Abstract
The epidemiological study of several multidrug-resistant Enterobacteriaceae isolated from five patients demonstrated in vivo dissemination of a 100-kb plasmid encoding the extended-spectrum β-lactamase TEM-24 from a clonal strain of Enterobacter aerogenes to different strains of Klebsiella pneumoniae, Escherichia coli, Proteus vulgaris, Proteus mirabilis, and Serratia marcescens.
In France, plasmid-mediated extended-spectrum beta-lactamases (ESBLs) have been mostly described from strains of Klebsiella pneumoniae (1, 2, 3, 6, 7, 15), but more recently infections caused by strains of Enterobacter spp. producing the TEM-24 ESBL have increased (5, 14, 24). The same phenomenon was observed in our University Hospital (2,000 beds, in Dijon, France). In 1996, 1997, and 1998 we isolated, respectively, 16, 37, and 50 Enterobacter aerogenes strains producing TEM-24 among totals of 78, 70, and 70 nonrepetitive ESBL-producing strains. All these strains were analyzed by pulsed-field gel electrophoresis (PFGE). During our continuous survey we found that five patients were cocolonized or coinfected with different multidrug-resistant species of enterobacteria. Following the use of imipenem, two strains of Proteus mirabilis and E. aerogenes resistant to this molecule were recovered from one patient. We report here the epidemiological study and the β-lactamase characterization of all the strains isolated from the five patients.
The origins of the strains are given in Table 1. The detection of ESBL production was performed by the double-disk synergy test (19) but with a quarter of the disk containing third-generation cephalosporin for Proteus sp. (9).
TABLE 1.
Origins of the strains
| Patient | Ward | Date of isolation | Source of isolate | Organisma | pI(s) of β-lactamase |
|---|---|---|---|---|---|
| 1 | Intensive care unit | 20 April 1998 | Sputum | EA (+) | 6.5, 8.3 |
| 22 May 1998 | Stool | EA (+) | 6.5, 8.3 | ||
| EC (+) | 6.5 | ||||
| 17 June 1998 | Sputum | EA (+) | 6.5, 8.3 | ||
| KP (+) | 6.5, 7.7 | ||||
| PV (+) | 6.5, >8.3 | ||||
| 17 June 1998 | Stool | EA (+) | 6.5, 8.3 | ||
| 2 | Urology surgery unit | 24 October 1998 | Stool | EA (+) | 6.5, 8.3 |
| 31 October 1998 | Surgical wound | EA (+) | 6.5, 8.3 | ||
| KP (+) | 6.5, 7.7 | ||||
| PV (+) | 6.5, >8.3 | ||||
| 3 | Dermatology unit | 24 August 1998 | Urine | EA (+) | 6.5, 8.3 |
| 18 November 1998 | Stool | EA (+) | 6.5, 8.3 | ||
| EC (+) | 6.5 | ||||
| KP (+) | 6.5, 7.7 | ||||
| 4 | Neurosurgery unit | 29 September 1999 | Sputum | EC (−) | |
| 3 October 1999 | Stool | EA (+) | 6.5, 8.3 | ||
| 20 November 1999 | Sputum | EA (+) | 6.5, 8.3 | ||
| EC (+) | 6.5 | ||||
| EC (−) | |||||
| 3 December 1999 | Sputum | EA (+) | 6.5, 8.3 | ||
| 28 February 2000 | Sputum | PM* (−) | 5.4 | ||
| 29 March 2000 | Surgical wound | PM* (+) | 5.4, 6.5 | ||
| EA* (+) | 6.5, 8.3 | ||||
| 5 | Rehabilitation unit | 24 February 2000 | Sputum | EA (+) | 6.5, 8.3 |
| 6 March 2000 | Sputum | EA (+) | 6.5, 8.3 | ||
| SM (+) | 6.5, >8.3 | ||||
| SM (−) | >8.3 |
EA, E. aerogenes; EC, E. coli; KP, K. pneumoniae; PV, P. vulgaris; PM, P. mirabilis; SM, S. marcescens; (+), ESBL-producing strain; (−), non-ESBL-producing strain; *, strain resistant to imipenem.
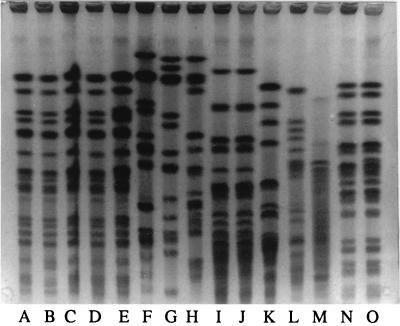
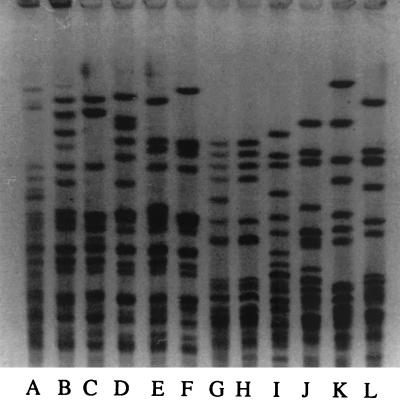
Analyses of chromosomal DNAs by PFGE were performed as described previously (15) but with a pulse range from 40 to 5 s for 20 h at 180 V for strains of E. aerogenes, K. pneumoniae, Serratia marcescens, and Escherichia coli. For P. mirabilis and Proteus vulgaris, we used a pulse range from 25 to 5 s for 20 h at 180 V (Fig. 1 and 2). A single profile was found for the strains of E. aerogenes, similar to that of the epidemic strain described in 1996 (24). For the other enterobacteria, strains from the same species (ESBL or not ESBL producing) isolated from the same patient shared concordant PFGE patterns, suggesting their clonal origin. Nevertheless, the strains of the same species isolated from the five patients were not related. This result excluded the possibility that resistant strains of K. pneumoniae, E. coli, or P. vulgaris were disseminated between the patients or that there was a common source of contamination.
FIG. 1.
PFGE of total DNAs from E. aerogenes (lanes A to E), K. pneumoniae (lanes F to H), S. marcescens (lanes I to K), and E. coli (lanes L to O) cut by XbaI. Lane A, patient 1 isolate; lane B, patient 3 isolate; lane C, patient 2 isolate; lane D, patient 4 isolate; lane E, patient 5 isolate; lane F, patient 1 isolate; lane G, patient 3 isolate; lane H, patient 2 isolate; lanes I and J, patient 5 isolate; lane K, unrelated strain; lane L, patient 1 isolate; lane M, patient 3 isolate; lanes N and O, patient 4 isolate.
FIG. 2.
PFGE of total DNAs from P. vulgaris (lanes A to F) and P. mirabilis (lanes G to L) cut by SmaI. Lane A, patient 1 isolate; lane B, patient 2 isolate; lanes C to F, unrelated strains; lanes G and H, patient 4 isolate; lanes I to L, unrelated strains.
A large plasmid of about 100 kb was isolated by the method of Birnboim and Doly from the ESBL-producing strains (4). The restriction patterns obtained after digestion of the plasmid by EcoRI were very similar. The plasmid was easily transferred from E. aerogenes, K. pneumoniae, P. vulgaris, P. mirabilis, and S. marcescens to E. coli K-12 C600, which is resistant to sodium azide (selection with 256 μg of sodium azide per ml and 8 μg of netilmicin per ml) or from E. coli to K. pneumoniae 10031, which is resistant to rifampin (selection with 100 μg of rifampin per ml and 8 μg of netilmicin per ml). Resistance to β-lactams was cotransferred with resistance to aminoglycosides (amikacin, kanamycin, netilmicin, tobramycin), sulfonamides, and chloramphenicol. The MICs of β-lactams (Table 2) were determined in Mueller-Hinton broth by a microdilution method for the clinical strains and their transconjugants. The levels of resistance were very similar among the transconjugants. For the P. mirabilis strains, extended-spectrum cephalosporin MICs were very low. This may explain why it was difficult to detect ESBL production in this species. P. mirabilis and E. aerogenes strains isolated from patient 4 in February and in March were resistant to imipenem (respectively, MICs of 8 and 16 to 32 μg/ml). Isoelectric focusing was performed as previously reported (24). The β-lactamase activity was located in the gels by an iodine starch procedure (20). A β-lactamase with a pI of 6.5 was detected in all the ESBL-producing strains as well as in their transconjugants. PCR was performed on plasmids extracted from the transconjugants and from the non-ESBL-producing P. mirabilis strain which was resistant to imipenem with primers J (forward, 5′-CTTATTCCCTTTTTTGCGGC-3′) and E (reverse, 5′-GGTCTGACAGTTACCAATGC-3′) (8) at positions 236 and 1079 of the TEM family gene β-lactamase according to Sutcliffe numbering (25). The sequence of the gene encoding the β-lactamase with a pI of 5.4 produced by the P. mirabilis strain was identical to that of TEM-1b (16, 25), and the sequence of the gene encoding the β-lactamase with a pI of 6.5 was identical to that of the extended-broad-spectrum β-lactamase TEM-24b (8, 17).
TABLE 2.
MICs of beta-lactam antibiotics for the clinical strains presented in Table 1
| Patient | Organisma | MIC (μg/ml)b
|
||||||
|---|---|---|---|---|---|---|---|---|
| TIC | PIP | CAZ | CTX | ATM | FEP | CEF | ||
| 1 | EA | >2,048 (>2,048) | 256 (16) | 512 (128) | 32 (1) | 64 (16) | 2 (0.5) | >1,024 (32) |
| EC | >2,048 (>2,048) | 256 (16) | 256 (64) | 4 (0.5) | 64 (8) | 2 (0.25) | 256 (32) | |
| KP | >2,048 (>2,048) | 32 (16) | 128 (128) | 1 (1) | 16 (16) | 0.5 (0.5) | 64 (32) | |
| PV | 256 (>2,048) | 8 (32) | 16 (128) | 1 (1) | 0.5 (16) | 1 (0.5) | 512 (32) | |
| 2 | EA | >2,048 (>2,048) | 256 (16) | 512 (128) | 32 (1) | 64 (16) | 2 (0.5) | >1,024 (32) |
| KP | >2,048 (>2,048) | 64 (32) | 256 (128) | 1 (1) | 32 (16) | 0.5 (0.5) | 64 (32) | |
| PV | 128 (>2,048) | 8 (32) | 16 (128) | 0.5 (1) | 0.25 (16) | 1 (0.5) | 256 (32) | |
| 3 | EA | >2,048 (>2,048) | 512 (16) | 512 (128) | 32 (0.5) | 64 (16) | 1 (0.5) | >1,024 (32) |
| EC | >2,048 (>2,048) | 64 (16) | 256 (64) | 2 (0.5) | 32 (8) | 1 (0.25) | 128 (32) | |
| KP | >2,048 (>2,048) | 64 (32) | 128 (128) | 2 (1) | 32 (16) | 1 (0.5) | 128 (64) | |
| 4 | EA | >2,048 (>2,048) | 256 (16) | 512 (128) | 32 (1) | 64 (16) | 2 (0.25) | >1,024 (32) |
| EC | >2,048 (>2,048) | 256 (16) | 128 (32) | 2 (0.5) | 32 (8) | 2 (0.25) | 32 (16) | |
| PM | 512 (>2,048) | 128 (16) | 8 (64) | 1 (1) | 0.25 (4) | 0.5 (0.25) | 16 (16) | |
| 5 | EA | >2,048 (>2,048) | 256 (16) | 512 (128) | 32 (1) | 64 (16) | 2 (0.5) | >1,024 (32) |
| SM | >2,048 (>2,048) | 64 (16) | 256 (128) | 1 (1) | 32 (8) | 1 (0.5) | 256 (128) | |
EA, E. aerogenes; EC, E. coli; KP, K. pneumoniae; PV, P. vulgaris; PM, P. mirabilis; SM, S. marcescens.
TIC, ticarcillin; PIP, piperacillin; CAZ, ceftazidime; CTX, cefotaxime; ATM, aztreonam; FEP, cefepime; CEF, cephalothin. Values in parentheses are MICs for the transconjugants.
These results demonstrate clearly that there has been an in vivo transfer of the plasmid encoding ESBL TEM-24 from E. aerogenes to K. pneumoniae, E. coli, P. mirabilis, S. marcescens, and P. vulgaris. In all cases, the ESBL was first detected in the strain of E. aerogenes and only later in other species in a site colonized by E. aerogenes. For patients 1, 2, and 3 we unfortunately did not keep the non-ESBL-producing species of Enterobacteriaceae isolated before the E. aerogenes strain. The transfer probably occurred in the wound of patient 2 because the resistant strains of K. pneumoniae and P. vulgaris were never found in the stools. For the two other patients, we isolated the non-ESBL-producing strain (E. coli and P. mirabilis for patient 4, S. marcescens for patient 5) from the same site as we did the identical TEM-24-producing strain. The transfer in vivo of plasmid has already been described (12, 21, 22, 24), but each report concerned only one patient. This study, the first one describing five patients, proves that the spread of plasmid is no longer exceptional and can concern species for which the ESBL TEM-24 had not yet been described, like P. vulgaris.
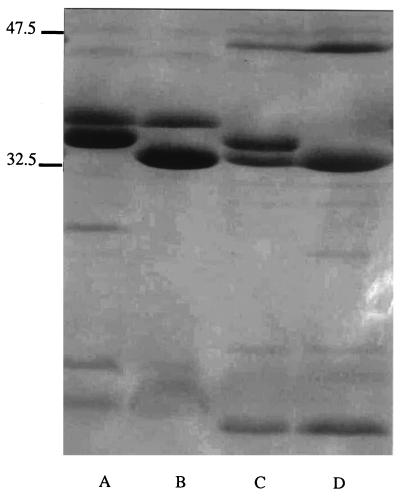
The analysis of the outer membrane proteins of the strains of P. mirabilis and E. aerogenes resistant to imipenem and isolated from patient 4 was carried out as previously reported (Fig. 3) (18, 23). A band of 40 kDa was not detected in the strain of E. aerogenes that is resistant to imipenem as already reported (10, 11, 13). In the P. mirabilis strain resistant to imipenem all the major bands were present. The resistance was probably due to some modifications in the penicillin-binding proteins, which we already described for this species (23). This report is the first description of the selection of two different species of Enterobacteriaceae resistant to imipenem in a single patient following treatment with imipenem. If such strains were to be more often isolated, there would soon be no medical therapies available.
FIG. 3.
Outer membrane protein profiles of P. mirabilis and E. aerogenes. Lane A, P. mirabilis ATCC 29906; lane B, P. mirabilis imipenem-resistant strain from patient 4; lane C, E. aerogenes imipenem-susceptible strain from patient 4; lane D, E. aerogenes imipenem-resistant strain from patient 4. Molecular mass standards in kilodaltons are given on the left.
In conclusion, the E. aerogenes strain producing TEM-24 isolated in our hospital represents a serious danger: it spreads very easily and is at the origin of plasmid dissemination among Enterobacteriaceae.
REFERENCES
- 1.Arlet C, Rouveau M, Casin I, Bouvet P J M, Lagrange P H, Philippon A. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J Clin Microbiol. 1994;32:2553–2558. doi: 10.1128/jcm.32.10.2553-2558.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermudes H, Arpin C, Jude F, El-Harrif Z, Bebear C, Quentin C. Molecular epidemiology of an outbreak due to extended-spectrum β-lactamase-producing enterobacteria in a French hospital. Eur J Clin Microbiol Infect Dis. 1997;16:523–527. doi: 10.1007/BF01708236. [DOI] [PubMed] [Google Scholar]
- 3.Bingen E H, Desjardins P, Arlet G, Bourgeois F, Kurkdjan P M, Lambert-Zechovsky N Y, Denamur E, Philippon A, Elion J. Molecular epidemiology of plasmid spread among extended broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J Clin Microbiol. 1993;31:179–184. doi: 10.1128/jcm.31.2.179-184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosi C, Davin-Regli A, Bornet C, Mallea M, Pages J M, Bollet C. Most Enterobacter aerogenes strains in France belong to a prevalent clone. J Clin Microbiol. 1999;37:2165–2169. doi: 10.1128/jcm.37.7.2165-2169.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun-Buisson C, Legrand P, Philippon A, Montralvers F, Ansquer M, Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;ii:302–306. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 7.Chanal C M, Sirot D L, Petit A, Labia R, Morand A, Sirot J L, Cluzel R A. Multiplicity of TEM-derived β-lactamase from Klebsiella pneumoniae strains isolated at the same hospital and relationships between the responsible plasmids. Antimicrob Agents Chemother. 1989;33:1915–1920. doi: 10.1128/aac.33.11.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanal C, Poupart M C, Sirot D, Labia R, Sirot J, Cluzel R. Nucleotide sequences of CAZ-2, CAZ-6, and CAZ-7 β-lactamase genes. Antimicrob Agents Chemother. 1992;36:1817–1820. doi: 10.1128/aac.36.9.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanal C, Sirot D, Romaszko J P, Bret L, Sirot J. Survey of prevalence of extended-spectrum β-lactamase among Enterobacteriaceae. J Antimicrob Chemother. 1996;38:127–132. doi: 10.1093/jac/38.1.127. [DOI] [PubMed] [Google Scholar]
- 10.Charrel R N, Pagès J M, De Micco P, Mallea M. Prevalence of outer membrane porin alteration in β-lactam antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow J W, Shlaes D M. Imipenem resistance associated with the loss of a 40 kDa outer membrane protein in Enterobacter aerogenes. J Antimicrob Chemother. 1991;28:499–504. doi: 10.1093/jac/28.4.499. [DOI] [PubMed] [Google Scholar]
- 12.d'Agata E, Venkataraman L, De Girolami P, Weigel L, Samore M, Tenover F. The molecular and clinical epidemiology of Enterobacteriaceae-producing extended-spectrum β-lactamase in a tertiary care hospital. J Infect. 1998;36:279–285. doi: 10.1016/s0163-4453(98)94171-8. [DOI] [PubMed] [Google Scholar]
- 13.de Champs C, Henquell C, Guelon D, Sirot D, Gazuy N, Sirot J. Clinical and bacteriological study of nosocomial infections due to Enterobacter aerogenes resistant to imipenem. J Clin Microbiol. 1993;31:123–127. doi: 10.1128/jcm.31.1.123-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Champs C, Sirot D, Chanal C, Bonnet R, Sirot J. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. The French Study Group. Antimicrob Agents Chemother. 2000;44:3177–3179. doi: 10.1128/aac.44.11.3177-3179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouby A, Neuwirth C, Bourg G, Bouzigues N, Carles-Nurit M J, Despaux E, Ramuz M. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32:301–305. doi: 10.1128/jcm.32.2.301-305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goussard S, Courvalin P. Sequence of the genes blaT-1B and blaT-2. Gene. 1991;102:71–73. doi: 10.1016/0378-1119(91)90540-r. [DOI] [PubMed] [Google Scholar]
- 17.Goussard S, Courvalin P. Updated sequence information for TEM β-lactamase genes. Antimicrob Agents Chemother. 1999;43:367–370. doi: 10.1128/aac.43.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkins J M, Towner K J. Enhanced resistance to cefotaxime and imipenem associated with outer membrane protein alterations in Enterobacter aerogenes. J Antimicrob Chemother. 1990;25:49–55. doi: 10.1093/jac/25.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Jarlier V, Nicolas M H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 20.Labia R, Barthélemy M, Masson J M. Multiplicité des bêta-lactamases: un problème d'isoenzymes? C R Acad Sci. 1976;283D:1597–1600. [PubMed] [Google Scholar]
- 21.Marchandin H, Carrière C, Sirot D, Jean-Pierre H, Darbas H. TEM-24 produced by four different species of Enterobacteriaceae, including Providencia rettgeri, in a single patient. Antimicrob Agents Chemother. 1999;43:2069–2073. doi: 10.1128/aac.43.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchandin H, Jean-Pierre H, de Champs C, Sirot D, Darbas H, Perigault P F, Carrière C. Production of a TEM-24 plasmid-mediated extended-spectrum β-lactamase by a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:213–216. doi: 10.1128/aac.44.1.213-216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuwirth C, Siébor E, Duez J M, Péchinot A, Kazmierczak A. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J Antimicrob Chemother. 1995;36:335–342. doi: 10.1093/jac/36.2.335. [DOI] [PubMed] [Google Scholar]
- 24.Neuwirth C, Siébor E, Lopez J, Péchinot A, Kazmierczak A. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum β-lactamase to other members of the family Enterobacteriaceae. J Clin Microbiol. 1996;34:76–79. doi: 10.1128/jcm.34.1.76-79.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]