ABSTRACT
Extracellular vesicles (EVs) long RNAs (exLRs) have been shown to be indicators for the diagnosis and prognosis of colorectal cancer (CRC); however, the dynamic changes of exLRs during perioperative period and their cellular sources in CRC remains largely unknown. In this study, exLR sequencing (exLR-seq) was performed on plasma samples from three CRC patients at four time points (before surgery [T0], after extubation [T1], 1 day after surgery [T2], and 3 days after surgery [T3]). Bioinformatics approaches were used to investigate the profile and biofunctions of exLRs and their cellular sources. Greater than 12,000 mRNAs and 2,000 lncRNAs were reliably detected in each exLR-seq sample. Compared with T0, there were 110 differentially expressed genes (DEGs) in T1, 60 DEGs in T2, and 50 DEGs in T3. A total of 11 DEGs were found at all three time points and were related to membrane potential. In addition, compared to T0, 22 differentially expressed lncRNAs (DELRs) were found in T1, 19 DELRs in T2, and 38 DELRs in T3. Moreover, only three DELRs were detected at all three time points. Interestingly, EVs from CD8 + T cells, CD4+ memory T cells and NK cells decreased after surgery and the absolute quantity of EVs from immune cells were reduced as well. In summary, this study was the first to characterize the dynamic changes of exLRs during perioperative period and the cellular sources. These findings established the foundation for further studies involving the effects of these dynamically changed exLRs on CRC.
KEYWORDS: Extracellular vesicles, long RNA sequencing, perioperative period, colorectal cancer
Graphical Abstract
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide [1,2]. According to the 2020 global cancer statistics, there are approximately 1.9 million newly diagnosed CRC patients and 935,000 CRC-related mortalities, accounting for 10% of global cancer cases and 9.4% of cancer-related deaths [3]. Despite the rapid development of advanced chemotherapy, targeted therapy and immunotherapy for tumors during the past decades, surgical resection remains the major treatment for CRC [4–7]. However, previous studies have demonstrated that surgical trauma causes long-term oncologic outcome by facilitating metastasis and recurrence of tumors [8,9]. During the perioperative period, a variety of factors participate in the metastasis and recurrence of primary tumors, such as dissemination of tumor cells, drugs used in anesthetic and analgesic procedures, destruction of the extracellular matrix, release of vascular endothelial growth factor (VEGF), post-operative immunosuppression [10]. Therefore, there is an urgent need to identify biomarkers involved in the postoperative metastasis and recurrence of tumors and evaluate the dynamic changes and biofunctions of them.
Extracellular vesicles (EVs) are lipid bilayer-enclosed, nanosized endocytic vesicles which could be secreted by most cell types [11,12]. EVs can modify the function of recipient cells by various bioactive contents, such as proteins (enzymes, extracellular matrix proteins, transcription factors, and receptors), DNAs, RNAs, and lipids [13]. It has been shown that EV long RNAs (exLRs), including circular RNA (circRNA), long non-coding RNA (lncRNA), and messenger RNA (mRNA), are abundant in human plasma [14–18]. ExLRs are considered to be valuable and functional [18,19] and play an important role in the progression of tumor development [20–22]. For example, Nabet et al. reported that an unshielded exosome RNA (RN7SL1) could act as a damage-associated molecular pattern (DAMP) to activate the pattern recognition receptor (PRR) RIG-I, driving anti-viral signaling when transferred to recipient breast cancer cells via an exosome, and ultimately leads to tumor growth and therapy resistance [16]. The CD274 mRNA in plasma-derived EVs is related to the response to anti-PD-1 antibodies in melanoma and non-small cell lung cancer [17]. In CRC, circulating EV microRNAs and lncRNAs are considered to be potential diagnostic biomarkers and related to mitomycin resistance [23–27]. These results showed that ExLRs could act as the cell-to-cell mediators of human cancers and promoted the progression of cancers. However, the dynamic changes of ExLRs during perioperative period and their biofunctions in the progression of CRC remains largely unknown.
In the current study, we first evaluated the expression profile of exLRs during the perioperative period. ExLRs sequencing (exLR-seq) was performed on plasma samples collected from three CRC patients at four specific timepoints (before surgery [T0], after extubation [T1], 1 day after surgery [T2], and 3 days after surgery [T3]) to detect the effects of surgical stress on the exLR expression profile. In addition, the biofunctions of the changed exLRs were also investigated to assess the effects of exLRs on CRC progression. Moreover, tracking the cellular source of circulating EVs provides biological information about the origin and the functional states [28]. However, the cellular origin of dynamic changes in circulating EVs during the post-operative period has not been thoroughly investigated. In this study, we also explored and compared the distinct cellular origins from plasma EV samples during the post-operative period.
Our study first evaluated the dynamic changes of exLRs during the perioperative period and their biological functions to find out appropriate biomarkers involved in the postoperative metastasis and recurrence of CRC. We also track the cellular sources of those dynamically changed exLRs to figure out the origin and functional states of these exLRs.
Material and methods
Patient specimens and clinical assessments
The present study recruited three CRC patients, all of whom underwent right hemicolectomy at Fudan University Shanghai Cancer Center by the same surgeon. All the participants were histologically confirmed to have colorectal adenocarcinoma (stage II) by two pathologists. Tumor staging was determined according to the AJCC Cancer Staging Manual. None of the patients received any other forms of therapy on the time of enrollment. This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center and informed written consent was obtained from all patients.
Isolation of plasma from blood
Peripheral blood samples were collected from three CRC patients at four times (before surgery, after extubation, 1 day after surgery, and 3 days after surgery) in 10-mL EDTA-coated vacutainer tubes. Plasma was then separated by centrifugation at 3000 rpm (~800 × g) for 10 min at 25°C within 2 h after blood collection. Then, samples were centrifuged at 13,000 rpm (~16,000 × g) for 10 min at 4°C to remove debris. The plasma samples were then stored at – 80°C until use, according to a previous publication [18].
Isolation of EVs and EV RNA
For every patient, 1 mL of plasma was used. EVs were isolated by affinity-based binding to spin columns via an exoRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, melted plasma was mixed with binding buffer and added to the exoEasy membrane affinity spin column. Samples were subjected to ultrafiltration using an Amicon Ultra-0.5 Centrifugal Filter 10 kDa (Merck Millipore, Germany) to reduce the eluate volume to 50 µL and exchange the buffer with phosphate buffer saline (PBS). For transmission electron microscopy (TEM), the size distribution measurement, and western blotting, the EVs were eluted with 400 μL of XE elution buffer, according to previous publications [29]. For TEM, ultrathin sections (100 nm) were cut using a LeicaUC6 ultra-microtome and post-stained with uranyl acetate for 10 min and with lead citrate for 5 min at room temperature before observation in a FEI Tecnai T20 TEM, operated at 120 kV. For EV RNA isolation, EVs were lysed on the column using QIAzol (Qiagen), and total RNA was then eluted and purified, as per other publications [30].
ExLR-seq analysis
The strategy for exLR-seq analysis includes plasma preparation, isolation of EV and EV RNAs, RNA-seq library construction, sequencing, and data analysis. Briefly, to remove DNA, total EV RNA isolated from 1 mL of plasma was treated with DNase I (NEB; Ipswich, Massachusetts, USA). RNA-seq libraries were generated using SMART technology (Clontech). ExLR-seq was performed on an Illumina sequencing platform (San Diego, California, USA) with 150 bp paired-end run metrics. Gene expression levels were calculated in transcripts per kilobase
million (TPM). Annotations of mRNAs and lncRNAs were retrieved from the GENCODE database, according to previous publications [20,31].
Identification of differentially expressed mRNAs and lncRNAs
Transcriptional profiles of EVs from the plasma of three CRC patients were evaluated during the perioperative period (before surgery [T0], after extubation [T1], 1 day after surgery [T2], and 3 days after surgery [T3]). Significantly differentially expressed genes (DEGs) and differentially-expressed lncRNAs (DELRs) were identified using the Limma R package (version 3.6.3) with a |logFC| > 1 and p < 0.1.
Functional enrichment analysis of DEGs and DELRs
The intersective hub genes during the perioperative period were selected for further analyses using a Venn diagram. A protein–protein interaction (PPI) network of hub genes was constructed using GeneMANIA (http://genemania.org/). Biological processes, cellular components, and molecular function of gene ontology (GO) functional analysis and Kyoto encyclopedia genes and genomes (KEGG) pathway were predicted using the Web-based Gene seT AnaLysis Toolkit (WebGestalt [http://www.webgestalt.org/]) and visualized using R software. In the functional analyses, the input parameters including gene names of all the DEGs, gene ontology (GO) and Kyoto encyclopedia genes and genomes (KEGG) pathways and the results can change depending on the query/input information.
Western blot analysis
Fifty milligrams of exosomes were extracted using 2X SDS lysis buffer, separated by 4%–12% SDS-PAGE, transferred to a PVDF membrane, blocked with 5% BSA in TBST, and probed with specific primary antibodies against Calnexin (1:1000 dilution; Abcam, Cambridge, UK), CD63 (1:1000 dilution; Abcam), and TSG101 (1:1500 dilution; Abcam). β-actin (1:5000 dilution; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) was used as a loading control. The chemiluminescent signals were detected with a chemiluminescence imaging system and quantified by Image J software (v1.37).
Data and statistical analyses
All statistical analyses were two-sided. A |logFC| > 1 and p < 0.05 were considered statistically significant. The following R software packages were used in this study: e1071, glmnet, varSelRF, pROC, and caret. Nonparametric T test was used in Limma Package and the comparison of immune cells producing EVs before and after surgery. Hypergeometric test was used in the functional enrichment analysis. Spearman’s rank correlation coefficient was utilized in the correlation analysis of different types of immune cells.
Results
Firstly, we evaluated the dynamic changes of mRNAs and lncRNAs during the perioperative period via ExLR-seq. Then, we detected the biological functions of these DEGs to find out appropriate biomarkers involved in the postoperative metastasis and recurrence of CRC. Finally, we track the cellular sources of those dynamically changed exLRs to figure out the origin and functional states of these exLRs.
Isolation and identification of EVs
To evaluate the integrity of isolated EVs from plasma, EV morphology was inspected by electron microscopy. As shown in Figure 1, the isolated vesicles in plasma were cup-shaped, rounded, and double membrane-bound vesicle-like (Figure 1a). Furthermore, flow cytometry exhibited a heterogeneous population of spherical nanoparticles, with abundant peaks ranging from 50 to 200 nm (Figure 1b). In addition, western blot analysis revealed characteristic exosomal marker (CD63 and TSG101) expression in isolated vesicles, but not in peripheral blood mononuclear cells (PBMCs). Calnexin, which is an intracellularly enriched protein in PBMCs and often used as a negative-control protein marker for EV identification, was detected in PBMCs, but not in isolated vesicles (Figure 1c). These data indicated that the isolated vesicles were composed mostly of exosomes.
Figure 1.
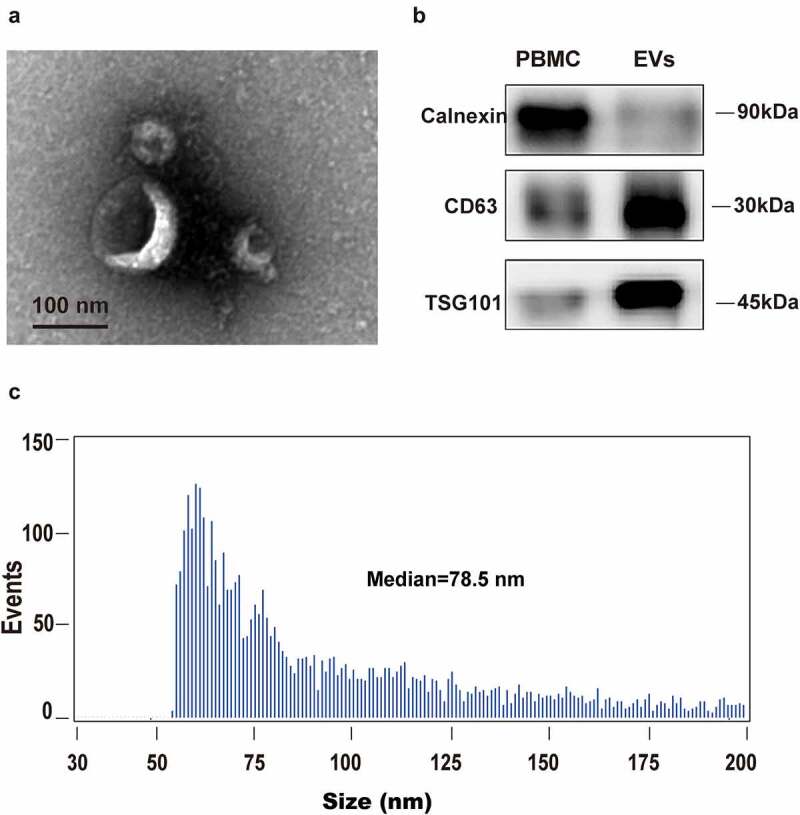
Human blood EVs confirmation. EVs were isolated and purified from plasma using membrane affinity spin columns. (a) Electron microscopy image of isolated vesicles. (b) Size distribution measurements of isolated vesicles. (c) Western blots of calnexin, which can be detected in PBMCs, but not in isolated vesicles, was used as a control. EV markers TSG101 and CD63 in isolated vesicles were detected in EVs, but not in PBMCs
Dynamic changes of mRNAs in EVs before and after surgery
ExLR-seq was conducted using plasma samples from three CRC patients at four timepoints. Approximately 12,924 mRNAs were reliably detected in each sample. Dynamic changes were observed in the expression profiles of mRNAs in EVs during the postoperative period. Briefly, as shown in Figures 2A, 110 DEGs in T1 were compared with those in T0. In addition, compared with T0, there were 60 DEGs in T2 (Figure 2b) and 50 DEGs in T3 (Figure 2c). Taking the intersection of DEGs in the three groups revealed that a total of 11 DEGs (hub DEGs), including DGKI, GRB14, KIAA1549, WT1, ACKR4, PLXNB3, KCNH8, TCTEX1D1, ILDR2, DYTN and CHRNA2, changed at all three timepoints compared to T0 (Figure 2d). Although it did not reach significance owing to the sample size, there existed an obvious trend (logFC| > 1 and p < 0.1) and details are shown in table 1 (Table 1, Fig S1).
Figure 2.
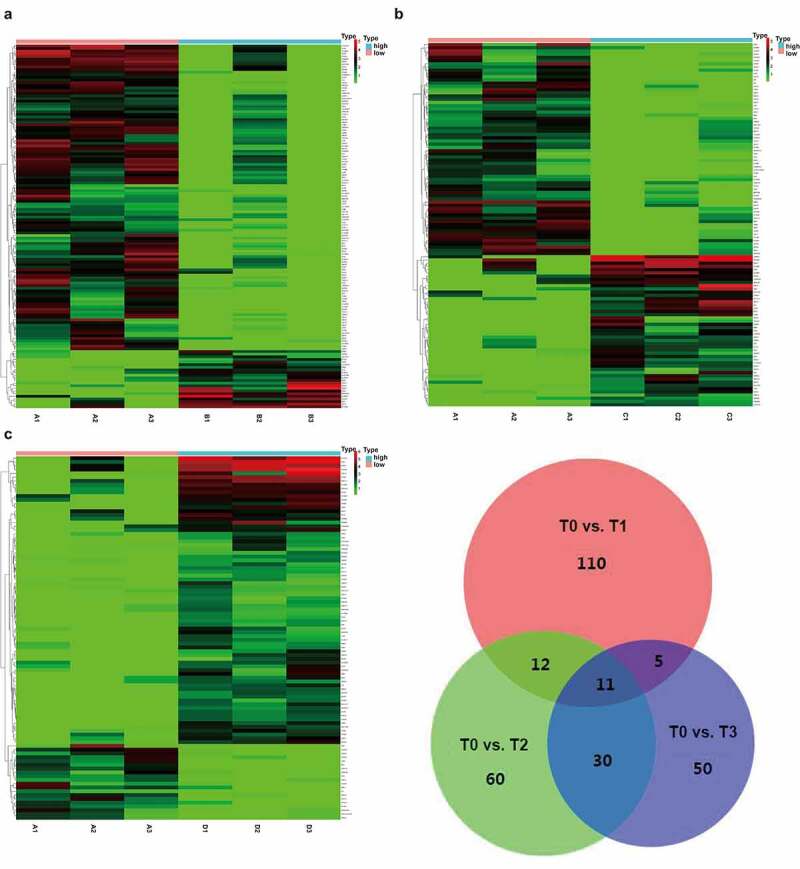
Comprehensive mRNAs in extracellular vesicles and functional annotations before and after surgery. (a-c) Significant DEGs in extracellular vesicles were screened and identified using the ‘Limma’ R package between samples before surgery and after extubation (Fig. A), or 1 day after surgery (Fig. B), or 3 days after surgery (Fig. C). (d) A total of 11 common DEGs were obtained in extracellular vesicles before and different time points after surgery using a Venn diagram
Functional annotations of mRNAs in EVs
Next, functional annotations of 11 altered mRNAs in EVs were determined. With respect to the biological process, nine of the 11 DEGs were involved in the biological regulation process and seven DEGs participated in the localization process and stimulus response. For the cellular component, nine DEGs were membrane components. In addition, molecular function analysis showed that six DEGs were protein-binding mRNAs (Figure 3a). To further determine the interactions among the 11 DEGs, the protein–protein interaction network was used (Figure 3b). Moreover, GO function analysis showed that the 11 hub DEGs most significantly involved in the regulation of membrane potential, cell chemotaxis, chemical synaptic transmission, and mesenchymal-epithelial transition (Figure 3c). KEGG pathway analysis revealed that the hub DEGs were enriched for some pathways, such as nicotinic acetylcholine receptors activities, Tie2 signaling, other semaphoring interactions, and choline and glycerolipid metabolism (Figure 3d).
Figure 3.
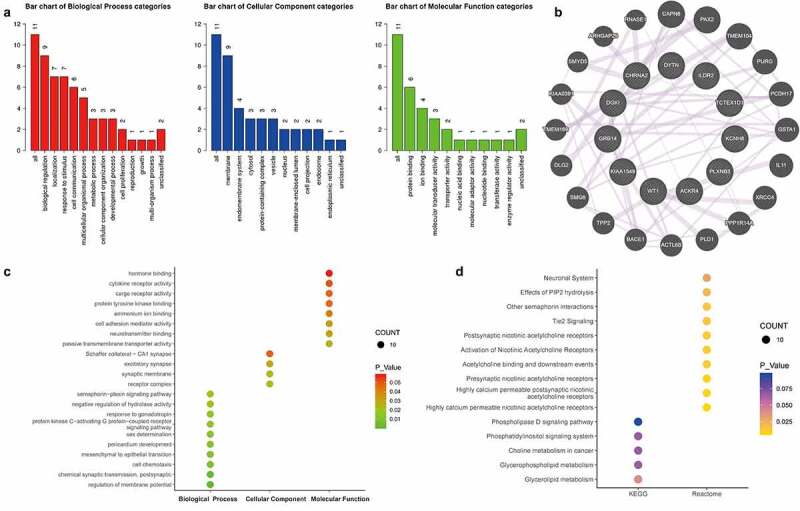
The functional annotations of mRNAs in EVs. (a) Biological processes, cellular components, and molecular function analysis from GO items of 11 hub genes were evaluated. (b) The protein–protein interaction network was used, showing direct interactions and potential associations between proteins. (c) The 11 hub genes most significantly involved in changed GO functions. (d) Significantly altered KEGG and Reactome pathways were predicted
Dynamic changes of lncRNAs in EVs before and after surgery
Except for mRNAs, approximately 2200 lncRNAs were detected in each sample. Dynamic changes were also observed in the expression profiles of lncRNAs in EVs during the post-operative period. As shown in Figure 4a, we identified 22 DELRs between T0 and T1. In addition, 19 DELRs were detected between T0 and T2 (Figure 4b) and 38 DELRs between T0 and T3 (Figure 4c). Furthermore, we found that only three DELRs (C15orf54, RP11-446N19.1, and RP11-87H9.4) changed at all three timepoints (Figure 4d). Although it did not reach significance owing to the sample size, there existed an obvious trend (logFC| > 1 and p < 0.1) and details are shown in table 2 (Table 2, Fig S2).
Figure 4.
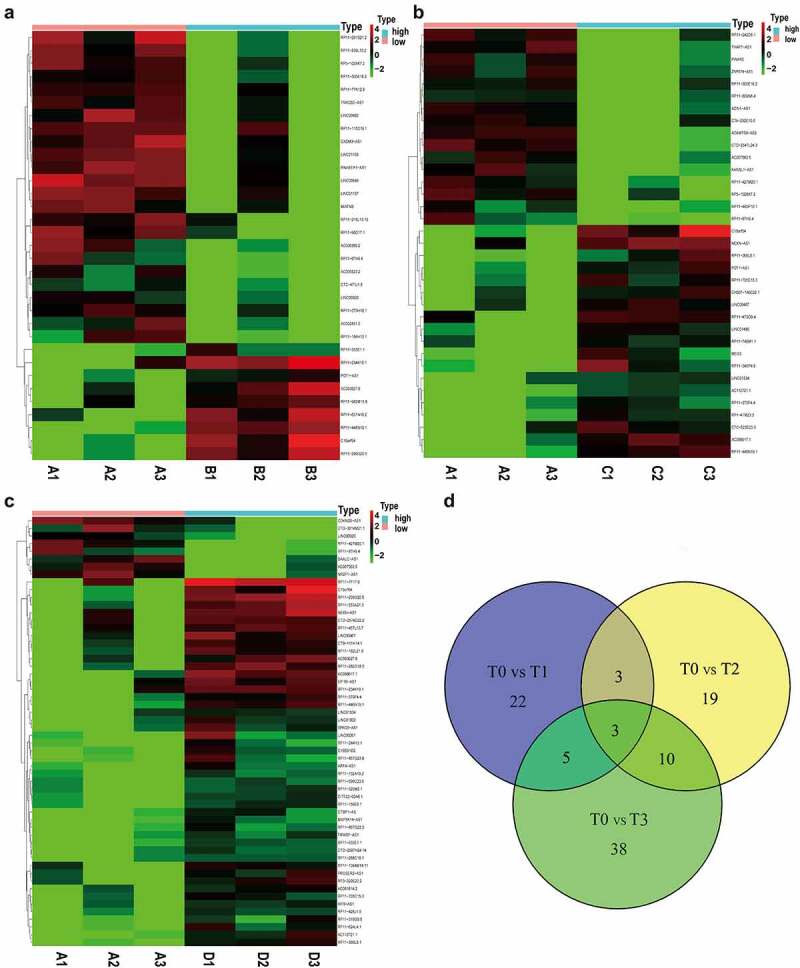
Comprehensive lncRNAs in EVs before and after surgery. (a-c) Significant lncRNAs in EVs were also screened and identified using the ‘Limma’ R package between samples before surgery and after extubation (Fig. A), or 1 day after surgery (Fig. B), or 3 days after surgery (Fig. C). (d) A total of 3 common DEGs were obtained in EVs before and different time points after surgery using a Venn diagram
Cell source analysis of EVs
Because blood EVs are derived from a variety of tissues, the xCell tool (http://xcell.ucsf.edu) was used to characterize the proportions of cell types derived from EVs. xCell is a webtool that performs cell-type enrichment analysis from gene expression data for 64 immune and stroma cell types. xCell is a gene signature-based method learned from thousands of pure cell types from various sources. xCell applies a novel technique for reducing associations between closely related cell types. We identified 67 immune and stroma cell types and evaluated the correlation between immune and stroma cells (Figure 5a). Dynamic changes of the origination of EVs during the postoperative period were also investigated (Figure 5b). Consensus clustering were utilized to explore potential clusters and Consensus clustering (or aggregated clustering) is a robust approach that relies on multiple iterations of the chosen clustering method on sub-samples of the dataset. Specifically, EVs derived from platelets were gradually reduced after surgery. Clinically, the immunosuppressive microenvironment caused by surgery can lead to tumor metastases and recurrence and the role of EVs in immune regulation has been intensively studied. Following infection, the release of EVs carrying immunomodulatory molecules by various immune cells can influence primary and secondary immune responses [32,33]. Therefore, we further investigated the changes in immune cells derived from EVs before and after surgery. As shown in Figure 5c, the total number of EVs derived from immune cells decreased after surgery. EVs derived from CD8 + T, CD4+ memory T, and natural killer (NK) cells decreased as well (Figure 5c).
Figure 5.
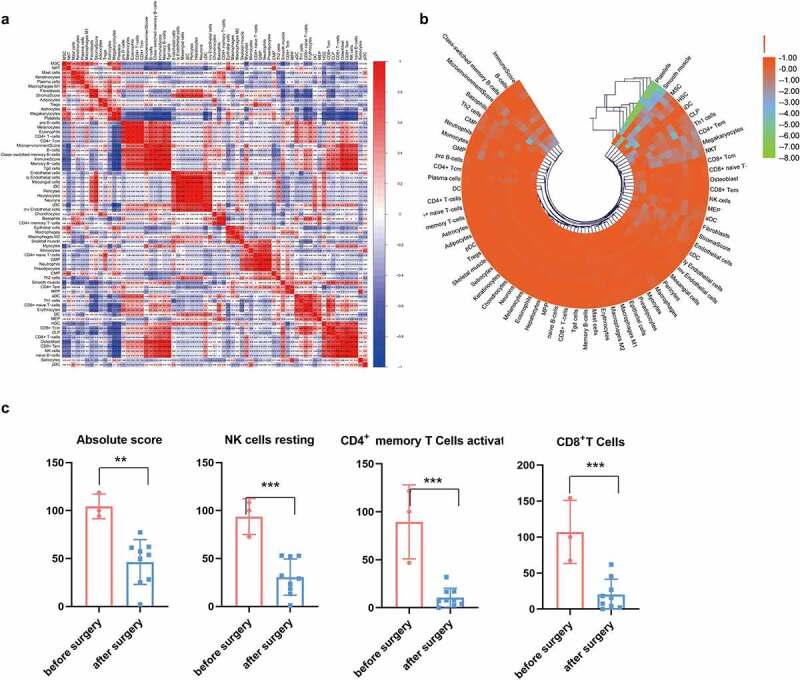
Cell source analysis of EVs. (a) The correlation of immune and stroma cell expression derived from EVs. (b) Dynamic changes of cells derived from EVs before and after surgery. (c) Comparison of immune cells producing EVs before and after surgery. * p < 0.05, ** p < 0.01, *** p < 0.01 vs before surgery
Discussion
Recently, EVs, as a molecular approach to analyze tumor diagnosis and progression, has attracted more and more attention [31,34]. While studies of EVs on cancer development have expanded rapidly, few studies have investigated the influence of surgical stress on exLRs and the cellular origins of exLRs. Our research showed that there are a large variety of exLRs in human blood. Given the high abundance and heterogeneity of blood exLRs, we intended to find the differences of expression profile and function in exLRs before and after surgery and their influence on CRC progression and prognosis.
As the size similarity between exosomes and other EVs, including ectosomes and MVs, has deeply hindered the development of isolation processes. It is critical to pay attention to the isolation of plasma, EV purification and preparation of EV RNA in order to obtain reliable exLR-seq data. In this study EVs were isolated via density gradient centrifugation (DG), which was intensively used in plasma and cell culture supernatants. Although the process is time-consuming and highly instrument-dependent, DG is easy to perform and yields exosomes with higher purity and size uniformity. The proportion of exosomes in the isolated EVs was confirmed from the aspect of morphology, particle size, and characteristic exosomal markers (CD63 and TSG101). Rapid development of EV isolation technology makes it possible to use EVs and exLRs for further cancer-related studies.
In this study, surgical stress caused significant changes to the expression profile of exLRs. 11 mRNAs, including DGKI, GRB14, KIAA1549, WT1, ACKR4, PLXNB3, KCNH8, TCTEX1D1, ILDR2, DYTN and CHRNA2, have dynamically changed at all three timepoints compared with T0. Several of these exLRs were associated with CRC progression. For example, WT1 expression in CRC primary tumors could be a novel independent marker for prognosis and tumor progression [35,36]. GRB14, which belongs to a small family of adapter proteins, could encode a growth factor receptor-binding protein that interacts with insulin receptors and insulin-like growth factor receptors (IGF-Rs) [37]. IGF-1 R expression is associated with tumor progression and poor prognosis in several cancer types, including gastrointestinal malignancies [38]. KIAA1549 belongs to the UPF0606 family and is related to oncogenic MAPK signaling [39]. ACKR4, which is a receptor for C–C type chemokines, has been shown to bind T cells and dendritic cell-activated chemokines and plays a significant role in controlling the migration of immune and cancer cells [40]. These findings indicated that mRNAs dynamically changed during the peri-operative period may play a role in the development of CRC. However, the exact effect of these mRNAs and the underlying mechanisms require further investigation.
Our data also showed that lncRNAs are enriched in EVs; however, only three common lncRNAs were detected at three time points compared with T0. This finding may due to our small sample size. In a corollary study, we will expand our samples to further explore the exact effect of surgery on EVs and the underlying mechanism.
Innate immune cells, such as NK cells, neutrophils, mast cells (MCs), macrophages, eosinophils and adaptive immune cells, including DCs, T cells, and B cells, derived from exosomes can directly interact with cancer cells and uptake by tumor cells inducing different types of immune responses [41,42]. Accumulating evidence has shown that EVs derived from immune cells can promote pro-tumor and anti-tumor immunity, which suggests a complex relationship between the immune cell-derived EVs and the immune system [43–46]. NK cells that were previously exposed to neuroblastoma (NB) can secrete exosomes containing NK cell receptors, such as CD56, KIR2DL2, and NKG2D receptors, which can subsequently stimulate normal NK cells, generating greater and more efficient cytotoxicity against NB tumor cells [45]. CD8 + T cell-derived exosomes with membrane expression of Fas ligand (FasL) can promote invasion and metastasis of Fas+ tumor cells through matrix metalloproteinase-9 (MMP-9)-mediated degradation of extracellular matrix proteins [43]. In our study, we found that total EVs derived from immune cells, especially from CD8 + T cells, CD4+ memory T cells and NK cells, decreased after surgery. Further studies are warranted to fully characterize EVs derived from immune cells, and to learn how to precisely engineer exosomes for therapeutic antitumor treatment.
Significant DELRs between T0 and T1
| LncRNA | Mean(T0) | Mean(T1) | logFC | pValue | FDR |
|---|---|---|---|---|---|
| AC002451.3 | 2.21713388 | 0.114178067 | −4.27934 | 0.0765225 | 0.683824 |
| AC005523.2 | 1.52729331 | 0.0228,35613 | −6.06355 | 0.0765225 | 0.683824 |
| AC006369.2 | 3.413328513 | 0.091342467 | −5.22375 | 0.0765225 | 0.683824 |
| AC093627.8 | 0.325408993 | 6.553166567 | 4.331866 | 0.0765225 | 0.683824 |
| C15orf54 | 0.081352247 | 11.77879127 | 7.177794 | 0.0765225 | 0.683824 |
| CADM3-AS1 | 6.056916993 | 0.548054667 | −3.46619 | 0.0765225 | 0.683824 |
| CTC-471J1.8 | 0.74328401 | 0.091342467 | −3.02456 | 0.0765225 | 0.683824 |
| LINC00649 | 8.314309343 | 0.548054667 | −3.9232 | 0.0765225 | 0.683824 |
| LINC00662 | 5.140611187 | 0.411041 | −3.64459 | 0.0765225 | 0.683824 |
| LINC00920 | 1.453565963 | 0.1370137 | −3.4072 | 0.0765225 | 0.683824 |
| LINC01133 | 5.082520677 | 0.479548 | −3.4058 | 0.0765225 | 0.683824 |
| LINC01137 | 5.426904483 | 0.662232667 | −3.03472 | 0.0765225 | 0.683824 |
| MIATNB | 4.27227625 | 0.433876667 | −3.29965 | 0.0765225 | 0.683824 |
| POT1-AS1 | 0.10169031 | 1.446493733 | 3.830306 | 0.0765225 | 0.683824 |
| RNASEH1-AS1 | 5.546831097 | 0.502383667 | −3.4648 | 0.0765225 | 0.683824 |
| RP11-115D19.1 | 4.29410445 | 1.141780667 | −1.91107 | 0.0765225 | 0.683824 |
| RP11-164H13.1 | 2.22593248 | 0.0228,35613 | −6.60698 | 0.0765225 | 0.683824 |
| RP11-216 L13.19 | 3.491072927 | 0.416797333 | −3.06625 | 0.0765225 | 0.683824 |
| RP11-234K19.1 | 0.750527333 | 11.01633047 | 3.875595 | 0.0765225 | 0.683824 |
| RP11-291B21.2 | 7.87761297 | 0.1370137 | −5.84537 | 0.0765225 | 0.683824 |
| RP11-299G20.5 | 0.081352247 | 6.911403467 | 6.408653 | 0.0765225 | 0.683824 |
| RP11-303E16.2 | 2.226079677 | 0.1826849 | −3.60708 | 0.0765225 | 0.683824 |
| RP11-333E1.1 | 0.041695967 | 1.0917443 | 4.710583 | 0.0765225 | 0.683824 |
| RP11-379H18.1 | 2.29573854 | 0.342534333 | −2.74464 | 0.0765225 | 0.683824 |
| RP11-446N19.1 | 0.062543933 | 5.9012772 | 6.560013 | 0.0765225 | 0.683824 |
| RP11-539L10.2 | 4.57341401 | 0.1370137 | −5.06088 | 0.0765225 | 0.683824 |
| RP11-631N16.2 | 0.245482033 | 6.750055167 | 4.78121 | 0.0765225 | 0.683824 |
| RP11-77K12.9 | 2.799639297 | 0.525219 | −2.41425 | 0.0765225 | 0.683824 |
| RP11-87H9.4 | 2.333000947 | 0.0228,35613 | −6.67476 | 0.0765225 | 0.683824 |
| RP11-95D17.1 | 4.896657887 | 0.269692367 | −4.18241 | 0.0765225 | 0.683824 |
| RP11-982M15.8 | 0.508451553 | 2.996266867 | 2.558984 | 0.0765225 | 0.683824 |
| RP5-1028K7.2 | 3.882608593 | 0.274027367 | −3.82463 | 0.0765225 | 0.683824 |
| TNRC6C-AS1 | 2.94829249 | 0.411041 | −2.84253 | 0.0765225 | 0.683824 |
Significant DELRs between T0 and T2
| LncRNA | Mean(T0) | Mean(T2) | logFC | pValue | FDR |
|---|---|---|---|---|---|
| AC007563.5 | 2.76586097 | 0.155973413 | −4.14836 | 0.0765225 | 0.621384 |
| AC098617.1 | 0.166783833 | 4.994414183 | 4.904264 | 0.0765225 | 0.621384 |
| AC112721.1 | 0.02084798 | 0.913731333 | 5.453791 | 0.0765225 | 0.621384 |
| ADAMTS9-AS2 | 4.195305597 | 0.038,993,353 | −6.7494 | 0.0765225 | 0.621384 |
| AZIN1-AS1 | 3.07585092 | 0.17547009 | −4.13169 | 0.0765225 | 0.621384 |
| C15orf54 | 0.081352247 | 23.33072509 | 8.163833 | 0.0765225 | 0.621384 |
| CH507-145 C22. | 0.38642318 | 2.695785687 | 2.802452 | 0.0765225 | 0.621384 |
| CTA-292E10.6 | 3.03295283 | 0.584900297 | −2.37446 | 0.0765225 | 0.621384 |
| CTC-523E23.3 | 0.542047333 | 3.586782283 | 2.726199 | 0.0765225 | 0.621384 |
| CTD-2547L24.3 | 5.05390719 | 0.038993353 | −7.01803 | 0.0765225 | 0.621384 |
| KANSL1-AS1 | 3.431506167 | 0.05849003 | −5.87451 | 0.0765225 | 0.621384 |
| LINC00467 | 0.406761243 | 3.144939917 | 2.950778 | 0.0765225 | 0.621384 |
| LINC01480 | 0.1022842 | 2.300809347 | 4.491486 | 0.0765225 | 0.621384 |
| LINC01534 | 0.271023733 | 1.089082163 | 2.006622 | 0.0765225 | 0.621384 |
| MEG3 | 0.020456837 | 2.239088517 | 6.774185 | 0.0765225 | 0.621384 |
| NEXN-AS1 | 0.833860547 | 9.482922053 | 3.507454 | 0.0765225 | 0.621384 |
| POT1-AS1 | 0.10169031 | 2.667381173 | 4.71317 | 0.0765225 | 0.621384 |
| PWAR5 | 3.006275807 | 0.136476737 | −4.46125 | 0.0765225 | 0.621384 |
| RP11-242D8.1 | 4.363404197 | 0.409430207 | −3.41376 | 0.0765225 | 0.621384 |
| RP11-303E16.2 | 2.226079677 | 0.38993353 | −2.51321 | 0.0765225 | 0.621384 |
| RP11-345P4.9 | 0.0613705 | 4.858066433 | 6.306693 | 0.0765225 | 0.621384 |
| RP11-366 L5.1 | 0.020338063 | 3.031805823 | 7.219851 | 0.0765225 | 0.621384 |
| RP11-379F4.4 | 0.125087867 | 1.32542017 | 3.405436 | 0.0765225 | 0.621384 |
| RP11-427M20.1 | 3.052397903 | 0.102365933 | −4.89814 | 0.0765225 | 0.621384 |
| RP11-446N19.1 | 0.062543933 | 3.929752793 | 5.973425 | 0.0765225 | 0.621384 |
| RP11-479O9.4 | 0.777359667 | 4.55295743 | 2.55015 | 0.0765225 | 0.621384 |
| RP11-495P10.1 | 1.398293977 | 0.077986707 | −4.1643 | 0.0765225 | 0.621384 |
| RP11-705 C15.3 | 0.162704497 | 2.89037666 | 4.150931 | 0.0765225 | 0.621384 |
| RP11-746M1.1 | 0.3273094 | 1.68878291 | 2.367257 | 0.0765225 | 0.621384 |
| RP11-809N8.4 | 1.43980743 | 0.253456793 | −2.50606 | 0.0765225 | 0.621384 |
| RP11-87H9.4 | 2.333000947 | 0.020473183 | −6.83231 | 0.0765225 | 0.621384 |
| RP1-47M23.3 | 0.041695967 | 1.749010003 | 5.390487 | 0.0765225 | 0.621384 |
| RP5-1028K7.2 | 3.882608593 | 0.368517333 | −3.39722 | 0.0765225 | 0.621384 |
| THAP7-AS1 | 4.052557717 | 0.136476737 | −4.89211 | 0.0765225 | 0.621384 |
| ZNF674-AS1 | 3.01449014 | 0.214463443 | −3.81311 | 0.0765225 | 0.621384 |
| LncRNA | Mean(T0) | Mean(T3) | logFC | pValue | FDR |
| AC007563.5 | 2.76586097 | 0.215818967 | −3.67984 | 0.0765225 | 0.355949 |
| AC091814.2 | 0.223718683 | 1.485052283 | 2.730756 | 0.0765225 | 0.355949 |
| AC093627.8 | 0.325408993 | 4.232574173 | 3.701209 | 0.0765225 | 0.355949 |
| AC098617.1 | 0.166783833 | 5.820427687 | 5.125074 | 0.0765225 | 0.355949 |
| AC112721.1 | 0.02084798 | 1.709872217 | 6.357837 | 0.0765225 | 0.355949 |
| ARF4-AS1 | 0.040913667 | 0.893591417 | 4.448961 | 0.0765225 | 0.355949 |
| BAALC-AS1 | 2.832459073 | 0.172655167 | −4.03609 | 0.0765225 | 0.355949 |
| C15orf54 | 0.081352247 | 12.67610939 | 7.283714 | 0.0765225 | 0.355949 |
| CDKN2B-AS1 | 4.44231614 | 0.36374714 | −3.6103 | 0.0765225 | 0.355949 |
| CITF22-92A6.1 | 0.081827333 | 0.85146136 | 3.379286 | 0.0765225 | 0.355949 |
| CTB-111H14.1 | 0.28473287 | 2.847558803 | 3.322045 | 0.0765225 | 0.355949 |
| CTBP1-AS | 0.041695967 | 0.94928833 | 4.508867 | 0.0765225 | 0.355949 |
| CTD-2574D22.2 | 0.833860547 | 3.866681563 | 2.213218 | 0.0765225 | 0.355949 |
| CTD-2587H24.1 | 0.1042399 | 0.609993183 | 2.548886 | 0.0765225 | 0.355949 |
| CTD-3014M21.1 | 3.569017227 | 0.161665397 | −4.46444 | 0.0765225 | 0.355949 |
| CYB561D2 | 0.040676123 | 1.130932207 | 4.797186 | 0.0765225 | 0.355949 |
| EIF1B-AS1 | 0.542047333 | 4.039256577 | 2.897599 | 0.0765225 | 0.355949 |
| KIF9-AS1 | 0.18304256 | 0.99462356 | 2.441971 | 0.0765225 | 0.355949 |
| LINC00261 | 0.040913667 | 2.71680064 | 6.053182 | 0.0765225 | 0.355949 |
| LINC00467 | 0.406761243 | 4.464962843 | 3.456394 | 0.0765225 | 0.355949 |
| LINC00920 | 1.453565963 | 0.080832697 | −4.16851 | 0.0765225 | 0.355949 |
| LINC01002 | 0.166783833 | 1.508721053 | 3.177275 | 0.0765225 | 0.355949 |
| LINC01534 | 0.271023733 | 1.066834697 | 1.976846 | 0.0765225 | 0.355949 |
| MAP3K14-AS1 | 0.02084798 | 0.650734907 | 4.96409 | 0.0765225 | 0.355949 |
| NEXN-AS1 | 0.833860547 | 8.247496673 | 3.306078 | 0.0765225 | 0.355949 |
| NR2F1-AS1 | 3.88181485 | 0.172655167 | −4.49077 | 0.0765225 | 0.355949 |
| PROSER2-AS1 | 0.2250252 | 1.895535413 | 3.074447 | 0.0765225 | 0.355949 |
| RP11-1094M14. | 0.3273094 | 1.935723837 | 2.564146 | 0.0765225 | 0.355949 |
| RP11-156E8.1 | 0.081827333 | 0.919915587 | 3.490847 | 0.0765225 | 0.355949 |
| RP11-182 L21.6 | 0.223718683 | 3.090707643 | 3.78818 | 0.0765225 | 0.355949 |
| RP11-234K19.1 | 0.750527333 | 4.799530327 | 2.676917 | 0.0765225 | 0.355949 |
| RP11-244H3.1 | 0.020456837 | 0.80383261 | 5.29624 | 0.0765225 | 0.355949 |
| RP11-282O18.3 | 0.162704497 | 4.722175677 | 4.859126 | 0.0765225 | 0.355949 |
| RP11-288C18.1 | 0.1042399 | 0.553462837 | 2.408579 | 0.0765225 | 0.355949 |
| RP11-299G20.5 | 0.081352247 | 9.772892627 | 6.90846 | 0.0765225 | 0.355949 |
| RP11-319G9.5 | 0.020338063 | 0.97249277 | 5.579433 | 0.0765225 | 0.355949 |
| RP11-320 M2.1 | 0.1022842 | 0.71546583 | 2.8063 | 0.0765225 | 0.355949 |
| RP11-333E1.1 | 0.041695967 | 0.642504777 | 3.945727 | 0.0765225 | 0.355949 |
| RP11-366 L5.1 | 0.020338063 | 1.697460663 | 6.383052 | 0.0765225 | 0.355949 |
| RP11-379F4.4 | 0.125087867 | 2.12802338 | 4.0885 | 0.0765225 | 0.355949 |
| RP11-427M20.1 | 3.052397903 | 0.028775863 | −6.72894 | 0.0765225 | 0.355949 |
| RP11-428J1.5 | 0.10169031 | 0.803844833 | 2.982735 | 0.0765225 | 0.355949 |
| RP11-446N19.1 | 0.062543933 | 2.991808677 | 5.580004 | 0.0765225 | 0.355949 |
| RP11-467L13.7 | 0.650817987 | 3.83507436 | 2.558929 | 0.0765225 | 0.355949 |
| RP11-553A21.3 | 0.18304256 | 6.535466787 | 5.158039 | 0.0765225 | 0.355949 |
| RP11-596 C23.6 | 0.1022842 | 0.54519963 | 2.414201 | 0.0765225 | 0.355949 |
| RP11-624L4.1 | 0.020338063 | 1.489609253 | 6.194608 | 0.0765225 | 0.355949 |
| RP11-705 C15.3 | 0.162704497 | 1.189071987 | 2.86951 | 0.0765225 | 0.355949 |
| RP11-732A19.2 | 0.081827333 | 0.596647493 | 2.866224 | 0.0765225 | 0.355949 |
| RP11-7F17.8 | 1.403326287 | 18.64460099 | 3.731836 | 0.0765225 | 0.355949 |
| RP11-867G23.3 | 0.041695967 | 0.716191363 | 4.102365 | 0.0765225 | 0.355949 |
| RP11-867G23.8 | 0.020338063 | 1.565923 | 6.266687 | 0.0765225 | 0.355949 |
| RP11-87H9.4 | 2.333000947 | 0.028775863 | −6.34118 | 0.0765225 | 0.355949 |
| RP3-329E20.2 | 0.204568367 | 2.282784317 | 3.48014 | 0.0765225 | 0.355949 |
| SPAG5-AS1 | 0.145935867 | 2.191379437 | 3.908433 | 0.0765225 | 0.355949 |
| TIPARP-AS1 | 0.083391933 | 0.705930717 | 3.081547 | 0.0765225 | 0.355949 |
Significant DEGs between T0 and T1
| GENE | Mean(T0) | Mean(T1) | logFC | pValue | FDR |
|---|---|---|---|---|---|
| TSPYL5 | 8.297698333 | 1 | −3.05271 | 0.0636026 | 0.381797 |
| NAGPA | 8.164393 | 1 | −3.02935 | 0.0636026 | 0.381797 |
| ZNF112 | 7.031611667 | 1 | −2.81386 | 0.0636026 | 0.381797 |
| GSTZ1 | 7.736256333 | 1 | −2.95164 | 0.0636026 | 0.381797 |
| SLC25A33 | 4.360061333 | 1 | −2.12435 | 0.0636026 | 0.381797 |
| C1orf186 | 5.760208 | 1 | −2.52612 | 0.0636026 | 0.381797 |
| CC2D2A | 5.340496 | 1 | −2.41697 | 0.0636026 | 0.381797 |
| DCLK1 | 7.265278667 | 1 | −2.86102 | 0.0636026 | 0.381797 |
| CXorf57 | 4.585493667 | 1 | −2.19708 | 0.0636026 | 0.381797 |
| IL2RA | 3.627303333 | 1 | −1.8589 | 0.0636026 | 0.381797 |
| C2orf81 | 6.706935333 | 1 | −2.74565 | 0.0636026 | 0.381797 |
| RPUSD2 | 8.42355 | 1 | −3.07443 | 0.0636026 | 0.381797 |
| RABL2A | 5.830274 | 1 | −2.54356 | 0.0636026 | 0.381797 |
| AC092835. | 6.381238667 | 1 | −2.67384 | 0.0636026 | 0.381797 |
| WT1 | 5.004191667 | 1 | −2.32314 | 0.0636026 | 0.381797 |
| GCNT4 | 4.203905 | 1 | −2.07173 | 0.0636026 | 0.381797 |
| TIGD2 | 5.404809 | 1 | −2.43424 | 0.0636026 | 0.381797 |
| MYT1 | 2.737559667 | 1 | −1.45289 | 0.0636026 | 0.381797 |
| C14orf80 | 4.038967333 | 1 | −2.01399 | 0.0636026 | 0.381797 |
| GOLGA7B | 5.057243333 | 1 | −2.33835 | 0.0636026 | 0.381797 |
| UNC13 C | 3.131791333 | 1 | −1.64699 | 0.0636026 | 0.381797 |
| ZNF705A | 5.314469 | 1 | −2.40993 | 0.0636026 | 0.381797 |
| POC1A | 3.826612667 | 1 | −1.93607 | 0.0636026 | 0.381797 |
| TIFAB | 7.848899333 | 1 | −2.97249 | 0.0636026 | 0.381797 |
| NLRP7 | 5.482675333 | 1 | −2.45488 | 0.0636026 | 0.381797 |
| CFAP53 | 5.940012 | 1 | −2.57047 | 0.0636026 | 0.381797 |
| TCTEX1D1 | 1 | 4.620729 | 2.20812 | 0.0636026 | 0.381797 |
| VNN3 | 1 | 10.84045167 | 3.438353 | 0.0636026 | 0.381797 |
| HTR2A | 1 | 3.366183667 | 1.751114 | 0.0636026 | 0.381797 |
| ILDR2 | 1 | 5.065631333 | 2.340742 | 0.0636026 | 0.381797 |
| EP400NL | 15.092232 | 2.712671 | −2.47602 | 0.0765225 | 0.381797 |
| DCPS | 14.96914533 | 2.027602667 | −2.88414 | 0.0765225 | 0.381797 |
| ZNF354 C | 11.624222 | 1.205520667 | −3.26941 | 0.0765225 | 0.381797 |
| FLT4 | 8.531011667 | 1.616561667 | −2.39979 | 0.0765225 | 0.381797 |
| PTPRO | 13.12034167 | 2.027602667 | −2.69396 | 0.0765225 | 0.381797 |
| RITA1 | 7.793430333 | 1.159849333 | −2.74832 | 0.0765225 | 0.381797 |
| KCNH8 | 6.572459333 | 1.159849333 | −2.5025 | 0.0765225 | 0.381797 |
| CCDC167 | 15.463013 | 2.210287667 | −2.80652 | 0.0765225 | 0.381797 |
| TRPV2 | 10.67998933 | 1.959095667 | −2.44665 | 0.0765225 | 0.381797 |
| A4GALT | 7.112375333 | 1.753575333 | −2.02003 | 0.0765225 | 0.381797 |
| GALK1 | 10.69892733 | 2.895356 | −1.88565 | 0.0765225 | 0.381797 |
| LTBP2 | 9.837398333 | 1.662232667 | −2.56515 | 0.0765225 | 0.381797 |
| TMEM234 | 6.152497 | 1.274027333 | −2.27178 | 0.0765225 | 0.381797 |
| DNALI1 | 7.875622 | 1.114178 | −2.82141 | 0.0765225 | 0.381797 |
| NEK3 | 10.27452633 | 1.760042 | −2.54539 | 0.0765225 | 0.381797 |
| MTFP1 | 9.792329333 | 1.433876667 | −2.77173 | 0.0765225 | 0.381797 |
| SELO | 8.185779 | 1.114178 | −2.87714 | 0.0765225 | 0.381797 |
| PCED1A | 9.752530333 | 1.959095667 | −2.31559 | 0.0765225 | 0.381797 |
| GSTCD | 10.69255167 | 1.159849333 | −3.2046 | 0.0765225 | 0.381797 |
| MMP23B | 5.119569333 | 1.479548 | −1.79087 | 0.0765225 | 0.381797 |
| FOXRED1 | 7.648389 | 1.639397333 | −2.22199 | 0.0765225 | 0.381797 |
| C9orf172 | 7.559422667 | 2.210287667 | −1.77404 | 0.0765225 | 0.381797 |
| FAM206A | 12.360496 | 1.913424667 | −2.69151 | 0.0765225 | 0.381797 |
| MIPEP | 12.83586 | 2.118945 | −2.59876 | 0.0765225 | 0.381797 |
| NUDT16L1 | 10.97319067 | 2.210287667 | −2.31168 | 0.0765225 | 0.381797 |
| PEMT | 8.485709667 | 1.890589 | −2.1662 | 0.0765225 | 0.381797 |
| GPSM1 | 4.377577333 | 1.122587333 | −1.9633 | 0.0765225 | 0.381797 |
| CDCA5 | 9.481528667 | 1.114178 | −3.08914 | 0.0765225 | 0.381797 |
| TRIM66 | 9.392559333 | 1.799246333 | −2.38413 | 0.0765225 | 0.381797 |
| EVC | 7.847389333 | 1.479548 | −2.40706 | 0.0765225 | 0.381797 |
| OSBPL7 | 7.09137 | 1.228356 | −2.52934 | 0.0765225 | 0.381797 |
| IL23A | 6.11104 | 1.388205333 | −2.1382 | 0.0765225 | 0.381797 |
| GYPE | 12.21936 | 2.552821667 | −2.259 | 0.0765225 | 0.381797 |
| ATG9B | 6.718192 | 1.269692333 | −2.40359 | 0.0765225 | 0.381797 |
| CHTF18 | 4.928835333 | 1.205520667 | −2.03159 | 0.0765225 | 0.381797 |
| NDOR1 | 5.756575333 | 1.593726 | −1.85281 | 0.0765225 | 0.381797 |
| TMEM150A | 6.262062 | 1.182685 | −2.40457 | 0.0765225 | 0.381797 |
| LRFN3 | 4.199165 | 1.525219 | −1.46109 | 0.0765225 | 0.381797 |
| WDR83 | 9.817685333 | 2.804013667 | −1.80789 | 0.0765225 | 0.381797 |
| HMCN1 | 5.950431667 | 1.433876667 | −2.05307 | 0.0765225 | 0.381797 |
| ASB13 | 9.439516 | 1.411041 | −2.74195 | 0.0765225 | 0.381797 |
| BCDIN3D | 5.399809 | 1.220657333 | −2.14525 | 0.0765225 | 0.381797 |
| S1PR2 | 5.867014667 | 1.045671333 | −2.4882 | 0.0765225 | 0.381797 |
| AMDHD1 | 5.082083667 | 1.525219 | −1.7364 | 0.0765225 | 0.381797 |
| SLC25A17 | 7.005707 | 1.593726 | −2.13613 | 0.0765225 | 0.381797 |
| ALG1 | 4.579064 | 1.045671333 | −2.13062 | 0.0765225 | 0.381797 |
| SSPN | 6.093317667 | 2.152322 | −1.50133 | 0.0765225 | 0.381797 |
| POGLUT1 | 6.763843667 | 1.662232667 | −2.02472 | 0.0765225 | 0.381797 |
| PARS2 | 4.567606333 | 1.0228,35667 | −2.15886 | 0.0765225 | 0.381797 |
| STK32B | 5.824938333 | 1.570890333 | −1.89066 | 0.0765225 | 0.381797 |
| FAM207A | 8.498970333 | 1.616561667 | −2.39436 | 0.0765225 | 0.381797 |
| LRWD1 | 9.251122333 | 1.365369667 | −2.76034 | 0.0765225 | 0.381797 |
| ERP27 | 4.538457667 | 1.319698667 | −1.78199 | 0.0765225 | 0.381797 |
| UBE3D | 6.706291 | 1.685068333 | −1.99271 | 0.0765225 | 0.381797 |
| RAG1 | 6.426811 | 1.342534333 | −2.25914 | 0.0765225 | 0.381797 |
| THNSL1 | 6.241041333 | 1.411041 | −2.14503 | 0.0765225 | 0.381797 |
| CHRNA2 | 2.411521667 | 16.41214167 | 2.766748 | 0.0765225 | 0.381797 |
| KLHDC8A | 3.710156667 | 1.416797333 | −1.38885 | 0.0765225 | 0.381797 |
| SDHAF4 | 6.697633333 | 2.096109333 | −1.67594 | 0.0765225 | 0.381797 |
| DDX28 | 5.240941667 | 1.867753333 | −1.48852 | 0.0765225 | 0.381797 |
| FLG | 5.70593 | 1.022835667 | −2.47989 | 0.0765225 | 0.381797 |
| GLYCTK | 3.64294 | 1.456712333 | −1.32239 | 0.0765225 | 0.381797 |
| MTERF2 | 8.744611 | 1.822082 | −2.26281 | 0.0765225 | 0.381797 |
| DDIAS | 7.581300667 | 1.137013667 | −2.7372 | 0.0765225 | 0.381797 |
| NIPAL2 | 7.583966667 | 1.822082 | −2.05736 | 0.0765225 | 0.381797 |
| QTRT1 | 6.379764 | 1.548054667 | −2.04305 | 0.0765225 | 0.381797 |
| SMYD5 | 9.282532667 | 1.822082 | −2.34893 | 0.0765225 | 0.381797 |
| AN-P2RY | 4.527150333 | 1.844917667 | −1.29505 | 0.0765225 | 0.381797 |
| SEC61A2 | 4.182814667 | 1.388205333 | −1.59125 | 0.0765225 | 0.381797 |
| ARL6 | 7.930565 | 1.039818 | −2.93109 | 0.0765225 | 0.381797 |
| FAM179A | 5.037166667 | 1.274027333 | −1.98322 | 0.0765225 | 0.381797 |
| SLC46A3 | 10.185989 | 1.091342333 | −3.22241 | 0.0765225 | 0.381797 |
| CAPN3 | 5.298772667 | 1.09807 | −2.27069 | 0.0765225 | 0.381797 |
| ITIH5 | 4.453339333 | 1.137013667 | −1.96964 | 0.0765225 | 0.381797 |
| FCN2 | 5.770094 | 1.525219 | −1.91958 | 0.0765225 | 0.381797 |
| ASB9 | 2.964853667 | 1.159849333 | −1.35402 | 0.0765225 | 0.381797 |
| ZNF90 | 4.634495667 | 1.068507 | −2.11682 | 0.0765225 | 0.381797 |
| MPP3 | 3.775197667 | 1.024517333 | −1.88161 | 0.0765225 | 0.381797 |
| JMJD7 | 3.819834333 | 1.068507 | −1.83791 | 0.0765225 | 0.381797 |
| PPIC | 7.103771 | 1.182685 | −2.58652 | 0.0765225 | 0.381797 |
| PAK3 | 4.371605 | 1.639397333 | −1.415 | 0.0765225 | 0.381797 |
| MAP9 | 6.329609667 | 1.416797333 | −2.15948 | 0.0765225 | 0.381797 |
| NR2C2AP | 3.038,413 | 1.388205333 | −1.1301 | 0.0765225 | 0.381797 |
| PIGV | 4.787995 | 1.479548 | −1.69427 | 0.0765225 | 0.381797 |
| TNFAIP8L3 | 4.463735333 | 1.525219 | −1.54924 | 0.0765225 | 0.381797 |
| HSD11B1L | 4.341861 | 1.159849333 | −1.90438 | 0.0765225 | 0.381797 |
| ACPP | 2.291559667 | 1.137013667 | −1.01108 | 0.0765225 | 0.381797 |
| RYR1 | 5.863586 | 1.296863 | −2.17676 | 0.0765225 | 0.381797 |
| ACKR4 | 6.268366 | 1.182685 | −2.40602 | 0.0765225 | 0.381797 |
| ZNF674 | 5.265564667 | 1.119454333 | −2.23379 | 0.0765225 | 0.381797 |
| SLC16A14 | 2.638718 | 1.228356 | −1.10311 | 0.0765225 | 0.381797 |
| NR6A1 | 1.286395667 | 5.917175 | 2.201574 | 0.0765225 | 0.381797 |
| ABCB9 | 1.225025333 | 2.864761 | 1.225603 | 0.0765225 | 0.381797 |
| GALNT18 | 1.204568333 | 3.584699 | 1.573336 | 0.0765225 | 0.381797 |
| PRRT2 | 1.122741 | 7.446128 | 2.729465 | 0.0765225 | 0.381797 |
| KHK | 1.771375333 | 5.577208333 | 1.654673 | 0.0765225 | 0.381797 |
| VSIG2 | 1.427099333 | 11.46590967 | 3.006193 | 0.0765225 | 0.381797 |
| DYTN | 2.382988333 | 13.68237767 | 2.521475 | 0.0765225 | 0.381797 |
| KIAA1549 | 1.041696 | 5.577024333 | 2.420561 | 0.0765225 | 0.381797 |
| NT5DC3 | 1.583743333 | 6.695642 | 2.079884 | 0.0765225 | 0.381797 |
| GRB14 | 1.041696 | 19.04747167 | 4.192593 | 0.0765225 | 0.381797 |
| SPHAR | 1.833860667 | 8.911786667 | 2.280831 | 0.0765225 | 0.381797 |
| CLDN12 | 1.437807667 | 4.590490667 | 1.674778 | 0.0765225 | 0.381797 |
| DGKI | 1.416959667 | 6.646423 | 2.229779 | 0.0765225 | 0.381797 |
| TAF6L | 1.142366333 | 4.517017333 | 1.983345 | 0.0765225 | 0.381797 |
| LIPC | 2.444002333 | 9.215349 | 1.914793 | 0.0765225 | 0.381797 |
| PPP1R13L | 1.166784 | 11.21800867 | 3.265207 | 0.0765225 | 0.381797 |
| PLXNB3 | 3.989695 | 15.27107967 | 1.936452 | 0.0765225 | 0.381797 |
Significant DEGs between T0 and T2
| GENE | Mean(T0) | Mean(T2) | logFC | pValue | FDR |
|---|---|---|---|---|---|
| CCHCR1 | 7.595336 | 1 | −2.92511 | 0.0636026 | 0.412752 |
| CBR3 | 11.61466033 | 1 | −3.53788 | 0.0636026 | 0.412752 |
| CCDC40 | 5.298982333 | 1 | −2.40572 | 0.0636026 | 0.412752 |
| CLEC4C | 4.453637333 | 1 | −2.15498 | 0.0636026 | 0.412752 |
| IL23A | 6.11104 | 1 | −2.61142 | 0.0636026 | 0.412752 |
| WT1 | 5.004191667 | 1 | −2.32314 | 0.0636026 | 0.412752 |
| OC4-APO | 3.817754 | 1 | −1.93272 | 0.0636026 | 0.412752 |
| APOC2 | 3.817754 | 1 | −1.93272 | 0.0636026 | 0.412752 |
| SMOC2 | 5.391121667 | 1 | −2.43059 | 0.0636026 | 0.412752 |
| APOC3 | 6.555501 | 1 | −2.71271 | 0.0636026 | 0.412752 |
| DTX3 | 4.302150667 | 1 | −2.10506 | 0.0636026 | 0.412752 |
| DHRS11 | 4.471552 | 1 | −2.16078 | 0.0636026 | 0.412752 |
| CCL20 | 3.247779667 | 1 | −1.69945 | 0.0636026 | 0.412752 |
| NLRP7 | 5.482675333 | 1 | −2.45488 | 0.0636026 | 0.412752 |
| GPA33 | 2.577333333 | 1 | −1.36588 | 0.0636026 | 0.412752 |
| ZBED6CL | 1 | 2.731812667 | 1.449859 | 0.0636026 | 0.412752 |
| PRRT3 | 1 | 2.731313667 | 1.449595 | 0.0636026 | 0.412752 |
| SAA1 | 1 | 5.330368667 | 2.414235 | 0.0636026 | 0.412752 |
| TCTEX1D1 | 1 | 5.456496 | 2.447975 | 0.0636026 | 0.412752 |
| CEBPE | 1 | 8.148108 | 3.026465 | 0.0636026 | 0.412752 |
| DIRAS1 | 1 | 4.648991667 | 2.216918 | 0.0636026 | 0.412752 |
| DEPDC4 | 1 | 5.40525 | 2.434361 | 0.0636026 | 0.412752 |
| VNN3 | 1 | 4.804029667 | 2.264245 | 0.0636026 | 0.412752 |
| WFDC1 | 1 | 11.938668 | 3.57757 | 0.0636026 | 0.412752 |
| ADGRG3 | 1 | 5.034594 | 2.331875 | 0.0636026 | 0.412752 |
| ILDR2 | 1 | 9.576249333 | 3.259461 | 0.0636026 | 0.412752 |
| IFI27L1 | 1 | 5.030390667 | 2.33067 | 0.0636026 | 0.412752 |
| OPLAH | 1 | 4.975158667 | 2.314743 | 0.0636026 | 0.412752 |
| TLR5 | 1 | 4.745244333 | 2.246482 | 0.0636026 | 0.412752 |
| RETN | 1 | 7.145587 | 2.837053 | 0.0636026 | 0.412752 |
| MAK | 1 | 5.000173 | 2.321978 | 0.0636026 | 0.412752 |
| GCKR | 1 | 6.585539 | 2.719302 | 0.0636026 | 0.412752 |
| FBXO40 | 1 | 4.339207 | 2.117431 | 0.0636026 | 0.412752 |
| MMP2 | 14.33027367 | 1.45041 | −3.30453 | 0.0765225 | 0.412752 |
| KCNH8 | 6.572459333 | 1.097483333 | −2.58223 | 0.0765225 | 0.412752 |
| PGAP2 | 7.833870333 | 1.311946667 | −2.57802 | 0.0765225 | 0.412752 |
| ZSWIM3 | 6.354159333 | 1.406925667 | −2.17516 | 0.0765225 | 0.412752 |
| KATNAL2 | 7.194896 | 1.194966667 | −2.59 | 0.0765225 | 0.412752 |
| GSTZ1 | 7.736256333 | 1.17547 | −2.7184 | 0.0765225 | 0.412752 |
| ARHGAP22 | 5.822747333 | 1.11698 | −2.3821 | 0.0765225 | 0.412752 |
| NEK3 | 10.27452633 | 1.857853667 | −2.46736 | 0.0765225 | 0.412752 |
| CYP2E1 | 9.410710667 | 1.214463333 | −2.95398 | 0.0765225 | 0.412752 |
| MMP23B | 5.119569333 | 1.429937 | −1.84007 | 0.0765225 | 0.412752 |
| ABCA9 | 9.573765 | 1.757507667 | −2.44556 | 0.0765225 | 0.412752 |
| SCARB1 | 9.162249333 | 2.033324 | −2.17186 | 0.0765225 | 0.412752 |
| KIF5C | 9.273256333 | 1.038993333 | −3.15789 | 0.0765225 | 0.412752 |
| SFT2D3 | 7.445747667 | 1.584900333 | −2.23202 | 0.0765225 | 0.412752 |
| C2orf81 | 6.706935333 | 1.389933667 | −2.27064 | 0.0765225 | 0.412752 |
| GZMK | 7.516827 | 1.11698 | −2.75052 | 0.0765225 | 0.412752 |
| FGFR3 | 4.588419667 | 1.019377333 | −2.17031 | 0.0765225 | 0.412752 |
| ADTRP | 4.90516 | 1.122839 | −2.12715 | 0.0765225 | 0.412752 |
| LRP5L | 4.650881333 | 1.020473333 | −2.18827 | 0.0765225 | 0.412752 |
| CCR7 | 5.365534 | 1.245678333 | −2.10679 | 0.0765225 | 0.412752 |
| CSRP2 | 7.636079333 | 1.194966667 | −2.67586 | 0.0765225 | 0.412752 |
| ERP27 | 4.538457667 | 1.11698 | −2.0226 | 0.0765225 | 0.412752 |
| PLA2R1 | 3.766903667 | 1.143312333 | −1.72016 | 0.0765225 | 0.412752 |
| CLEC1A | 5.842795333 | 1.506913667 | −1.95506 | 0.0765225 | 0.412752 |
| RSPO3 | 11.824911 | 1.389933667 | −3.08874 | 0.0765225 | 0.412752 |
| C1orf56 | 3.535961667 | 1.019496667 | −1.79425 | 0.0765225 | 0.412752 |
| IDUA | 3.500385 | 1.17547 | −1.57428 | 0.0765225 | 0.412752 |
| FOLR2 | 4.340185667 | 1.370437 | −1.66312 | 0.0765225 | 0.412752 |
| CHRNA2 | 2.411521667 | 25.994894 | 3.430213 | 0.0765225 | 0.412752 |
| HSD17B3 | 2.288780667 | 6.393580667 | 1.482045 | 0.0765225 | 0.412752 |
| C14orf80 | 4.038967333 | 1.584900333 | −1.34959 | 0.0765225 | 0.412752 |
| APOBEC3H | 3.586465667 | 1.327571 | −1.43377 | 0.0765225 | 0.412752 |
| ADAMTSL3 | 3.572188 | 1.019496667 | −1.80895 | 0.0765225 | 0.412752 |
| C19orf44 | 5.222961667 | 1.682383667 | −1.63436 | 0.0765225 | 0.412752 |
| CERCAM | 4.230867 | 1.573249 | −1.42721 | 0.0765225 | 0.412752 |
| FBLN1 | 8.063541 | 1.467920333 | −2.45764 | 0.0765225 | 0.412752 |
| FAM184A | 7.253860333 | 1.584900333 | −2.19436 | 0.0765225 | 0.412752 |
| ZNF630 | 4.970839333 | 1.467920333 | −1.75972 | 0.0765225 | 0.412752 |
| C15orf65 | 2.889942 | 1.370437 | −1.0764 | 0.0765225 | 0.412752 |
| VPS9D1 | 1.838730333 | 5.310771667 | 1.530212 | 0.0765225 | 0.412752 |
| ALG14 | 4.971241667 | 1.194966667 | −2.05664 | 0.0765225 | 0.412752 |
| ID4 | 7.315216667 | 1.081892667 | −2.75734 | 0.0765225 | 0.412752 |
| SPINT1 | 3.621315333 | 1.019377333 | −1.82883 | 0.0765225 | 0.412752 |
| MAP10 | 4.540429 | 1.584900333 | −1.51844 | 0.0765225 | 0.412752 |
| ABI3BP | 7.148441 | 1.214463333 | −2.55731 | 0.0765225 | 0.412752 |
| RBKS | 3.832568333 | 1.184258667 | −1.69433 | 0.0765225 | 0.412752 |
| DCAF4 | 1.490964 | 4.681461667 | 1.650714 | 0.0765225 | 0.412752 |
| CIB2 | 2.768122667 | 1.05849 | −1.3869 | 0.0765225 | 0.412752 |
| GREB1L | 4.893538333 | 1.467920333 | −1.7371 | 0.0765225 | 0.412752 |
| HSD11B1L | 4.341861 | 1.467920333 | −1.56454 | 0.0765225 | 0.412752 |
| RYR1 | 5.863586 | 1.29245 | −2.18167 | 0.0765225 | 0.412752 |
| SNTG1 | 3.070525 | 1.389933667 | −1.14347 | 0.0765225 | 0.412752 |
| ACKR4 | 6.268366 | 1.429937 | −2.13214 | 0.0765225 | 0.412752 |
| ZNF835 | 4.583497333 | 1.23396 | −1.89315 | 0.0765225 | 0.412752 |
| TIFAB | 7.848899333 | 1.102366 | −2.83189 | 0.0765225 | 0.412752 |
| NCBP2L | 1.061370667 | 5.636411 | 2.408848 | 0.0765225 | 0.412752 |
| FKBP10 | 1.040913667 | 2.451751333 | 1.235962 | 0.0765225 | 0.412752 |
| DYTN | 2.382988333 | 9.130906 | 1.937986 | 0.0765225 | 0.412752 |
| MMP1 | 1.020848 | 4.238790333 | 2.053885 | 0.0765225 | 0.412752 |
| SEC14L2 | 1.569465667 | 6.796782 | 2.114578 | 0.0765225 | 0.412752 |
| MCEMP1 | 6.165867667 | 27.79493433 | 2.172446 | 0.0765225 | 0.412752 |
| FICD | 1.10424 | 5.152503 | 2.22222 | 0.0765225 | 0.412752 |
| KIAA1549 | 1.041696 | 3.803068 | 1.868229 | 0.0765225 | 0.412752 |
| ADORA2B | 2.313422667 | 7.665795333 | 1.728407 | 0.0765225 | 0.412752 |
| ARMC2 | 1.549127667 | 3.928262667 | 1.342435 | 0.0765225 | 0.412752 |
| GRB14 | 1.041696 | 18.48755433 | 4.149548 | 0.0765225 | 0.412752 |
| CEL | 1.020848 | 10.00939533 | 3.293515 | 0.0765225 | 0.412752 |
| SRPK3 | 1.325409 | 4.096300333 | 1.627884 | 0.0765225 | 0.412752 |
| KCNMB1 | 1.711832333 | 12.59957767 | 2.879762 | 0.0765225 | 0.412752 |
| DNAH2 | 1.750527333 | 8.483400667 | 2.276853 | 0.0765225 | 0.412752 |
| DGKI | 1.416959667 | 9.680691667 | 2.772311 | 0.0765225 | 0.412752 |
| SDCBP2 | 1.062544 | 4.338208 | 2.029577 | 0.0765225 | 0.412752 |
| WDSUB1 | 2.146639 | 8.531141667 | 1.990659 | 0.0765225 | 0.412752 |
| WASF1 | 3.338877 | 15.118132 | 2.178845 | 0.0765225 | 0.412752 |
| CRP | 1.488113333 | 4.649315667 | 1.643534 | 0.0765225 | 0.412752 |
| ZDHHC15 | 1.083392 | 3.372840667 | 1.638409 | 0.0765225 | 0.412752 |
| ACCSL | 1.610142 | 4.907580667 | 1.607824 | 0.0765225 | 0.412752 |
| LRRC69 | 1.312719667 | 2.734634667 | 1.058789 | 0.0765225 | 0.412752 |
| PLXNB3 | 3.989695 | 17.17698433 | 2.106126 | 0.0765225 | 0.412752 |
| DGAT2 | 1.101690333 | 4.806447667 | 2.125252 | 0.0765225 | 0.412752 |
| PIGV | 4.787995 | 1.479548 | −1.69427 | 0.0765225 | 0.381797 |
| TNFAIP8L3 | 4.463735333 | 1.525219 | −1.54924 | 0.0765225 | 0.381797 |
| HSD11B1L | 4.341861 | 1.159849333 | −1.90438 | 0.0765225 | 0.381797 |
| ACPP | 2.291559667 | 1.137013667 | −1.01108 | 0.0765225 | 0.381797 |
| RYR1 | 5.863586 | 1.296863 | −2.17676 | 0.0765225 | 0.381797 |
| ACKR4 | 6.268366 | 1.182685 | −2.40602 | 0.0765225 | 0.381797 |
| ZNF674 | 5.265564667 | 1.119454333 | −2.23379 | 0.0765225 | 0.381797 |
| SLC16A14 | 2.638718 | 1.228356 | −1.10311 | 0.0765225 | 0.381797 |
| NR6A1 | 1.286395667 | 5.917175 | 2.201574 | 0.0765225 | 0.381797 |
| ABCB9 | 1.225025333 | 2.864761 | 1.225603 | 0.0765225 | 0.381797 |
| GALNT18 | 1.204568333 | 3.584699 | 1.573336 | 0.0765225 | 0.381797 |
| PRRT2 | 1.122741 | 7.446128 | 2.729465 | 0.0765225 | 0.381797 |
| KHK | 1.771375333 | 5.577208333 | 1.654673 | 0.0765225 | 0.381797 |
| VSIG2 | 1.427099333 | 11.46590967 | 3.006193 | 0.0765225 | 0.381797 |
| DYTN | 2.382988333 | 13.68237767 | 2.521475 | 0.0765225 | 0.381797 |
| KIAA1549 | 1.041696 | 5.577024333 | 2.420561 | 0.0765225 | 0.381797 |
| NT5DC3 | 1.583743333 | 6.695642 | 2.079884 | 0.0765225 | 0.381797 |
| GRB14 | 1.041696 | 19.04747167 | 4.192593 | 0.0765225 | 0.381797 |
| SPHAR | 1.833860667 | 8.911786667 | 2.280831 | 0.0765225 | 0.381797 |
| CLDN12 | 1.437807667 | 4.590490667 | 1.674778 | 0.0765225 | 0.381797 |
| DGKI | 1.416959667 | 6.646423 | 2.229779 | 0.0765225 | 0.381797 |
| TAF6L | 1.142366333 | 4.517017333 | 1.983345 | 0.0765225 | 0.381797 |
| LIPC | 2.444002333 | 9.215349 | 1.914793 | 0.0765225 | 0.381797 |
| PPP1R13L | 1.166784 | 11.21800867 | 3.265207 | 0.0765225 | 0.381797 |
| PLXNB3 | 3.989695 | 15.27107967 | 1.936452 | 0.0765225 | 0.381797 |
| NR6A1 | 1.286395667 | 3.191675333 | 1.310979 | 0.0765225 | 0.209152 |
| DKK1 | 1.225025333 | 4.089077333 | 1.738964 | 0.0765225 | 0.209152 |
| SCGB1C2 | 1.143198 | 5.987795333 | 2.38895 | 0.0765225 | 0.209152 |
| GLIS3 | 1.122741 | 4.273733 | 1.928472 | 0.0765225 | 0.209152 |
| NCBP2L | 1.061370667 | 2.974784333 | 1.486856 | 0.0765225 | 0.209152 |
| FKBP10 | 1.040913667 | 2.901915667 | 1.479155 | 0.0765225 | 0.209152 |
| PPP1R14 C | 1.062544 | 3.190217 | 1.586132 | 0.0765225 | 0.209152 |
| GRTP1 | 2.972792 | 8.086871667 | 1.443763 | 0.0765225 | 0.209152 |
| VSIG2 | 1.427099333 | 7.269909333 | 2.348,852 | 0.0765225 | 0.209152 |
| DYTN | 2.382988333 | 9.401901 | 1.980181 | 0.0765225 | 0.209152 |
| MMP1 | 1.020848 | 7.486689333 | 2.87456 | 0.0765225 | 0.209152 |
| SEC14L2 | 1.569465667 | 13.37922333 | 3.091649 | 0.0765225 | 0.209152 |
| JSRP1 | 1.284733 | 3.192776333 | 1.313343 | 0.0765225 | 0.209152 |
| TRIM16L | 1.894874667 | 8.569771 | 2.177154 | 0.0765225 | 0.209152 |
| FKBP14 | 1.166784 | 2.745736333 | 1.234656 | 0.0765225 | 0.209152 |
| KIAA1549 | 1.041696 | 5.596469 | 2.425583 | 0.0765225 | 0.209152 |
| TMEM17 | 1.081352333 | 2.765306333 | 1.354603 | 0.0765225 | 0.209152 |
| SH3D21 | 1.081352333 | 2.318455333 | 1.100327 | 0.0765225 | 0.209152 |
| ADORA2B | 2.313422667 | 6.558939333 | 1.503434 | 0.0765225 | 0.209152 |
| GRB14 | 1.041696 | 22.80897233 | 4.452595 | 0.0765225 | 0.209152 |
| KIAA1211 L | 1.10424 | 2.301469667 | 1.059502 | 0.0765225 | 0.209152 |
| DYNC1I1 | 2.342312 | 21.485078 | 3.19733 | 0.0765225 | 0.209152 |
| CEL | 1.020848 | 4.892537667 | 2.260815 | 0.0765225 | 0.209152 |
| SUCNR1 | 3.725300333 | 39.45302667 | 3.404707 | 0.0765225 | 0.209152 |
| CRYM | 1.162704333 | 6.564460333 | 2.497192 | 0.0765225 | 0.209152 |
| SRPK3 | 1.325409 | 7.290892 | 2.459658 | 0.0765225 | 0.209152 |
| KCNMB1 | 1.711832333 | 15.167389 | 3.147359 | 0.0765225 | 0.209152 |
| SPHAR | 1.833860667 | 5.496761 | 1.583698 | 0.0765225 | 0.209152 |
| DNAH2 | 1.750527333 | 9.255194667 | 2.402474 | 0.0765225 | 0.209152 |
| DGKI | 1.416959667 | 4.966386333 | 1.809398 | 0.0765225 | 0.209152 |
| WASF1 | 3.338877 | 33.65391 | 3.333339 | 0.0765225 | 0.209152 |
| PRSS50 | 1.040676 | 11.52869267 | 3.469636 | 0.0765225 | 0.209152 |
| HBE1 | 2.098255333 | 45.805628 | 4.448263 | 0.0765225 | 0.209152 |
| PPP1R13L | 1.166784 | 2.609403333 | 1.161182 | 0.0765225 | 0.209152 |
| ACCSL | 1.610142 | 12.29111 | 2.932355 | 0.0765225 | 0.209152 |
| PLXNB3 | 3.989695 | 46.17292067 | 3.532697 | 0.0765225 | 0.209152 |
| HOOK2 | 1.416959667 | 3.573092 | 1.334374 | 0.0765225 | 0.209152 |
| DGAT2 | 1.101690333 | 2.642294333 | 1.262072 | 0.0765225 | 0.209152 |
| FAM229A | 1.061014333 | 3.219210333 | 1.601263 | 0.0765225 | 0.209152 |
| ADORA2B | 2.313422667 | 7.665795333 | 1.728407 | 0.0765225 | 0.412752 |
| ARMC2 | 1.549127667 | 3.928262667 | 1.342435 | 0.0765225 | 0.412752 |
| GRB14 | 1.041696 | 18.48755433 | 4.149548 | 0.0765225 | 0.412752 |
| CEL | 1.020848 | 10.00939533 | 3.293515 | 0.0765225 | 0.412752 |
| SRPK3 | 1.325409 | 4.096300333 | 1.627884 | 0.0765225 | 0.412752 |
| KCNMB1 | 1.711832333 | 12.59957767 | 2.879762 | 0.0765225 | 0.412752 |
| DNAH2 | 1.750527333 | 8.483400667 | 2.276853 | 0.0765225 | 0.412752 |
| DGKI | 1.416959667 | 9.680691667 | 2.772311 | 0.0765225 | 0.412752 |
| SDCBP2 | 1.062544 | 4.338208 | 2.029577 | 0.0765225 | 0.412752 |
| WDSUB1 | 2.146639 | 8.531141667 | 1.990659 | 0.0765225 | 0.412752 |
| WASF1 | 3.338877 | 15.118132 | 2.178845 | 0.0765225 | 0.412752 |
| CRP | 1.488113333 | 4.649315667 | 1.643534 | 0.0765225 | 0.412752 |
| ZDHHC15 | 1.083392 | 3.372840667 | 1.638409 | 0.0765225 | 0.412752 |
| ACCSL | 1.610142 | 4.907580667 | 1.607824 | 0.0765225 | 0.412752 |
| LRRC69 | 1.312719667 | 2.734634667 | 1.058789 | 0.0765225 | 0.412752 |
| PLXNB3 | 3.989695 | 17.17698433 | 2.106126 | 0.0765225 | 0.412752 |
| DGAT2 | 1.101690333 | 4.806447667 | 2.125252 | 0.0765225 | 0.412752 |
| PIGV | 4.787995 | 1.479548 | −1.69427 | 0.0765225 | 0.381797 |
| TNFAIP8L3 | 4.463735333 | 1.525219 | −1.54924 | 0.0765225 | 0.381797 |
Significant DEGs between T0 and T3
| GENE | Mean(T0) | Mean(T3) | logFC | pValue | FDR |
|---|---|---|---|---|---|
| WT1 | 5.004191667 | 1 | −2.32314 | 0.0636026 | 0.209152 |
| CLEC1A | 5.842795333 | 1 | −2.54666 | 0.0636026 | 0.209152 |
| APOC3 | 6.555501 | 1 | −2.71271 | 0.0636026 | 0.209152 |
| XG | 2.106219667 | 1 | −1.07466 | 0.0636026 | 0.209152 |
| EYA1 | 9.177906 | 1 | −3.19817 | 0.0636026 | 0.209152 |
| PRRT3 | 1 | 3.799291667 | 1.92573 | 0.0636026 | 0.209152 |
| P4HTM | 1 | 2.007482 | 1.005387 | 0.0636026 | 0.209152 |
| OR2B6 | 1 | 10.21555833 | 3.352696 | 0.0636026 | 0.209152 |
| CAPN8 | 1 | 3.670159 | 1.875843 | 0.0636026 | 0.209152 |
| GLDC | 1 | 3.748804 | 1.90643 | 0.0636026 | 0.209152 |
| TCTEX1D1 | 1 | 3.947338 | 1.98088 | 0.0636026 | 0.209152 |
| RNF215 | 1 | 2.582703333 | 1.368882 | 0.0636026 | 0.209152 |
| DIRAS1 | 1 | 4.19538 | 2.068801 | 0.0636026 | 0.209152 |
| KCNQ4 | 1 | 2.161521333 | 1.112047 | 0.0636026 | 0.209152 |
| DEPDC4 | 1 | 3.900750333 | 1.963752 | 0.0636026 | 0.209152 |
| GRIK5 | 1 | 3.591186 | 1.84446 | 0.0636026 | 0.209152 |
| GFAP | 1 | 2.811727 | 1.491457 | 0.0636026 | 0.209152 |
| SLC29A2 | 1 | 2.196367667 | 1.13512 | 0.0636026 | 0.209152 |
| NKAIN2 | 1 | 4.292667667 | 2.101874 | 0.0636026 | 0.209152 |
| HTR2A | 1 | 4.016981333 | 2.006112 | 0.0636026 | 0.209152 |
| WFDC1 | 1 | 3.885309667 | 1.95803 | 0.0636026 | 0.209152 |
| ZMYND12 | 1 | 3.471236667 | 1.79545 | 0.0636026 | 0.209152 |
| PGA3 | 1 | 2.335415 | 1.223679 | 0.0636026 | 0.209152 |
| KBTBD12 | 1 | 2.296444667 | 1.199402 | 0.0636026 | 0.209152 |
| ILDR2 | 1 | 10.93074 | 3.450319 | 0.0636026 | 0.209152 |
| IFI27L1 | 1 | 2.527466 | 1.337692 | 0.0636026 | 0.209152 |
| OPLAH | 1 | 3.064273667 | 1.615545 | 0.0636026 | 0.209152 |
| IGSF21 | 1 | 2.188809 | 1.130146 | 0.0636026 | 0.209152 |
| KCNA2 | 1 | 3.154114333 | 1.657235 | 0.0636026 | 0.209152 |
| TLR5 | 1 | 3.060803667 | 1.613911 | 0.0636026 | 0.209152 |
| VIPR2 | 1 | 6.112661333 | 2.611801 | 0.0636026 | 0.209152 |
| HIST1H1T | 1 | 4.333762667 | 2.11562 | 0.0636026 | 0.209152 |
| SLC4A11 | 1 | 3.405952333 | 1.768058 | 0.0636026 | 0.209152 |
| MAK | 1 | 2.680692333 | 1.422606 | 0.0636026 | 0.209152 |
| CDHR1 | 1 | 25.677904 | 4.682456 | 0.0636026 | 0.209152 |
| PPP1R32 | 1 | 4.505728667 | 2.17176 | 0.0636026 | 0.209152 |
| GCKR | 1 | 3.954844667 | 1.983621 | 0.0636026 | 0.209152 |
| FBXO40 | 1 | 2.491368333 | 1.316938 | 0.0636026 | 0.209152 |
| KCNH8 | 6.572459333 | 1.080832667 | −2.60429 | 0.0765225 | 0.209152 |
| DZIP1 | 7.629405 | 1.424371667 | −2.42124 | 0.0765225 | 0.209152 |
| ARHGAP22 | 5.822747333 | 1.086327667 | −2.42224 | 0.0765225 | 0.209152 |
| NKX2-3 | 4.342062 | 1.383955333 | −1.64958 | 0.0765225 | 0.209152 |
| PLA2R1 | 3.766903667 | 1.060624667 | −1.82846 | 0.0765225 | 0.209152 |
| SLC2A4 | 9.014120333 | 1.014388 | −3.15158 | 0.0765225 | 0.209152 |
| OC4-APO | 3.817754 | 1.071939667 | −1.8325 | 0.0765225 | 0.209152 |
| APOC2 | 3.817754 | 1.071939667 | −1.8325 | 0.0765225 | 0.209152 |
| CRTAC1 | 6.897075667 | 1.201323 | −2.52136 | 0.0765225 | 0.209152 |
| CHRNA2 | 2.411521667 | 13.60272067 | 2.49588 | 0.0765225 | 0.209152 |
| LAYN | 6.237966 | 1.028776 | −2.60015 | 0.0765225 | 0.209152 |
| SMOC2 | 5.391121667 | 1.083884667 | −2.31437 | 0.0765225 | 0.209152 |
| FAM155A | 4.772379 | 1.101041 | −2.11584 | 0.0765225 | 0.209152 |
| IFNG | 3.556483333 | 1.028776 | −1.78952 | 0.0765225 | 0.209152 |
| VPS9D1 | 1.838730333 | 11.01438633 | 2.582607 | 0.0765225 | 0.209152 |
| SLC35D3 | 1.572791333 | 4.819860333 | 1.615664 | 0.0765225 | 0.209152 |
| LRFN4 | 3.209380667 | 1.115103333 | −1.52512 | 0.0765225 | 0.209152 |
| ACKR4 | 6.268366 | 1.101041 | −2.50922 | 0.0765225 | 0.209152 |
| CYS1 | 1.38868 | 5.606796333 | 2.013462 | 0.0765225 | 0.209152 |
| HSD11B1L | 4.341861 | 1.159849333 | −1.90438 | 0.0765225 | 0.381797 |
| ACPP | 2.291559667 | 1.137013667 | −1.01108 | 0.0765225 | 0.381797 |
| RYR1 | 5.863586 | 1.296863 | −2.17676 | 0.0765225 | 0.381797 |
| ACKR4 | 6.268366 | 1.182685 | −2.40602 | 0.0765225 | 0.381797 |
| ZNF674 | 5.265564667 | 1.119454333 | −2.23379 | 0.0765225 | 0.381797 |
| SLC16A14 | 2.638718 | 1.228356 | −1.10311 | 0.0765225 | 0.381797 |
| NR6A1 | 1.286395667 | 5.917175 | 2.201574 | 0.0765225 | 0.381797 |
| ABCB9 | 1.225025333 | 2.864761 | 1.225603 | 0.0765225 | 0.381797 |
| GALNT18 | 1.204568333 | 3.584699 | 1.573336 | 0.0765225 | 0.381797 |
| PRRT2 | 1.122741 | 7.446128 | 2.729465 | 0.0765225 | 0.381797 |
| KHK | 1.771375333 | 5.577208333 | 1.654673 | 0.0765225 | 0.381797 |
| VSIG2 | 1.427099333 | 11.46590967 | 3.006193 | 0.0765225 | 0.381797 |
| DYTN | 2.382988333 | 13.68237767 | 2.521475 | 0.0765225 | 0.381797 |
| KIAA1549 | 1.041696 | 5.577024333 | 2.420561 | 0.0765225 | 0.381797 |
| NT5DC3 | 1.583743333 | 6.695642 | 2.079884 | 0.0765225 | 0.381797 |
| GRB14 | 1.041696 | 19.04747167 | 4.192593 | 0.0765225 | 0.381797 |
| SPHAR | 1.833860667 | 8.911786667 | 2.280831 | 0.0765225 | 0.381797 |
| CLDN12 | 1.437807667 | 4.590490667 | 1.674778 | 0.0765225 | 0.381797 |
| DGKI | 1.416959667 | 6.646423 | 2.229779 | 0.0765225 | 0.381797 |
| TAF6L | 1.142366333 | 4.517017333 | 1.983345 | 0.0765225 | 0.381797 |
| LIPC | 2.444002333 | 9.215349 | 1.914793 | 0.0765225 | 0.381797 |
| PPP1R13L | 1.166784 | 11.21800867 | 3.265207 | 0.0765225 | 0.381797 |
| PLXNB3 | 3.989695 | 15.27107967 | 1.936452 | 0.0765225 | 0.381797 |
In short, this report presented abundant exLRs in human plasma and the exLR dynamic changes. ExLRs originating from CD8 + T and CD4+ memory T cells were reduced during the perioperative period. Future studies will focus on the specific exLRs dynamically changed during the perioperative period and their origins to explore the impact on the occurrence and progression of CRC and the potential underlying mechanism.
Conclusions
To the best of our knowledge, we investigated the effects of surgical stress on the expression profile and cellular sources of blood exLR by exLR sequencing of CRC patients at four time points before and after surgery. In addition, we also investigated the function of these changed exLRs during the perioperative period. These findings open an avenue for the investigation of EVs at different time points and lay foundation to find out exLRs involved in the postoperative metastasis and recurrence of tumors.
Supplementary Material
Acknowledgements
We thank PI Shenglin Huang for the technical guidance. This work was supported by grants from National Natural Science Foundation of China (No. 81471852) and Shanghai Natural Science Foundation Program (20ZR1412900).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by grants from National Natural Science Foundation of China (No. 81471852); Natural Science Foundation of Shanghai (No. 20ZR1412900).
Highlights
Eleven DEGs and three DELRs dynamically changed during perioperative period.
The dynamically changed exLRs were related to various biofunctions.
EVs originating from immune cells decreased after surgery.
Disclosure statement
The authors declare that they have no conflict of interest.
Supplemental material
Supplemental data for this article can be accessed here
Consent for publication
We would like to submit the enclosed manuscript entitled ‘Dynamic plasma extracellular vesicle long RNA profiling changes during the peri-operative period’, which we wish to be considered for publication in ‘Bioengineered’. Manuscript is approved by all authors for publication. All the authors listed have approved the manuscript that is enclosed.
Ethics approval and consent to participate
All research and procedures involving human subjects were approved by the ethics committee of Fudan University Shanghai Cancer Center. The methods in this study were carried out in accordance with the approved guidelines by the ethics committee of Fudan University Shanghai Cancer Center. All donors provided written informed consents for participation
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- [3].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- [4].Addae JK, Gani F, Fang SY, et al. A comparison of trends in operative approach and postoperative outcomes for colorectal cancer surgery. J Surg Res. 2017;208:111–120. [DOI] [PubMed] [Google Scholar]
- [5].Labianca R, Nordlinger B, Beretta GD, et al. Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi64–72. [DOI] [PubMed] [Google Scholar]
- [6].Martinez-Useros J, Garcia-Carbonero N, Li W, et al. UNR/CSDE1 Expression Is Critical to Maintain Invasive Phenotype of Colorectal Cancer through Regulation of c-MYC and Epithelial-to-Mesenchymal Transition. J Clin Med. 2019;8(4):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Benson ABB-ST, Chan E, Chen YJ, et al. National Comprehensive Cancer Network, Localized colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11(5):519–528. [DOI] [PubMed] [Google Scholar]
- [8].Gustafsson UO, Oppelstrup H, Thorell A, et al. Adherence to the ERAS protocol is Associated with 5-Year Survival After Colorectal Cancer Surgery: a Retrospective Cohort Study. World J Surg. 2016;40(7):1741–1747. [DOI] [PubMed] [Google Scholar]
- [9].van der Bij GJ, Oosterling SJ, Beelen RH, et al. The perioperative period is an underutilized window of therapeutic opportunity in patients with colorectal cancer. Ann Surg. 2009;249(5):727–734. [DOI] [PubMed] [Google Scholar]
- [10].Hiller JG, Perry NJ, Poulogiannis G, et al. Perioperative events influence cancer recurrence risk after surgery. Nat Rev Clin Oncol. 2018;15(4):205–218. [DOI] [PubMed] [Google Scholar]
- [11].Raposo G, Stoorvogel W.. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. [DOI] [PubMed] [Google Scholar]
- [13].Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–289. [DOI] [PubMed] [Google Scholar]
- [14].Zhou R, Chen KK, Zhang J, et al. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer. 2018;17(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Del Re M, Biasco E, Crucitta S, et al. The detection of androgen receptor splice variant 7 in Plasma-derived Exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017;71(4):680–687. [DOI] [PubMed] [Google Scholar]
- [16].Nabet BY, Qiu Y, Shabason JE, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170(2):352–66 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Del Re M, Marconcini R, Pasquini G, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer. 2018;118(6):820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Zhao J, Yu S, et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem. 2019;65(6):798–808. [DOI] [PubMed] [Google Scholar]
- [19].Li C, Sun YD, Yu GY, et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell. 2020;38(5):734–47 e9. [DOI] [PubMed] [Google Scholar]
- [20].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Z, Zhu X, Huang S. Extracellular vesicle long non-coding RNAs and circular RNAs: biology, functions and applications in cancer. Cancer Lett. 2020;489:111–120. [DOI] [PubMed] [Google Scholar]
- [22].Wang W, Hong G, Wang S, et al. Tumor-derived exosomal miRNA-141 promote angiogenesis and malignant progression of lung cancer by targeting growth arrest-specific homeobox gene (GAX). Bioengineered. 2021;12(1):821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Desmond BJ, Dennett ER, Danielson KM. Circulating Extracellular Vesicle MicroRNA as Diagnostic Biomarkers in Early Colorectal Cancer-A Review. Cancers (Basel). 2019;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen X, Liu Y, Zhang Q, et al. Exosomal Long Non-coding RNA HOTTIP Increases Resistance of Colorectal Cancer Cells to Mitomycin via Impairing MiR-214-Mediated Degradation of KPNA3. Front Cell Dev Biol. 2020;8:582723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu J, Dong W, Liang J. Extracellular Vesicle-TransportedLong Non-Coding RNA (LncRNA) X Inactive-Specific Transcript (XIST) in Serum is a Potential Novel Biomarker for Colorectal Cancer Diagnosis. Med Sci Monit. 2020;26:e924448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen M, Xu R, Ji H, et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep. 2016;6(1):38397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xie L, Circular PZ. RNA circ_0000467 regulates colorectal cancer development via miR-382-5p/EN2 axis. Bioengineered. 2021;12(1):886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li Y, He X, Li Q, et al. EV-origin: enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput Struct Biotechnol J. 2020;18:2851–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu L, Sun HT, Wang S, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yu S, Li Y, Liao Z, et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69(3):540–550. [DOI] [PubMed] [Google Scholar]
- [31].Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Admyre C, Bohle B, Johansson SM, et al. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J Allergy Clin Immunol. 2007;120(6):1418–1424. [DOI] [PubMed] [Google Scholar]
- [33].Bhatnagar S, Shinagawa K, Castellino FJ, et al. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110(9):3234–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Aslan A, Erdem H, Celik MA, et al. Investigation of Insulin-Like Growth Factor-1 (IGF-1), P53, and Wilms’ Tumor 1 (WT1) Expression Levels in the Colon Polyp Subtypes in Colon Cancer. Med Sci Monit. 2019;25:5510–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ichinohasama R, Oji Y, Yokoyama H, et al. Sensitive immunohistochemical detection of WT1 protein in tumors with anti-WT1 antibody against WT1 235 peptide. Cancer Sci. 2010;101(5):1089–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ding X, Iyer R, Novotny C, et al. Inhibition of Grb14, a negative modulator of insulin signaling, improves glucose homeostasis without causing cardiac dysfunction. Sci Rep. 2020;10(1):3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene. 2016;35(20):2634–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Usta D, Sigaud R, Buhl JL, et al. A Cell-Based MAPK Reporter Assay Reveals Synergistic MAPK Pathway Activity Suppression by MAPK Inhibitor Combination BRAF-driven BRAF-Driven Pediatric Low-Grade Glioma Cells. Mol Cancer Ther. 2020;19(8):1736–1750. [DOI] [PubMed] [Google Scholar]
- [40].Whyte CEOM, Kara EE, Abbott C, et al. ACKR4 restrains antitumor immunity by regulating CCL21. J Exp Med. 2020;217(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wei R, Xiao Y, Song Y, et al. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dong Gao LJ. Exosomes in cancer therapy a novel experimental strategy. Am J Cancer Res. 2018;8(11):2165–2175. [PMC free article] [PubMed] [Google Scholar]
- [43].Cai Z, Yang F, Yu L, et al. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J Immunol. 2012;188(12):5954–5961. [DOI] [PubMed] [Google Scholar]
- [44].Hong CS, Sharma P, Yerneni SS, et al. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myeloid leukemia. Sci Rep. 2017;7(1):14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lugini L, Cecchetti S, Huber V, et al. Immune surveillance properties of human NK cell-derived exosomes. J Immunol. 2012;189(6):2833–2842. [DOI] [PubMed] [Google Scholar]
- [46].Aah AS-H, Maryam Behfar W, Sam-t RM, et al. NK Cell–derived Exosomes From NK Cells Previously Exposed to Neuroblastoma Cells Augment the Antitumor Activity of Cytokine-activated NK Cells. J Immunother. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


