ABSTRACT
MicroRNAs (miRNAs) have been proved to be involved in many biological processes during tumorigenesis and progression, including cell proliferation and cell cycle progression. However, the potential role of miR-26b-5p in tongue squamous cell carcinoma (TSCC) remains unclear. In the present study, we demonstrated that miR-26b-5p was decreased in TSCC tissues in both TCGA-TSCC subset and eight paired samples from TSCC patients, while Proline Rich 11 (PRR11) was obviously increased. Transfection of miR-26b-5p mimics inhibited CALL7 cell proliferation by arresting the cells at the S/G2 transition. Meanwhile, miR-26b-5p inhibitor had the opposite biological functions. The results of luciferase activity and RNA-pulldown assays indicated that miR-26b-5p directly targeted the PRR11 3ʹ -untranslated region in CAL27 cells. Furthermore, the effects of miR-26b-5p on cell cycle regulation were reversed after treatment with siRNA against PRR11. In summary, our findings indicate that miR-26b-5p induce cell cycle arrest in TSCC by targeting PRR11. Hence, targeting miR-26b-5p could be a promising therapeutic strategy for the treatment of TSCC.
KEYWORDS: Mir-26b-5p, prr11, cell cycle, tongue squamous cell carcinoma, proliferation
Introduction
Tongue squamous cell carcinoma (TSCC) is the most prevalent malignancy of the oral cavity worldwide [1–3]. Despite advances in detection and treatment, the outcome of patients with TSCC remains unsatisfactory in recent years [4]. In normal cells, cellular growth and proliferation are stringently regulated, while derangements of the cell cycle can lead to uncontrolled proliferation and provide tumor cells with growth advantages. Hence, it is essential to dissect the molecular mechanism supporting the growth advantage of TSCC cells, which might contribute to providing novel therapeutic targets for TSCC treatment.
Accumulating evidence supports the involvement of microRNAs (miRNAs) in the regulation of cell proliferation, apoptosis, invasion, migration and other phenotypes by binding to the 3ʹ- untranslated region (UTR) of its target genes. Previous studies have demonstrated that miR-26b-5p plays important roles in the development and progression of various cancers, including lung cancer, liver cancer and myeloma [5–7]. For instance, miR-26b-5p inhibites cell proliferation and induces apoptosis in multiple myeloma cells by targeting JAG1, and maintains the stemness of hepatocellular carcinoma cells by inhibiting HSC71/HSPA8. miR-26b-5p inhibition promoted the growth of Burkitt lymphoma cells by repressing the KPNA2 expression [8]. Additionally, miR-26b-5p upregulation restricted the malignant features of human apillary thyroid cancer by degrading beta-catenin [9]. To data, The involvement of miR-26b-5p in TSCC remain unclear.
Recent high-throughput studies have facilitated an integrative understanding of the molecular mechanisms underlying carcinogenesis, metastasis, and chemoresistance in cancer research [10–13]. Using bioinformatics tools, we were able to understand miRNA functions by identifying miRNA targets. Proline-rich 11 (PRR11) protein, which has been implicated in the regulation of cell cycle progression [14], was predicted as a candidate target of miR-26b-5p in the current study. Several studies have indicated that PRR11 is overexpressed in various cancers, including lung, ovarian, esophageal and pancreatic cancers [15–18]. However, the potential roles of PRR11 in TSCC remain unclear.
In the present study, miR-26b-5p was found to be downregulated in TSCC, and contributed to the inhibition of cell proliferation. Together with the prediction of the miR-26b-5p binding site within the PRR11 3ʹ-UTR, we hypothesized that miR-26b-5p could suppress TSCC cell proliferation via targeting PRR11. Our results shed new light on the mechanism that provides TSCC cells with growth advantages.
Materials and methods
Tissues and cell line
Eight pairs of frozen samples were collected from TSCC patients, and informed consent was obtained from the Hunan Cancer Hospital. All experiments were approved by the ethics committee of the Hunan Cancer Hospital (KYJJ-2020-222), Changsha, China. The TSCC cell line CAL27 was purchased from Shanghai Genechem Co., Ltd, and routinely cultured in DMEM with 10% FBS (Gibco, Gaithersburg, MD).
Bioinformatics and statistical analysis
The gene expression data (FPKM) of TSCC patients were downloaded and filtered from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov), and the candidate gene data were subsequently extracted to form a new matrix. Differences in gene expression between groups were assessed using the Student’s t-test. Using ENCORI [19], candidate miRNA-target pairs were selected, and the Pearson correlation coefficient (PCC) values between the expressions of two genes in each pair were subsequently calculated. The pairs with PCC< −0.2 and corrected p-value < 0.05 were considered statistically significantly correlated, and the correlation between candidate genes in different datasets was visualized as scatter diagrams. Because TSCC is always considered as a subset of head and neck squamous cell carcinoma, overall survival (OS) analysis of candidate genes was performed using the web tool OncoLnc (http://www.oncolnc.org) in the TCGA-HNSC dataset [20]. The Kaplan–Meier method was used to estimate OS.
Quantitative real-time PCR
Each siRNA, miRNA mimic or inhibitor (GenePharm, Shanghai, China) was transfected into CAL27 cells for 48 h using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA), and then reverse transcribed to cDNA using PrimeScriptTM RT-PCR Kit (Takara, Dalian, China) according to the manufacturer’s instructions. For miRNA quantitation, reverse transcription was performed using the PrimeScript RT Reagent Kit (Takara, Dalian, China) with specific stem-loop primers. Quantitative Real-Time PCR (qRT-PCR) was performed using SYBR® Premix DimerEraser™ (Takara, Dalian, China) in a Roche LightCycler 480 II Real-Time PCR system (Roche, Basel, Switzerland). The threshold cycle value (Ct) of each product was determined and normalized against that of the internal control GAPDH or U6 (for miRNA), and the differences were compared by t-test using SPSS version 23.0, the statistical significance set at P < 0.05.
Cell counting kit-8 assay
After treatment, the cells were seeded into 96-well plates at 2 × 103 cells/well. The Cell counting kit-8 (CCK-8, Beyotime, China, C0041) reagent was injected into the wells after 0-, 12-, 24-, 48- and 72 h of culturing. Finally, the optical density at 450 nm was recorded using a microplate reader after a 2 h incubation.
Cell cycle analysis
Cell cycle analysis was performed by using propidium iodide staining and flow cytometry. After washing once with cold PBS, cell pellets (approximately 1.0 × 106 cells) were resuspended in 200 μL of cold PBS, and fixed in 4 mL of 70% ethanol overnight at −20 °C. Samples were subsequently collected by centrifugation and resuspended in 500 μL of buffer containing 40 μg/ml propidium iodide (Beyotime, China, C1052) and 100 μg/mL RNase A (Beyotime, China, C1052). All samples were incubated for 30 min at 37 °C before analysis on a CytoFLEX flow cytometer (Beckman Coulter). Data were analyzed using the CytExpert (Beckman Coulter, Version 2.0) software.
EdU staining
EdU (5-ethynyl-2ʹ-deoxyuridine) staining was carried out using Cell-Light EdU Apoll®567 In Vitro Kit (RiboBio, Guangzhou, China, C10310). The CAL27 cells were seeded and transfected with these molecules. Then, 48 h after transfection, the cells were washed and incubated with 10 μM EdU for 30 min. Fixation and penetration of cells were performed, followed by DAPI (Thermo, USA, 62,248) staining. After washing with PBS, the plates were observed and photographed under a microscope (Olympus, Tokyo, Japan, CX41-72C02) at 200× magnification.
Dual-luciferase reporter gene assay
Vectors, pmirGLO-PRR11 3ʹ-UTR-wt (wild-type) and pmirGLO-PRR11 3ʹ-UTR-mut (miR-26b-5p binding site mutated), for the luciferase reporter assay were generated based on the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI). The plasmids were then co-transfected with miR-26b-5p or negative control (NC) mimics into CAL27 cells using Lipofectamine 2000 according to the manufacturer’s guidelines, respectively. Relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
RNA pulldown
Biotinylated probes complementary to PRR11 mRNA were synthesized (GenePharm, Shanghai, China) using a random probe as the control. The M-280 streptavidin magnetic beads (Sigma-Aldrich, St. Louis, MO) were coated by incubation with the probes. The cell lysates of CAL27 transfected with PRR11 overexpression vectors, with wild-type or mutated miR-26b-5p binding sites, were prepared after 48 h. Lysates were then incubated with the probe-coated beads at 4°C overnight, and the molecules interacting with PRR11 mRNA were captured after washing. The bound RNAs were subsequently purified using TRIzol and the miR-26b-5p abundance was measured by qRT-PCR.
Western blot
The harvested cells were lysed in RIPE buffer at 4°C for 30 min and centrifuged at 15,000 × g for 15 min to obtain the protein sediment. Equal amounts of total proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS–PAGE) before transferring to PVDF membranes (Millipore, Billerica, MA). Subsequently, the membrane was incubated with primary antibodies against PRR11 (Invitrogen, CA, USA, MA5-26,460, 1:2,000 dilution), cyclin D1 (Bioss, Beijing, China, bs-0623 R, 1:500 dilution), p-RbSer807 (abcam, CA, UK, ab131264, 1:1,000 dilution) and β-actin (ProteinTech, IL, USA, 66,009, 1:2,000 dilution) overnight at 4°C. After incubation with the corresponding secondary antibody (ProteinTech Group Inc., Chicago, IL, 1:6,000 dilution) for 1 h at room temperature, the signals were measured using an enhanced chemiluminescence (ECL) kit (Pierce, Rockford, IL).
Results
This study was designed to investigate the role of the miR-26b-5p/PRR11 axis in TSCC. We studied the expression pattern of miR-26b-5p in TSCC, as well as its role in cell proliferation and cell cycle progression. Together with the prediction of the miR-26b-5p binding site within the PRR11 3ʹ-UTR, we hypothesized and verified that miR-26b-5p could suppress TSCC cell proliferation by targeting PRR11.
miR-26b-5p was downregulated in TSCC tissues
Using the data from the TCGA-TSCC subset, we found that miR-26b-5p was significantly downregulated (p = 0.0207) in TSCC tissues as compared with the normal tongue tissues (Figure 1(a)). To validate the results of high-throughout sequencing, the expression levels of miR-26b-5p were subsequently examined in collected tissues by qRT-PCR. Compared with the matched para-cancer tissues, the expression of miR-26b-5p was lower (p = 0.0089) in TSCC tissues (Figure 1(b)). Moreover, the Kaplan-Meier survival curve suggests poor OS with lower expression of miR-26b-5p than those in the high miR-26b-5p express ion group (Figure 1(c)).
Figure 1.
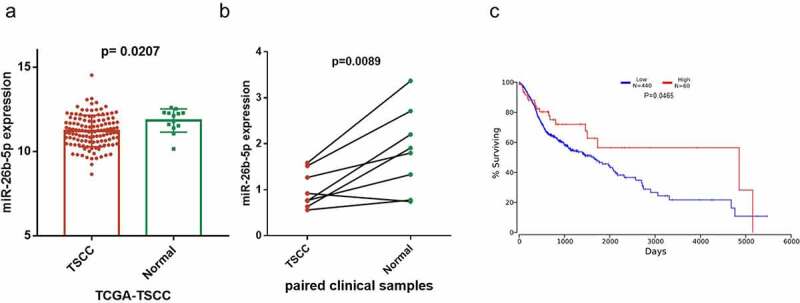
Expression and prognostic value of miR-26b-5p in TSCC. (a) the level of miR-26b-5p in TSCC tissues in TCGA dataset compared with normal tongue tissues. (b) comparison of miR-26b-5p expression in 8 paired samples collected from the TSCC patients. (c) kaplan-meier plots for overall survival (OS) in TCGA-HNSC patients, grouped by low and high expression of miR-26b-5p. P values were obtained using log-rank test
miR-26b-5p inhibits cell proliferation of CAL27
Because miR-26b-5p was significantly downregulated in TSCC, we evaluated the effect of miR-26b-5p on cell proliferation of CAL27 cells. miR-26b-5p expression in CAL27 cells was regulated by transfection with miR-26b-5p mimics and inhibitors (Figure 2(a)). As indicated in Figure 2(b)), miR-26b-5p mimics inhibited the growth of CAL27 cells, whereas the inhibitor enhanced the growth of CAL27 cells. We further examined the effects of miR-26b-5p on cell cycle modulation in CAL27 cells. Compared with the control, miR-26b-5p mimics caused cycle arrest at the S-phase, and restrained CAL27 cell transit to the G2/M phase (Figure 2(c)). Further EdU staining indicated that miR-26b-5p mimics led to reductions in S phase cells, but miR-26b-5p inhibitor significantly increased the number of proliferating cells (Figure 2(d)), indicating the suppressive role of miR-26b-5p on cell proliferation. To decipher the potential mechanism of miR-26b-5p in regulating CAL27 cell proliferation, we predicted the downstream target of miR-26b-5p using the ENCORI database (http://starbase.sysu.edu.cn/), and found that there was a binding site for miR-26b-5p in the PRR11 mRNA 3ʹ-UTR. Consequently, the expression of PRR11 and cell cycle-related proteins, including CDK1 and p-RbSer807, was detected by western blotting. Our results showed that miR-26b-5p mimic decreased the protein levels of PRR11, cyclinD1 and p-RbSer807, which are the indicators of enhanced cell cycle progression, while miR-26b-5p inhibitor elevated the expression levels of these proteins (Figure 2(e)). Collectively, these findings suggest that miR-26b-5p suppresses CAL27 cell proliferation by arresting the cells at the S/G2 transition.
Figure 2.
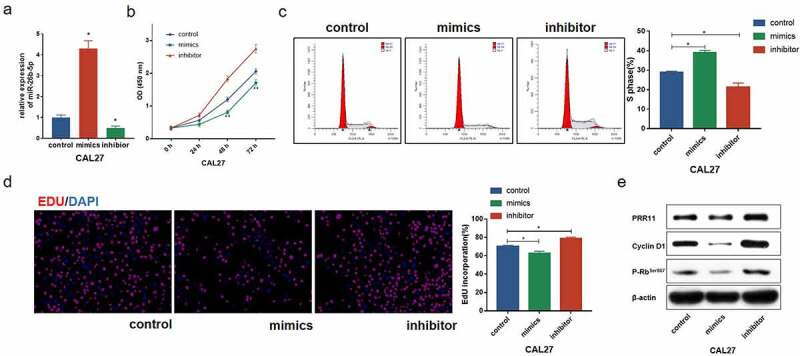
Effect of miR-26b-5p on cell cycle progression in CAL27 cells. (a) effects of miR-26b-5p mimics and inhibitor on miR-26b-5p expression in CAL27 cells. (b) the viability of each group of cells was detected by CCK-8 assay. (c) for each group, cell cycle distribution was detected by flow cytometry analysis. (d) EdU staining (red) was performed to check the proliferating cells. cell nucleus were stained with DAPI (blue). (e) levels of PRR11, cyclinD1, and p-RbSer807 were detected by western blotting analysis in each group, with β-actin as the reference protein. *, p < 0.05
PRR11 is a target of miR-26b-5p in CAL27
We found that PRR11 was significantly upregulated in TSCC tissues as compared with the normal tongue tissues in the TCGA-TSCC subset (Figure 3(a)) and collected paired samples (Figure 3(b)) (p = 0.0207 and p = 0.0207). Moreover, the Kaplan-Meier survival curve suggests poor OS with high expression of PRR11 compared with those in the low PRR11 expression population (Figure 3(c)). A negative correlation between the expression of miR-26b-5p and PRR11 was validated in the TCGA-TSCC subset and collected samples (Figure 4(a-b)). Luciferase reporter assays and RNA pull-down assays were also performed to further investigate the interaction between these two molecules. Transfection with miR-26b-5p mimics significantly decreased the luciferase activity of the PRR11 3ʹ-UTR wild-type reporter gene, but had no effect on that of the PRR11 3ʹ-UTR mutated reporter gene (Figure 5(a)). These results suggest that miR-26b-5p inhibits PRR11 dependent on 3ʹ-UTR binding. Furthermore, mutation of PRR11 3ʹ-UTR significantly decreased the abundance of miR-26b-5p captured by PRR11 probes (Figure 5(b), right panel), further validating the binding interaction between miR-26b-5p and PRR11.
Figure 3.
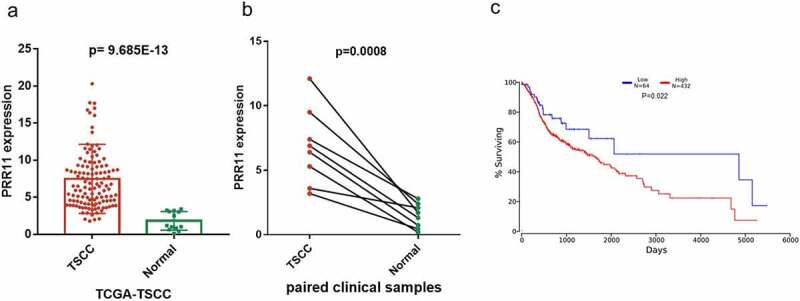
Expression and prognostic value of PRR11 in TSCC. (a) the level of PRR11 in TSCC tissues in TCGA dataset compared with normal tongue tissues. (b) comparison of PRR11 expression in 8 paired samples collected from TSCC patients. (c) kaplan-meier plots for overall survival (OS) in TCGA-HNSC patients, grouped by low and high expression of PRR11. P values were obtained using log-rank test
Figure 4.
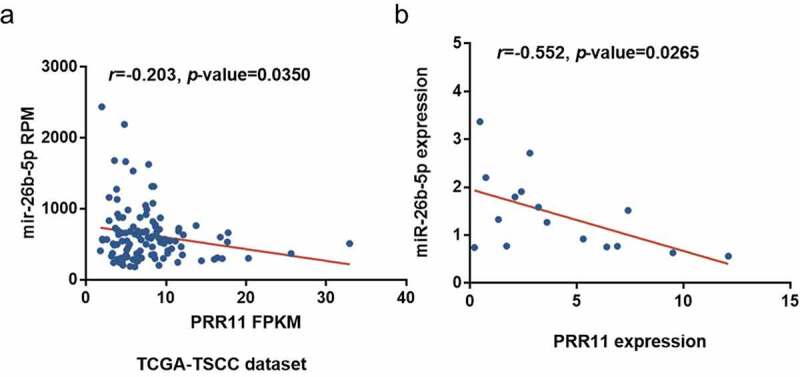
Correlation of miR-26b-5p and PRR11 expression in TSCC. pearson correlation analysis shows a negative correlation between miR-26b-5p and PRR11 mRNA level in the (a) TCGA-TSCC subset (n = 110) and (b) 8 paired samples
Figure 5.
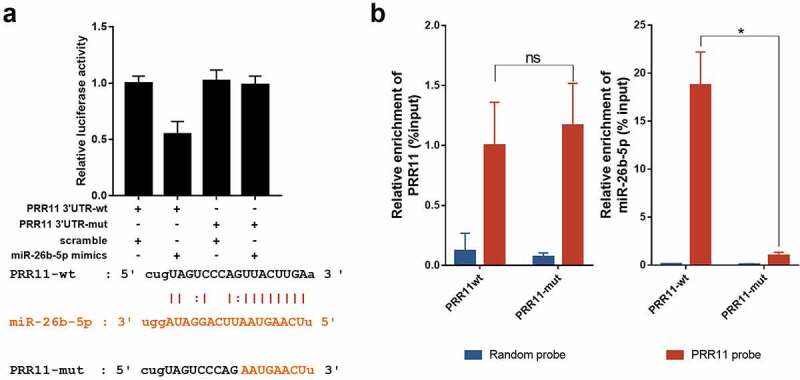
miR-26b-5p repressed PRR11 by binding to the 3ʹ-UTR of PRR11. (a) effects of miR-26b-5p on the luciferase activity of the reporter gene inserted downstream of the wildtype and mutated PRR11 3ʹ-UTR in CAL27 cells. the sequence of the miR-26b-5p binding site in the 3ʹUTR of PRR11 mRNA and its corresponding mutation were indicated. (b) RNA pulldown assay was performed in CAL27 cells transfected with wildtype and mutated PRR11, followed by qRT-PCR to detect the abundance of PRR11 and miR-26b-5p. data are presented as the mean ± standard deviation (SD). ns, not significant; *, p < 0.05
miR-26b-5p inhibited cell cycle progression via targeting PRR11 in CAL27
To determine the specific role of the miR-26b-5p/PRR11 axis in TSCC, we co-transfected miR-26b-5p inhibitor and si-PRR11 into CAL27 cells. The CCK-8 assay results indicated that PRR11 knockdown attenuated the effect of miR-26b-5p inhibitor on cell proliferation (Figure 6(a)). Moreover, western blotting revealed that transfection of si-PRR11 downregulated the expression of PRR11 in CAL27 cells, and weakened the facilitation effect of miR-26b-5p inhibitor on the expression of cell cycle-related genes (Figure 6(b)). These results highlight that the miR-26b-5p/PRR11 axis is involved in modulating the cell cycle progression of CAL27 cells.
Figure 6.
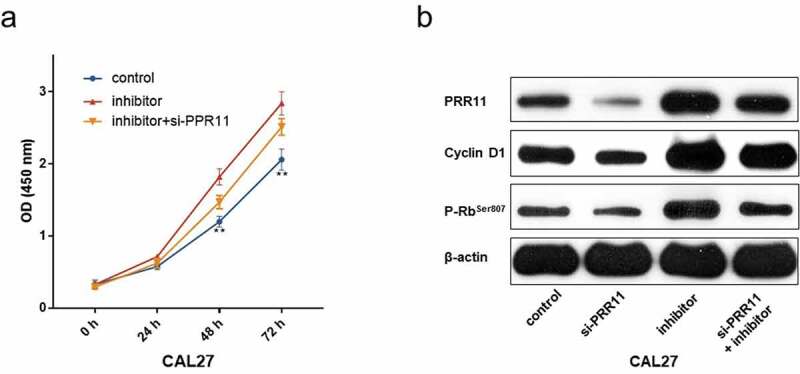
PRR11 knockdown attenuates the repression effects of miR-26b-5p on CAL27cell proliferation. (a) the viability of each group of cells was detected by CCK-8 assay. (b) western blot analysis of the impact of si-PRR11 on PRR11, cCyclinD1 and p-RbSer807 expression. data are presented as the mean ± SD
Discussion
The treatment of TSCC patients remains unsatisfactory in recent years, facilitating research on the molecular mechanisms of malignant phenotypes in TSCC. Cell cycle control provides cancer cells with a growth advantage, and can be investigated as a promising therapeutic target for TSCC treatment. In the present study, we found that the dysfunction of the miR-26b-5p/PRR11 axis were involved in the regulation of cell cycle progression. Moreover, the expression of the miR-26b-5p/PRR11 axis was significantly associated with OS in TSCC patients.
Recently, accumulating evidence has indicated that miRNAs are involved in the regulation of cell cycle progression in cancer [21,22]. However, few studies have focused on the tumorigenesis and progression of tongue cancer. In the last decade, several studies have identified miR-26b-5p to be crucial in the development and progression of cancers. As reported, miR-26b-5p is a tumor suppressor and modulator in cell cycle regulation that targets different genes, including CCND2, PLOD2, JAG1, MAP3K9 and KPN2 [6,8,23–25]. Notably, the ceRNA hypothesis sparked another miRNA-mediated mechanism, in which miRNA acts as the key modulator linking competing endogenous RNAs, including long non-coding RNA (lncRNA), circular RNA (circRNA), pseudogenes and protein-coding genes. For instance, circRNA_000203 exacerbates cardiac hypertrophy via the miR-26b-5p/Gata4 axis [26]. LINC00657 represses miR-26b-5p and enhances COMMD8 expression to promote NSCLC progression [27]. In addition, lncRNA HCG11 participates in the regulation of HUVEC proliferation by suppressing miR-26b-5p on QKI-5 expression [28]. These results support the idea that miR-26b-5p is implicated in the negative regulation of cell growth, which is consistent with our findings. In the current study, we identified PRR11, a promising oncogene, as a novel target of miR-26b-5p in TSCC. The expression of PRR11 is closely related to tumorigenesis, progression and poor prognosis in cancers, including lung, gastric, pancreatic, breast, esophageal and ovarian cancers [15,29–33]. PRR11 was also demonstrated to promote anti-estrogen resistance in breast cancer by amplifying the PI3K signaling pathway [34]. Additionally, PRR11 activated the Akt/mTOR autophagy signaling pathway to facilitate tumorigenesis in non-small cell lung cancer, suggesting that this gene may affect cancer cells through different signal transduction pathways. All the aforementioned results support the results of our study. To further validate the roles of the miR-26b-5p/PRR11 axis in TSCC, we performed rescue experiments, which indicated that miR-26b-5p inhibited cell cycle progression by targeting PRR11 in CAL27 cells. Consistent with our results, a previous study has indicated that PRR11 promotes TSCC cell proliferation by regulating the expression of cell cycle-related proteins, and facilitating S/G2 phase transition [35].
Conclusions
Altogether, we have uncovered a novel miR-26b-5p/PRR11 axis and elaborated its involvement in cell cycle regulation in TSCC. Moreover, our study provides novel insights into future understanding of the molecular mechanisms of cell cycle progression in TSCC.
Acknowledgements
We acknowledge TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Funding Statement
The present study was supported in part by the National Natural Science Foundation of China (grant no. 81703005), the Natural Science Foundation of Hunan Province (grant nos. 2017JJ3195, 2018JJ3317 and 2021JJ30427), the Hunan Provincial Science and Technology Department (grant nos. 2018SK2120, 2018SK2122, 2018SK50907 and 2018SK7005), the Research Project of Health and Family Planning Commission of Hunan Province (grant no. B2013-097, B2019092, C2019080, 20201650, 20201653 and 202102082037), Changsha Science and Technology Bureau Foundation (grant no. kq1706044 and kq1901074) and the Key Clinical Specialty Construction Project (Head&Neck Surgery) of the Provincial Health Commission of Hunan Province.
Authors’ contributions
YLong, LY, YLiu, YX and XZ conceived and designed the study; LY, AX, CL, JD and ZL collected and prepared the samples; LY, YLiu, AX, SL, HZ, MP and HR performed data research and analysis; YLong, LY and YLiu wrote the manuscript. All authors reviewed the manuscript and approved the final version.
Availability of data and materials
The datasets analyzed in the current study are available from the TCGA (cancergenome.nih.gov) repository. TCGA allows researchers to download relevant data for research and publish relevant articles.
Disclosure statement
The authors declare that they have no competing interests.
Ethics approval
The present study was approved by the Ethics Committee of Hunan Cancer Hospital (KYJJ-2020-222), Changsha, China.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. [DOI] [PubMed] [Google Scholar]
- [3].Gulland A. Oral cancer rates rise by two thirds. BMJ. 2016;355:i6369. [DOI] [PubMed] [Google Scholar]
- [4].Jansen L, Buttmann-Schweiger N, Listl S, et al. Differences in incidence and survival of oral cavity and pharyngeal cancers between Germany and the United States depend on the HPV-association of the cancer site. Oral Oncol. 2018;76:8–15. [DOI] [PubMed] [Google Scholar]
- [5].Khosla R, Hemati H, Rastogi A, et al. miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC. Liver Int. 2019;39(9):1692–1703. [DOI] [PubMed] [Google Scholar]
- [6].Jia CM, Tian YY, Quan LN, et al. miR-26b-5p suppresses proliferation and promotes apoptosis in multiple myeloma cells by targeting JAG1. Pathol Res Pract. 2018;214(9):213–221. [DOI] [PubMed] [Google Scholar]
- [7].Wu T, Chen W, Liu S, et al. Huaier suppresses proliferation and induces apoptosis in human pulmonary cancer cells via upregulation of miR-26b-5p. FEBS Lett. 2014;588(12):2107–2114. [DOI] [PubMed] [Google Scholar]
- [8].Niu F, Kazimierska M, Nolte IM, et al. The miR-26b-5p/KPNA2 Axis Is an Important Regulator of Burkitt Lymphoma Cell Growth. Cancers (Basel). 2020;12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhou A, Pan H, Sun D, et al. miR-26b-5p Inhibits the Proliferation, Migration and Invasion of Human Papillary Thyroid Cancer in a beta-Catenin-Dependent Manner. Onco Targets Ther. 2020;13:1593–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun W, Qiu Z, Huang W, et al. Gene expression profiles and proteinprotein interaction networks during tongue carcinogenesis in the tumor microenvironment. Mol Med Rep. 2018;17(1):165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tian F, Zhao J, Fan X, et al. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the cancer genome atlas database. J Thorac Dis. 2017;9(1):42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dai P, He Y, Luo G, et al. Screening candidate microRNA-mRNA network for predicting the response to chemoresistance in osteosarcoma by bioinformatics analysis. J Cell Biochem. 2019;120(10):16798–16810. [DOI] [PubMed] [Google Scholar]
- [13].Zhao Y, Huang J, Chen J.. The integration of differentially expressed genes based on multiple microarray datasets for prediction of the prognosis in oral squamous cell carcinoma. Bioengineered. 2021;12(1):3309–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ji Y, Xie M, Lan H, et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int J Biochem Cell Biol. 2013;45(3):645–656. [DOI] [PubMed] [Google Scholar]
- [15].Zhu J, Hu H, Wang J, et al. PRR11 Overexpression Facilitates Ovarian Carcinoma Cell Proliferation, Migration, and Invasion Through Activation of the PI3K/AKT/beta-Catenin Pathway. Cell Physiol Biochem. 2018;49(2):696–705. [DOI] [PubMed] [Google Scholar]
- [16].Chen J, Yang HM, Zhou HC, et al. PRR11 and SKA2 promote the proliferation, migration and invasion of esophageal carcinoma cells. Oncol Lett. 2020;20(1):639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lin C, Xia J, Gu Z, et al. Downregulation of USP34 inhibits the growth and migration of pancreatic cancer cells via Inhibiting the PRR11. Onco Targets Ther. 2020;13:1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang L, Zhang Y, Lei Y, et al. Proline-rich 11 (PRR11) drives F-actin assembly by recruiting the actin-related protein 2/3 complex in human non-small cell lung carcinoma. J Biol Chem. 2020;295(16):5335–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].J A. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67. [Google Scholar]
- [21].Wang L, Ge S, Zhou F. MicroRNA-487a-3p inhibits the growth and invasiveness of oral squamous cell carcinoma by targeting PPM1A. Bioengineered. 2021;12(1):937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu H, Chi Z, Jin H, et al. MicroRNA miR-188-5p as a mediator of long non-coding RNA MALAT1 regulates cell proliferation and apoptosis in multiple myeloma. Bioengineered. 2021;12(1):1611–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Y, Liu WB, Liu KJ, et al. Overexpression of miR-26b-5p regulates the cell cycle by targeting CCND2 in GC-2 cells under exposure to extremely low frequency electromagnetic fields. Cell Cycle. 2016;15(3):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Miyamoto K, Seki N, Matsushita R, et al. Tumour-suppressive miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by regulating PLOD2 in bladder cancer. Br J Cancer. 2016;115(3):354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu S, Li L, Li M, et al. Effect of miR-26b-5p on cis-diamine dichloroplatinum-induced ovarian granulosa cell injury by targeting MAP3K9. Vitro Cell Dev Biol Anim. 2020;56(3):213–221. [DOI] [PubMed] [Google Scholar]
- [26].Li H, Xu JD, Fang XH, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. 2020;116(7):1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qi J, Luo X, Ma Z, et al. Downregulation of miR-26b-5p, miR-204-5p, and miR-497-3p expression facilitates exercise-induced physiological cardiac hypertrophy by augmenting autophagy in rats. Front Genet. 2020;11:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Du J, Han R, Li Y, et al. LncRNA HCG11/miR-26b-5p/QKI5 feedback loop reversed high glucose-induced proliferation and angiogenesis inhibition of HUVECs. J Cell Mol Med. 2020;24(24):14231–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Song Z, Liu W, Xiao Y, et al. PRR11 Is a prognostic marker and potential oncogene in patients with gastric cancer. PLoS One. 2015;10(8):e0128943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sakai Y, Ohbayashi C, Yanagita E, et al. PRR11 immunoreactivity is a weak prognostic factor in non-mucinous invasive adenocarcinoma of the lung. Pathologica. 2017;109(3):133–139. [PubMed] [Google Scholar]
- [31].Tan S, Jiang Z, Hou A, et al. Expression of PRR11 protein and its correlation with pancreatic cancer and effect on survival. Oncol Lett. 2017;13(6):4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Y, Zhang C, Mai L, et al. PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep. 2019;52(2):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou L, Deng ZZ, Li HY, et al. Overexpression of PRR11 promotes tumorigenic capability and is associated with progression in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:2677–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee KM, Guerrero-Zotano AL, Servetto A, et al. Proline rich 11 (PRR11) overexpression amplifies PI3K signaling and promotes antiestrogen resistance in breast cancer. Nat Commun. 2020;11(1):5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang C, Yu L, Ren X, et al. The oncogenic potential of PRR11 gene in tongue squamous cell carcinoma cells. J Cancer. 2019;10(11):363–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the current study are available from the TCGA (cancergenome.nih.gov) repository. TCGA allows researchers to download relevant data for research and publish relevant articles.


