard and the delineation predicted by the model to the gold standard. The expert (B.H.) was arranged to review the delineation predicted by the model because it was sometimes difficult to mark the entire lesion boundary precisely under WLI. Correct delineation performance was defined as the intersection region over the gold standard exceeding the threshold overlap ratio of 0.6. Meanwhile, early ESCC videos were collected from WCH between January 2019 and December 2020 to assess the ability of the model in identifying lesions and delineating margins.
Comparing the delineation performance of the model with that of endoscopists
For comparing the delineation performance of the model with that of endoscopists, another data set (one-fifth from the internal data set) was randomly collected for further evaluation. Delineation performance was estimated by the model and endoscopists (7 senior endoscopists with >4 years of experience in endoscopy and 7 expert endoscopists with >8 years of experience in endoscopy), respectively. Endoscopists were required to delineate images only under WLI in the comparison data set. We then compared delineation performance of both endoscopists and model under the overlap ratio of 0.6, and the experts' (C.C.W. and B.H.) delineation was used as gold standards.
Outcome measures
Evaluation of the detection performance of the model: The accuracy, sensitivity, and specificity of the model in detecting lesions under WLI were calculated as follows: Accuracy = true predictions/total number of cases, sensitivity = true positive/positive, specificity = true negative/negative, positive predictive value (PPV) = true positive/(true positive + false positive [FP]), negative predicted value (NPV) = true negative/(true negative + false negative [FN]).
Evaluation of the delineation performance of the model and endoscopists per-image: The accuracy, mean intersection over union (mIoU), sensitivity, and specificity were used to assess the performance of the model and endoscopists on delineating lesions under WLI.
The overlap ratio was defined as the ratio of intersection between the delineation region of the gold standard and that of the AI model or region marked by endoscopists to the gold standard. The accuracy of the model or endoscopists on delineating lesion margins was interpreted as exceeding the threshold overlap ratio of 0.6. Accuracy = true predictions/total number of cases.
For each image, the areas where the AI model or endoscopists' delineation performance coincided with the gold standard were true positives. The absence of the AI model or endoscopists' delineation performance on the gold standard was a FP. The absence of the gold standard on the model or endoscopists' delineation performance was a FN. Intersection over union = true positive/(true positive + FP + FN). Sensitivity = true positive/positive, specificity = true negative/negative.
Statistical analysis
Least significant difference-test with a significance level of 0.05 was used to compare mIoU, sensitivity, and specificity per-image between model and endoscopists. The interobserver agreement of the model or endoscopists was calculated using the Fleiss kappa statistics. The statistical significance was set to P < 0.05. All continuous variables are expressed as the mean within a range. Statistical analyses were conducted using SPSS software, version 22.0 (SPSS Inc, Chicago, IL).
RESULTS
Characteristics of patients and lesions in test image sets
A total of 1,479 WLI images from 262 patients were selected as the internal validation data set to evaluate the model's detection performance. Next, another 563 WLI images from other 96 patients treated at 4 other hospitals were selected as an external validation data set to evaluate the model's detection performance. The demographics of the selected patients and lesions are summarized in Table 1 and Figure 1. Then, we collected 1,114 cancer images (from the internal data set) and another 211 cancer images (from the external data set) to evaluate the delineation performance of the model.
Table 1.
Characteristics of patients and lesions in test image sets
| Characteristics | WCH data set (n = 1,239) | External test data set (n = 96) | |
| Training (n = 977) | Internal test data set(n = 262) | ||
| Patient characteristics | |||
| Age, yr | 61 (36–84) | 63 (39–82) | 65 (40–85) |
| Sex | |||
| Male | 649 | 186 | 62 |
| Female | 328 | 76 | 34 |
| Lesion characteristics | |||
| Size (mm), mean (range) | 29 (5–88) | 27 (5–85) | 26 (5–70) |
| Location (Ut/Mt/Lt) | 142/572/263 | 55/167/40 | 26/49/21 |
| Macroscopic type (IIa/IIb/IIc/IIa + IIc) | 15/557/368/37 | 4/177/53/28 | 0/59/32/5 |
| Invasion depth (EP-LPM/MM/SM/uncertain) | 483/235/222/37 | 146/67/40/9 | 51/32/8/5 |
Values are median (range).
EP-LPM, epithelium-lamina propria; Lt, lower esophagus; MM, muscularis mucosa; Mt, middle esophagus; SM, submucosa; Ut, upper esophagus; WCH, West China Hospital.
Detection and delineation performance of the model
The AI model achieved an accuracy of 85.7% in per-image (95% confidence interval [CI]: 83.9%–87.5%), while corresponding sensitivity, specificity, PPV, and NPV in per-image for detecting early ESCC were 92.6% (95% CI, 90.6%–94.5%), 80.0% (95% CI, 77.1%–82.9%), 81.8% (95% CI, 79.2%–84.4%), and 91.3% (95% CI, 89.1%–93.5%) at internal validation. At external validation, the AI model achieved an accuracy of 84.5% in per-image (95% CI: 81.5%–87.5%), while corresponding sensitivity, specificity, PPV, and NPV in per-image for detecting early ESCC were 89.5% (95% CI, 86.0%–93.0%), 79.0% (95% CI, 74.1%–83.9%), 82.6% (95% CI, 78.5%–86.7%), and 87.7% (95% CI, 83.6%–91.8%) (Table 2 and Figure 2).
Table 2.
Diagnostic performance of the model for lesions in per-image
| Internal validation set (n = 1,479) | External validation set (n = 563) | |
| Accuracy (95% CI) | 85.7 (83.9–0.87.5) | 84.5 (81.5–87.5) |
| Sensitivity (95% CI) | 92.6 (90.6–0.94.5) | 89.5 (86.0–93.0) |
| Specificity(95% CI) | 80.0 (77.1–0.82.9) | 79.0 (74.1–83.9) |
| PPV (95% CI) | 81.8 (79.2–0.84.4) | 82.6 (78.5–86.7) |
| NPV(95% CI) | 91.3 (89.1–0.93.5) | 87.7 (83.6–91.8) |
Values are given in percentages.
CI, confidence interval; NPV, negative predicted value; PPV, positive predictive value.
Figure 2.
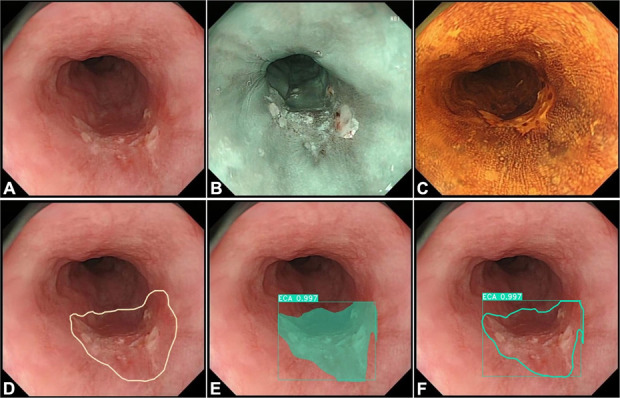
Evaluating the detection and delineation performance of the model. (a) A case of cancer in the esophagus with WLI. (b and c) Same case of cancer in the esophagus with narrow-band imaging and iodine staining, respectively. (d) Margins of the same lesion under WLI were manually delineated (white polygonal frames) by an expert who took the margins of the lesions under NBI, iodine staining, and resection specimen as reference. (e) The AI model correctly detected the lesion by indicating it with a square frame and a polygonal frame colored by dark cyan. (f) The AI model delineated the margin of the lesion (a dark cyan polygonal frame). AI, artificial intelligence; NBI, narrow-band imaging; WLI, white light imaging.
For delineation analysis, the accuracy, mIoU, sensitivity, and specificity of the AI model for early ESCC were 93.4% (95% CI, 91.9%–94.9%), 70.3% (95% CI, 69.3%–71.4%), 86.6% (95% CI, 85.6%–87.5%), and 81.4% (95% CI, 80.3%–82.5%), respectively, at internal validation. At external validation, the AI model achieved an accuracy of 95.7% in per-image (95% CI: 93.0%–98.4%), while corresponding mIoU, sensitivity, and specificity in per-image for delineating margins of early ESCC were 71.0% (95% CI, 68.3%–73.2%), 91.1% (95% CI, 89.3%–92.8%), and 78.0% (95% CI, 75.2%–80.3%) (Table 3 and Figure 2).
Table 3.
Delineation performance of the model for lesions in per-image
| Internal validation set (n = 1,114) | External validation set (n = 211) | |
| Accuracy (95% CI) | 93.4 (91.9–94.9) | 95.7 (93.0–98.4) |
| mIoU (95% CI) | 70.3 (69.3–71.4) | 71.0 (68.3–73.2) |
| Sensitivity(95% CI) | 86.6 (85.6–87.5) | 91.1 (89.3–92.8) |
| Specificity(95% CI) | 81.4 (80.3–82.5) | 78.0 (75.2–80.3) |
Values are given in percentages. The accuracy of delineating lesions margins was interpreted as exceeding the threshold overlap ratio of 0.6. Accuracy = true predictions/total number of cases.
CI, confidence interval; mIoU, mean intersection over union.
Testing of the model in videos
We successfully tested the model in 20 real WLI videos from patients with early ESCC. In clip1 presenting an early ESCC, the model successfully identified the lesion and accurately delineated lesion margins during the examination (see Supplementary Video, Supplementary Digital Content 1, http://links.lww.com/CTG/A729). Meanwhile, we have developed a website to provide free access to our AI model (http://huaxi-ai.innovsight.com/).
Comparison of the delineation performance between the AI model and endoscopists per-image
The delineating margins of the lesions determined by the AI model were compared with the values obtained by senior and expert endoscopists. The average time to detect and delineate 1 image was 17 ms for the model, 113 seconds for senior endoscopists, and 92 seconds for expert endoscopists. The delineation performance measures are summarized in Table 4. The AI model demonstrated better performance in accuracy compared with senior endoscopists (98.1% vs 78.6%, P = 0.002), and it has a similar performance when compared with that of the expert endoscopists (98.1% vs 95.3%, P = 0.608) on delineating margins under an overlap ratio of 0.60. The kappa value of 2 times repeated of the model reads was 1. However, both senior and expert endoscopists had a low interobserver agreement (kappa score, 0.198 and 0.140, respectively). In addition, the mIoU of the AI model was significantly greater than that of senior (76.2% vs 60.5%, P < 0.001) and expert endoscopists (76.2% vs 64.0%, P < 0.001). Further analysis of sensitivity revealed that the AI model was superior to that of the senior endoscopists (88.5% vs 76.3%, P < 0.001); yet, there was no significant difference between the AI model and expert endoscopists (88.5% vs89.6%, P = 0.425). Moreover, it was demonstrated that the AI model achieved significantly higher specificity than the senior (85.8% vs 79.3%, P < 0.001) and expert endoscopists (85.8% vs 71.0%; P < 0.001) (Table 4, Figure 3; see Supplementary Figure 2, Supplementary Digital Content 3, http://links.lww.com/CTG/A731).
Table 4.
Performance of the model and endoscopists in delineating lesions in per-image
| Metrics | AI model | Senior endoscopists | Expert endoscopists |
| Accuracy (95% CI) | 98.1 (96.3–99.9) | 78.6 (65.3–89.7) | 95.3 (93.0–97.5) |
| mIoU (95% CI) | 76.2 (74.4–77.9) | 60.5 (59.5–61.5) | 64.0 (63.1–65.0) |
| Sensitivity (95% CI) | 88.5 (87.0–90.0) | 76.3 (75.1–77.5) | 89.6 (88.9–90.3) |
| Specificity (95% CI) | 85.8 (83.9–87.3) | 79.3 (78.2–80.4) | 71.0 (70.0–72.1) |
Values are given in percentages. The accuracy of delineating lesions margins was interpreted as exceeding the threshold overlap ratio of 0.6. Accuracy = true predictions/total number of cases.
CI, confidence interval; mIoU, mean intersection over union.
Figure 3.
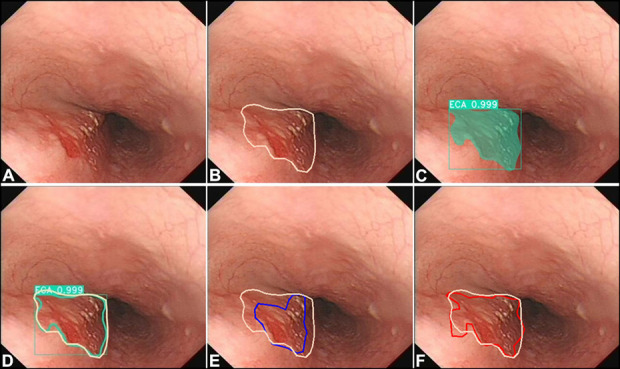
Comparing the delineation performance of the model with that of endoscopists. (a) A case of cancer in the esophagus with white light imaging. (b) Margins of the same lesion under WLI were manually delineated (white polygonal frames, used as the gold standard) by an expert who took the margins of the lesions under NBI, iodine staining, and resection specimen as reference. (c) The AI model correctly detected the lesion by indicating it with a square frame and a polygonal frame (dark cyan). (d) Margins of the same lesion under WLI were delineated AI model (a dark cyan polygonal frame) and the gold standard (white polygonal frames). (e) Margins of the same lesion under WLI were delineated by a senior endoscopist (a blue polygonal frame) and the gold standard (white polygonal frames). (f) Margins of the same lesion under WLI were delineated by an expert endoscopist (a red polygonal frame) and the gold standard (white polygonal frames). AI, artificial intelligence; NBI, narrow-band imaging; WLI, white light imaging.
DISCUSSION
In the present study, we successfully developed a DCNN-based AI model to detect and delineate margins of early ESCC under WLI. Our data demonstrated the AI model had good accuracy in detecting and delineating margins of early ESCC at internal and external validation data sets. To the best of our knowledge, this is the first study that evaluated the delineation performance of the model of early ESCC under WLI. Moreover, in unprocessed videos, the model achieved real-time identification and delineation of early ESCC under WLI.
With the rapid progress in the development of AI, especially in the advancement of computer-aided diagnosis for detection of early ESCC, several studies have shown that AI can significantly improve the diagnosis of such lesions (16–19). However, most previous models were developed based on NBI/ME-NBI. Fukuda et al. (16) have proposed that a DCNN is capable of diagnosing early ESCC. They reported that the detection rate of early ESCC under NBI was 86.4%, thus demonstrating the promising diagnostic performance of DCNNs. Furthermore, in combination with DCNNs, analysis of IPCL under ME-NBI can be applied to further evaluate the depth of the lesion (20).
In addition, WLI is the most basic and available endoscopy diagnostic modality for early ESCC. Therefore, the application of AI to WLI might be beneficial; however, studies focusing on early diagnosis of ESCC under WLI are limited, possibly because it is difficult to achieve high accuracy. Luo et al. (21) developed an AI model to identify early ESCC and gastric cancer under WLI with an accuracy of 91.5%–97.7%. However, most of the patients were diagnosed with advanced-stage cancers in their validation set. Although Cai et al. (22). reported that the accuracy of their model in the screening of early ESCC was 86.4%, their validation data set contained only 187 images from 52 patients. Ohmori et al. (18) developed a computer-aided diagnosis model for early ESCC under WLI. However, their diagnostic accuracy was only 81%; thus, further investigation is required to prove the diagnostic accuracy of early ESCC under WLI. In the present study, our internal and external data sets contained more images compared with previous studies (18). Meanwhile, further results indicated that the accurate rate of our model in identifying early ESCC under WLI was 85.7% and 84.5% in internal and external validation, respectively.
Meanwhile, although endoscopy treatment of early ESCC has achieved a very high cure rate, there are also some complications such as bleeding, perforations, and stricture formation (6). As one of the main adverse effects, strictures formation can be difficult to manage. Postendoscopic submucosal dissection strictures usually require repeated endoscopy dilatation or even additional surgical treatment (23,24). Therefore, it is crucial to accurately delineate early ESCC to establish rational treatment and improve patients' quality of life. Various diagnostic techniques are used to measure the margins of early ESCC such as WLI, ME-NBI, and iodine staining (25). Still, because some of the lesion margins are not clear, delineating the margins of early ESCC under WLI endoscopy is difficult even for expert endoscopists because they require large amounts of experience and time (26). Meanwhile, WLI is typically used to evaluate the extent of early ESCC because NBI and ME-NBI are not always available, especially in economically underdeveloped areas (27). Until recently, no studies were focusing on the delineation of early ESCC under WLI by using an AI model. In this study, the model showed an accuracy of 93.4% and 95.7% in delineating lesions in internal and external validation, respectively. The sensitivity of delineation performance is higher than the specificity of the model for lesions in per-image. It suggested that our AI model can help ensure complete delineation of the lesion under WLI. In addition, we further compared the delineating accuracy of our AI model with that of expert endoscopists. The accuracy and sensitivities of the AI model in delineating the margins of early ESCC exceeded those of the nonexpert endoscopists. Besides, the delineating accuracy and sensitivities of our AI model were comparable with that of expert endoscopists. Meanwhile, the mIoU and specificities of the AI model in delineating the margins of early ESCC were both significantly higher than those of the senior and expert endoscopists. Moreover, the time required by the model for delineation of the lesions was shorter than that required by the senior and expert endoscopists. Therefore, these findings suggested that the developed model may not only enhance the performance of nonexpert endoscopists to delineate early ESCC margins but also quickly assist expert endoscopists in delineating the margins of early ESCC under WLI.
There are several limitations in this study. First, the training data set mainly contained early ESCC and noncancer endoscopy images under WLI, whereas it lacked inflammation, ulcers, advanced cancer, and other types of esophageal lesions. Therefore, this may profoundly influence the specificity of the model in clinical practice. Second, this study lacked rebuilding the pathological result on the endoscopic image of the lesions regarding the selection of the gold standard during the evaluation of delineation performance evaluation. The gold standard was defined as delineation of the lesions under WLI marked by the expert endoscopists who took iodine staining and NBI as a reference. Third, we used only video clips to show the identification and delineation performance of our model. Statistical analysis was not conducted on video tests because it is difficult for experts to evaluate the delineation performance of the model based on each frame. Next, we will conduct a further man-machine experiment to compare the detecting accuracy of our AI model with that of endoscopists. Meanwhile, the delineation performance of the model should be further evaluated by comparing it with other advanced diagnostic techniques such as NBI, magnified endoscopy, and iodine staining.
In conclusion, our AI model was successfully constructed to help determine the diagnosis of early ESCC. Meanwhile, the successful development of this model is helpful for the delineation of early ESCC to establish a rational treatment plan.
CONFLICTS OF INTEREST
Guarantor of the article: Bing Hu, MD.
Specific author contributions: B.H., W.L., and X. Y.: study design. W.L., X.Y., and L.G.: analysis and interpretation of data. Y.H. and J.F.: training and testing models. W.L. and X.Y.: drafting of the manuscripts. B.H., F.P., C.W., Z.S., F.T., C.Y., W.Z., and S.B.: critical revision of the manuscript.
Financial support: This work was supported by National Natural Science Foundation of China (Grant No: 82,170,675) and 1·3·5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC21011).
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ Screening for early esophageal squamous cell carcinoma (ESCC) is generally performed by standard endoscopic observation with white light imaging (WLI).
✓ It is difficult to detect and delineate the margins of early ESCC using WLI.
WHAT IS NEW HERE
✓ An artificial intelligence (AI) model was successfully constructed to help determine the diagnosis of early ESCC.
✓ This is the first evaluation of AI in delineating margins of early ESCC under WLI.
✓ The AI model was effective in detecting and delineating margins of early ESCC under WLI.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A729, http://links.lww.com/CTG/A730, http://links.lww.com/CTG/A731, http://links.lww.com/CTG/A732
Wei Liu and Xianglei Yuan contributed equally to this work.
Contributor Information
Wei Liu, Email: lwym117@163.com.
Xianglei Yuan, Email: yuanxianglei94@163.com.
Linjie Guo, Email: guolinjie1985@126.com.
Feng Pan, Email: fengliupan@126.com.
Chuncheng Wu, Email: hxwucc@163.com.
Zhongshang Sun, Email: nice22hh@163.com.
Feng Tian, Email: lcw261@163.com.
Cong Yuan, Email: 709564580@qq.com.
Wanhong Zhang, Email: 412854774@qq.com.
Shuai Bai, Email: 327263946@qq.com.
Jing Feng, Email: 596283512@qq.com.
Yanxing Hu, Email: huyanxing@hotmail.com.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ, RE T. Esophageal cancer. In: Thun MJ, Linet MS, Cerhan JR, et al. (eds). Cancer Epidemiology and Prevention. 4th edn, Oxford University Press 2017: New York, 2018, pp 579–92. [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics 2020. CA Cancer J Clin 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4.Lagergren J, Smyth E, Cunningham D, et al. Oesophageal cancer. Lancet 2017;390(10110):2383–96. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb-Vedi E, Kauppila JH, Malietzis G, et al. Long-term survival in esophageal cancer after minimally invasive compared to open esophagectomy: A systematic review and meta-analysis. Ann Surg 2019;270(6):1005–17. [DOI] [PubMed] [Google Scholar]
- 6.di Pietro M, Canto MI, Fitzgerald RC. Endoscopic management of early adenocarcinoma and squamous cell carcinoma of the esophagus: Screening, diagnosis, and therapy. Gastroenterology 2018;154(2):421–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebi M, Shimura T, Yamada T, et al. Multicenter, prospective trial of white-light imaging alone versus white-light imaging followed by magnifying endoscopy with narrow-band imaging for the real-time imaging and diagnosis of invasion depth in superficial esophageal squamous cell carcinoma. Gastrointest Endosc 2015;81(6):1355.e2. [DOI] [PubMed] [Google Scholar]
- 8.Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol 2012;9(5):278–87. [DOI] [PubMed] [Google Scholar]
- 9.Lee CT, Chang CY, Lee YC, et al. Narrow-band imaging with magnifying endoscopy for the screening of esophageal cancer in patients with primary head and neck cancers. Endoscopy 2010;42(8):613–9. [DOI] [PubMed] [Google Scholar]
- 10.Kitagawa Y, Uno T, Oyama T, et al. Esophageal cancer practice guidelines 2017 edited by the Japan Esophageal Society: Part 1. Esophagus 2018;16(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono S, Fujishiro M, Niimi K, et al. Long-term outcomes of endoscopic submucosal dissection for superficial esophageal squamous cell neoplasms. Gastrointest Endosc 2009;70:860–6. [DOI] [PubMed] [Google Scholar]
- 12.Zeki SS, Bergman JJ, Dunn JM. Endoscopic management of dysplasia and early oesophageal cancer. Best Pract Res Clin Gastroenterol 2018;36-37:27–36. [DOI] [PubMed] [Google Scholar]
- 13.Lui TKL, Tsui VWM, Leung WK. Accuracy of artificial intelligence–assisted detection of upper GI lesions: A systematic review and meta-analysis. Gastrointest Endosc 2020;92(4):821–30. [DOI] [PubMed] [Google Scholar]
- 14.Chahal D, Byrne MF. A primer on artificial intelligence and its application to endoscopy. Gastrointest Endosc 2020;92(4):813.e4. [DOI] [PubMed] [Google Scholar]
- 15.Bang CS, Lee JJ, Baik GH. Computer-aided diagnosis of esophageal cancer and neoplasms in endoscopic images: A systematic review and meta-analysis of diagnostic test accuracy. Gastrointest Endosc 2020;93(5):1006–15. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda H, Ishihara R, Kato Y, et al. Comparison of performances of artificial intelligence vs expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc 2020;92(4):848–55. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, Xiao X, Wu C, et al. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc 2020;91(1):41–51. [DOI] [PubMed] [Google Scholar]
- 18.Ohmori M, Ishihara R, Aoyama K, et al. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc 2020;91(2):301.e1. [DOI] [PubMed] [Google Scholar]
- 19.Tokai Y, Yoshio T, Aoyama K, et al. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus 2020;17(3):250–6. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y-Y, Xue D-X, Wang Y-L, et al. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy 2018;51(4):333–41. [DOI] [PubMed] [Google Scholar]
- 21.Luo H, Xu G, Li C, et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: A multicentre, case-control, diagnostic study. Lancet Oncol 2019;20(12):1645–54. [DOI] [PubMed] [Google Scholar]
- 22.Cai SL, Li B, Tan WM, et al. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc 2019;90(5):745.e2. [DOI] [PubMed] [Google Scholar]
- 23.Bhatt A, Mehta NA. Stricture prevention after esophageal endoscopic submucosal dissection. Gastrointest Endosc 2020;92(6):1187–9. [DOI] [PubMed] [Google Scholar]
- 24.Abe S, Iyer PG, Oda I, et al. Approaches for stricture prevention after esophageal endoscopic resection. Gastrointest Endosc 2017;86(5):779–91. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara R, Arima M, Iizuka T, et al. Endoscopic submucosal dissection/endoscopic mucosal resection guidelines for esophageal cancer. Dig Endosc 2020;32(4):452–93. [DOI] [PubMed] [Google Scholar]
- 26.Park CH, Yang DH, Kim JW, et al. Clinical practice guideline for endoscopic resection of early gastrointestinal cancer. Clin Endosc 2020;53(2):142–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes AB. Esophageal squamous cell carcinoma - precursor lesions and early diagnosis. World J Gastrointest Endosc 2012;4(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]