ABSTRACT
To identify how circular RNA circRNA_0082835 impacts melanoma cells and lymphatic metastasis to observe whether it exerts effects through its action mechanism of sponging microRNA miR-429. Clinical baseline information was collected, and clinical samples were used for detection on circRNA_0082835 and EZH2. The expression of circRNA_0082835, EZH2, and miR-429 was detected by quantitative real-time PCR (RT-qPCR). Cell proliferation was tested with cell counting kit-8 (CCK-8). Flow cytometry was applied to examination of cell cycle levels. Cell invasion and migration were observed by transwell and wound healing. The expression of Wnt/β-catenin pathway, cell cycle and epithelial–mesenchymal transition (EMT) marker proteins was analyzed by western blot. Dual-luciferase determined the binding of miR-429 and circ_0082835. As a result, the expression of circRNA_0082835 was increased and that of miR-429 was decreased with the increase in lymphatic metastasis level. CircRNA_0082835 expression was downregulated by circ_0082835 interference, upregulated by EZH2 interference and also downregulated after transfection of both shRNA-circ_0082835 and shRNA-EZH2. Inhibiting circ_0082835 and EZH2 suppressed the proliferation, invasion and migration, regulated the cell cycle levels, inhibited Wnt/β-catenin and attenuated EMT in melanoma cells. Inhibition of circ_0082835 and/or EZH2 elevated miR-429 expression. The binding among miR-429 and circ_0082835 was verified. MiR-429 inhibitor reversed the effect of circ_0082835 interference while having no significant impact on EZH2. In conclusion, circRNA_0082835 sponges miR-429 to affect the anti-tumor effect of miR-429 in primary melanoma and lymphatic metastasis.
KEYWORDS: CircRNA_0082835, miR-429, primary melanoma, EZH2, Wnt/β-catenin
Graphical Abtstract
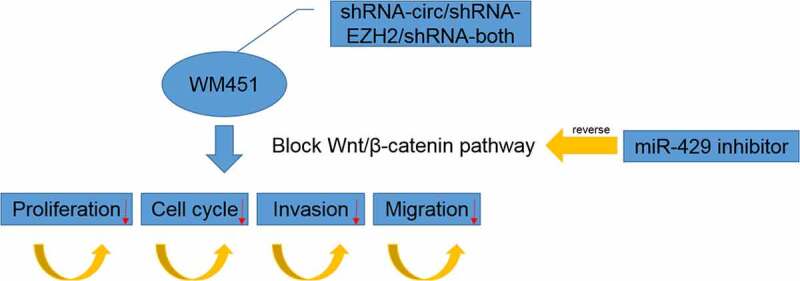
Introduction
Melanoma usually refers to malignant melanoma, a type of skin cancer originated from melanocytes, which often develops on the skin and sometimes on the mucosal site or even viscera including the intracranial site[1,2]. The main metastasis route of melanoma is lymphatic metastasis which is often occurred after stage III [3]. While this disease can be accurately diagnosed with biopsy, it is still a challenge to treat patients with melanoma as surgical margin has only limited preciseness and the efficacy largely depends on cancer stages [4]. Furthermore, progression assessment in lack of accuracy and diagnostic biopsy inadequate for differentiation between normal nevi and melanoma are also considered impediments for the melanoma treatment [5]. Thus, it is necessary to broaden the knowledge of melanoma and to develop more effective treatment for this horrible disease.
The implication of circular RNAs (circRNAs) in cancer has been reported in many studies. For instance, Shang et al. elucidated the oncogenicity of circMTUS1 in conjunctival melanoma [6]; Wei et al. demonstrated the promotive effect of circ_0020710 on melanoma progression via regulation of miR-370-3p/CXCL12 pathway [7]. According to the high throughput screening (HTS) of circRNAs in a research on melanoma [8], circRNA_0082835 is one of the circRNAs and found to be significantly overexpressed in melanoma tissue. Furthermore, it is noted on circBase (http://circrna.org/) that circRNA_0082835 is located in the EZH2 gene (chr7: 148,511,050–148,511,229). High levels of EZH2 was shown to be associated with more malignant forms of melanoma [9,10]. High EZH2 expression was related to the increased proliferation, thicker primary melanomas, and increased invasion [9]. Knockdown of EZH2 suppressed proliferation, restored a senescent-like phenotype of melanoma cells, and inhibited the growth of xenografts in mice [11]. Inhibition of EZH2 negatively regulated the proliferation of human melanoma cells [12]. Therefore, why we intended to find out how circRNA_0082835 affects melanoma cells.
ENCORI database (http://starbase.sysu.edu.cn/) was consulted for potential miRNAs that interact with cicrRNA_0082835, one of which was miR-429. It is well substantiated that miR-429 plays an anti-tumor role in various cancer types. miR-429 overexpression obviously inhibited proliferation and promoted apoptosis of gastric cancer cells, which was reversed by miR-429 inhibitor [13]. Upregulation of miR-429 suppressed the increase of tumor size and weight in nude mice [14]. miR-429 expression was obviously decreased in glioblastoma tissues and human glioma cells and miR-429 played inhibition effects on the proliferation and invasion of glioblastoma cells [15]. The tumor-specific expression of miR-429 is also shown on ENCORI website. However, the role of miR-429 in regulation of melanoma cells remains unknown and needs us to explore.
Therefore, we assume that circRNA_0082835 may be able to influence the cellular process of melanoma cells through interaction with miRNA-429, which alters the effects of miRNA-429. We detected the expression of circRNA_0082835 and miRNA-429 in melanoma tissues and expression of circRNA_0082835 and EZH2 in melanoma cells. Subsequently, cell counting kit-8 (CCK-8), flow cytometry, transwell and wound healing assays were used to explore the effects of inhibiting circRNA_0082835 and/or EZH2 on the biological function of melanoma cells and reversible effect of inhibiting miRNA-429 on the effects of inhibiting circRNA_0082835 and/or EZH2. Our study is expected to provide a possible channel to the development of effective treatment for malignant melanoma in the future.
Materials and methods
Clinical sample collection
Ten melanoma patients (five males and five females, aged between 44 and 67) from The First Affiliated Hospital of Nanjing Medical University diagnosed with malignant melanoma (metastasis or no metastasis) were chosen and melanoma tissues from metastasis or no metastasis groups were surgically resected from 10 melanoma patients. The clinical baseline information of 10 melanoma patients is shown in Table 1. Informed consent forms were signed by all patients. The ethics committee has approved this study of The First Affiliated Hospital of Nanjing Medical University between December 2018 and December 2020 in our department (2020-SR-563).
Table 1.
TNM stage and lymphatic metastasis of malignant melanoma
| Age | Gender | Tumor size (mm) | TNM stage | Lymphatic metastasis |
|---|---|---|---|---|
| 47 | Male | 1 | IIA | No |
| 63 | Female | 0.1 | IA | No |
| 44 | Female | 0.3 | IA | No |
| 67 | Male | 0.4 | IA | No |
| 63 | Female | 0.2 | IA | No |
| 56 | Male | 0.1 | IV | No |
| 64 | Female | 0.1 | IIIC | Yes |
| 49 | Female | 0.3 | IIIA | Yes |
| 53 | Male | 0.1 | IIIA | Yes |
| 58 | Male | 0.2 | IV | Yes |
Cell lines and treatments
Human melanoma cell lines available in our laboratory include cell strains A375 (cat. no. CRL-1619), WM451 (cat. no. CRL-2813), SK-MEL-24 (cat. no. HTB-71), and WM35 (cat. no. CRL-2807), all sourced from American Type Culture Collection (ATCC). Normal human melanocyte cell line HM (cat. no. MZ-2700) was purchased from Mingzhou Biological Technology Co., Ltd (Ningbo, China). The cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) or Eagle’s Minimum Essential Medium (EMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 15% FBS. For subculture, the complete growth medium was removed, and the cell layers were rinsed with 0.25% (w/v) Trypsin-0.53 mM ethylene diamine tetraacetic acid solution (EDTA; GIBCO BRL, Grand Island, NY, USA). Next, 3.0 mL of Trypsin-EDTA was added to the flask for dispersion of the cells. 8.0 mL of complete growth medium was then added, and the cells were aspirated by gently pipetting. Cell suspension in new culture vessels was incubated at 37°C. The medium was renewed every 2–3 days.
shRNA plasmids of circ_0082835 and EZH2 as well as miRNA-429 inhibitor were constructed or synthesized by GenePharma (Shanghai, China).
Quantitative real-time PCR (RT-qPCR)
Total RNA was extracted using TRIzolTM reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed to synthesize cDNA, according to the manufacturer’s protocol of the PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan). RT-qPCR was performed with SYBR Premix Ex Taq (TaKaRa) on a CFX96 Realtime System (Bio-Rad Laboratories, Inc., Hercules, CA). The procedure was 30 s at 95°C for the holding stage, 5 s at 95°C and 30 s at 60°C in total 40 cycles for the cycling stage. Relative quantification was calculated with the application of 2−ΔΔCt method [16]. The expression levels of circRNA_0082835 and EZH2 were normalized to GAPDH and miR-429 expression levels to U6.
Cell Counting Kit-8 (CCK-8)
Cell proliferation was examined using CCK-8 (Beyotime, Nanjing, China) referring to a previous study [17]. Melanoma cells were added into the plate at a density of 100 μl 2 × 103 cells per well. After treatments in different groups, 10 μl of CCK-8 solution was added to each well for incubation with the cells for 2 h. Absorbance was measured using a Multiskan™ Go microplate spectrophotometer (Thermo Fisher Scientifc, Inc.) at 450 nm, respectively, after incubation for 24 h, 48 h, and 72 h.
Flow cytometry
Melanoma cell cycles were detected with flow cytometry. Cells in the number of approximately 1 × 106 were centrifuged, suspended in 0.3 ml PBS buffer (Aladdin, Shanghai, China) containing 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) and fixed at −20°C for 24 h after addition of 0.7 ml absolute ethyl alcohol. The cells were then centrifuged at 3000 r/min for 30 min and resuspended by 1 ml PBS (Aladdin, Shanghai, China) after the supernatant was discarded. After centrifugation once again, the cells were suspended in 100 μl 1 mg/ml RNase A (Beyotime, Nanjing, China) and placed at 37°C for 30 min to achieve sedimentation of the cells. A 400 μl 50 μg/ml propidium iodine (PI; Sigma-Aldrich, St. Louis, MO, USA) was used to stain the cells in the dark for 10 min before detection by a Beckman Coulter CytoFLEX and BD FLOWJO-V10 software (Beckman Coulter, Inc.).
Transwell assay
The invasion ability of the cells was assayed by transwell assay referring to a previous study [18]. A 100 μl of Matrigel (Corning Incorporated, Corning, NY, USA) diluted in serum-free medium (Sigma-Aldrich, St. Louis, MO, USA) at 4°C and then used to precoat the membrane of the upper chamber. The chamber was placed in a 37°C environment for about 1 h to allow gel formation. Cells growing in the logarithmic stage were washed with PBS (Aladdin, Shanghai, China) and suspended in serum-free medium. The density of the cells was adjusted to 5 × 105/ml. Next, 500 μl of culture medium containing 5% FBS was added to the lower chamber of the 24-well plate. Transwell chamber was put in the plate by a tweezer. A 150 μl of cell suspension was added to the upper chamber followed by incubation for 24 h. Subsequently, transwell chamber was taken out, and the culture medium was absorbed. Matrigel and cells in the upper chamber were gently wiped with a cotton swab. The chamber was then put in a new 24-well plate and fixed with 600 μl of 4% paraformaldehyde (Aladdin, Shanghai, China) for 30 min, followed by staining with 0.1% crystal violet (Solarbio, Beijing, China) for 10 min. After air drying, cells were observed and counted with a high-power microscope from five different visual angles.
Wound healing assay
Cell migration was detected by wound healing assay referring to a previous study [19]. A horizontal line was evenly drawn across the hole on the back of the 6-well plate using a ruler, about every 0.5 ~ 1 cm. 5 × 105 cells were added to well and grown overnight. The tip of a pipette was used to create wound on the cell surface the next day. The cells were then washed 3 times with PBS (Aladdin, Shanghai, China) and incubated in serum-free medium in a 37°C, 5% CO2 incubator. Images were taken at 0 h, 6 h, 12 h and 24 h of incubation.
Western blot analysis
Gene expression of target proteins was detected by western blot referring to a previous study [17]. Protein samples were prepared using RIPA lysis buffer (Beyotime, Nanjing, China). And protein concentration was determined by bicinchoninic acid (BCA) kit (Abcam, Cambridge, MA, USA). SurePAGE™ precast sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (GenScript, Nanjing, China) was used for electrophoresis. SDS-PAGE buffer was added to the protein samples, which were then denatured by boiling water bath at 100°C for 5 min. The samples after cooldown were loaded into the well and electrophorized until bromophenol blue reached the bottom of the gel. After the samples were transferred to polyvinylidene fluoride (PVDF) membrane (Corning Incorporated, Corning, NY, USA) and fixed with 5% bovine serum albumin (BSA; Thermo Fisher Scientific, Waltham, MA, USA), they were incubated against Wnt5a (ab282153; dilution, 1:1000; Abcam), β-catenin (ab68183; dilution, 1:1000; Abcam), CyclinD1 (ab16663; dilution, 1:200; Abcam), CyclinE1 (ab33911; dilution, 1:1000; Abcam), CRKL (ab32018; dilution, 1:1000; Abcam), E-cadherin (E-cad) (ab40772; dilution, 1:10,000; Abcam), N-cadherin (N-cad) (ab76011; dilution, 1:5000; Abcam) and GAPDH (ab9485; dilution, 1:2500; Abcam) at 4°C overnight and then with diluted goat Anti-Rabbit IgG (H + L) secondary antibody (ab6721 dilution, 1:2000; Abcam) at room temperature for 1 h. The protein bands were visualized on a Tanon Chemiluminescence Imaging system with an ECL regent (Thermo Fisher Scientific). The density of the protein bands was quantified using Alphalmager™ 2000 Imaging System (Alpha Innotech).
Dual-luciferase reporter assay
The interaction between miR-429 and circ_0082835 or EZH2 was verified by dual-luciferase reporter assay referring to a previous study [20]. Transfection of WT-circ_0082835/EZH2, MUT-circ_0082835/EZH2 and miR-429 mimic was performed using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) when the cells converged about 70%. 48 h after transfection, the culture medium was absorbed. The cells were lysed and centrifuged at 10,000 × g for 5 min, and the supernatant was collected. The firefly and Renilla luciferase activities were detected according to the guidelines of the dual-luciferase reporter assay kits (Abcam, Cambridge, MA, USA) using Dual-Luciferase Reporter Assay System (Promega Biotech Co., Madison, USA). The ratio of the firefly and Renilla luciferase activities represented the relative luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation (SD). Two-group comparisons were analyzed with Student’s t test and one-way ANOVA followed by the Turkey’s test was applied to the comparison between multiple groups. GraphPad Prism 6 analyzed the data and graphed the figures. P value less than 0.05 suggests significant statistical difference.
Results
Increased circRNA_0082835 expression and decreased miR-429 expression in clinical melanoma samples
The detection of circRNA_0082835 and miR-429 expressions in clinical melanoma samples was conducted by RT-qPCR. The result showed rising circRNA_0082835 expression level and declining miR-429 expression level as the number of metastatic lymph nodes increased (Figure 1(a-B)). circRNA_0082835 expression was negatively related to the miR-429 expression (Figure 1(c)). Therefore, increased circRNA_0082835 expression and decreased miR-429 expression are ascertained in clinical melanoma samples.
Figure 1.
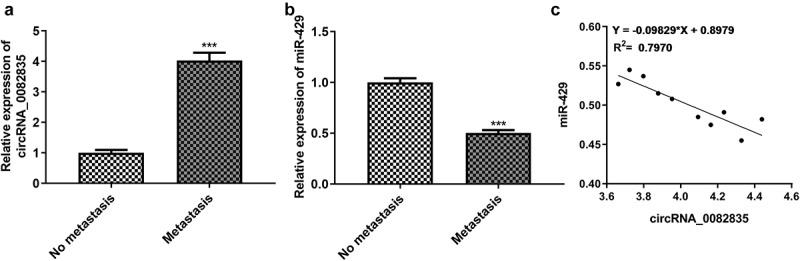
Increased circRNA_0082835 expression and decreased miR-429 expression in clinical melanoma samples. (a-b) Relative expressions of circRNA_0082835 and miR-429 in metastatic melanoma, detected by qPCR. ***P < 0.001 vs. No metastasis group. (c) correlation analysis between circRNA_0082835 and miR-429
Inhibiting circRNA_0082835 and/or EZH2 suppresses melanoma cell proliferation
The expressions of circRNA_0082835 and EZH2 in different melanoma cell strains were detected first by RT-qPCR, which found that WM451 cells had the highest expression of both (Figure 2(a-B)). Thus, WM451 was selected for the subsequent experiments. shRNA plasmids of circRNA_0082835 (shRNA-circ) and EZH2 (shRNA-EZH2) were constructed and, respectively, transfected or co-transfected into WM451 cells. The transfection effects of above shRNA plasmids were determined by RT-qPCR. Greatly downregulated circRNA_0082835 and upregulated EZH2 were shown in shRNA-circ group; Increased circRNA_0082835 expression and greatly decreased EZH2 expression were shown in shRNA-EZH2 group; The expressions of these two were both downregulated in shRNA-both group (Figure 2(c-d)). CCK-8 examined WM451 cell proliferation after transfection and found that transfection of shRNA-circ, shRNA-EZH2 and/or weakened cell proliferation to different degrees (Figure 2(e)), among which co-transfection of shRNA-circ and shRNA-EZH2 inhibited cell proliferation the most. The above results tell us that Inhibiting circRNA_0082835 and/or EZH2 suppresses melanoma cell proliferation.
Figure 2.
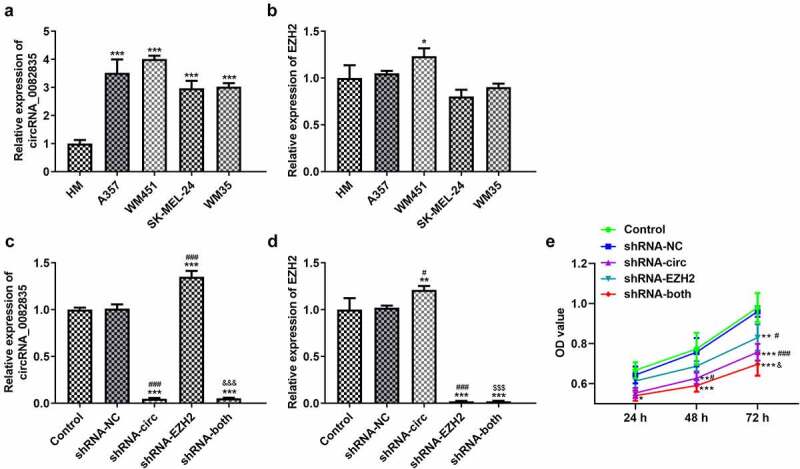
Inhibiting circRNA_0082835 and/or EZH2 suppresses melanoma cell proliferation. (a-b) Relative expressions of circRNA_0082835 and EZH2 in different melanoma cell strains, detected by qPCR. *P < 0.05 and ***P < 0.001 vs. HM group. (c-d) Relative expressions of circRNA_0082835 and EZH2 in WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by qPCR. (e) OD value of WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by CCK-8. *P < 0.05, **P < 0.01 and ***P < 0.01 vs. Control group. #P < 0.05 and ###P < 0.001 vs. shRNA-NC group. $$$P < 0.001 vs. shRNA-circ group. &&&p < 0.001 vs. shRNA-EZH2 group
Inhibiting circRNA_0082835 and/or EZH2 regulates cell cycle and suppresses the invasion and migration of melanoma cells
The flow cytometry, transwell assay and wound healing assay were used to detect the effects of inhibiting circRNA_0082835 and/or EZH2 on cell cycle, invasion and migration of melanoma cells. The percentage of G1 phase in the full cell cycle of WM451 cells increased, while that of S phase decreased after transfection of shRNA-circ and/or shRNA-EZH2. The percentage of G2 phase increased compared to the NC after shRNA-circ transfection, while its decline was observed in shRNA-EZH2 group and shRNA-both group (Figure 3(a)). Additionally, the invasion ability of WM451 cells was largely weakened by shRNA-circ, slightly weakened by shRNA-EZH2 and significantly weakened by shRNA-both (Figure 3(b)). The migration ability of WM451 cells, evidenced by much wider scratch in shRNA-circ, shRNA-EZH2 and shRNA-both groups (Figure 3(c)). These results indicate that melanoma cell cycle can be regulated, and its invasion and migration abilities can be suppressed by inhibition of circRNA_0082835 and/or EZH2.
Figure 3.
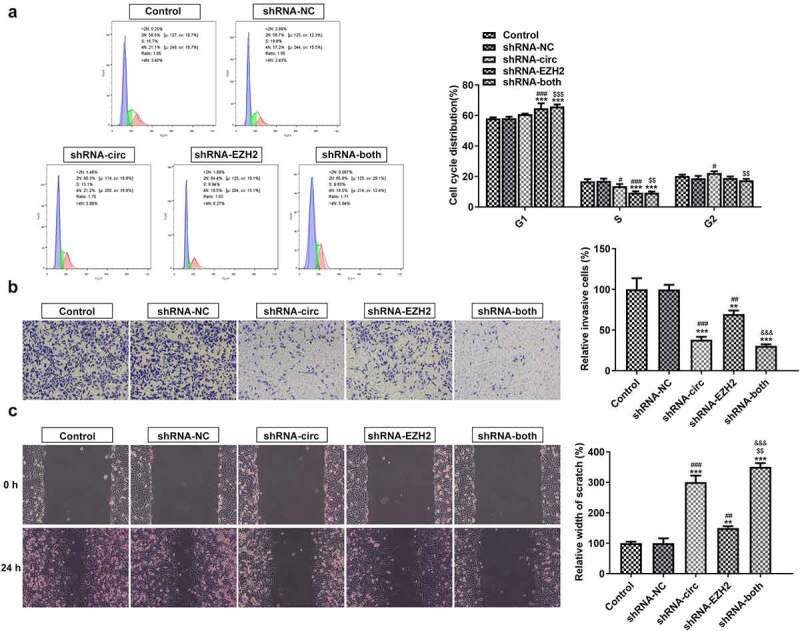
Inhibiting circRNA_0082835 and/or EZH2 regulates cell cycle and suppresses the invasion and migration of melanoma cells. (a) WM451 cell cycle distribution before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by flow cytometry. (b) Relative invasive WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by transwell. (c) Relative wound width of WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by wound healing. **P < 0.01 and ***P < 0.01 vs. Control group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. shRNA-NC group. $$P < 0.01 and $$$P < 0.001 vs. shRNA-circ group. &&&p < 0.001 vs. shRNA-EZH2 group
Inhibiting circRNA_0082835 and/or EZH2 blocks Wnt/β-catenin, suppresses cell cycle-related protein expression and attenuates epithelial–mesenchymal transition (EMT) in melanoma cells
The expressions of Wnt/β-catenin signaling-related proteins, cell cycle-related proteins, and EMT-related proteins were determined by western blot analysis. It was found that Wnt/β-catenin expression and cell cycle-related protein expression were both downregulated by contrast with the NC after shRNA-circ, shRNA-EZH2 or shRNA – both were transfected into WM351 cells (Figure 4(a)). Meanwhile, transfection of shRNA-circ and/or shRNA-EZH2 noticeably reduced the expression of CRKL and N-cad, while elevating that of E-cad to varying degrees (Figure 4(b)). These data suggest that inhibiting circRNA_0082835 and/or EZH2 blocks Wnt/β-catenin pathway, suppresses cell cycle-related protein expression and attenuates EMT in melanoma cells.
Figure 4.
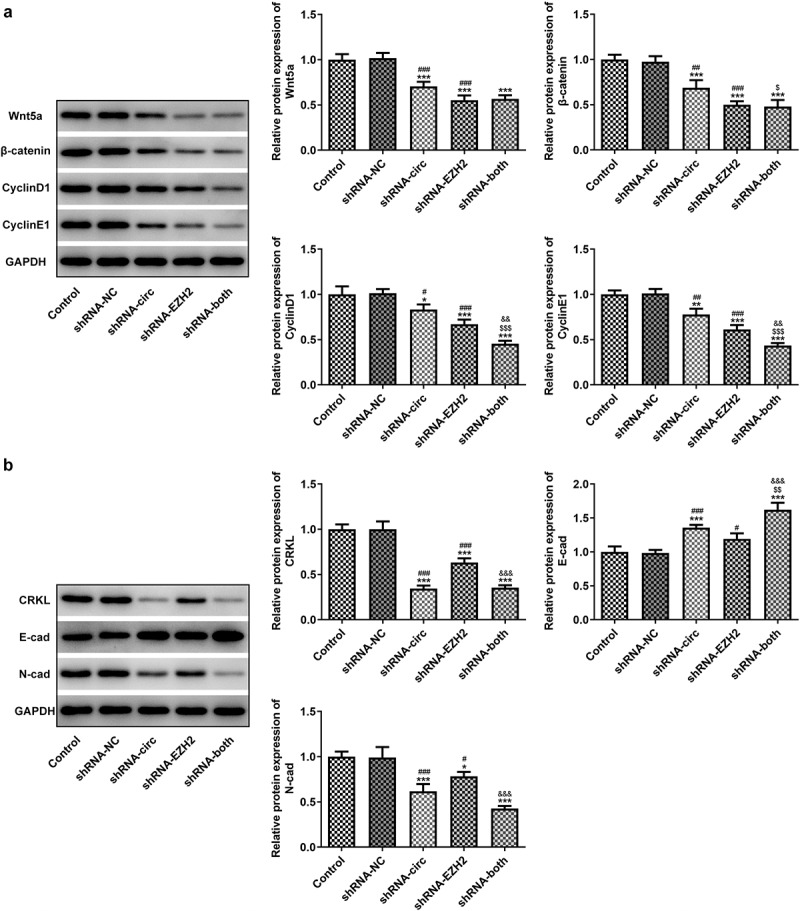
Inhibiting circRNA_0082835 and/or EZH2 blocks Wnt/β-catenin, suppresses cell cycle-related protein expression and attenuates EMT in melanoma cells. (a) Relative protein expressions of Wnt/β-catenin signaling-related Wnt5a and β-catenin, cell cycle-related CyclinD1 and CyclinE1 in WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by western blot. (b) Relative protein expressions of EMT-related CRKL, E-cad and N-cad in WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by western blot. *P < 0.05, **P < 0.01 and ***P < 0.01 vs. Control group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. shRNA-NC group. $P < 0.05, $$P < 0.01 and $$$P < 0.001 vs. shRNA-circ group. &&p < 0.01 and &&&p < 0.001 vs. shRNA-EZH2 group
MiRNA-429 binds to circRNA_0082835 and its inhibition reverses the anti-proliferation effect of cicrRNA_0082835 knockdown
The transfection effects, interaction between miR-429 and circ_0082835 and the reversible effect of miR-429 inhibitor on cicrRNA_0082835 knockdown for proliferation of melanoma cells were respectively detected by RT-qPCR, Dual-luciferase reporter assay and CCK-8 assay. miR-429 expression increased notably after transfection of shRNA-circ and co-transfection of shRNA-circ and shRNA-EZH2, whereas only minorly increased miR-429 was observed in shRNA-EZH2 group (Figure 5(a)). Dual-luciferase reporter activity confirmed the binding of miRNA-429 to circRNA_0082835, while no significant change of the relative luciferase activity was observed in EZH2 group (Figure 5(b-c)). Subsequently, miR-429 inhibitor and shRNA-circ were transfected into WM351 cells for further exploration on the interaction between miR-429 and circRNA_0082835. Decreased miR-429 expression was detected in WM351 cells co-transfected with shRNA-circ/shRNA-EZH2 and miR-429 inhibitor (Figure 5(d-e)). The proliferation level of WM351 cells was higher in shRNA-circ/shRNA-EZH2 + miR-429 inhibitor group than that in shRNA-circ/shRNA-EZH2 group (Figure 5(f)). These results demonstrate that miR-429 binds to circRNA_0082835 and that its inhibition reverses the anti-proliferation effect of circRNA_0082835 knockdown. It is also worth notice that the reversal in shRNA-EZH2 group was not as obvious as that in shRNA-circ group, which also shows the direct interaction between miR-429 and circRNA_0082835.
Figure 5.
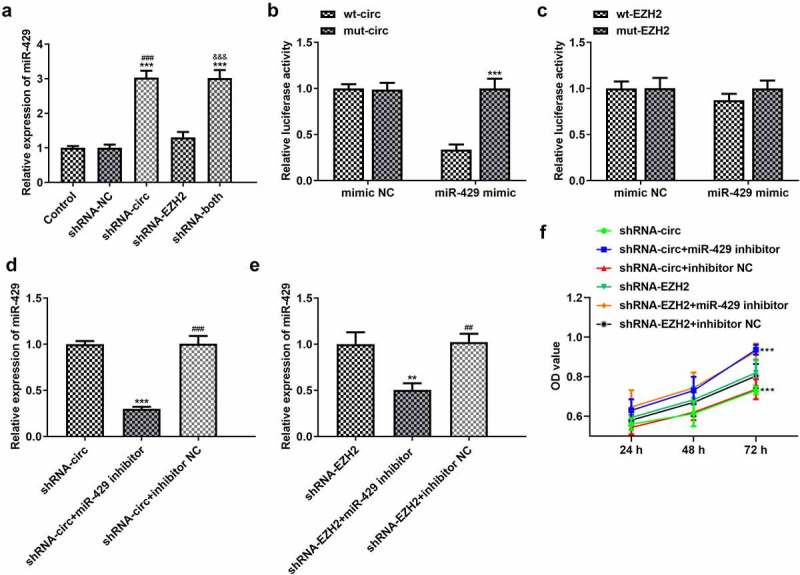
MiRNA-429 binds to circRNA_0082835 and its inhibition reverses the anti-proliferation effect of cicrRNA_0082835 knockdown. (a) Relative expression of miR-429 in WM451 cells before and after transfection of shRNA-circ and/or shRNA-EZH2, detected by qPCR. ***P < 0.001 vs. Control group. ###P < 0.001 vs. shRNA-NC group. &&&P < 0.001 vs. shRNA-EZH2 group. (b-c) Relative luciferase activity in wt-circ + mimic NC, mut-circ + mimic NC, wt-circ + miR-429 mimic, wt-EZH2 + mimic NC, mut-EZH2 + mimic NC, wt-EZH2 + miR-429 mimic and mut-EZH2 + miR-429 mimic, detected by dual-luciferase reporter assay. ***P < 0.001 vs. wt-circ group. (d-e) Relative expression of miR-429 in WM451 cells before and after the transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups was detected by qPCR. **P < 0.01 and ***P < 0.001 vs. shRNA-circ group. ##P < 0.01 and ###P < 0.001 vs. shRNA-circ +miR-429 inhibitor group. (f) OD values of WM451 cells before and after the transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups were detected by CCK-8. ***P < 0.001 vs. shRNA-circ group
MiR-429 inhibitor affects cell cycle regulation and reverses the anti-invasion and anti-migration effects of circRNA_0082835 knockdown
Cell cycle distribution, invasion level and migration ability of WM451cells were examined again after transfection of miR-429 inhibitor, respectively, by flow cytometry, transwell and wound healing. Figure 6(a) shows that compared to the NC, the percentage of G1 phase decreased and that of S phase increased in shRNA-circ group after transfection of miR-429 inhibitor. The percentage of G2 phase was reduced in shRNA-EZH2 group, compared to the NC, after transfection of miR-429 inhibitor. Meanwhile, the percentages of G1 phase and S phase exhibited no significant changes in sh-EZH2 group, neither did that of G2 phase in shRNA-circ group. Moreover, miR-429 inhibitor enlarged significantly the number of relative invasive WM451 cells in shRNA-circ group in comparison to the NC, while shRNA-EZH2 groups only exhibited exiguous changes without statistical significance (Figure 6(b)). Additionally, the wound width of WM451 cells was much narrower after transfection of miR-429 inhibitor compared to the NC in shRNA-circ group, whereas shRNA-EZH2 groups again exhibited no significant difference (Figure 6(c)). It can be concluded based on these results that miR-429 inhibitor affects cell cycle regulation and reverses the anti-invasion and anti-migration effects of circRNA_0082835 knockdown.
Figure 6.
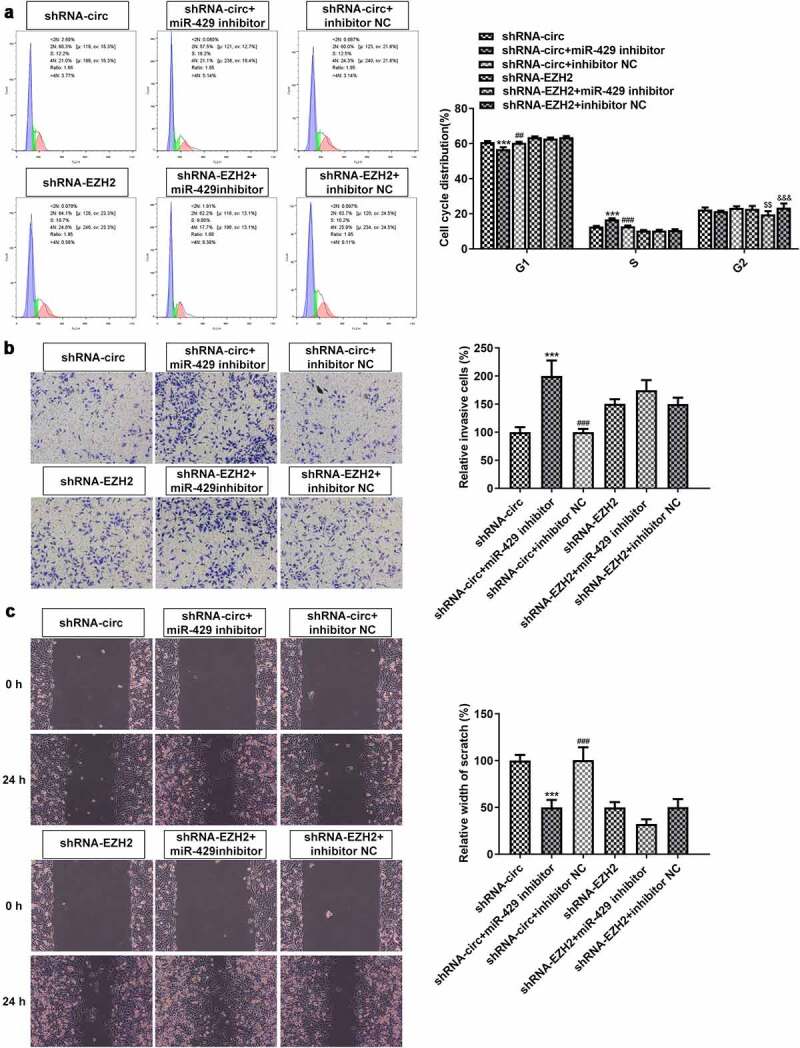
MiR-429 inhibitor affects cell cycle regulation and reverses the anti-invasion and anti-migration effects of circRNA_0082835 knockdown. (a) WM451 cell cycle distribution before and after transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups, detected by flow cytometry. (b) Relative invasive WM451 cells before and after transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups, detected by transwell. (c) Relative wound width of WM451 cells before and after transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups, detected by wound healing. ***P < 0.001 vs. shRNA-circ group. ##P < 0.01 and ###P < 0.001 vs. shRNA-circ +miR-429 inhibitor group. $$P < 0.01 vs. shRNA-EZH2 group. &&&P < 0.001 vs. shRNA-EZH2 + miR-429 inhibitor group
MiR-429 inhibitor reverses the effects of circRNA_0082835 knockdown on Wnt/β-catenin, cell cycle-related protein expressions and EMT in melanoma cells
The expressions of Wnt/β-catenin signaling-related proteins, cell cycle-related proteins, and EMT-related proteins were affected by the inhibitory effects of shRNA-circ transfection, which was reversed by miR-429 inhibitor. The expressions of Wnt5a, β-catenin, CyclinD1 and CyclinE1 were upregulated in WM451 cells (Figure 7(a)). Meanwhile, in shRNA-EZH2 groups, miR-429 inhibitor slightly increased Wnt5a expression and had no significant impact on the expressions of the rest. Furthermore, increased expression of CRKL and N-cad and decreased expression of E-cad were observed in shRNA-circ + miR-429 inhibitor group compared to the NC (Figure 7(b)). Generally speaking, shRNA-EZH2 groups were not significantly affected by miR-429 inhibitor, even though N-cad expression was elevated by co-transfection of miR-429 inhibitor and shRNA-EZH2. The results suggest that miR-429 inhibitor reverses the effects of circRNA_0082835 knockdown on Wnt/β-catenin, cell cycle-related protein expressions as well as EMT in melanoma cells.
Figure 7.
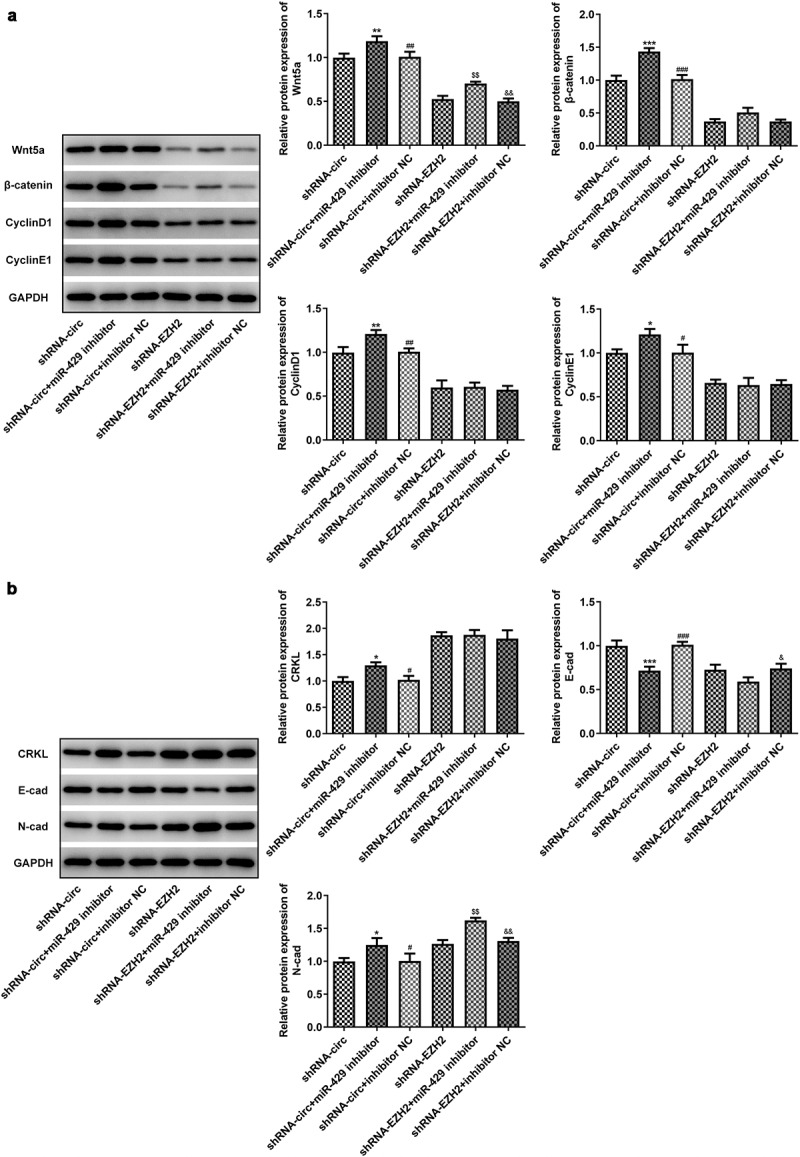
MiR-429 inhibitor reverses the effects of circRNA_0082835 knockdown on Wnt/β-catenin, cell cycle-related protein expressions and EMT in melanoma cells. (a) Relative expressions of Wnt/β-catenin signaling-related proteins and cell cycle-related proteins in WM451 cells before and after transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups, detected by western blot. (b) Relative expressions of and EMT-related proteins in WM451 cells before and after transfection of miR-429 inhibitor in shRNA-circ groups and shRNA-EZH2 groups, detected by western blot. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. shRNA-circ group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. shRNA-circ +miR-429 inhibitor group. $$P < 0.01 vs. shRNA-EZH2 group. &P < 0.05 and &&P < 0.01 vs. shRNA-EZH2 + miR-429 inhibitor group
Discussion
Primary melanoma is a type of skin cancer with high malignant level and high metastatic level, which can also arise on other sites such as the mucosal layer and the intracranial region [21]. This cancer is indeed once incurable and has now become a preventable and curable cancer, with the premise of early screening and diagnosis [22]. The initial form of melanoma can be just similar to a mole or is a mole (cancerized under long-term exposure to the sun), due to which early diagnosis tends to be easily overlooked by the patients [23,24]. While this disease can be largely prevented by daily sunscreen application, patients with diagnosed malignant melanoma will have to go through treatment including surgical intervention and immunotherapy, both being able to result in significant adverse symptoms [25,26]. Thus, the quest for a more effective and less harmful approach to melanoma treatment is still a long way to go.
In recent years, an explosion of studies has suggested that circular RNAs (circRNAs) point to the dysregulation of certain biological processes, which further reveals its close implication in cancer incidence and development [27,28]. A study conducted high-throughput sequencing on circRNA types and expressions in malignant melanoma tissues, where circRNA_0082835 overexpression was shown to be a significant one [8]. CircBase annotation of circRNA_0082835 shows its site on EZH2, whose expression exhibits no significant difference in cutaneous melanoma, according to GEPIA database.
Therefore, we carried out a series of experiments to identify the role of circRNA_0082835 and EZH2 in melanoma cells.
We transfected the shRNA plasmids of circRNA_0082835 and/or EZH2 into melanoma cells WM451 to knockdown circRNA_0082835 and EZH2 expressions. It was then found that the proliferation, invasion and migration levels of WM451 cells were conspicuously reduced after shRNA-circ or shRNA-EZH2 transfection and were reduced most prominently after co-transfection of shRNA-circ and shRNA-EZH2. CircRNA_0082835 and/or EZH2 knockdown also changed the distribution of different cell cycle phases in WM451 cells. The expressions of marker proteins that are related to Wnt/β-catenin signaling pathway and EMT were assayed as well in this study to explore more potentials of circRNA_0082835. Wnt/β-catenin is a classic pathway that has been extensively studied in various cellular progresses and is abnormally activated in many cancer types to exert a carcinogenic effect [29,30]. Inhibited Wnt/β-catenin expression was demonstrated in our study by circRNA_0082835 and/or EZH2 knockdown. Epithelial–mesenchymal transition (EMT) is another participant in cancer progression that contributes to cancer cell invasion and resistance to anti-cancer drugs [31]. We observed decreased EMT-related N-cad expression and increased E-cad expression in WM451 cells after transfection of shRNA-circ and/or shRNA-EZH2. Thus, it is validated here that inhibiting circRNA_0082835 and/or EZH2 may suppress melanoma cell growth by suppressing proliferation, invasion and migration, regulating cell cycle, blocking Wnt/β-catenin pathway and attenuating EMT.
According to ENCORI database, there exist several miRNAs that circRNA_0082835 may interact with, one of which is miRNA-429. This miRNA caught our attention for reports on its anti-tumor property and how it prevents melanoma cells from aggravated progression [20,32,33]. Therefore, we carried out more experiments to identify the relationship between miRNA-429 and circRNA_0082835 and the role of it in melanoma cells. We first found noticeably upregulated miRNA-429 expression after circRNA_0082835 inhibition and then verified the binding of miRNA-429 to circRNA_0082835 by performing dual-luciferase reporter assay. We further examined whether miRNA-429 inhibition affects the effect of circRNA_0082835 knockdown on melanoma cells. Our results showed that miR-429 inhibitor reversed the anti-proliferation, anti-migration and anti-invasion effects of shRNA-circ interference in melanoma cells. Furthermore, the expressions of Wnt/β-catenin signaling-related proteins and cell cycle-related proteins inhibited by circRNA_0082835 knockdown were further elevated after transfection of miR-429 inhibitor, which also reversed the effect of circRNA_0082835 knockdown on EMT-related protein expressions. Meanwhile, impact on the effects of EZH2 knockdown was scarce in shRNA-EZH2 + miR-429 inhibitor groups.
Conclusion
Our study is the first to describe the role of circRNA_0082835 in primary melanoma and lymphatic metastasis. It supports that circRNA_0082835 sponges miR-429 and that its overexpression inhibits the anti-tumor effect of miR-429 in melanoma cells. Data in our study may inspire a new approach to the therapeutic strategies for this malignant disease.
Acknowledgements
The authors thank all for participating in this study and laboratory assay. We also thank Han Chen MD, Ph.D. for her terrific advice, critical comments, and technical assistance with this manuscript.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure statement
The author(s) declare that they have no competing interests. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval
All procedures performed were in accordance with the declaration of the ethical standards of the institutional research committee and with the 1964 Helsinki 387 Declaration and its later amendments. The ethics committee has approved this study of the First Affiliated Hospital of Nanjing Medical University between December 2018 and December 2020 in our department (No. 2020-SR-563).
Consent to participate
All the patients provided informed consent upon this study.
Consent for publication
Written informed was obtained from the patient for the publication of this study and any accompanying images. A copy of written consent is available for review by the Editor-in-Chief of this journal.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Authors’ contributions
YTS collect the clinical data, complete the manuscript and translation. ZQH helped to draft the manuscript and participated in the experiments. BLL, CJL, JFL, and JLL supported collecting the data, participated in the experiments and follow-up patients. JT and GY designed and supervised this study also revised the manuscript for important intellectual content and translation. All authors read and approved the final manuscript.
References
- [1].Rastrelli M, Tropea S, Rossi CR, et al. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- [2].Kudchadkar RR, Lowe MC, Khan MK, et al. Metastatic melanoma. CA Cancer J Clin. 2020;70(2):78–85. [DOI] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [4].Pavri SN, Clune J, Ariyan S, et al. Malignant melanoma: beyond the basics. Plast Reconstr Surg. 2016;138(2):330e–40e. [DOI] [PubMed] [Google Scholar]
- [5].Abbas O, Miller DD, Bhawan J.. Cutaneous malignant melanoma: update on diagnostic and prognostic biomarkers. Am J Dermatopathol. 2014;36(5):363–379. [DOI] [PubMed] [Google Scholar]
- [6].Shang Q, Li Y, Wang H, et al. Altered expression profile of circular RNAs in conjunctival melanoma. Epigenomics. 2019;11(7):787–804. [DOI] [PubMed] [Google Scholar]
- [7].Wei CY, Zhu MX, Lu NH, et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol Cancer. 2020;19(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29. [DOI] [PubMed] [Google Scholar]
- [9].Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24(2):268–273. [DOI] [PubMed] [Google Scholar]
- [10].McHugh JB, Fullen DR, Ma L, et al. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol. 2007;34(8):597–600. [DOI] [PubMed] [Google Scholar]
- [11].Fan T, Jiang S, Chung N, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res. 2011;9(4):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tiffen JC, Gunatilake D, Gallagher SJ, et al. Targeting activating mutations of EZH2 leads to potent cell growth inhibition in human melanoma by derepression of tumor suppressor genes. Oncotarget. 2015;6(29):27023–27036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu D, Xia P, Diao D, et al. MiRNA-429 suppresses the growth of gastric cancer cells in vitro. J Biomed Res. 2012;26:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Su Z, Jiang G, Chen J, et al. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting AKT1 in renal cell carcinoma. Mol Clin Oncol. 2020;12:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peng G, Liao Y, Shen C. miRNA-429 inhibits astrocytoma proliferation and invasion by targeting BMI1. Pathol Oncol Res. 2017;23(2):369–376. [DOI] [PubMed] [Google Scholar]
- [16].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [17].Yang X, Jiang J, Zhang C, et al. Baicalein restrains proliferation, migration, and invasion of human malignant melanoma cells by down-regulating colon cancer associated transcript-1. Braz J Med Biol Res = Rev Bras Pesqui Med Biol. 2019;52(12):e8934. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [18].Chen XE, Chen P, Chen S, et al. Long non-coding RNA FENDRR inhibits migration and invasion of cutaneous malignant melanoma cells. Biosci Rep. 2020;40(3):BSR20191194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [19].Chen G, Xie Y. miR-495 inhibits proliferation, migration, and invasion and induces apoptosis via inhibiting PBX3 in melanoma cells. Onco Targets Ther. 2018;11:1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Z, Chen J, Wa Q, et al. Knockdown of circ_0084043 suppresses the development of human melanoma cells through miR-429/tribbles homolog 2 axis and Wnt/β-catenin pathway. Life Sci. 2020;243:117323. [DOI] [PubMed] [Google Scholar]
- [21].Kibbi N, Kluger H, Choi JN. Melanoma: clinical presentations. Cancer Treat Res. 2016;167:107–129. [DOI] [PubMed] [Google Scholar]
- [22].Testori AAE, Blankenstein SA, van Akkooi ACJ. Primary melanoma: from history to actual debates. Curr Oncol Rep. 2019;21(12):112. [DOI] [PubMed] [Google Scholar]
- [23].Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–358. [DOI] [PubMed] [Google Scholar]
- [24].Pampena R, Lai M, Piana S, et al. Nevus-associated melanoma: facts and controversies. G Ital Dermatol Venereol. 2020;155(1):65–75. [DOI] [PubMed] [Google Scholar]
- [25].Joyce D, Skitzki JJ. Surgical management of primary cutaneous melanoma. Surg Clin North Am. 2020;100(1):61–70. [DOI] [PubMed] [Google Scholar]
- [26].Namikawa K, Yamazaki N. Targeted therapy and immunotherapy for melanoma in Japan. Curr Treat Options Oncol. 2019;20(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- [29].Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shang S, Hua F, Hu ZW. The regulation of beta-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–33989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164:257–264. [DOI] [PubMed] [Google Scholar]
- [32].Huang D, Wang F, Wu W, et al. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting the AKT1 in melanoma. Cancer Biomark. 2019;26:63–68. [DOI] [PubMed] [Google Scholar]
- [33].Sheng H, Guo YH, Cao DS, et al. MiR-429-5p attenuates the migration and invasion of malignant melanoma by targeting LIMK1. Eur Rev Med Pharmacol Sci. 2020;24:2625–2631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


