ABSTRACT
Glioma is a common life-threatening tumor with high malignancy and high invasiveness. LncRNA ZFPM2 antisense RNA 1 (ZFPM2-AS1) was confirmed to be implicated in numerous tumors, while its biological function and mechanism have not been thoroughly understood in glioma. The gene expression was measured by RT-qPCR. Cell proliferation, cell cycle, and cell apoptosis of glioma cells were validated by CCK-8, colony formation, flow cytometry and TUNEL assays. The effect of ZFPM2-AS1 on tumor growth was verified by in vivo assay. The exploration on ZFPM2-AS1-mediated mechanism was carried out via ChIP, luciferase reporter, and RIP assays. In the present study, ZFPM2-AS1 was demonstrated as a highly-expressed lncRNA in glioma tissues and cells. ZFPM2-AS1 silencing suppressed cell proliferation and cell cycle, but facilitated cell apoptosis. In addition, the inhibitive effect of silenced ZFPM2-AS1 was also observed in tumor growth. Furthermore, we found that SP1 interacted with ZFPM2-AS1 promoter to transcriptionally activate ZFPM2-AS1 expression. Moreover, ZFPM2-AS1 was identified as a competing endogenous RNA (ceRNA) for miR-515-5p to target SOD2. Rescue assays verified that SOD2 overexpression partially abolished the suppressive impact of ZFPM2-AS1 silencing on glioma cell growth. In conclusion, this study corroborated the regulatory mechanism of SP1/ZFPM2-AS1/miR-515-5p/SOD2 axis in glioma, indicating that targeting ZFPM2-AS1 might be an effective way to treat glioma.
KEYWORDS: ZFPM2-AS1, SP1, miR-515-5p, SOD2, glioma
Introduction
Glioma, a common brain tumor derived from the central nervous system, exhibits numerous heterogeneous partitions [1]. Among which, high-grade glioma (such as glioblastoma) is fatal and accompanied with unfavorable prognosis; moreover, it also constitutes a majority of deaths related to brain tumors [2]. Glioblastoma leads to pathological changes in the brain vasculature, which, in turn, facilitates glioblastoma development, enhances tumor aggressiveness, and exacerbates therapeutic resistance [3]. Although great advances in glioma treatment have been achieved in recent years, the improvement in therapeutic outcomes is still limited [4]. Patients with advanced-stage glioma face poor prognosis because of its drug resistance and infiltrative nature [5,6]. Therefore, further research is quite necessary to identify novel molecular mechanisms that might be useful to improve glioma diagnosis and prognosis, thereby prolonging the long-term survival [7,8].
Long non-coding RNAs (lncRNAs), exceeding 200 nucleotides, are identified as heterogeneous transcripts that are widely participate in pathophysiological processes [9]. Extensive studies suggested that lncRNAs exerted essential functions in the occurrence and development of glioma. For example, lncRNA NEAT1 sponged miR-132 to target SOX2, thus facilitating glioma cell migration and invasion [10]. LncRNA HOTAIR was recognized as a diagnostic and prognostic biomarker for glioma [11]. In addition, lncRNA MIR22HG contributed to cell growth in glioma by activating Wnt/β-catenin signaling [12]. Previously, lncRNA ZFPM2 antisense RNA 1 (ZFPM2-AS1) has been reported to accelerate cell migration and invasion of hepatocellular carcinoma through the miR-139/GDF10 axis [13]. Furthermore, ZFPM2-AS1 contributed to epithelial-mesenchymal transition (EMT) and cell migration by regulating the miR-511-3p/AFF4 axis in lung adenocarcinoma [14]. However, the function and mechanism of ZFPM2-AS1 have not yet been elucidated in glioma.
MicroRNAs (miRNAs) are a class of single-stranded RNAs (20–23 nucleotides), which can act as either oncogene or tumor suppressor in various cancers, including glioma [15,16]. For example, miR-451 inhibited glioma cell proliferation, invasion, and apoptosis by regulating the PI3K/AKT signaling pathway [17]. MiR-4516 predicted unfavorable prognosis and acted as an oncogene in glioblastoma by targeting PTPN14 [18]. miR-515-5p has been reported to act as a tumor suppressor in various human cancers, such as prostate cancer [19], non-small cell lung cancer [20], and breast cancer [21]. Nevertheless, the biological role of miR-515-5p in glioma remains unclear.
The present study aimed to explore the clinical significance of ZFPM2-AS1 expression in glioma, and to investigate the biological function and underlying mechanism of ZFPM2-AS1 in regulating the malignant phenotypes of glioma.
Materials and methods
Clinical specimens
The glioma tissues and adjacent normal tissues were obtained from 30 patients who were newly diagnosed at The Affiliated Sir Run Run Hospital of Nanjing Medical University. Besides, serum samples were also collected from 30 healthy volunteers as a healthy control group. The informed consent was signed by each patient, and this study was permitted by the Ethics Committee of the Affiliated Sir Run Run Hospital of Nanjing Medical University. The collected specimens were immediately frozen by liquid nitrogen and kept at −80°C after surgical resection.
Cell lines
Human glioma cell lines (A172, LN229, U87, T98G) and normal human astrocytes (NHA) used in our study were obtained from ATCC (Manassas, VA). The cell lines mentioned above were cultured in the DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Gibco, Waltham, MA) under a humid atmosphere with 5% CO2 at 37°C.
Cell transfection
The shRNA targeting ZFPM2-AS1 (sh-ZFPM2-AS1#1 and sh-ZFPM2-AS1#2), the negative control (sh-NC), overexpression vector (oe) targeting SP1 (oe-SP1) or SOD2 (oe-SOD2), negative control (oe-NC), miR-515-5p mimics, and NC mimics were offered by GenePharma (Shanghai, China). A172 and LN229 cell lines were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen).
RT-qPCR
Total RNAs from tissues and cells were extracted by TRIzol (Invitrogen) and used for cDNA synthesis performed with Reverse Transcription Kit (Toyobo, Osaka, Japan). The qPCR experiment was performed via SYBR Green Super Mix (Bio-Rad, Hercules, CA). MiR-515-5p expression was standardized to a U6 transcript, and GAPDH was set as the negative control for ZFPM2-AS1, SP1 and SOD2. The 2−ΔΔCT method [22] was applied to calculate the relative expression.
CCK-8 assay
Transfected cells (1 × 104 cells/well) were inoculated in 96-well plates. Under the indicated condition (5% CO2, 37°C), 10 μl CCK8 was added, and the cells were incubated for another 2 h. Cell viability was analyzed at 0, 24, 48, 72 h by determining the absorbance at 450 nm via a microplate reader [23].
Colony formation assay
A172 and LN229 cells (1000 cells/well) were planted in the 6-well plates and incubated for 14 days at 37°C. Then, colonies (over 50 cells) were immobilized in 4% formaldehyde (Sigma-Aldrich) for sequential staining via 0.5% crystal violet (Sigma-Aldrich). After staining, the colonies were manually counted.
Flow cytometry analysis
Briefly, transfected A172 and LN229 cells were collected and immobilized with 75% ethanol overnight at 4°C. After that, cells were rinsed and re-suspended with PBS. Then, the cells were incubated with RNase (10 mg/ml) and PI (1 mg/ml) for half an hour at 37°C in the dark. The analysis of cell cycle was conducted by flow cytometry [24].
TUNEL assay
After transfection, A172 and LN229 cells were treated with paraformaldehyde (4%) for 15 min and Triton-X 100 (0.25%) for 20 min. Then, TUNEL detection kit (Roche, Basel, Switzerland) was used to treat the cells. After DAPI staining, the cells were observed using a fluorescent microscope [25].
Xenograft tumors
Male nude mice (18–22 g, 4 weeks old) were obtained from SJA Laboratory Animal Co., Ltd. (Hunan, China) and randomly divided into two groups. A172 cells stably transfected with sh-ZFPM2-AS1#1 and sh-NC were hypodermically injected into the mice. Every 7 days, tumor volume was estimated through measuring the width and length of tumors by caliper. Four weeks later, every mouse was sacrificed by dislocation, and Xenografted tumors were collected for weighting [26]. The study was approved by the Ethics Committee of the Affiliated Sir Run Run Hospital of Nanjing Medical University.
Immunohistochemistry (IHC)
After fixation and paraffin-embedment, the tissues acquired from xenografts were cut into 4-µm sections. Then, anti-Ki67 (ab16667; Abcam) was used to incubate the sections. Later, the sections were washed with PBS, incubated with HRP-conjugated secondary antibody (Abcam), and finally photographed using a light microscope [27].
ChIP assay
Following the manufacturer’s protocol, ChIP Assay Kit (Beyotime) was applied for ChIP assay in transfected A172 and LN229 cells [28]. The cells were collected, immobilized in formaldehyde, and then lysed in ChIP lysis buffer. After the cross-linking of DNAs and proteins, the formaldehyde was quenched in glycine buffer. Then, sonication was used to generate DNA fragments (200–400 bp). Afterward, the fragmented DNAs were precipitated by anti-SP1 (Abcam). Anti-IgG (Abcam) was applied as the negative control. Finally, precipitated DNAs were determined by qPCR.
Luciferase reporter assay
To confirm the binding of SP1 to ZFPM2-AS1 promoter, ZFPM2-AS1 promoter fragment with SP1 wild-type (WT) or mutant (Mut) binding site was inserted into pGL3-basic vector (Promega, Madison, WI, USA), and the plasmids were named as ZFPM2-AS1 promoter-WT and ZFPM2-AS1 promoter-Mut. Then, the recombinant constructs were co-transfected into A172 and LN229 cells, together with oe-SP1.
To confirm the interaction of miR-515-5p with ZFPM2-AS1 (or SOD2), full-length sequence of ZFPM2-AS1 (or SOD2) with WT or Mut miR-515-5p binding site were inserted into pmirGLO vector (Promega) to construct pmirGLO-ZFPM2-AS1-WT/Mut (or pmirGLO-SOD2-WT/Mut) reporter. Then, the reporters were co-transfected with NC mimics or miR-515-5p mimics into A172 and LN229 cells. Two days later, Dual-luciferase Reporter Assay System (Promega) was used to determine relative luciferase activity.
RNA pull-down assay
RNA pull-down assay was conducted through applying biotin-labeled ZFPM2-AS1 (Bio-ZFPM2-AS1) as probe and examining the potential miRNAs by qPCR analysis later [29]. Biotin-control (Bio-NC) was utilized as control biotin-labeled lncRNA. Briefly, lysis buffer (650 μl) was added into the collected cells to obtain cell extracts, and then the cell lysates (2 μg) were respectively mixed with Bio-ZFPM2-AS1 (ZFPM2-AS1 biotin probe) or Bio-NC (ZFPM2-AS1 no-biotin probe). Afterward, 100 μl of pierce streptavidin agarose beads (Baili Biotech) was added for each binding reaction, and then the complexes were incubated for 45 min at room temperature. Later, the beads were gathered and the precipitated RNAs were eluted and detected by RT-qPCR.
RIP assay
RIP experiments were using the Magna RIP kit (Millipore, Bedford, USA) according to the manufacturer’s protocol [30]. Briefly, A172 and LN229 cells were lysed in RIP lysis buffer, and then incubated with magnetic beads conjugated with anti-Ago2 or anti-IgG (control). After purification, the immunoprecipitated RNA was subjected to RT-qPCR.
Statistical analysis
SPSS 20.0 (SPSS, Chicago, IL, USA) was used for all statistical analysis, and results were expressed as mean ± SD. The differences between two or among multiple groups were examined via Student’s t-test or one-way ANOVA, and considered as statistical significance when P < 0.05. In vitro experiments were repeated at least three times. Pearson’s correlation analysis was applied to evaluate gene expression correlation.
Results
ZFPM2-AS1 knockdown inhibited the tumorigenesis of glioma
At first, the ZFPM2-AS1 expression level in glioma tissues was measured via RT-qPCR. Compared with the control tissues, ZFPM2-AS1 was highly expressed in glioma tissues (Figure 1a). Then, we analyzed the clinical potential of ZFPM2-AS1 in glioma patients. As presented in Table 1, the expression of ZFPM2-AS1 was associated with the tumor size, kamofsky performance score (KPS), and WHO stage. In addition, we assessed the diagnostic value of serum ZFPM2-AS1, and the results showed that the area under curve (AUC) was 0.9214, implying that ZFPM2-AS1 might be used as an indicator for glioma diagnosis (Figure 1b). Moreover, ZFPM2-AS1 expression was highly expressed in glioma cell lines (A172, LN229, U87 and T98G) compared to NHA cell line (Figure 1c). Then, A172 and LN229 cell lines were selected for the following experiments due to the higher expression of ZFPM2-AS1 in A172 and LN229 cell lines. To examine the biological function of ZFPM2-AS1 in glioma, loss-of-function assays were performed. RT-qPCR indicated that ZFPM2-AS1 expression was decreased in A172 and LN229 cells after sh-ZFPM2-AS1#1/2 transfection (Figure 1d). CCK-8 assay indicated that the viability of A172 and LN229 cells was hindered by silencing ZFPM2-AS1 (Figure 1e). Furthermore, the decreased colonies were found in ZFPM2-AS1-knockdown group by colony formation assay (figure 1f). In addition, the flow cytometry results implied that ZFPM2-AS1 depletion boosted cell ratio in G0/G1 phase, whereas reduced cells in S phase (Figure 1g and Supplementary Figure S1). TUNEL assay suggested that apoptosis rate was enhanced in A172 and LN229 cells upon ZFPM2-AS1 deficiency (Figure 1h). All data indicated that ZFPM2-AS1 silencing restrained cell growth in glioma.
Figure 1.
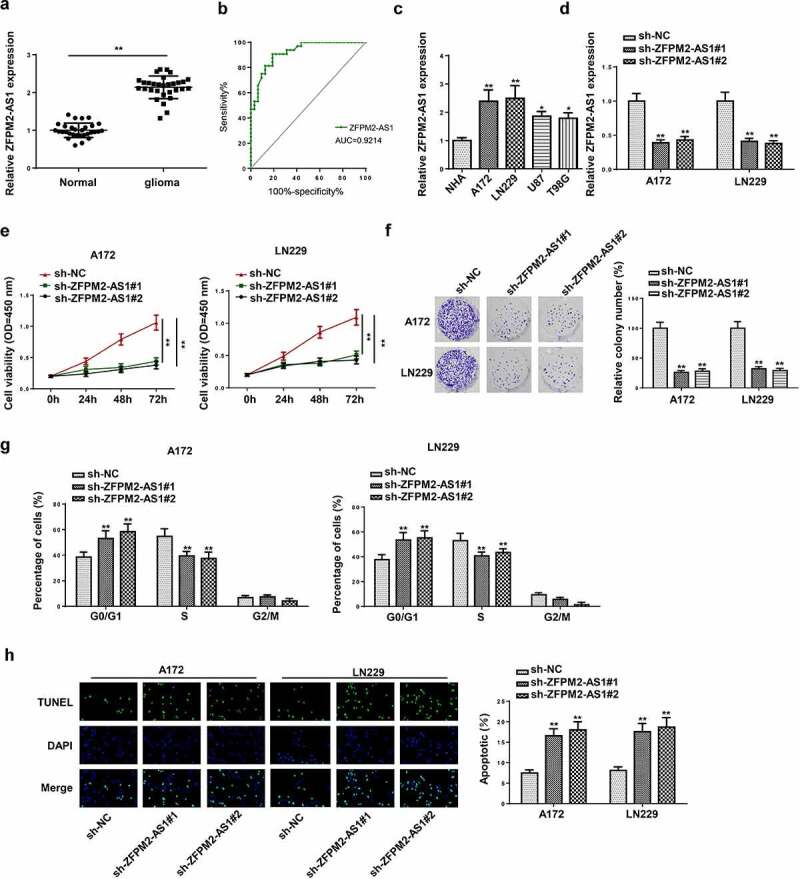
ZFPM2-AS1 knockdown inhibited the tumorigenesis of glioma. (a) ZFPM2-AS1 expression in glioma tissues and matched control tissues by RT-qPCR. (b) ROC curve analysis was used to assess the diagnostic values of the circCSNK1G1. (c) ZFPM2-AS1 expression in glioma cell lines and control cell line (NHA) by RT-qPCR. (d) RT-qPCR analysis of ZFPM2-AS1 expression in A172 and LN229 cells transfected with sh-ZFPM2-AS1#1 and sh-ZFPM2-AS1#2. (e) CCK-8 assay was performed to detect A172 and LN229 cell viability upon ZFPM2-AS1 knockdown. (f) The proliferation of ZFPM2-AS1-downregulated A172 and LN229 cells was evaluated via colony formation assay. (g) Flow cytometry analysis for cell cycle detection in sh-ZFPM2-AS1#1/2 group or sh-NC group. (h) Cell apoptosis after ZFPM2-AS1 deficiency was testified through TUNEL assay. *p < 0.05, **p < 0.01
Table 1.
The association between ZFPM2-AS1 expression and clinicopathologic parameters in glioma patients
| ZFPM2-AS1 expression |
|||
|---|---|---|---|
| Variable | Low | High | p-value |
| Age | 0.186 | ||
| < 50 | 10 | 15 | |
| ≥ 50 | 3 | 2 | |
| Gender | 0.112 | ||
| Male | 12 | 14 | |
| Female | 1 | 3 | |
| Tumor size | 0.004 | ||
| < 5 cm | 10 | 7 | |
| ≥ 5 cm | 3 | 10 | |
| KPS | 0.002 | ||
| ≥ 70 | 10 | 6 | |
| < 70 | 3 | 11 | |
| WHO stage | 0.003 | ||
| I+ II | 9 | 7 | |
| III+IV | 4 | 10 | |
ZFPM2-AS1 depletion hindered tumor growth in glioma
In this section, the effect of ZFPM2-AS1 on glioma tumor growth was further analyzed in vivo. As demonstrated in Figure 2a, tumors harvested from the mice in sh-ZFPM2-AS1#1 group were smaller than those from the mice in sh-NC group. Moreover, the tumor volume and weight were also decreased by knocking down ZFPM2-AS1 expression (Figure 2b and c). Additionally, the coloration intensity of Ki-67 (proliferation antigen) was significantly inhibited with sh-ZFPM2-AS1#1 transfection (Figure 2d). Collectively, these results showed that ZFPM2-AS1 deficiency impaired tumorigenesis in glioma.
Figure 2.
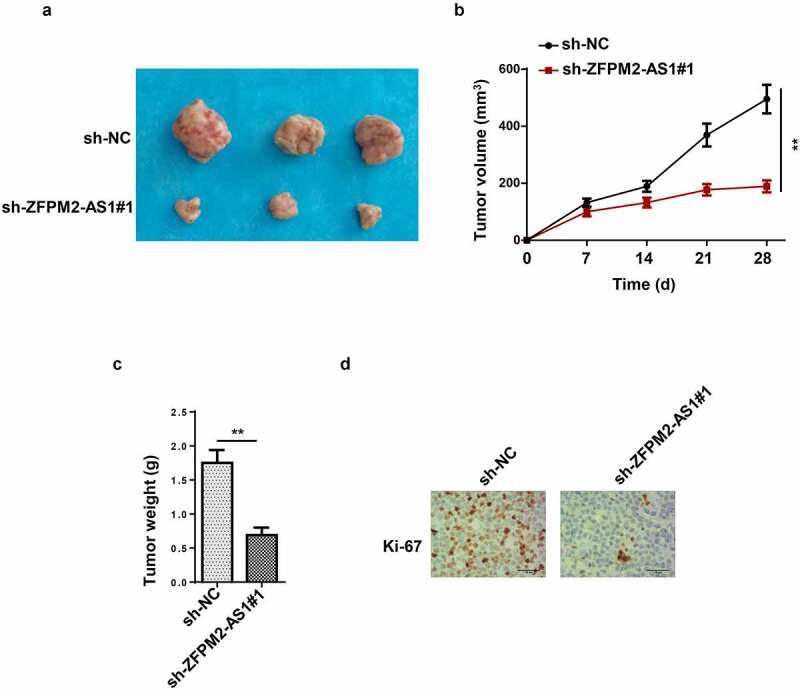
Effect of ZFPM2-AS1 in glioma tumorigenesis. (a) The tumors excised from mice in sh-ZFPM2-AS1#1 group or sh-NC group. (b and c) The tumor volume and weight were confirmed in ZFPM2-AS1-knockdown group and control group. (d) Function of silenced ZFPM2-AS1 on Ki-67 expression was confirmed by IHC assay. **p < 0.01
ZFPM2-AS1 was transcriptionally induced by SP1
Subsequently, we inspected the upstream mechanism that caused the upregulation of ZFPM2-AS1. Previously, numerous reports have proved that transcription factors could transcriptionally activate the expression of lncRNAs [31,32]. Then, we used UCSC website (http://genome.ucsc.edu/) to predict the promising transcription factor for ZFPM2-AS1, and SP1 was identified. According to JASPAR (http://jaspar.genereg.net/), the DNA motif and binding sequence between ZFPM2-AS1 promoter and SP1 were obtained (Figure 3a). Data from ChIP assay further validated the interactivity of SP1 with ZFPM2-AS1 promoter (Figure 3b). Subsequently, we transfected oe-SP1 vectors into A172 and LN229 cells, and found the up-regulated expression of SP1 in such cells (Figure 3c). Luciferase reporter assay uncovered that SP1 overexpression increased the luciferase activity of ZFPM2-AS1 promoter-WT, while that of ZFPM2-AS1 promoter-Mut displayed no remarkable differences (Figure 3d). Then, we detected ZFPM2-AS1 expression in A172 and LN229 cells with SP1 overexpression, and data displayed that ZFPM2-AS1 was upregulated in SP1-overexpressed A172 and LN229 cells (Figure 3e). These data revealed that SP1 activated ZFPM2-AS1 expression through transcriptional regulation.
Figure 3.
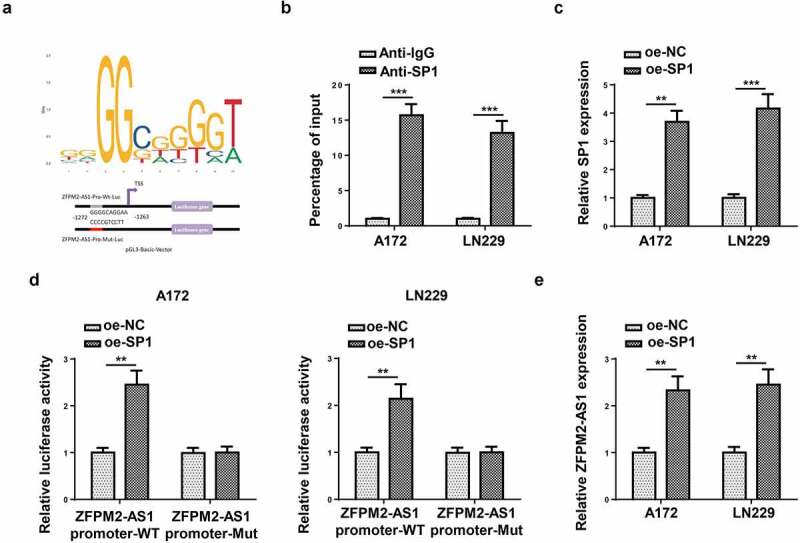
SP1 transcriptionally activated ZFPM2-AS1 expression. (a) The DNA motif of SP1 and its binding site with ZFPM2-AS1 promoter. (b) ChIP assay was used to confirm the enrichment of ZFPM2-AS1 promoter in the beads conjugated with anti-SP1 or anti-IgG. (c) RT-qPCR was applied to detect oe-SP1 transfection efficiency. (d) Luciferase reporter assay was adopted to prove the interaction of SP1 with ZFPM2-AS1 promoter. (e) ZFPM2-AS1 expression in SP1-overexpressed cells was assessed by RT-qPCR. **p < 0.01, ***p < 0.001
ZFPM2-AS1 sponged miR-515-5p in glioma
Next, we inspected the downstream mechanism mediated by ZFPM2-AS1. Considering that lncRNA was widely reported to involve in ceRNA network, we used starBase (http://starbase.sysu.edu.cn/) to hypothesize potential miRNAs for ZFPM2-AS1. As a result, 10 candidate miRNAs (miR-650, miR-3612, miR-4428, miR-519e-5p, miR-515-5p, miR-653-5p, miR-4735-3p, miR-18a-5p, miR-18b-5p, miR-511-3p) were found. Among which, miR-515-5p had the highest binding capacity with ZFPM2-AS1 (Figure 4a). A previous study indicated that ZFPM2-AS1 could serve as a sponge of miR-515-5p in retinoblastoma [33], but their interplay was not explored in glioma. Subsequently, miR-515-5p expression was found to be downregulated in glioma tissues and cells (Figure 4b). For further analysis, we introduced miR-515-5p mimics into glioma cells, and found miR-515-5p was overexpressed (Figure 4c). Then, as demonstrated by luciferase reporter assay, miR-515-5p overexpression decreased the luciferase activity of ZFPM2-AS1-WT rather than that of ZFPM2-AS1-Mut (Figure 4d). In addition, compared to complex captured by anti-IgG, ZFPM2-AS1 and miR-515-5p were abundant in compound precipitated by anti-Ago2 (Figure 4e). Moreover, ZFPM2-AS1 expression was negatively correlated with miR-515-5p expression in glioma tissues (figure 4f). Taken together, ZFPM2-AS1 acted as a sponge of miR-515-5p.
Figure 4.
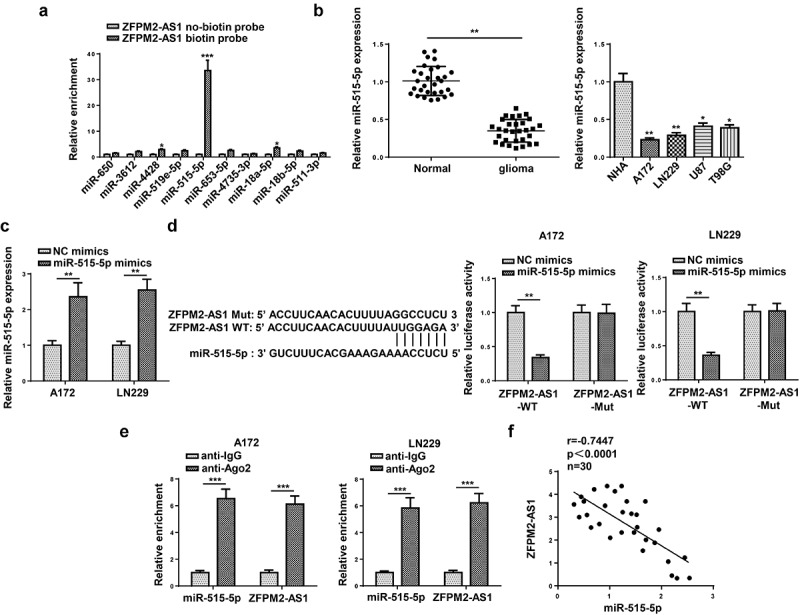
ZFPM2-AS1 interacted with miR-515-5p. (a) Potential miRNAs that had complementary base for ZFPM2-AS1 were obtained from starBase, and the interaction between ZFPM2-AS1 and these miRNAs was validated by RNA pull-down assay. (b) MiR-515-5p expression in glioma tissues and cells. (c) The transfection efficiency of miR-515-5p was confirmed by RT-qPCR. (d) Luciferase reporter assay was performed to verify the binding site between miR-515-5p and ZFPM2-AS1. (e) RIP assay was conducted to testify the interaction between miR-515-5p and ZFPM2-AS1. (f) Expression association between miR-515-5p and ZFPM2-AS1 was validated by Pearson’s association analysis. *p < 0.05, **p < 0.01, ***p < 0.001
SOD2 was targeted by miR-515-5p
To investigate the downstream target gene of miR-515-5p, 12 potential targets of miR-515-5p were predicted using StarBase (Supplementary Figure S2). Then, the expression of these genes was examined in miR-515-5p-overexpressed glioma cells, and the results showed that SOD2 expression was significantly lower than other genes (Figure 5a). In addition, it was detected that SOD2 was highly expressed in glioma tissues and cells (Figure 5b and c). Luciferase reporter assay revealed the decreased luciferase activity of SOD2-WT in miR-515-5p-overexpressed cells, while that of SOD2-Mut remained unchanged (Figure 5d). By RIP assay, we found that miR-515-5p and SOD2 were both abundant in anti-Ago2-precipitated complex (Figure 5e). Finally, it was found that SOD2 expression was negatively correlated with miR-515-5p and positively correlated with ZFPM2-AS1 in glioma tissues (figure 5f). Conclusively, miR-515-5p targeted SOD2 in glioma.
Figure 5.
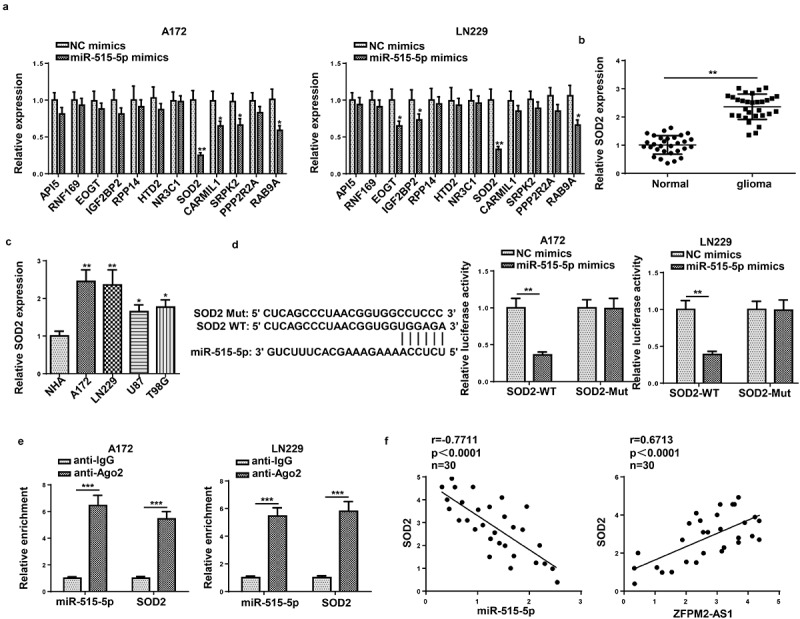
SOD2 was the target gene of miR-515-5p. (a) Effect of miR-515-5p upregulation on the expression of the predicted targets. (b and c) RT-qPCR analysis of SOD2 expression in glioma tissues and cells. (d and e) The binding of SOD2 to miR-515-5p was proved by luciferase reporter and RIP assays. (f) Pearson’s association analysis was used to validate the expression correlation between SOD2 and miR-515-5p (or ZFPM2-AS1). *p < 0.05, **p < 0.01, ***p < 0.001
ZFPM2-AS1 accelerated glioma cell growth via increasing SOD2 expression
Finally, rescue assays were conducted to verify whether SOD2 impacted the regulatory effect of ZFPM2-AS1 on cellular processes in glioma. At first, the overexpression efficiency of SOD2 was confirmed by RT-qPCR (Figure 6a). As revealed in Figure 6b, the cell viability of glioma inhibited by ZFPM2-AS1 knockdown was rescued via overexpressing SOD2. Colony formation assay indicated that SOD2 overexpression counteracted ZFPM2-AS1 silencing-mediated inhibitive effect on cell colonies (Figure 6c). The knockdown of ZFPM2-AS1 promoted cell cycle arrest, which was recovered by SOD2 upregulation (Figure 6d and Supplementary Figure S3). Furthermore, the accelerating effect of ZFPM2-AS1 silencing on cell apoptosis was neutralized by SOD2 augment (Figure 6e). Generally, ZFPM2-AS1 facilitated glioma cell growth by upregulating SOD2.
Figure 6.
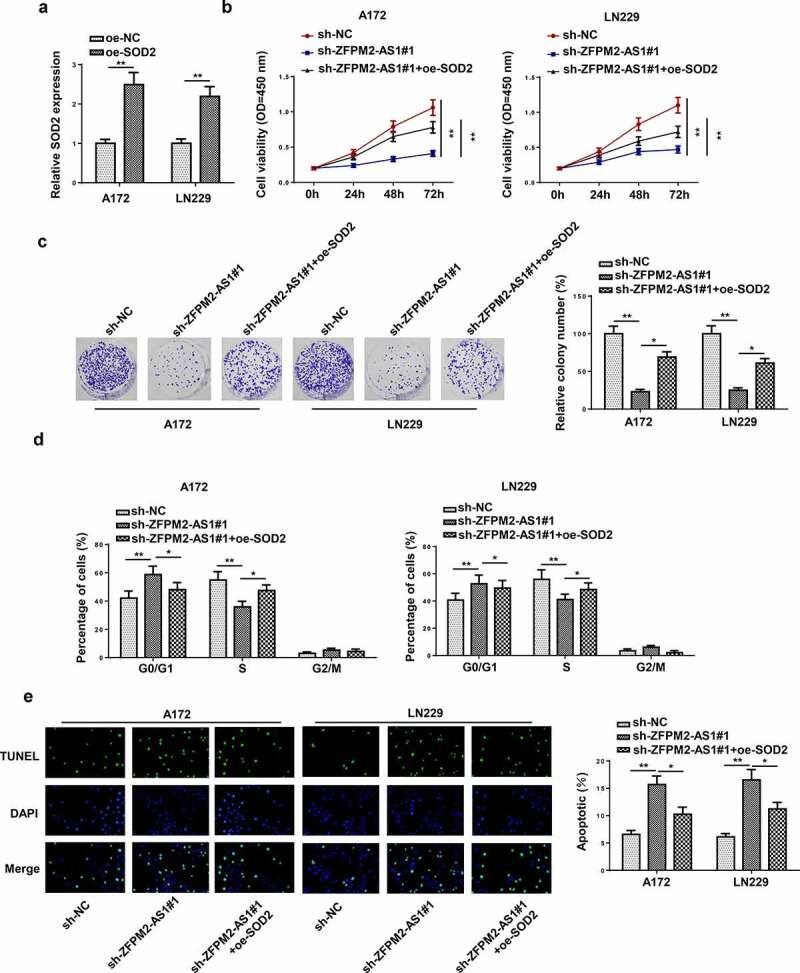
SOD2 participated in ZFPM2-AS1-mediated cell growth in glioma. (a) RT-qPCR analysis confirmed the upregulation of SOD2 in glioma cells transfected with SOD2 overexpression plasmid. (b and c) Cell proliferation in each group was evaluated by CCK-8 and colony formation assay. (d) Flow cytometry analysis was conducted to determine the cell cycle in each group. (e) The cell apoptosis was assessed via TUNEL assay. *p < 0.05, **p < 0.01
Discussion
High proliferative potential and aggressive angiogenesis are typical characteristics of glioma and cause frequent recurrence and unfavorable prognosis [34]. Understanding the specific molecular events underpinning glioma development could be beneficial for earlier detection and better prognosis. Previous studies have indicated that lncRNAs participate in the progression of glioma. For example, Li et al. reported that LINC00319 promoted tumorigenesis of glioma and was associated with poor prognosis in glioma [35]. LINC01116 facilitates the proliferation, migration, and invasion of glioma cells by targeting VEGFA [36]. Herein, we inspected the level and functional potential of lncRNA ZFPM2-AS1 in glioma. It was found that ZFPM2-AS1 was highly expressed in glioma. In vitro and in vivo assays exhibited that ZFPM2-AS1 deficiency diminished cell proliferation and cell cycle, enhanced cell apoptosis, and retarded tumor growth in nude mice. Importantly, accumulating studies have demonstrated that the upregulation of lncRNAs could be attributed to the function of transcription factor [37,38]. As a key transcription factor, SP1 was widely recognized to control the expression of lncRNAs [39,40]. Herein, our experiments confirmed the interaction between SP1 and ZFPM2-AS1 promoter. All the data confirmed the oncogenic role of ZFPM2-AS1 in glioma, and ZFPM2-AS1 was transcriptionally induced by SP1.
Numerous studies have revealed that lncRNA and miRNA can form a control network to exert regulatory functions in human cancers [41]. This novel mechanism known as ceRNA network was emerged as an important modulator involved in epigenetic modification, in which lncRNAs altered tumor-related genes by sponging miRNAs [42,43]. Therefore, we assumed that ZFPM2-AS1 might act as a ceRNA to participate in glioma progression. Through bioinformatics, we found ten promising miRNAs, and then miR-515-5p was identified with the highest possibility. Previous studies uncovered that miR-515-5p served as a cancer-inhibitor by suppressing cell migration and invasion in hepatocellular carcinoma [44]. MiR-515-5p was also reported to diminish cell proliferation in lung squamous cell carcinoma [45]. In our study, we observed that miR-515-5p was downregulated in glioma tissues and cells. In addition, miR-515-5p was found to interact and negatively associate with ZFPM2-AS1. Overall, these results implied that ZFPM2-AS1 displayed functional importance in glioma via serving as a ceRNA to regulate miR-515-5p.
Superoxide dismutase 2 (SOD2) has been reported as an oncogenic regulator in non-small cell lung cancer [46]. Moreover, SOD2 was found to improve liver cell detoxification capability [47]. In addition, SOD2 was also confirmed to play an important part in HER2-positive breast cancer [48]. Our research revealed that SOD2 was targeted by miR-515-5p, and rescued the inhibitive effect of silenced ZFPM2-AS1 on glioma cell growth.
Conclusions
Our study was the first to explore the function and mechanism of ZFPM2-AS1 in glioma, and discovered that ZFPM2-AS1 upregulated by SP1 promoted glioma cell growth via targeting miR-515-5p/SOD2 axis. This discovery indicated that ZFPM2-AS1 might be a prospective biomarker for glioma treatment.
Supplementary Material
Funding Statement
This work was supported by Nanjing Health Science and Technology Development Special Fund Project (YKK19166 and YKK19167).
Highlights
ZFPM2-AS1 knockdown inhibits glioma progression
ZFPM2-AS1 is transcriptionally induced by SP1
ZFPM2-AS1 promotes glioma progression via miR-515-5p/SOD2 axis
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Bakas S, Akbari H.. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Venkatesh HS, Tam LT, Woo PJ, et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature. 2017;549(7673):533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kane JR. The role of brain vasculature in glioblastoma. Mol Neurobiol. 2019;56(9):6645–6653. [DOI] [PubMed] [Google Scholar]
- [4].Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. [DOI] [PubMed] [Google Scholar]
- [5].Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9(7):717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lu QR, Qian L. Developmental origins and oncogenic pathways in malignant brain tumors. Wiley Interdiscip Rev Dev Biol. 2019;8(4):e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zachariah MA, Oliveira-Costa JP, Carter BS, et al. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018;20(9):1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kiran M, Chatrath A, Tang X, et al. A prognostic signature for lower grade gliomas based on expression of long non-coding RNAs. Molecular Neurobiology. 2019;56(7):4786–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Glen H. Lenvatinib therapy for the treatment of patients with advanced renal cell carcinoma. Future Oncol. 2016;12(19):2195–2204. [DOI] [PubMed] [Google Scholar]
- [10].Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tan SK, Pastori C, Penas C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Han M, Wang S, Fritah S, et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain. 2020;143(2):512–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].He H, Wang Y, Ye P, et al. Long noncoding RNA ZFPM2-AS1 acts as a miRNA sponge and promotes cell invasion through regulation of miR-139/GDF10 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li J, Ge J, Yang Y, et al. Long noncoding RNA ZFPM2-AS1 is involved in lung adenocarcinoma via miR-511-3p/AFF4 pathway. J Cell Biochem. 2020;121(3):2534–2542. [DOI] [PubMed] [Google Scholar]
- [15].Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. [DOI] [PubMed] [Google Scholar]
- [16].Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. [DOI] [PubMed] [Google Scholar]
- [17].Nan Y, Han L, Zhang A, et al. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. [DOI] [PubMed] [Google Scholar]
- [18].Cui T, Bell EH, McElroy J, et al. miR-4516 predicts poor prognosis and functions as a novel oncogene via targeting PTPN14 in human glioblastoma. 2019;38(16):2923–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang X, Zhou J, Xue D, et al. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. [DOI] [PubMed] [Google Scholar]
- [20].Li J, Tang Z, Wang H, et al. CXCL6 promotes non-small cell lung cancer cell survival and metastasis via down-regulation of miR-515-5p. Biomed Pharmacother. 2018;97:1182–1188. [DOI] [PubMed] [Google Scholar]
- [21].Qiao K, Ning S, Wan L, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(1):418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [23].Hong Y, Liang H, Uzair-Ur-Rehman, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang L, Kang W, Lu X, et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–1900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [25].Liu B, Wu S, Ma J, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang H, Liu S, Tang L, et al. Long non-coding RNA (LncRNA) MRPL23-AS1 promotes tumor progression and carcinogenesis in osteosarcoma by activating Wnt/β-catenin signaling via inhibiting microRNA miR-30b and upregulating myosin heavy chain 9 (MYH9). Bioengineered. 2021;12(1):162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen L, Hu N, Wang C, et al. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17(3):319–329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [28].Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [29].Yu S, Wang D, Shao Y, et al. SP1-induced lncRNA TINCR overexpression contributes to colorectal cancer progression by sponging miR-7-5p. Aging (Albany NY). 2019;11(5):1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782. [DOI] [PubMed] [Google Scholar]
- [31].Zhou T, Wu L, Ma N, et al. SOX9-activated FARSA-AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis. 2020;11(12):1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang X-W, Li Q-H, Xu Z-D, et al. STAT1-induced regulation of lncRNA ZFPM2-AS1 predicts poor prognosis and contributes to hepatocellular carcinoma progression via the miR-653/GOLM1 axis. Cell Death Dis. 2021;12(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lyv X, Wu F, Zhang H, et al. Long noncoding RNA ZFPM2-AS1 knockdown restrains the development of retinoblastoma by modulating the MicroRNA-515/HOXA1/Wnt/β-Catenin axis. Invest Ophthalmol Vis Sci. 2020;61(6):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li Q, Wu Q, Li Z, et al. LncRNA LINC00319 is associated with tumorigenesis and poor prognosis in glioma. Eur J Pharmacol. 2019;861:172556. [DOI] [PubMed] [Google Scholar]
- [36].Ye J, Zhu J, Chen H, et al. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. International Journal of Cancer. 2020;146(1):248–261. [DOI] [PubMed] [Google Scholar]
- [37].Huang T, Wang G, Yang L, et al. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol. 2017;36(11):947–958. [DOI] [PubMed] [Google Scholar]
- [38].Zhang W, et al. A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives tumor growth and metastasis in hepatocellular carcinoma through the Wnt/β-catenin pathway. Mol Oncol. 2019;13(10):2194–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang Z-Q, Cai Q, Hu L, et al. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis. 2017;8(6):e2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu T-P, Liu -X-X, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. [DOI] [PubMed] [Google Scholar]
- [41].Poliseno L, Pandolfi PP. PTEN ceRNA networks in human cancer. Methods. 2015;77-78:41–50. [DOI] [PubMed] [Google Scholar]
- [42].Zhang H, Liu S, Tang L, et al. Long non-coding RNA (LncRNA) MRPL23-AS1 promotes tumor progression and carcinogenesis in osteosarcoma by activating Wnt/β-catenin signaling via inhibiting microRNA miR-30b and upregulating myosin heavy chain 9 (MYH9). Bioengineered. 2021;12(1):162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zheng S, Guo Y, Dai L, et al. Long intergenic noncoding RNA01134 accelerates hepatocellular carcinoma progression by sponging microRNA-4784 and downregulating structure specific recognition protein 1. Bioengineered. 2020;11(1):1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ni JS, Zheng H, Ou Y-L, et al. miR-515-5p suppresses HCC migration and invasion via targeting IL6/JAK/STAT3 pathway. Surg Oncol. 2020;34:113–120. [DOI] [PubMed] [Google Scholar]
- [45].Ye P, Lv X, Aizemaiti R. H3K27ac-activated LINC00519 promotes lung squamous cell carcinoma progression by targeting miR-450b-5p/miR-515-5p/YAP1 axis. 2020;53(5):e12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu J, Yao L, Zhang M, et al. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY). 2019;11(18):7830–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Li M, Peng L, et al. SOD2 promotes the expression of ABCC2 through lncRNA CLCA3p and improves the detoxification capability of liver cells. Toxicol Lett. 2020;327:9–18. [DOI] [PubMed] [Google Scholar]
- [48].Zhong BL, Bian L-J, Wang G-M, et al. Identification of key genes involved in HER2-positive breast cancer. Eur Rev Med Pharmacol Sci. 2016;20(4):664–672. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


