ABSTRACT
The extracellular matrix (ECM) shows an essential effect during the occurrence and procession of human cancers. Type III collagen is a crucial component of ECM. Collagen Type III Alpha 1(COL3A1) is aberrantly expressed in a variety of cancers. Nevertheless, the role of COL3A1 in pan-cancer stays unidentified. In this study, we explored public databases, including Cancer Genome Atlas (TCGA), GTEx, GEPIA, cBioPortal, Oncommine, TIMER and GENEMANIA databases to identify the differential expression of COL3A1 in human cancer tissues and normal samples, followed by its prognostic value for patient survival. In addition, we explore the association between COL3A1 expression and immune infiltration. Further, we used the GeneMANIA database and Gene Set Enrichment Analysis (GSEA) to investigate Protein–Protein Interaction (PPI) and gene functional enrichment. Results show that COL3A1 expressed higher in tumor samples than in normal samples. Upregulation of COL3A1 is associated with a worse prognosis and a more advanced cancer stage. COL3A1 expression shows significant positive correlations with tumor-infiltrating immune cells (TIICs), including neutrophils, macrophages, CD8 + T cells, CD4 + T cells, dendritic cells, and B cells. Markers of TIICs demonstrated distinct patterns of COL3A1-related immune infiltration. COL3A1 expression was associated with ECM receptor interaction, regulation of actin cytoskeleton and focal adhesion pathways via GSEA analysis. In conclusion, COL3A1 may be a molecular biomarker for prognosis and immune infiltration in pan-cancer. It might act as a potential target for a new insight of human cancers management.
KEYWORDS: COL3A1, pan-cancer, tumor microenvironment, immune analysis, prognosis
graphical
Introduction
Worldwide, the incidence and mortality of cancer remain high and a serious public health problem [1]. In 2015, it was reported that close to 4,300,000 people were diagnosed with cancer, and about 2,800,000 people died from cancer in China [2]. And the number continues to grow. It is believed that cancer diagnosis and treatment have imposed a huge economic burden on countries all over the world [3]. Despite the tremendous advances in neoadjuvant chemoradiation therapy, targeted therapy, and immunotherapy in recent years, the overall survival for cancer remains unsatisfactory. Because of the limited understanding of tumorigenesis, especially for those which are indiscoverable for diagnosis or stubborn for present treatment. Pancreatic cancer, for example, currently has an overall five-year survival rate of only about 10% [4]. Therefore, there is an urgent demand to sequentially investigate the mechanism underlying tumorigenesis and identifying novel markers of early diagnosis, prognosis, and management.
In the past decade, immunotherapy has made breakthroughs in the treatment of a variety of cancers [5]. However, in cancers such as pancreatic cancer, the effectiveness of immunotherapy is frustrating [6]. Hence, it is crucial to find new targets to improve the efficacy of immunotherapy to optimize clinical treatment strategies. In recent years, the research on tumor microenvironment (TME) for advanced immunotherapy is in full swing [7–9]. TME plays an essential element in the development, metastasis, and chemotherapy resistance of cancer [7,8]. However, the underlying mechanism of interaction between TME and immune cells remains unclear. The extracellular matrix (ECM) is defined as the non-cellular component of the intercellular compartment that provides basic structural support to cells. Recent studies have found that ECM plays an important role in tumorigenesis [10]. Alterations in ECM can promote malignant phenotypes of cancer, such as invasion, metastasis [11,12].
Type III collagen, encoded by COL3A1, is an integral ECM protein that was discovered in 1971 [13]. Type III collagen has various important physiological functions. For example, mutations of COL3A1 can induce aneurysms, intestinal rupture [14]. It has been reported that the aberrant overexpression of COL3A1 occurs in several cancers [15–19]. For instance, COL3A1 overexpression is related to a worse survival and it might be a potential marker for early diagnosis in ovarian cancer [15]. Liu and his colleagues demonstrated that COL3A1 was upregulated in both primary and metastatic brain tumors [16]. And in rectal cancer, COL3A1 expression can be a predictor of response to neoadjuvant therapy [17]. The evidence above shows the potential of COL3A1 in pan-cancer research. However, little research focuses on the effect of COL3A1 in pan-cancer.
In the present study, we run an integrated bioinformatics analysis by mining data from diverse databases. We aim to comprehensively investigate the COL3A1 expression in cancer tissues and normal tissues. We are also intended to explore the prognostic role of COL3A1 in a variety of tumors and to identify the correlation between COL3A1 and immune cells. We assume to find that COL3A1 expression is elevated and acts as a cancer promoter in pan-cancer. In addition, by interacting with tumor-infiltrating immune cells, overexpression of COL3A1 is related to the poor survival of cancer patients. Therefore, COL3A1 could be considered as a promising biomarker of prognosis and improving immunotherapy.
Methods
Raw data collection and analysis
Gene expression data of cancers were collected from TCGA research network (https://portal.gdc.cancer.gov/), in which the method of acquisition and application complied with the guidelines and regulations. TCGA database contains clinical and genetic data of over 10,000 tumor samples across 33 types of human cancers, which could be acquired via UCSC Xena browser (https://xena.ucsc.edu/). The expression of normal samples was retrieved from Genotype-Tissue Expression (GTEx) (https://gtexportal.org/home/datasets). TIMER (https://cistrome.shinyapps.io/timer) was applied to analyze the COL3A1 expression in human cancers [20]. Further information on tumor types is shown in Supplementary Text S1.
COL3A1 gene alterations in human cancers
The cBioPortal (http://cbioportal.org) is a publicly accessible, online database for exploring cancer genomic datasets [21,22]. We used the quick search module to investigate the modification of COL3A1 gene in human cancers, including mutation, fusion, amplification, deep deletion, and multiple alterations.
Association of COL3A1 with survival and clinical stage
Overall survival (OS), disease-specific survival (DSS), and progression-free survival (PFI) were adopted to identify the association of COL3A1 with survival in various types of cancer. P-values and hazard ratio (HR) with 95% confidence interval (CI) were generated for Kaplan–Meier curves by log-rank and univariate Cox proportional hazards regression. Clinical information such as age and stage was included for multivariate Cox regression. GEPIA database (http://gepia.cancer-pku.cn) [23] is an online platform for dissecting the RNA sequencing expression data from the TCGA and the GTEx projects by using a standard processing approach. The stage survival plots module was applied to explore the interaction between COL3A1 expression and clinical stage, respectively.
Expression of COL3A1 in relation to immune cells
We explored the relationship between COL3A1 expression and immune cells through the TIMER database. The abundances of six immune infiltrate (CD4 + T cells, CD8 + T Cells, neutrophils, dendritic cells, macrophages, and B cells) are estimated by TIMER algorithm. And we next explored the purity of cancers via TIMER. We also evaluated the correlation between the expression of COL3A1 and gene markers of tumor-infiltrating immune cells (TIICs) in human cancers.
Tumor mutation burden (TMB) is becoming one novel biomarker for immune checkpoint blockade [24]. And the role of microsatellite instability (MSI) in tumor immunotherapy is also gradually being recognized [25]. In the present study, Sangerbox tools platform (http://www.sangerbox.com/tool) was applied to value the ESTIMATEScore for investigating TMB-MSI association. The related procedure on Sangerbox tools platform is based on R script of ESTIMATE algorithm, which theoretically could also be performed by R software.
Growing evidence suggests that the tumor immune microenvironment shows a crucial role in cancer development. Sangerbox tool was used to estimate the ratio of the immune-stromal component in TME to establish the relationship between the estimated proportions of immune and matrix with COL3A1 expression. Furthermore, the results are presented in the form of ImmuneScore, StromalScore, and ESTIMATEScore.
Protein–protein interaction network
We applied GeneMANIA online tool (http://www.genemania.org) [26,27] to build a protein–protein interaction (PPI) network of COL3A1. GeneMANIA is an interactive and publicly available online database for building PPI networks, generating hypotheses for gene function prediction, and detecting functionally similar genes. This network integration algorithm is characterized by the following bioinformatic approaches: site prediction, the interaction of physiology, co-expression, co-localization, analysis of gene enrichment and genetic interaction.
Gene set enrichment analysis
We applied GSEA to investigate the biological signaling pathways involved under the high and low expressions of COL3A1. We displayed the top three terms of KEGG pathways and the top four terms of HALLMARK analysis. Net enrichment score (NES), gene ratio, and P value were adopted to screen the KEGG pathway enrichment results. Pathways with FDR < 0.05, |NES|>1 and NOM p < 0.05 were considered to be significant [28].
Statistical analysis and Visualization
The comparisons of COL3A1 expression between tumor and normal tissues were tested by Wilcoxon rank sum test. The Spearman’s correlation analysis was adopted between COL3A1 level and immune cells scores in TIMER. All analytical methods above and R packages (survminer, survival, ggplot2, and clusterProfiler) were performed using R software version v4.0.3 (The R Foundation for Statistical Computing, 2020). p < 0.05 was defined as statistically significant.
Results
In this study, we intended to explore the association between COL3A1 and immunity and to provide its prognostic value as a promising biomarker in human cancers. Multiple databases were used to study genomic alterations, expression patterns, survival analysis of COL3A1 expression and its interaction with tumor immune infiltration in pan-cancer. Finally, we investigated Protein–Protein Interaction (PPI) and gene functional enrichment.
COL3A1 expression and genetic alterations in human cancers
We evaluated the expression of COL3A1 in various tumors and adjacent normal samples to investigate the relationship between COL3A1 and cancer. The data originally from TCGA and GTEx database were explored. The result indicates that compared to that in normal samples, mRNA expression of COL3A1 was significantly higher in BRCA, CHOL, COAD, ESCA, GBM, HNSC, KICH, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PRAD, READ, SKCM, STAD, TGCT, THCA, UCEC, and UCS cancer samples, suggesting that COL3A1 may act as an oncogene in the progression of a variety of cancers (Figure 1a). It is believed that genomic mutations are strongly related to tumorigenesis and tumor progression. Then, we performed comparative analysis to investigate genomic mutation of COL3A1 in cancers through cBioPortal. Genetic alteration profiling revealed that mutation of COL3A1 is one of the most common single alteration factors in SKCM, LUAC, UCEC, LUSC, and BLCA (Figure 1b). The expression data obtained through the TIMER database can be obtained from Supplementary Figure S1.
Figure 1.
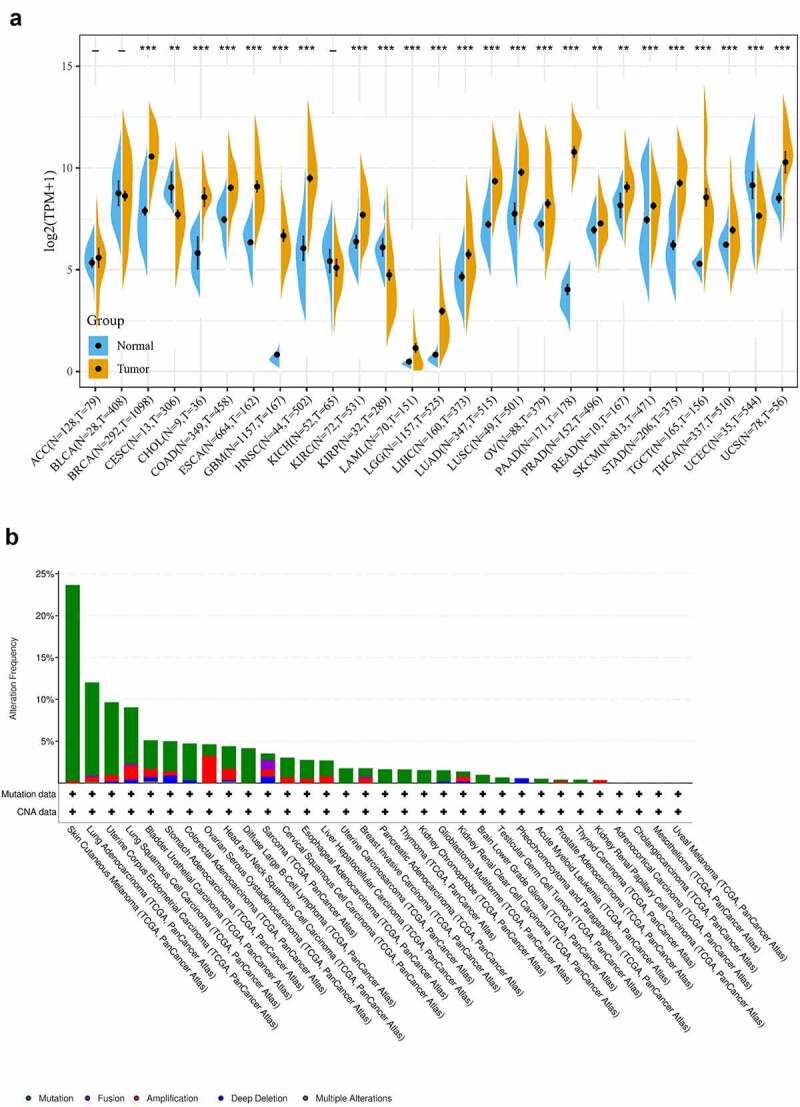
(a) COL3A1 expression levels in pan-cancer from TGCA and GTEx data. Tumor tissues are displayed with a yellow spindle and the normal tissues are displayed with a blue spindle, respectively. *, p < 0.05, **, p < 0.01, and ***, p < 0.001. (b) Type and rate of genetic alterations in COL3A1 in different cancers. We used the cBioPortal to explore the COL3A1 modifications in human cancers. These results are shown as histograms of the frequency of COL3A1 alterations in different cancers. Color images can be obtained from the online database
COL3A1 as a prognostic factor in human cancers
Next, the prognostic effect of COL3A1 for pan-cancer [OS, DSS, and PFI] was further evaluated. Upregulation of COL3A1 was significantly associated with a poor prognosis. In particular, in MESO and LGG, a high level of COL3A1 predicts a worse OS, DSS, and PFI (Figure 2a-F, Supplementary Table S1-6). These results indicate that COL3A1 is an important factor that affects cancer prognosis. Furthermore, stage plots in GEPIA database indicate that COL3A1 expression is strongly related to advanced tumor stage (P < 0.001, Figure 2g). The above results suggest that COL3A1 may be an important factor in the progression and metastasis of cancer.
Figure 2.
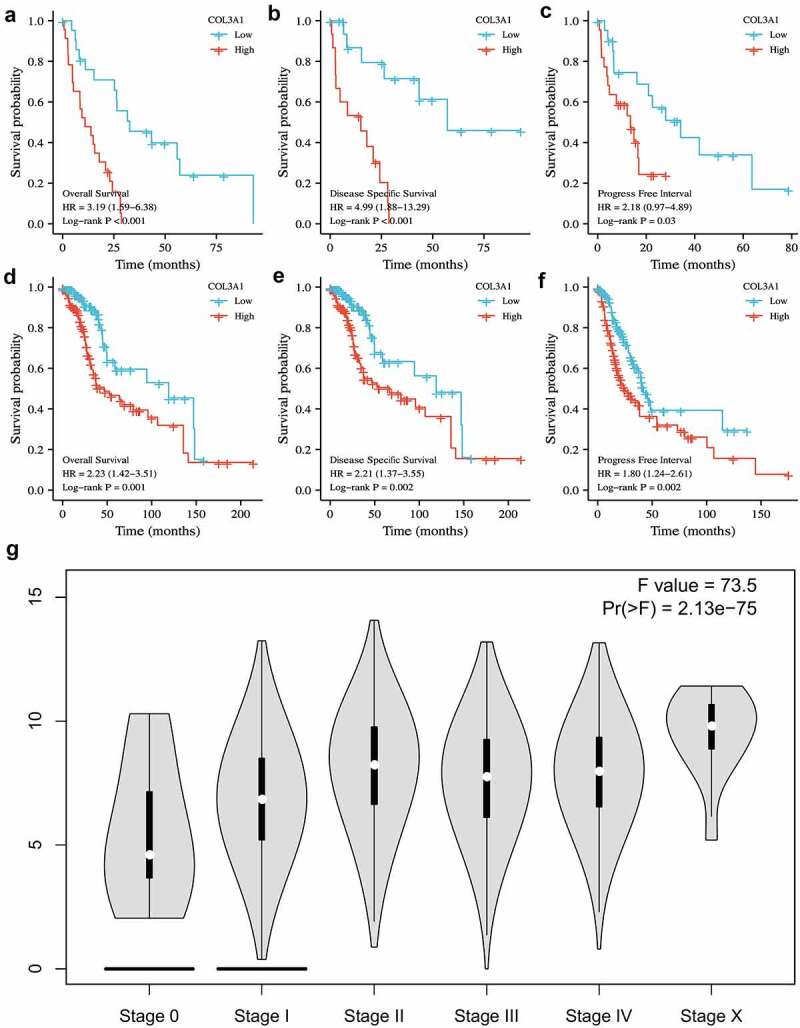
The role of COL3A1 in the prognosis of different cancers in TCGA database and the relationship with clinical stage in GEPIA online database. (a-c) OS, DSS, PFI in MESO and (d-f) in LGG. (g) COL3A1 level was positively associated with advanced cancer stage
COL3A1 expression and immune cells infiltration analysis
Despite the prior results confirming the prognostic value of COL3A1 in human tumors, its functions need to be further explored. TIICs are important components of the TME and play an essential effect in the regulation of cancer migration and metastasis in various cancers [29]. Tumor-infiltrating lymphocytes have an important survival predictive value in cancers [30]. Here, the association between COL3A1 expression and immune cells was investigated via TIMER database. The scores of B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells, and tumor purity were calculated separately. The results showed that COL3A1 expression was positively correlated with tumor purity in 23 kinds of cancer. Furthermore, the expression of COL3A1 was significantly associated with CD4 + T cells infiltration in 20 kinds of cancer, B cells in 15 types of cancer, CD8 + T cell in 20 kinds of cancer, macrophages in 27 types of cancer, dendritic cells in 24 cancer types and neutrophils in 24 kinds of cancer. And BRCA, BLCA, and COAD were the top three types of cancer associated with COL3A1 expression level in immune cells infiltration.
The COL3A1 expression was negatively associated with tumor purity (r = −0.53, P = 6.6e-29) and B cell (r = −0.13, P = 0.009), and was positively correlated with CD4 + T cell (r = 0.16, P = 0.0019), CD8 + T cell (r = 0.207, P = 6.53e- 05), macrophage (r = 0.493, P = 9.54e-24), neutrophil (r = 0.221, P = 1.98e-5) and dendritic cell (r = 0.208, P = 6.29e-5) in BLCA. The level of COL3A1 was negatively associated with tumor purity (r = – 0.300, P = 3.53e-22), and was positively correlated with CD8 + T cell (r = 0.307, P = 8.54e-23), CD4 + T cell (r = 0.238, P = 7.25e-14), macrophage (r = 0.506, P = 3.8e-65), neutrophil (r = 0.276, P = 4.56e-18), dendritic cell (r = 0.292, P = 3.6e-20) in BLCA. In COAD, the expression level of COL3A1 was notably correlated with CD8 + T cell (r = 0.241, P = 8.21e-7), CD4 + T cell (r = 0.481, P = 1.03e-24), macrophage (r = 0.607, P = 3.53e-42), neutrophil (r = 0.564, P = 5.25e-35) and dendritic cell (r = 0.576, P = 7.42e-37), and was negatively associated with tumor purity (r = −0.378, P = 3.08e-15). (Figure 3) Further information was available in the Supplementary Text S2.
Figure 3.
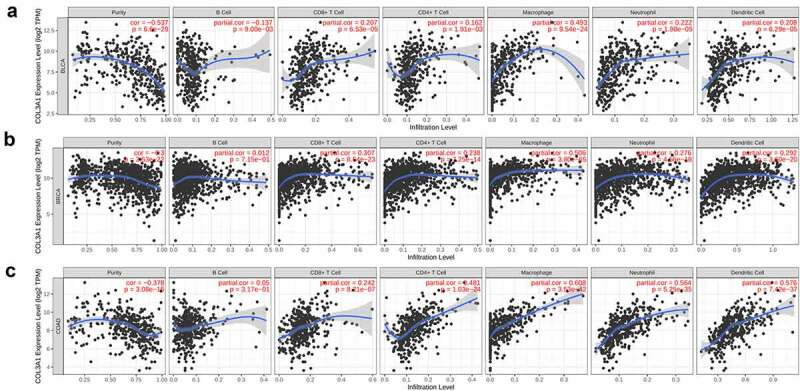
Association between COL3A1 expression and the level of immune infiltration in cancer. (a-c) Correlation between COL3A1 expression and immune cells infiltration in BLCA, BRCA and COAD
High ImmuneScore and StromalScore are positively correlated with the proportion of immune and substrate. ESTIMATEScore is a combined score that represents the combined ratio of the two components in TME. The expression of COL3A1 is most associated with StromalScore in BLCA (R = 0.878), BRCA (R = 0.776) and CESE (R = 0.707). The expression of COL3A1 is most associated with ImmuneScore in BLCA (R = 0.878), COAD (R = 0.864) and LGG (R = 0.497). The expression of COL3A1 is most associated with ESTIMATEScore in BLCA (R = 0.878), BRCA (R = 0.776) and COAD (R = 0.864). (Figure 4a) Correlations between other types of tumors and StromalScore, ImmuneScore and ESTIMATEScore can be acquired from Supplementary Figures S2-4.
Figure 4.
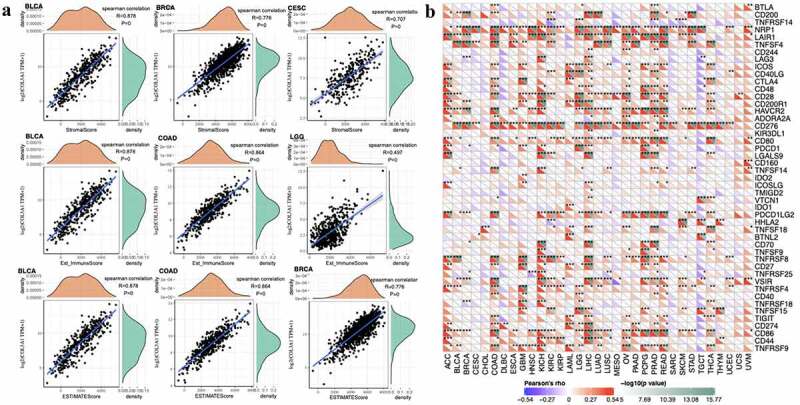
(a) Top 3 cancers related with ImmuneScore, StromalScore and ESTIMATEScore. (b) Association between COL3A1 expression and immune marker sets
The correlation between COL3A1 and Immune Marker Sets, TMB, MSI
Immune surveillance and immune escape are issues that must be faced in tumor immunotherapy. And these mechanisms may have important implications for cancer patient survival. Cancer cells could utilize immune checkpoint genes such as PD-1, CTLA-4 to evade immune surveillance. We examined the association between COL3A1 and immune checkpoint genes followed by further analysis, which is the correlation between COL3A1 and the degree of immune infiltration in human cancers. In LIHC, COL3A1 expression was positively associated with the expression of CD200, NRP1, LAIR1, CD244, LAG3, ICOS, CD40LG, CTLA4, CD48, CD28, CD200R1, HAVCR2, ADORA2A, CD276, CD80, PDCD1, LGALS9, ICOSLG, TMIGD2, VTCN1, PDCD1LG2, TNFSF18, CD70, TNFRSF8, CD27, VSIR, TNFSF15, TIGIT, CD274, CD86, CD44, TNFRSF9 (Figure 4b). High COL3A1 expression has a significantly important effect in the mediation of immune evasion based on the above findings. Moreover, COL3A1 was positively related with TMB in THYM, LGG, LAML, LUAD, OV, and SARC, as well as negatively associated with TMB in KIRP, GBM, ESCA, CESE, and BRCA (Figure 5a). COL3A1 was positively associated with MSI in COAD, TGCT, and was negatively correlated with HNSC, BRCA, SKCM, and LUSC (Figure 5b).
Figure 5.
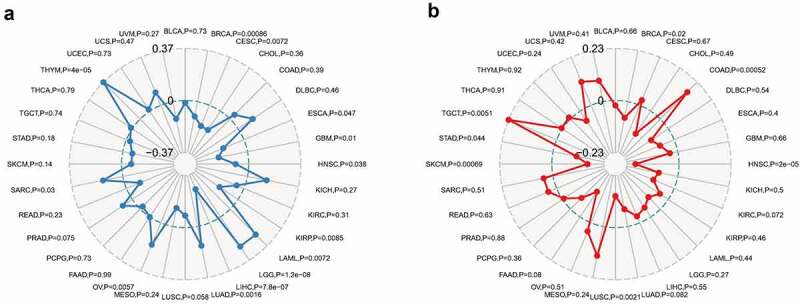
Association between COL3A1 expression and tumor mutation burden (TMB) and microsatellite instability (MSI) in human cancers. (a) Association of COL3A1 expression with TMB in human cancers. (b) Association of COL3A1 expression with MSI in human cancers. Spearman’s correlation coefficients and p values are shown in the radar map
PPI network of COL3A1 and GSEA
Subsequently, we constructed a PPI network for COL3A1 to investigate the underlying mechanisms that COL3A1 involved in the tumorigenesis of cancer using GeneMANIA online database (Figure 6). The result showed that COL3A1 had a significant correlation with DDR1, which participates in cancer metastasis and therapy resistance [31]. Furthermore, COL3A1 was predicted to be significant associated with SPARC. We used GSEA to identify functional enrichment of high COL3A1 expression and low COL3A1 expression (Figure 7). The results suggested that high expression of COL3A1 was mainly correlated with regulation of actin cytoskeleton, focal adhesion, and ECM receptor interaction via KEGG enrichment. However, there was no similar enrichment in HALLMARK terms.
Figure 6.
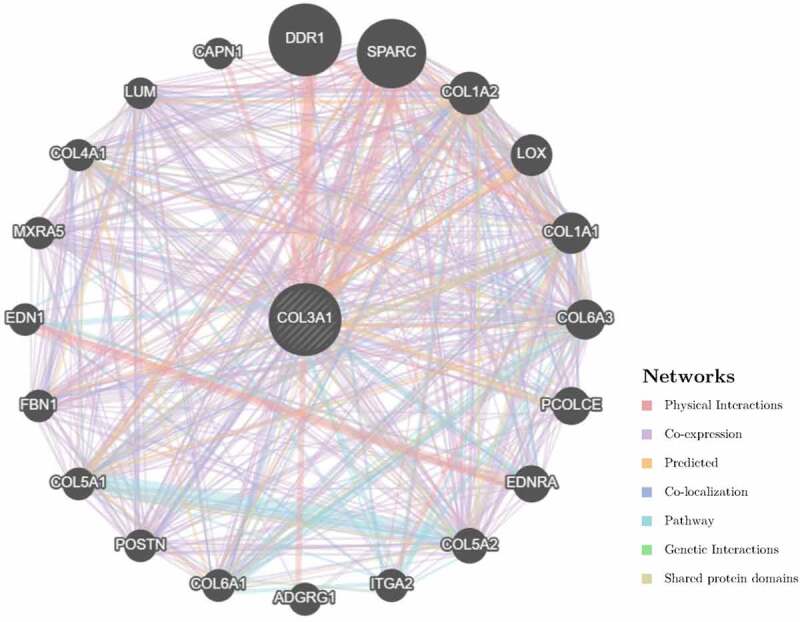
The COL3A1 PPI network was performed in GeneMANIA. COL3A1 has a strong association with DDR1 and SPARC in physical interactions. COL3A1 also correlates with other collagen genes. PPI, protein–protein interaction
Figure 7.
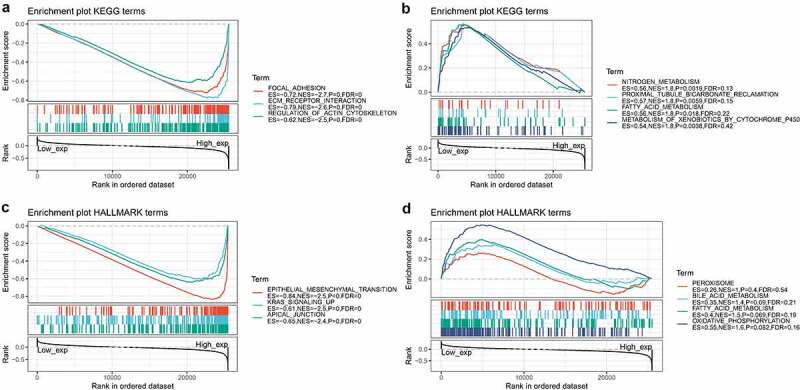
GSEA in high COL3A1 group and low COL3A1 group. (a) Enrichment analysis of KEGG pathway in the high COL3A1 expression group. (b) Enrichment analysis of KEGG pathway in the low COL3A1 expression group. (c) Pathway analysis of HALLMARK by the high COL3A1 group. (d) Enrichment in HALLMARK by samples with low COL3A1 expression
Discussion
COL3A1 is one of the members of the collagen family and is mainly expressed in extensible connective tissues including skin and vessels. The mutation of COL3A1 may cause vascular Ehlers–Danlos syndrome (vEDS), which is a life-threatening genetic disease [14]. COL3A1 transcription level was increased from adenoma to carcinoma [32], indicating involvement of COL3A1 in carcinogenesis. The COL3A1 was shown to promote malignant progression and drug resistance in a variety of cancers [33–35]. Despite the above findings, the functional roles and mechanism of COL3A1 in pan-cancer remains elucidative. The relationship between COL3A1 overexpression and clinical parameters or prognosis requires further explorations.
In the present study, we analyzed COL3A1 pan-cancer expression and investigated the relationship between the abnormal expression and prognosis. The findings indicated that the COL3A1 expression was upregulated in a variety of cancers and may hamper the survival of cancer patients. We found COL3A1 was significantly overexpressed in PAAD and BRCA, compared to the adjacent normal tissues. On the contrary, the COL3A1 expression was downregulated in UCEC and CESC. COL3A1 may serve different functions in different types of cancers. Mutations were considered as one of the most frequent alterations in human cancers. Mutations of COL3A1 were most common in SKCM, followed by LUSC and UCEC. Survival analysis revealed that overexpression of COL3A1 was associated with poor OS and DFS. High expression of COL3A1 was associated with poor survival in MESO, UVM, and LGG. In addition, high COL3A1 expression was correlated with advanced cancer stage, indicating that COL3A1 may promote tumor progression. The results show that high COL3A1 expression may hamper cancer patients’ survival.
Normally, the immune system could identify and eradicate malignant cells in TME. Nonetheless, tumors may adopt different strategies to evade monitoring by the immune system, leading to further growth and invasion. Cancer immunotherapy such as checkpoint inhibitors, cell therapy, and therapeutic antibodies may repair the normal anti-tumor effects. TIICs are of great value on the therapeutic effects in various types of tumors [36,37]. In the current study, common immune checkpoints genes were collected to explore the association between COL3A1 expression and checkpoint genes in a variety of tumor types. Overexpression of PD-L1 and PD-1 was associated with worse histological grade and prognosis in multiple cancers [38]. High expression of COL3A1 was associated with poor survival in a variety of cancers. These results showed that the COL3A1 expression was related to the degree of TIICs. MSI is associated with a higher risk of cancer and with clinicopathological features such as increased TMB and higher quality of tumor-infiltrating lymphocytes [39]. TMB can be used as a marker of response to immune checkpoint inhibitor therapy [24]. Thus, our work elucidates the underlying effect of COL3A1 in cancer immunology and its prognostic values for human cancers.
Furthermore, COL3A1 plays a vital role in cell adhesion, migration, proliferation, and differentiation via interactions with cell-surface receptor integrins [40]. In this study, pathway enrichment analysis demonstrated that high expression of COL3A1 was correlated with focal adhesion and ECM receptor interaction. The two pathways showed a crucial function in the migration and metastasis of cancers [41,42]. And they may be related to the sensitivity of the chemo- or radio-therapy [43]. PPI network revealed that COL3A1 showed a significant association with DDR1. Ambrogio et al. reported that DDR1 could be a therapeutic target for KRAS-driven non-small-cell lung cancer [44]. Another primary study revealed that in bladder cancer upregulation of DDR1 could promote invasion and migration via the EMT pathway [45]. Considering the above research results which can be used as the verification of our bioinformatics results, it may indicate that COL3A1 may act as a molecular biomarker mediating in several pathways of human cancer development.
Despite our exploration and integration of information from various databases, we acknowledge that our study has certain limitations. Firstly, although we obtained some meaningful insights into COL3A1 in human cancer from data mining, validation by conducting these findings to cellular or animal experiments would be in favor of clinical practicality. Further mechanism investigations will be of benefit in clarifying the effects of COL3A1 in vivo and in vitro. Second, post-translational modifications are important for regulating intracellular signal transduction and regulatory factors activity, though no related information about COL3A1 could be found in the databases. In addition, despite COL3A1 expression was found to correlate with immune and clinical survival in pan-cancer, whether COL3A1 is affecting clinical survival through the associated immune pathway still requires further investigation.
Taken together, the results from the present research illuminate the strong association between COL3A1 expression and the prognosis of various cancers. Since COL3A1 is overexpressed in multiple human cancers and associated with poorer survival, it could serve as a potential target for cancer management. Furthermore, our study yields key insights into the significant effect of COL3A1 in oncogenesis and metastasis, offering potential mechanisms by which COL3A1 expression may regulate tumor immunity and EMT in cancers. Future basic and clinical studies based on COL3A1 and tumor microenvironment may bring benefits to our clinical strategies for optimizing cancer treatment.
Conclusion
In conclusion, we herein report that COL3A1 is heterogeneously expressed in diverse cancers and its expression is correlated with the tumor immune microenvironment and pan-cancer prognosis. COL3A1 is promising to be further investigated as a biomarker for practical application in cancer prognosis and related cancer immunotherapy. Despite the relevance of COL3A1 in cancer studies, this seems to be the first pan-cancer analysis carried out to date.
Supplementary Material
Acknowledgements
We are grateful to TCGA and GTEx databases for their availability and contributors for providing their valuable datasets. We appreciate the aforementioned online databases and platforms for their selfless sharing and technical support.
Funding Statement
This research was financially sponsored by the project of National Natural Science Foundation of China [81903150];National Natural Science Foundation of China [82073238];
Disclosure statement
The authors do not have a conflict of interest.
Data availability statement
This study analyzed publicly available datasets. All raw data were available from TCGA(https://portal.gdc.cancer.gov/).
Author contributions
D-MH performed the design of this research. Z-HY and DC drafted the manuscript and analyzed the results. DC, L-YT, CX, and W-SD acquired and analyzed the data. C-LX, Z-HY, L-PY, and Y-ZP helped discuss the results. D-MH and Z-HY revised the manuscript. All authors contributed to this article and gave approval for the submission.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021. Feb 4;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016. Mar-Apr;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- [3].McGuire S. World cancer report 2014. geneva, switzerland: world health organization, international agency for research on cancer, who press, 2015. Adv Nutr. 2016. Mar 15;7(2):418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021. Jan;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- [5].Becht E, Giraldo NA, Dieu-Nosjean MC, et al. Cancer immune contexture and immunotherapy. Curr Opin Immunol. 2016Apr;39:7–13. [DOI] [PubMed] [Google Scholar]
- [6].Ho WJ, Jaffee EM, Zheng L.. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol. 2020. Sep;17(9):527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang S, Li Y, Xing C, et al. Tumor microenvironment in chemoresistance, metastasis and immunotherapy of pancreatic cancer. Am J Cancer Res. 2020. Jul 1;10(7):1937–1953. [PMC free article] [PubMed] [Google Scholar]
- [8].Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017. Feb 28;387: 61–68. [DOI] [PubMed] [Google Scholar]
- [9].Irvine DJ, Dane EL. Enhancing cancer immunotherapy with nanomedicine. Nat Rev Immunol. 2020. May;20(5):321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014. Dec;15(12):786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sharma P, Ng C, Jana A, et al. Aligned fibers direct collective cell migration to engineer closing and nonclosing wound gaps. Mol Biol Cell. 2017. Sep 15;28(19):2579–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang T, Hamilla S, Cam M, et al. Extracellular matrix stiffness and cell contractility control RNA localization to promote cell migration. Nat Commun. 2017. Oct 12;8(1):896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Miller EJ, Epstein EH Jr, Piez KA. Identification of three genetically distinct collagens by cyanogen bromide cleavage of insoluble human skin and cartilage collagen. Biochem Biophys Res Commun. 1971. Mar 19;42(6):1024–1029. [DOI] [PubMed] [Google Scholar]
- [14].Byers PH, Belmont J, Black J, et al. Diagnosis, natural history, and management in vascular Ehlers-Danlos syndrome. Am J Med Genet C Semin Med Genet. 2017. Mar;175(1):40–47. [DOI] [PubMed] [Google Scholar]
- [15].Engqvist H, Parris TZ, Kovács A, et al. Immunohistochemical validation of COL3A1, GPR158 and PITHD1 as prognostic biomarkers in early-stage ovarian carcinomas. BMC Cancer. 2019. Sep 18;19(1):928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu Y, Carson-Walter EB, Cooper A, et al. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J Neurooncol. 2010. Aug;99(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gonçalves-Ribeiro S, Sanz-Pamplona R, Vidal A, et al. Prediction of pathological response to neoadjuvant treatment in rectal cancer with a two-protein immunohistochemical score derived from stromal gene-profiling. Ann Oncol. 2017. Sep 1;28(9):2160–2168. [DOI] [PubMed] [Google Scholar]
- [18].Tian Y, Ke Y, Ma Y. High expression of stromal signatures correlated with macrophage infiltration, angiogenesis and poor prognosis in glioma microenvironment. PeerJ. 2020. May 20;8: e9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shi Y, Zheng C, Jin Y, et al. Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6a methylation-mediated COL3A1 up-Regulation. Front Oncol. 2020;10:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017. Nov 1;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012. May;2(5):401–404. Erratum in: Cancer Discov2012 Oct;2(10):960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013. Apr 2;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017. Jul;3(45(W1)):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019. Jan 1;30(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chang L, Chang M, Chang HM, et al. Biomarker for Cancer Immunotherapy. Appl Immunohistochem Mol Morphol. 2018. Feb;26(2):e15–e21. [DOI] [PubMed] [Google Scholar]
- [26].Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010. Jul;3(suppl_2):W214–20. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018. Jul;2(46(W1)):W60–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005. Oct 25;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015. Mar 31;16(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018. Jan;19(1):40–50. [DOI] [PubMed] [Google Scholar]
- [31].Gao Y, Zhou J, Li J. Discoidin domain receptors orchestrate cancer progression: a focus on cancer therapies. Cancer Sci. 2021. Mar;112(3):962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Paper W, Kroeber M, Heersink S, et al. Elevated amounts of myocilin in the aqueous humor of transgenic mice cause significant changes in ocular gene expression. Tamm ER Exp Eye Res. 2008. Sep;87(3):257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Januchowski R, Świerczewska M, Sterzyńska K, et al. Increased Expression of Several Collagen Genes is Associated with Drug Resistance in Ovarian Cancer Cell Lines. J Cancer. 2016. Jun 25;7(10):1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srour MK, Gao B, Dadmanesh F, et al. Gene expression comparison between primary triple-negative breast cancer and paired axillary and sentinel lymph node metastasis. Breast J. 2020. May;26(5):904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen Y, Pan Y, Ji Y, et al. Network analysis of differentially expressed smoking-associated mRNAs, lncRNAs and miRNAs reveals key regulators in smoking-associated lung cancer. Exp Ther Med. 2018. Dec;16(6):4991–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang Z, Guo X, Gao L, et al. The immune profile of pituitary adenomas and a novel immune classification for predicting immunotherapy responsiveness. J Clin Endocrinol Metab. 2020. Sep 1;105(9):e3207–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kikuchi T, Mimura K, Ashizawa M, et al. Characterization of tumor-infiltrating immune cells in relation to microbiota in colorectal cancers. Cancer Immunol Immunother. 2020. Jan;69(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Giraldo NA, Becht E, Pagès F, et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin Cancer Res. 2015. Jul 1;21(13):3031–3040. [DOI] [PubMed] [Google Scholar]
- [39].Cohen R, Hain E, Buhard O, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019. Apr 1;5(4):551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen Z, Soutto M, Rahman B, et al. Integrated expression analysis identifies transcription networks in mouse and human gastric neoplasia. Genes Chromosomes Cancer. 2017. Jul;56(7):535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev. 2011. Jul 18;63(8):610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yeh MH, Tzeng YJ, Fu TY, et al. Extracellular matrix-receptor interaction signaling genes associated with inferior breast cancer survival. Anticancer Res. 2018. Aug;38(8):4593–4605. [DOI] [PubMed] [Google Scholar]
- [43].Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015. Apr;(31):65–75. DOI: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- [44].Ambrogio C, Gómez-López G, Falcone M, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med. 2016. Mar;22(3):270–277. [DOI] [PubMed] [Google Scholar]
- [45].Xie X, He H, Zhang N, et al. Overexpression of DDR1 promotes migration, invasion, though EMT-related molecule expression and COL4A1/DDR1/MMP-2 signaling axis. Technol Cancer Res Treat. 2020Jan-Dec;19:1533033820973277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study analyzed publicly available datasets. All raw data were available from TCGA(https://portal.gdc.cancer.gov/).


