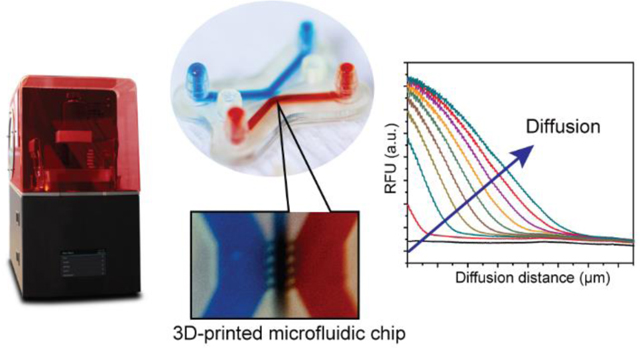
Abstract
Hydrogels allow for controlling the diffusion rate and amount of solute according to the hydrogel network and thus have found many applications in drug delivery, biomaterials, toxicology, and tissue engineering. This paper describes a 3D-printed microfluidic chip for the straightforward partitioning of hydrogel barriers between microchannels. We use a previously-reported 3-channel architecture whereby the middle channel is filled with a hydrogel – acting like a porous barrier for diffusive transport – and the two side channels act as sink and source; the middle channel communicates with the side channels via orthogonal, small capillary channels that are also responsible for partitioning the hydrogel during filling. Our 3D-printed microfluidic chip is simple to fabricate by stereolithography (SL), inexpensive, reproducible, and convenient, so it is more adequate for transport studies than a microchip fabricated by photolithographic procedures. The chip was fabricated in a resin made of poly(ethylene glycol) diacrylate (PEG-DA) (MW = 258) (PEG-DA-258). The SL process allowed us to print high aspect ratio (37:1) capillary channels (27 μm-width and 1 mm-height) and enable the trapping of liquid-phase hydrogels in the hydrogel barrier middle channel. We studied the permeability of hydrogel barriers made of PEG-DA (MW = 700) (PEG-DA-700, 10% polymer content by wt. in water) – as a model of photopolymerizable barriers – and agarose (MW = 120,000, 2% polymer content by wt. in water) – as a model of thermally-gelled barriers. We measured the diffusion of fluorescein, 10k-dextran-Alexa 680 and BSA-Texas Red through these barriers. Fluorescein diffusion was observed through both 10% PEG-DA-700 and 2% agarose barriers while 10k-dextran-Alexa 680 and BSA-Texas Red diffused appreciably only through the 2% agarose hydrogel barrier. Our microfluidic chip facilitates the tuning of such barriers simply by altering the hydrogel materials. The straightforward trapping of selective barriers in 3D-printed microchannels should find wide applicability in drug delivery, tissue engineering, cell separation, and organ-on-a-chip platforms.
Graphical Abstract
We demonstrated a stereolithographically 3D-printed chip for the straightforward injection of hydrogel barriers in microchannels and the facile measurement of molecular diffusivities through these hydrogels.
Introduction
Exploring mass transport across semi-permeable materials is critical for developing analytical platforms in a variety of fields, such as protein synthesis1, microphysiological cell culture models2, epigenetics3, immunoisolation4, and selective protein filters5. Hydrogels are a particular class of porous materials that, due to their tunable transport properties, excellent biocompatibility6, and ease of manufacture, have been widely applied to drug delivery7, biomaterials research8,9, tissue engineering10, and toxicology11. Researchers have integrated hydrogel materials into microfluidic devices for developing biosensors and cell culture systems with complex biofunctionalities.12–17 However, traditional microfluidic technologies have used photolithography- and molding-based processes (including soft lithography) that are labor- and time-intensive and are restricted to 2D planar/layered designs.18–21 In addition, microfluidic devices often require the intervention of a trained specialist for bonding of each layer and additional extra-steps such as the fabrication of inlets/outlets and the integration of functional materials including hydrogels, porous membranes, and microbeads.22–25
3D-printing technology overcomes many of the fabricating limitations of conventional microfluidic devices.26–29 As selected examples, Miri et al. presented a stereolithography (SL)-based bioprinting platform for fabricating a 3D heterogeneous hydrogel microfluidic device.30 This platform provides a multi-material fabrication capability at high spatial resolution by using a digital micromirror device based bioprinter using gelatin methacryloyl (GelMA) and poly(ethylene glycol) diacrylate (PEG-DA) (MW = 700) (PEG-DA-700); the biocompatibility was validated by introducing cell-laden GelMA into a microfluidic device for fabricating cellularized constructs. Zhang and Larsen reported SL printing of PEG-DA-700 hydrogel printing for fabricating biofunctionalized complex 3D perfusion networks.31 PEG-DA-700 resin was employed to construct an in vitro vascular network-like hydrogel device with 3D complexity, facile perfusion setup, and vascular networks and biocompatibility for culturing cells. However, the molecule size which can diffuse across PEG-DA-700 hydrogel is very restricted due to the small pore size of the hydrogel. Our group has studied SL-based high-resolution printing to fabricate biomicrofluidic devices utilizing a formulation based on PEG-DA (MW = 258) (PEG-DA-258) and a UV absorber.32–34 Recently, we developed an SL co-printing process to build a selective porous hydrogel barrier using PEG-DA (MW = 575) (PEG-DA-575) or 40% PEG-DA-700 in the microfluidic channel in order to control the diffusion of small ions or molecules; the technique reduces overall fabricating costs and shortens the duration of design iteration.35 However, the types of photopatternable hydrogels that can be used as porous barriers are limited (for ex., PEG-DA at an arbitrary MW is not commercially available) and it is difficult to optimize production conditions for each hydrogel material.
In order to simplify the fabrication of porous barriers, here we adopted a different strategy that consists of trapping a porous hydrogel barrier (between a sink and a source microchannel) in a 3D-printed chamber. This strategy, which was previously demonstrated in PDMS devices20, drastically simplifies the process for fabricating a porous barrier in a microfluidic environment and offers a virtually unlimited choice of hydrogels for building porous barriers. Our method starts by using a PEG-DA-258 based resin to 3D-print high aspect-ratio, high-resolution microchannels; the design is based on a previously-reported PDMS 3-channel design, whereby the center channel is filled with hydrogel (acting like a porous barrier) while the side channels act as source and sink channels.20 The center channel communicates with the source/sink channel via capillary channels that are too small for the hydrogel to penetrate (i.e. causing “capillary pinning”36), effectively trapping the hydrogel in the center channel during hydrogel filling and allowing for diffusion of solutes into the hydrogel during device operation. The porosity and chemical structure of the hydrogels is used to select the size and nature of molecules that can be transported across the porous barriers. We demonstrate size-based selective transport (and exclusion) of molecules through porous barriers made from photocured 10% PEG-DA-700 and thermally-gelled 2% agarose, both entrapped in the center channel of our 3D-printed chip.
Experimental
Resins for stereolithographic 3D-printing
For 3D-printing the microfluidic device, we used a photocurable resin that consisted of an acrylate macromer, PEG-DA-258 (Sigma Aldrich), a photoinitiator, 0.6% (w/w) Irgacure 819 (IRG) (BASF Corporation), and a photosensitizer, 0.6% (w/w) 2-isopropyl thioxanthone (ITX). We introduced the three resin components into a polypropylene tube wrapped with aluminium foil to prevent photopolymerization by ambient light. We vigorously mixed the resin using a mini vortexer and heated the mixture in a 70 oC oven for 30 min to completely dissolve the ITX and IRG. For preparing the hydrogels, we used two different formulations – a 10% PEG-DA-700 (w/w in water) resin containing 0.6% IRG, and a 2% (w/w) agarose (Sigma Aldrich) solution. The 10% PEG-DA-700 hydrogel resin was prepared similar to the PEG-DA-258 resin. The 2% agarose was prepared by heating the solution in a microwave oven (for about a minute) until the agarose powder was completely dissolved.
3D-printer setup and fabrication of the 3D-printed chip for partitioning hydrogels
The 3D-printed hydrogel partitioning microfluidic chip was designed using Inventor® (Autodesk, San Rafael, CA) and printed with the DLP-SL printer Asiga Pico2 HD (Asiga, Sydney, Australia) fitted with a 385 nm-wavelength LED (intensity: 32.21 mW cm−1) projector. The Pico2 HD printer has a projected pixel (XY) resolution of 27 μm and a Z-layer resolution of 10 μm. The high absorbance of our photocurable resin at the wavelength of the projector (385 nm), together with the optical and mechanical features of the printer, enable the 3D-printing of high-resolution biomicrofluidic devices.33 Prior to printing, the 3D CAD file was converted to STL format and then sliced using Asiga Composer which is an STL support and build preparation package from Asiga. The slice images and the UV exposure settings for each layer were loaded to the 3D-printer for fabricating the device. Calibrating the zero position of the build plate of the 3D-printer before starting the print is necessary for successful printing.
To fabricate a transparent microfluidic chip, glass slides (75 mm (L) × 50 mm (W) × 1.0 mm (T)) were used as a substrate for building the print.32 Before printing, the glass slides were sequentially rinsed with acetone, isopropyl alcohol, and de-ionized (D.I.) water and then completely dried in a 70 oC oven overnight. The surface of the glass slide was derivatized with 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) (Sigma-Aldrich) at 85 oC for 8 hrs after activating with oxygen plasma (Deiner Zepto, Thierry Corporation) at 60 W and 670 mTorr pressure for 180 s. The silanized glass slides were coated on one side with uncured PEG-DA-258 resin and attached to the aluminum build plate by brief exposure to UV light. For fabricating the 3D-printed hydrogel partitioning chip, the Asiga Pico2 HD build tray was filled with ~50 mL of the PEG-DA-258 resin. After printing the object, the glass slide with the printed chip was removed from the build plate and the cured PEG-DA-258 residue scratched off from the back side of the glass slide using a razor blade. Uncured PEG-DA-258 resin on the surface, and inside the microchannels of the 3D-printed chip were cleaned using D.I. water and dried with pressurized air.
To produce the hydrogel barrier in between the sink and source microchannels, two different methods were applied depending on the hydrogel material. For the 10% PEG-DA-700 UV curable hydrogel, the resin was directly added to the hydrogel reservoir and flowed into the hydrogel barrier channel due to capillary action. After filling the hydrogel barrier channel, the microfluidic channels were filled with mineral oil to prevent the 10% PEG-DA-700 resin from encroaching into the flanking channels during the UV curing process. The 10% PEG-DA-700 resin in the hydrogel barrier channel was photo-cured in a UV transilluminator box for 1 hr to make the porous barrier. The mineral oil in the microfluidic channels was removed by sequential aspiration and flushing with acetonitrile to eliminate any oil residue. For the 2% agarose hydrogel fabrication, the heated (liquid) agarose solution was quickly injected into the hydrogel barrier channel and allowed to cool down to form a gel. We used a Nikon SMZ-1500 stereomicroscope fitted with a Canon Rebel DSLR camera to take images of the 3D-printed devices. The microfluidic chips with the hydrogel barriers were stored in a water bath to prevent drying of the hydrogel due to evaporation.
Diffusion analysis
To analyze the diffusion of molecules through the hydrogel barriers, three fluorescent molecules of different molecular weights were utilized - 0.1 mM fluorescein (M.W. = 332 Da), 0.2 mM 10k-dextran labeled with Alexa 680 (M.W. = 10 kDa) and 1.0 mM bovine serum albumin (BSA) labeled with Texas Red (M.W. = 66.5 kDa). The fluorescent molecule solutions were injected into the source channel of the 3D-printed chip while PBS buffer was inserted into the sink channel. Time-lapse fluorescent images were captured for 90 min using an inverted epifluorescence microscope (Nikon Eclipse Ti, Plan Apo λ 2× objective). The images were analyzed using ImageJ (NIH, Bethesda, MD) to characterize the diffusion process.
Results and discussion
Resin selection
A detailed characterization of a PEG-DA-258 based resin for 3D-printing transparent high-resolution microfluidic devices and the rationale behind selecting the different resin components have been described in our earlier publications.32,33 Briefly, the high crosslinking density of low molecular weight PEG-DA (M.W. = 258) makes UV-polymerized PEG-DA-258 structures water-impermeable and therefore, suitable for fabricating microfluidic channel walls.32,37 The high absorption and photochemical efficiency of the photoinitiator Irgacure 819 (IRG) at the wavelength (385 nm) of the UV-LED projector of the Pico2 HD 3D-printer make it an effective photoinitiator, even at low (0.6%) concentrations.32 The high molar extinction coefficient of the photosensitizer 2-isopropyl thioxanthone (ITX) at 385 nm reduces the penetration depth of the UV-light into the resin26 and enables the printing of high-resolution channels by preventing cross-linking of uncured resin inside the microchannel voids.33 Although IRG and ITX are yellow-colored solids, their high molecular absorptivity at 385 nm (846.9 M−1cm−1 for IRG and 5155 M−1 cm−1 for ITX) allows them to be added to the resin at relatively low concentrations (0.6%), thereby minimizing the coloration of the final 3D-printed devices.26,33
Fabrication of the 3D-printed molecule diffusion chip
A schematic of the stereolithographic fabrication process of the microfluidic chip for partitioning hydrogels is shown in Fig. 1. Prior to printing, the designed device was sliced into 10 μm-thick layers. The PEG-DA-258 resin tray was loaded onto the Asiga Pico2 HD 3D-printer and a silanized glass slide was attached to the build plate using a few drops of PEG-DA-258 which was polymerized with a UV lamp. The silane, 3-(trimethoxysilyl)propyl methacrylate or TMSPMA, ends in an acrylate group. Therefore, adhesion of the acrylate resin (PEG-DA-258) is promoted by the acrylate group in TMSPMA on the surface when photopolymerization of the first layer of PEG-DA-258 starts by exposure of the resin to UV light. The first two layers were printed with a long (4.5 s) UV exposure to ensure strong adhesion of the printed parts to the silanized glass plate. After fabricating a ~100 μm-thick base, the channels, roofs, inlets, and reservoirs were printed with a short (0.15 s) exposure time for each layer (10 μm slices).
Fig. 1.
Cross-sectional schematic of the process for 3D-printing the microfluidic chip by stereolithography. The fabrication process entails step-by-step building of (i) the bottom layer, (ii) the channel layer, (iii) the roof layer, and (iv) the inlets and reservoirs. In reality, fabrication of the 3D-printed microchip from (i) bottom layer to (iv) inlets and reservoirs proceed upside down, however, we show the process upside up for clarity. The SL printing process is followed by a post-printing process (outside of the 3D-printer) consisting of (v) channel washing, (vi) hydrogel solution loading, (vii) porous barrier fabrication with UV-curing or thermal gellation, and (viii) completion of the 3D-printed chip.
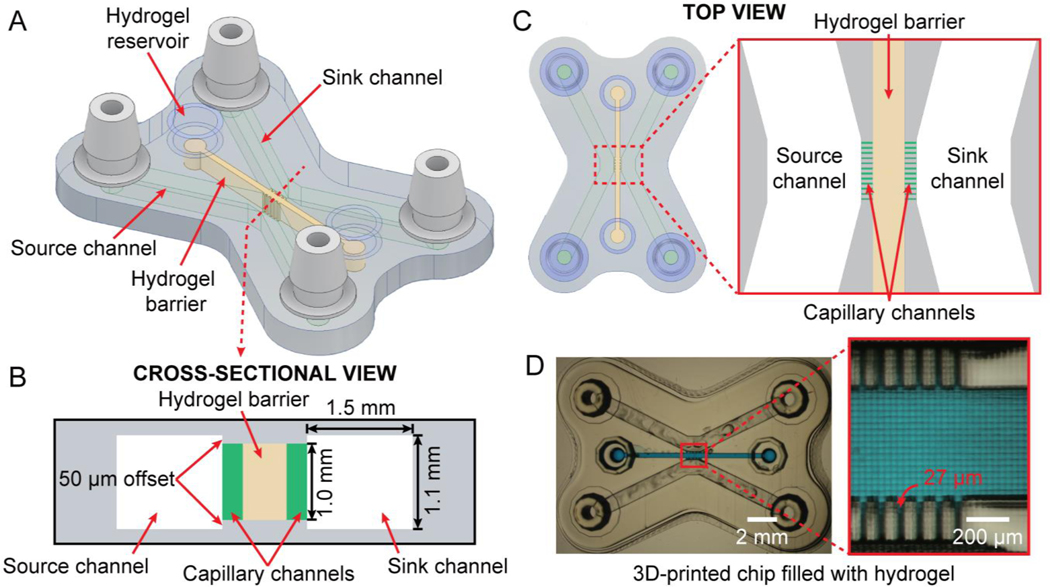
The architecture of our microfluidic chip is explained in Fig. 2. The 3D CAD design, comprising the central channel and the two flanking channels (sink/source), with capillary channels communicating the central and flanking channels, is illustrated in Fig. 2A. Fig. 2B displays a cross-sectional view of the device. Integrated barb-connectors on either side serve as inlets and outlets. The central hydrogel barrier channel (1.0 mm-height × 500 μm-width) is connected to the source and sink channels through a comb-like high aspect-ratio, single-pixel wide capillary channels. Optimizing the geometry of the interface between the capillary and the flanking channels can increase the capillary burst pressure required by an expanding hydrogel meniscus to leak into the adjacent compartment. The capillary burst pressure can be increased by ensuring that the capillary channels expand into the source and sink channels in all directions. Therefore, the roof and floor of the flanking compartments are offset by 50 μm to the top and bottom of the capillary channels, respectively (that is, while the capillary channels are 1 mm-high, the source and sink channels are 1.1 mm-high) – a feature that would be laborious to achieve by photolithography or molding, requiring several levels of photolithographic patterning. Fig. 2C shows a top view of the 3D-printed chip depicting the hydrogel barrier (brown) and the comb-like capillary channels (green). The 200 μm-long capillary channels (1.0 mm-height × 27 μm-width) pin the hydrogel within the barrier due to surface tension and prevents it from spilling into the source and sink channels.20 Fig. 2D shows a micrograph of the 3D-printed chip with an enlarged view of the capillary channel part. The capillary channels in our 3D-printed molecule diffusion chip have a 37:1 aspect ratio, again a ratio that is very difficult to achieve by photolithography or soft lithography.38 The width of the capillary channels (27 μm) corresponds to the width of a single pixel of the printer’s projector, and the width of the walls in Fig. 2D corresponds to the width of two pixels (54 μm). Interestingly, high aspect ratio channels are both necessary for 3D-printing (the farther the roof is from the floor, the easier it is to avoid channel clogging in stereolithography) and beneficial for establishing gradients (more surface area for mass transport).
Fig. 2.
Structure of the 3D-printed hydrogel partitioning chip. (A) Oblique view of the 3D CAD design containing three channels: two flanking microfluidic channels (source and sink) and a central channel filled with a hydrogel barrier, connected to the source/sink channels via 27 μm-wide comb-like capillary channels. (B) Cross-sectional schematic of the 3D-printed chip; the capillary channels are depicted in green for clarity. (C) Top-view of the CAD design of the chip and a close-up view of the source/sink channels and the hydrogel barrier connected via capillary channels (green). (D) Micrograph of the 3D-printed hydrogel partitioning chip and a magnified image of the hydrogel barrier entrapped by the capillary channels. The hydrogel barrier channel is filled with blue dye for visualization purposes.
Before injecting the hydrogel barrier resin, the printed device was thoroughly washed with D.I. water to remove residual uncured resin from the microchannels in a dark room and completely dried using compressed air. To fabricate the 10% PEG-DA-700 hydrogel barrier, the 10% PEG-DA-700 resin was loaded into one of the reservoirs and allowed to flow into the hydrogel barrier channel by capillary force. Mineral oil was inserted into the flanking microfluidic channels (acting as source and sink) to prevent leakage of the 10% PEG-DA-700 resin during UV irradiation. Because of the low amount of UV crosslink-able groups in the 10% PEG-DA-700 resin, we need a long UV exposure (1 hr) to completely cure the hydrogel barrier. Mineral oil was removed from the channels by aspiration and the channels were washed using acetonitrile and D.I. water. A 2% agarose hydrogel barrier was fabricated by injecting the liquefied agarose solution at 95 oC. The liquid-phase 2% agarose also flowed into the hydrogel barrier channel due to capillary force, and after cooling down, the 2% agarose hydrogel was formed in the center channel. The 3D-printed chips with the porous barriers were stored in D.I. water to ensure the integrity of the hydrogel structure.
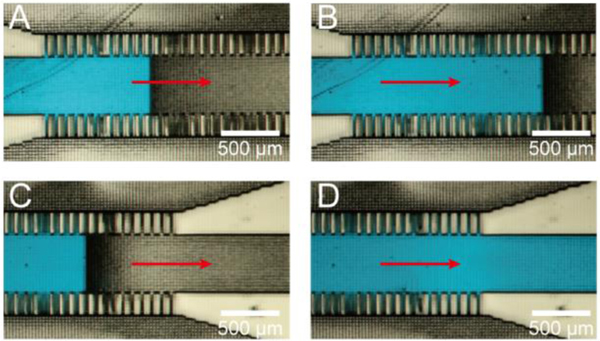
Hydrogel introduction is in itself a remarkable process when observed under the microscope. Fig. 3 displays a hydrogel resin being introduced into the hydrogel barrier channel and capillary channels. We added a few droplets of an aqueous solution of a blue food-coloring dye (FD&C Blue) to the resin for visualization purposes. From Fig. 3A–D, the blue dye slowly moves from the left to the right by first filling the capillary channels and then the hydrogel barrier channel. Interestingly, the advancing liquid profile in the hydrogel barrier channel is rectangular due to the phase-guiding effect of the pixelated channel surfaces.39 Supplementary video 1 shows the hydrogel introduction into hydrogel barrier channel and capillary channel.
Fig. 3.
Hydrogel precursor (colored in blue) introduction into the hydrogel channel. (A-D) Sequence of images depicting the stepwise advancement of fluid with a flat front due to phase guiding by the pixelated channel walls and roof. Note how capillary forces tend to draw fluid into the side capillary channels before fluid advances into the wider hydrogel barrier channel. See supplementary video 1.
Compared to the previous work by our group35, this design affords great freedom to select a hydrogel. By fabricating a channel that can contain any hydrogel barrier in between the source and the sink channels, it is possible to fabricate various types of hydrogel barriers between the flanking channels, such as extracellular matrix (ECM) and collagen, as well as precursors capable of generating hydrogels through UV curing. Hydrogel compositions that are cytocompatible and physiologically relevant can thus be easily incorporated into experiments.
Diffusion measurements using a 10% PEG-DA-700 hydrogel barrier
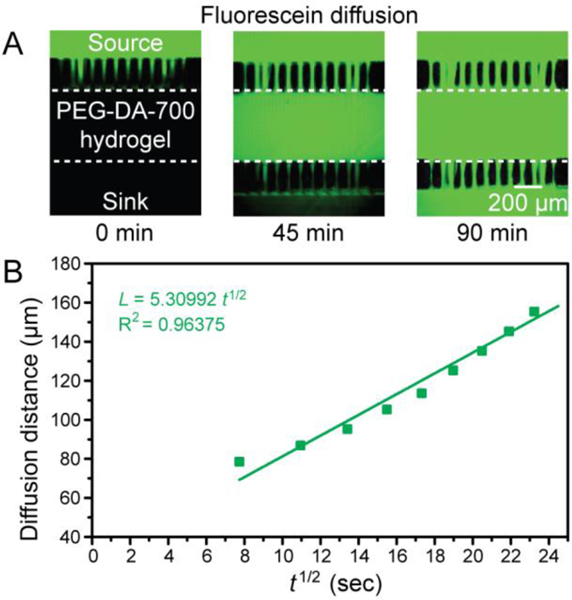
Using our chip, we measured the diffusion of different solutes across a 10% PEG-DA-700 hydrogel barrier introduced into the central channel. We compared the transport of fluorescein (0.1 mM), Alexa 680 labeled 10k-dextran (0.2 mM), and Texas Red labeled BSA (1 mM) using an inverted epifluorescence microscope. These fluorescent compounds were inserted into the source channel and PBS buffer was inserted into the sink channel. The changes in the fluorescence intensity profile as the molecules were allowed to diffuse from the source to the sink channel were observed for 90 min. When the molecules were loaded into the source channel, green signal (fluorescein), magenta signal (10k-dextran-Alexa 680), and red signal (BSA-Texas Red) were initially observed only in the source channel, while no signal was detected at 0 min in the hydrogel barrier and the sink channel. Only fluorescein was seen to diffuse through the 500 μm-wide 10% PEG-DA-700 hydrogel barrier after 45 mins. After 90 mins, fluorescein was observed in the hydrogel barrier and in the sink channel (Fig. 4A) while 10k-dextran and BSA stayed in the source channel (Fig. S1). We provide a video of fluorescein diffusion through the 10% PEG-DA-700 hydrogel barrier in Supplementary video 2. From the measured data, we were able to determine the diffusivity of fluorescein in 10% PEG-DA-700 as follows.
Fig. 4.
Fluorescein diffusion analysis using the 3D-printed hydrogel partitioning chip with a 10% PEG-DA-700 hydrogel barrier. (A) Fluorescence image of fluorescein (colored in green) diffusion at 0 min and 90 min through the 10% PEG-DA-700 hydrogel barrier. The dotted line depicts the boundary of the hydrogel barrier channel. (B) A plot of the characteristic diffusion length of fluorescein across the 10% PEG-DA-700 hydrogel barrier as a function of the square root of time (dots), the green line is a linear fit of the dots, L = 5.31 t1/2 (adjusted R2 = 0.96).
To calculate the diffusivity of a solute across a hydrogel barrier, we applied Fick’s second law of diffusion:40
| (1) |
where C(x,t) indicates the time-evolving concentration of solute in water along the x axis (which is defined as an axis parallel to the capillary channels) and D represents the diffusivity of the molecule in the hydrogel barrier.
A one-dimensional analytical solution to equation (1) can be written as follows,
| (2) |
where indicates the solute concentration at the junction of the source channel and the hydrogel barrier and indicates the initial solute concentration in the PEG-DA-700 barrier.40 In this experiment, we assume that and are constant.
The characteristic diffusion length () at a given time () is defined as the distance from the source boundary where the concentration of the diffusing molecule is 50% of the source concentration () and can be approximated as
| (3)40 |
To calculate the diffusivity of fluorescein in the 10% PEG-DA-700 hydrogel barrier, time-lapse images of fluorescein diffusion through the hydrogel barrier was taken from 0 min to 90 min using an inverted epifluorescence microscope and analyzed with ImageJ to obtain the diffusion profiles. The experimentally observed characteristic diffusion length () of fluorescein as a function of the square root of time () was plotted and a linear regression analysis carried out to estimate the diffusivity (). The fluorescein molecule diffusivity in the 10% PEG-DA-700 was evaluated to be 2.82 × 10−7 cm2 s−1 from the slope () of the fitted curve (L = 5.31 , adjusted R2 = 0.96) in Fig. 4B.
Diffusion measurements using a 2% agarose barrier
The advantage of our 3D-printed hydrogel partitioning chip design is that we can easily change the hydrogel material, which changes the pore size of the diffusion barrier and enables selective transport of different solutes across the barrier. We altered the hydrogel barrier material to 2% agarose and measured again the diffusivity of fluorescein, Alexa 680 labeled-10k-dextran, and Texas Red labeled-BSA across an agarose barrier. The high viscosity of the 2% agarose solution allowed us to increase the width of the capillary channels to 100 μm, which also provides 63.7% more area of contact between the hydrogel and the sink/source solutions compared to the design used for the PEG-DA-700 barrier. To monitor the transport of the three molecules through the 2% agarose hydrogel barrier in the 3D-printed chip, time-lapse images were obtained from 0 min to 10 min with the inverted epifluorescence microscope and examined using ImageJ. As shown in Fig. 5A, fluorescence signals were detected only in the source channel at t=0 min while no signal was detected initially in the 2% agarose hydrogel barrier and the sink channel. Fluorescein, 10k-dextran-Alexa 680, and BSA-Texas Red gradually diffused through the 2% agarose hydrogel barrier and spread into the sink channel (Fig. 5B). To determine the diffusivity of the three molecules in the 2% agarose hydrogel barrier, we applied Fick’s second law of diffusion as above. The diffusivities of fluorescein, 10k-dextran-Alexa 680, and BSA-Texas Red in the 2% agarose hydrogel barrier were calculated by plotting the experimentally observed characteristic diffusion length () as a function of the square root of time () (Fig. 5C). The diffusivity of fluorescein, 10k-dextran-Alexa 680, and BSA-Texas Red in 2% agarose were estimated to be 1.06 × 10−5 cm2 s−1, 2.09 × 10−6 cm2 s−1, and 1.61 × 10−6 cm2 s−1, respectively, from the slope () of the fitted curves in Fig. 5C. This methodology can be straightforwardly generalized to measuring the diffusivity of any fluorescently-labelled molecule in virtually any hydrogel.
Fig. 5.
Diffusion analysis of fluorescein (green), 10k-dextran-Alexa 680 (purple), and BSA-Texas Red (red) using the 3D-printed hydrogel partitioning chip with a 2% agarose hydrogel barrier. (A-B) Fluorescence image of fluorescein (colored in green), 10k-dextran-Alexa 680 (colored in magenta), and BSA-Texas Red (colored in red) diffusion at 0 min and 10 min through the 2% agarose hydrogel barrier. (C) A plot of the characteristic diffusion length of fluorescein, 10k-dextran-Alexa 680, and BSA-Texas Red across the 2% agarose hydrogel barrier as a function of the square root of time (dots). The green line is a linear fit of the green dots (diffusion length of fluorescein), L = 32.62729 t1/2 (adjusted R2 = 0.99584). The magenta line is a linear fit of the magenta dots (diffusion length of 10k-dextran-Alexa 680), L = 14.44622 t1/2 (adjusted R2 = 0.99815). The red line is a linear fit of the red dots (diffusion length of BSA-Texas Red), L = 12.68639 t1/2. (adjusted R2 = 0.98712).
3D-printed porous barriers could be used to mimic various dynamic biological and physicochemical processes that have been, until now, mimicked by traditional photolithographic or soft lithographic methods. Dynamic biological processes such as cancer metastasis41, angiogenesis42, cell to cell interactions43, blood brain barrier44, biomolecules/cells capture and transport45, and organ-on-a-chip46,47 has been carried out by using a porous hydrogel barrier. In addition, hydrogel-based chips have been applied to the programmable delivery and sustained release of mock drugs48, osmotic pumping toward wearable human-device interface49, and impedance sensing platform for cell viability assay or drug toxicity screening50.
Conclusions
In summary, we have developed a DLP-SL printing method to fabricate a device that can partition injectable hydrogels into microfluidic compartments. The microchannels of the 3D-printed device, which consist of a central hydrogel compartment flanked on either side by source and sink channels, were fabricated using a PEG-DA-258 resin that can be easily prepared in any laboratory. High aspect-ratio (37:1) single-pixel wide comb-like capillary channels orthogonal to the axis of the source and sink channels enabled the trapping of the hydrogel in the central channel. Our simple 3D-printed chip architecture prevents cross-channel convective transport but permits selective diffusive transport of molecules across the barrier. The molecular selectivity of the porous barrier can be tuned by the nature and structure of the hydrogel entrapped within the central channel. We selected 10% PEG-DA-700 and 2% agarose as the hydrogel barrier material to analyze the diffusion – and, crucially, measure the diffusivity – of differently sized molecules in the hydrogel of choice. A low-MW molecule (fluorescein) could diffuse through both 10% PEG-DA-700 and 2% agarose barrier, whereas large-MW molecules (10k-dextran-Alexa 680 and BSA-Texas Red) only passed through the 2% agarose hydrogel barrier. The 3D-printed chip described here will enable simple, reproducible, inexpensive, and convenient fabricating of microfluidic devices with integrated hydrogel barriers that can be used in the fields of drug delivery, tissue engineering, cell separation, epigenomics research, protein synthesis, and organ-on-a-chip platforms, among others.
Supplementary Material
Acknowledgements
This research was partially supported by the National Institutes of Health (Grant number: 5R01CA181445), the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2018R1C1B5085897), and the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT (MSIT) of Korea as Global Frontier Project (Grant number: H-GUARD_2014M3A6B2060302).
Footnotes
Electronic Supplementary Information (ESI) available: hydrogel precursor introduction into hydrogel barrier and fluorescein diffusion across hydrogel. See supplementary information.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Timm AC, Shankles PG, Foster CM, Doktycz MJ, and Retterer ST, Small, 2016, 12, 810. [DOI] [PubMed] [Google Scholar]
- 2.Mondrinos M, Yi Y–S, Wu N–K, Ding X, and Huh D, Lab Chip 2017, 17, 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma S, Revenga MF, Sun Z, Sun C, Murphy TW, Xie H, González-Maeso J, and Lu C, Nat. Biomed. Eng, 2018, 2, 183. [PMC free article] [PubMed] [Google Scholar]
- 4.La Flamme KE, Popat KC, Leoni L, Markiewicz E, La Tempa TJ, Roman BB, Grimes CA, and Desai TA, Biomaterials, 2007, 28, 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom AJ, Sipe DM, Shishkin Y, Ke Y, Devaty RP, and Choyke WJ, Biomed. Microdevices, 2004, 6, 261. [DOI] [PubMed] [Google Scholar]
- 6.Seliktar D, Science, 2012, 336, 1124. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Y, and Park K, Adv. Drug. Deliv. Rev, 2001, 53, 321. [DOI] [PubMed] [Google Scholar]
- 8.Lutolf MP, Nat. Mater, 2009, 8, 451. [DOI] [PubMed] [Google Scholar]
- 9.Caliari SR, and Burdick JA, Nat. Methods, 2016, 13, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KY, and Mooney DJ, Chem. Rev, 2001, 101, 1869. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen EH, Daly WT, Le NNT, Farnoodian M, Belair DG, Schwartz MP, Lebakken CS, Ananiev GE, Saghiri MA, Knudsen TB, Sheibani N, and Murphy WL, Nat. Biomed. Eng, 2017, 1, 0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skardal A, Shupe T, and Atala A, Drug Discov. Today, 2016, 21, 1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, and Radisic M, Lab Chip, 2017, 17, 2395. [DOI] [PubMed] [Google Scholar]
- 14.Sackmann EK, Fulton AL, and Beebe DJ, Nature, 2014, 507, 181. [DOI] [PubMed] [Google Scholar]
- 15.Sontheimer-Phelps A, Hassell BA, and Ingber DE, Nat. Rev. Cancer, 2019, 19, 65. [DOI] [PubMed] [Google Scholar]
- 16.Deligkaris K, Tadele TS, Olthuis W, and van den Berg A, Sens. Actuators B Chem, 2010, 147, 765. [Google Scholar]
- 17.Chung BG, Lee K–H, Khademhosseini A, and Lee S–H, Lab Chip, 2012, 12, 45. [DOI] [PubMed] [Google Scholar]
- 18.Huang CP, Lu J, Seon H, Lee AP, Flanagan AL, Kim H–Y, Putnam AJ, and Jeon NL, Lab Chip, 2009, 9, 1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung S, Sudo R, Mack PJ, Wan C–R, Vickerman V, and Kamm RD, Lab Chip, 2009, 9, 269. [DOI] [PubMed] [Google Scholar]
- 20.Mosadegh B, Huang C, Park JW, Shin HS, Chung BG, Whang S–K, Lee K–H, Kim HJ, Brody J, and Jeon NL, Langmuir, 2007, 23, 10910. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Li L, and Luo C, Lab Chip, 2016, 16, 1757. [DOI] [PubMed] [Google Scholar]
- 22.Kim YT, Lee D, Heo HY, Sim JE, Woo KM, Kim DH, Im SG, and Seo TS, Biosens. Bioelectron, 2016, 78, 489. [DOI] [PubMed] [Google Scholar]
- 23.Fan X, Jia C, Yang J, Mao H, Jin Q, and Zhao J, Biosens. Bioelectron, 2015, 71, 380. [DOI] [PubMed] [Google Scholar]
- 24.Benam KH, Villenave R, Lucches C, Varone A, Hubeau C, Lee H–H, Alves SE, Salmon M, Ferrante TC, Weaver JC, Bahinski A, Hamilton GA, and Ingber DE, Nat. Methods, 2016, 13, 151. [DOI] [PubMed] [Google Scholar]
- 25.Kim YT, Lee D, Heo HY, Kim DH, and Seo TS, Lab Chip, 2015, 15, 4148. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee N, Parra-Cabrera C, Kim YT, Kuo AP, and Folch A, Adv. Mater, 2018, 30, 1800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharjee N, Urrios A, Kang S, and Folch A, Lab Chip, 2016, 16, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Au AK, Huynh W, Horowitz LF, and Folch A, Angew. Chem. Int. Ed, 2016, 55, 3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macdonald NP, Cabot JM, Smejkal P, Guijt RM, Paull B, and Breadmore MC, Anal. Chem, 2017, 89, 3858. [DOI] [PubMed] [Google Scholar]
- 30.Miri AK, Nieto D, Iglesias L, Hosseinabadi HG, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen S, Shin SR, Zhang YS, and Khademhosseini A, Adv. Mater, 2018, 30, 1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, and Larsen NB, Lab Chip, 2017, 17, 4273. [DOI] [PubMed] [Google Scholar]
- 32.Urrios A, Parra-Cabrera C, Bhattacharjee N, Gonzalez-Suarez AM, Rigat-Brugarolas LG, Nallapatti U, Samitier J, DeForest CA, Posas F, Garcia-Cordero J, and Folch A, Lab Chip, 2016, 16, 2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo AP, Bhattacharjee N, Lee Y–S, Castro K, Kim YT, and Folch A, Adv. Mater. Technol, 2019, 1800395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y–S, Bhattacharjee N, and Folch A, Lab Chip, 2018, 18, 1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YT, Castro K, Bhattacharjee N, and Folch A, Micromachines, 2018, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gumuscu B, Bomer JG, van den Berg A, and Eijkel JCT, Lab Chip, 2015, 15, 664. [DOI] [PubMed] [Google Scholar]
- 37.Gong H, Bickham BP, Woolley AT, and Nordin GP, Lab Chip, 2017, 17, 2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folch A, Ayon A, Hurtado O, Schmidt MA, Toner M, J. Biomech. Eng 2009, 121, 28. [DOI] [PubMed] [Google Scholar]
- 39.Vulto P, Podszun S, Meyer P, Hermann C, Manz A, and Urban GA, Lab Chip, 2011, 11, 1561. [DOI] [PubMed] [Google Scholar]
- 40.B. J. Kirby, Cambridge University Press: Cambridge, UK, 2010, ISBN 978–0-521–11903-0.
- 41.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, and Kamm RK, Proc. Natl. Acad. Sci. USA, 2012, 109, 13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, and Kamm RD, Proc. Natl. Acad. Sci. USA, 2015, 112, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai J, Adriani G, Dang T–M, Tu T–Y, Penny H–XL, Wong S–C, Kamm RD, and Thiery J–P, Oncotarget, 2015, 6, 25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bang S, Lee S–R, Ko J, Son K, Tahk D, Ahn J, Im C, Jeon NL, Sci. Rep, 2017, 7, 8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HS, Chu WK, Zhang K, and Huang X, Lab Chip, 2013, 13, 3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh D, Hamilton GA, and Ingber DE, Trends Cell Biol, 2011, 21, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huh D, Torisawa Y, Hamilton GA, Kim HJ, and Ingber DE, Lab Chip, 2012, 12, 2156. [DOI] [PubMed] [Google Scholar]
- 48.Lin S, Yuk H, Zhang T, Parada GA, Koo H, Yu C, and Zhao X, Adv. Mater, 2016, 28, 4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shay T, Dickey MD, and Velev OD, Lab Chip, 2017, 17, 710. [DOI] [PubMed] [Google Scholar]
- 50.Tran TB, Cho S, and Min J, Biosens. Bioelectron, 2013, 50, 453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.