ABSTRACT
Osteoporosis is defined as a bone condition characterized by bone mass reduction, bone micro-architectural and quality deterioration, leading to compromised strength and increased chances of fracture. Evidence have shown an essential role of microRNAs (miRNAs) in various osteogenic differentiation processes. However, the function of miR-15a-5p in the differentiation of osteogenic cells and possible mechanisms remains unclear. The present study explored the expression of miR-15a-5p in human osteoporosis specimens and during the osteogenic differentiation of MC3T3-E1 cells. Functions of miR-15a-5p were determined using miR-15a-5p mimics and inhibitors. Luciferase assay was used to verify the binding of miR-15a-5p and PDCD4 3ʹUTR. Alizarin Red Staining (ARS) and Alkaline phosphatase (ALP) activity were used to determine the miR-15a-5p role in osteogenic differentiation. Finally, Wnt pathway inhibitor was used to determine the miR-15a-5p/PDCD4/Wnt signaling pathway in regulating osteogenic differentiation. We found miR-15a-5p expression was increased in human osteoporosis specimens and during differentiation of MC3T3-E1 cells. PDCD4 was also identified as a target of miR-15a-5p and was found to be involved in osteogenic differentiation. Further, miR-15a-5p mimics attenuated the effects of PDCD4 overexpression. Finally, use of XAV939 (Wnt pathway inhibitor) downregulated osteogenic differentiation in miR-15a5p/PDCD4/Wnt-dependent signaling pathway. In conclusion, miR-15a-5p induced differentiation of osteoblasts and mineralization by modulating osteoblast differentiation factors, mainly OSX, ALP, OCN, and RUNX2, by inhibiting PDCD4 and Wnt signaling pathways. This study provides a modality for the future use of miR-15a-5p in the treatment and prevention of osteoporosis.
KEYWORDS: miR-15a-5p, osteoporosis, programmed cell death 4 (PDCD4), Wnt/β-catenin pathway
1. Introduction
Osteoporosis (OP) is a bone condition marked by bone mass reduction, weakened bone microarchitecture and quality, leading to hindered strength and increased risks of fracture [1,2]. The disease also has socio-economic effects with prevalence of approximately 6.6% in men and 22.1% in women above 50 years [3,4]. Fundamental interactions of osteoclasts and osteoblasts regulate bone metabolism. Osteoclasts accelerate resorption through bone break-down, whereas osteoblasts induce bone production through regulation of differentiation and proliferation of osteoblasts precursors [5]. Osteoblast differentiation is controlled through essential factors of transcription, like runt-related transcription factor 2 (Runx2) and osterix (Osx), and an upregulated bone matrix proteins expression, for instance, type 1 collagen (ColA1), osteopontin (OPN), and alkaline phosphatase (ALP) [6]. The differentiation of these factors induces mineralization, and leads to bone formation.
MicroRNA are epigenetic regulators that interfere with expression of mRNA by binding the target’s 3′-untranslated region (3′-UTR), consequently changing the biological activity [7]. According to evidence, microRNAs have a role in osteoclast behavior and osteoblast function during development, metastasis, and degeneration of bones. For instance, in women, serum miR-22-3p and miR-328-3p levels are linked to osteoporotic fracture [8]. In menopausal diabetic women, serum miR-188-3p, miR-550a-5p, and miR-382-3p are specific skeletal fracture signatures [9]. In in vivo osteoporosis models, miR-188 deficient mice demonstrate minor responses to overproduction of marrow fat mediated by age and loss of bone [10]. MiR-503 silencing ramps up the formation of CD14-positive peripheral blood mononuclear cells (PBMCs) by the osteoclast in human, hence worsening osteoporosis development in ovariectomized mice. Severe loss and increased bone resorption are displayed in osteoclast-specific miR-34a knockout mice, while skin cancer-mediated bone metastasis and estrogen deficiency-mediated osteoporosis are compromised in mice with miR-34a overexpression [11].
MiR-15a-5p plays important functions in cancer regulation [12]. For instance, miR-15a-5p inhibits hepatocellular carcinoma through BDNF targeting [13]and fibrosis through vascular endothelial growth factor A (VEGFA) targeting [14]. MiR-15a/16-2 has been associated with the WNT5A interference to regulate mesenchymal stem cells osteogenesis [15]. MiR-15a-5p has been reported to modulate viability and degradation of osteoarthritis chondrocytes matrix through VEGFA targeting [16]
The programmed cell death 4 (PDCD4) is a tumor inhibitor and its aberrant expression has been reported in various types of cancers, such as pancreatic [17], lung [18]and breast cancers [19]. In osteogenesis, PDCD4 suppress mesenchymal cells differentiation and proliferation and subsequently induce their apoptosis through Wnt/B-catenin pathway suppression [20]. Wnt/β-catenin pathway positively influences bone mass by replicating pre-osteoblast enhancement, stimulating osteoblastogenesis, inhibiting osteoblast, and eventually inducing osteocyte’s apoptosis [21] However, the role of miR-15a-5p in osteogenic differentiation is still unclear.
The present investigation hypothesized that MicroRNA-15a-5p could induce osteogenic MC3T3-E1 cells differentiation by targeting PDCD4 via Wnt/β-catenin dependent signaling pathway. This study aimed at assessing the role of MicroRNA-15a-5p and PDCD4 in osteogenic differentiation of MC3T3-E1 cells. Further, the study investigated whether miR-15a-5p overexpression attenuates the effects of PDCD4 overexpression in osteogenic differentiation of MC3T3-E1 cells. Finally, the effects of XAV939 (Wnt pathway inhibitor) on osteogenic differentiation of MC3T3-E1 cells was also investigated.
2. Materials and methods
2.1. Human clinical samples
During the period from March 2016 to June 2018, bone tissue specimens were extracted from the transcervical region of the femoral neck from 20 patients with OP and 20 normal subjects having osteoarthritis at the Department of Orthopedic Surgery of Changshu Hospital Affiliated to Nanjing University of Chinese Medicine. The study approval was obtained from Changshu Hospital Affiliated to Nanjing University of Chinese Medicine Ethical Committee of the Hospital, which adhered to the Helsinki Declaration. Written informed consents were obtained from the patients or their guardians.
2.2. Cell culture and differentiation
MC3T3-E1 cells were used in the investigation. Cells were acquired from the Chinese Academy of Science biobank and maintained in α-MEM media containing 10% FBS. The media was also supplemented with 100 μg/mL of streptomycin and 100 units/mL of penicillin A. The cells were cultured at 37°C and 5% CO2. MC3T3-E1 cells’ differentiation was induced by cultivation in osteogenic medium (OM), made by supplementing α-MEM media with 0.05 mmol/L L-ascorbic acid-2-phosphate, 100 nmol/L dexamethasones, and 10 mmol/L β-glycerophosphate.
2.3. Cell transfection
MC3T3-E1 (3 × 105) cells were plated into 6-well plates for 24 hours prior to transfection. The MC3T3-E1 cells were then transfected using miR-15a-5p mimic or inhibitors and PDCD4-siRNA. For the PDCD4 overexpression (OE), PDCD4-OE plasmid was constructed by cloning the full-length coding sequence of PDCD4into the pcDNA3.1 plasmid (Invitrogen). All the oligonucleotide sequences were purchased from Genepharma (Shanghai). The transient cells transfection was done with Lipofectamine 2000TM (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s guidelines.
2.4. Cell proliferation
Cell proliferation assessment was done using MTT assay. Experimental MC3T3-E1 cells (103 cells/well) were inoculated in a 96-well culture plate and grown for 16 days in α-MEM supplemented with 10% FBS in the presence or lack of an osteogenic media (OM). The sampling and counting of cells were done every 4 days, during which 0.5 mg/mL of MTT was added. Cells were then cultured for 3 hours at 37°C to allow for the formation of MTT-formazan. Supernatant was later discarded followed by an addition of dimethyl sulfoxide (DMSO). Finally, the absorbance was determined spectrophotometrically in a microplate reader at an optical density of 490 nm.
2.5. Alkaline phosphatase (ALP) activity
ALP activity was spectrophotometrically assessed [22]. The α-MEM culture media was eliminated and cells subsequently washed thrice using PBS. The cells were then lysed using a lysis buffer made of 10 mM Tris-HCL at pH 7.5 and 0.1% Triton X 100 (Sigma–Aldrich), at 4°C for 1 h. Cells were later incubated for 30 min at 37°C using 10mMp-nitrophenyl phosphate (p-NP) in alkaline buffer (100 Mm diethanolamine and 0.5mMMgCl2, pH 10.5). The reaction was then blocked in 0.2 M NaOH and the absorbance was assessed at 405 nm using a Spectrophotometric ELISA reader. The ALP activity, U/cell number, was determined on the 7th, 14th and 31s day. Normalization of ALP activity to the total content of deoxyribonucleic acid (DNA) was done, and results finally presented in µM/(min × µg DNA). Three independent assays were performed and data presented as means ±SD.
2.6. Alizarin red S (ARS) staining
Deposition of calcium was assessed using alizarin red S (ARS) [23]. Approximately 2 × 104 cells /cm2 were seeded in a 24-well culture plates and stimulated using medium (OM) containing 50 µg/mLL-AA and 10mMβ-GP for 7 and 14 days. Fixation of cells was done for 10 minutes in 10% formalin in PBS at a room temperature, followed by washing in distilled water. Subsequently, the cells were stained for 10 min at 37°C using 2% ARS (pH 4.2). The ARS solution were then removed, and the cell samples were later washed thrice in deionized water. Calcium quantification was done by treatment with cetylpyridinium chloride (CPC) (Sigma). The samples stained by ARS were treated for 1 h using 10% CPC solution (1 ml), to desorb calcium ions. Later, 100 ul of the eluted stain was then added to a 96-well plate. The solution’s absorbance was determined at using multifunction microplate scanner at 590 nm. A standard curve was made using the RS and CPC. The calcium deposition expression was done as molar equivalent of calcium. One mole of ARS binds to two moles of calcium in the Alizarin Red S-calcium complex.
2.7. RT-quantitative polymerase chain reaction
Extraction of total RNA from MC3T3-E1 cells was done by RT-qPCR RNAiso Plus (TaKaRa Biotechnology, China), according to the manufacturers’ instructions. Total concentration and purity of RNA were detected using Nanodrop Assay (Thermofisher). Reverse transcription was carried out with Prime Script™ RT Master Mix, to synthesize the first strand cDNA. qPCR was also done with SYBR Premix Ex Taq II (TaKaRa Biotechnology, China). The assay primer sequence was as follows:
RUNX2, F: 5ʹ- CCCAGTATGAGAGTAGGTGTCC −3ʹ, R: 5ʹ- GGGTAAGACTGGTCATAGGACC −3ʹ;
ALP, F: 5ʹ-ACACCTTGACTGTGGTTACTG-3, R: 5ʹ-CCATATAGGATGGCCGTGAAG-3ʹ;
OCN, F: 5ʹ-ACACCATGAGGACCATCTTTC-3, R: 5ʹ-CGGAGTCTGTTCACTACCTTATT-3;
PDCD4, F 5ʹ-TCG TCGTTACGATTGGTTAGTC-3, R5ʹ-GAAAAATCTCTA ACCCTTCTCGC-3ʹ;
β-actin, F:5ʹ-AATGTGGCTGAGGACTTTG-3, R: 5ʹ-GGGACTTCCTGTAACCACTTATT-3ʹ.
U6: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; R, 5ʹ-TGGTGTCGTGGAGTCG-3ʹ.
The system was then run through Applied Biosystems 7500 Real-Time PCR (Applied Biosystems, Canada). The amplification of qPCR was done as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min. The relative gene expression levels were assessed using 2−∆∆Ct method.
2.8. Bioinformatics analysis
TargetScan (http://www.targetscan. org), and miRanda (http://www.microrna.org) and PicTar (http://pictar.mdc-berlin.de) were used for the analysis of potential miR-15a-5p binding sites on the target genes. The analysis consistency and predictions through these websites confirms their reliability.
2.9. Luciferase reporter gene assay
Luciferase reporter gene experiment was done by an overnight inoculation of approximately 5 × 105 HEK293T cells into a 24-well plate. We created pmirGLO-PDCD4-WT or PDCD4-Mut plasmids having wildtype PDCD4 3ʹUTR or mutant PDCD4 3ʹUTR, respectively. Then, co-transfected pmirGLO-miR-15-5pWT reporter plasmids or its mutant vector (at 150 ng each) into MC3T3-E1 cells with 50 nM miR-15a-5p mimics using Lipofectamine 2000 reagent. Cells were cultured further for 48 hours. The firefly and renilla luciferase activity were assessed by Dual-Luciferase Reporter Analysis System following the manufacturer’s guideline (Promega). The relative luciferase activity was finally determined based on the ratio of the fluorescence of firefly/renilla.
2.10. Western blot analysis
SDS-PAGE was carried out on Bio-Rad mini-protean electrophoresis system. For every sample, 20 µg of total protein was extracted and mixed using 6X SDS sample buffer, and heated for 5 min at 90°C. The proteins were later transferred into the membrane of polyvinylidene fluoride (PVDF), which was then blocked for 1 hour at room temperature using 5% nonfat milk (in 1 X PBS). It was then incubated overnight, at 4 °C using specific rabbit primary antibodies against various proteins as follows: OCN (1:1000), RUNX2 (1:1000), ALP (1:1000), PDCD4 (1:1000), and β− actin (1:1000). The membrane was later washed and then incubated at a room temperature for 2 h in diluted horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibodies, in 5% nonfat milk (in 1 X PBS) in the ratio 1:5,000. The membrane was rewashed in for three times in 0.5% PBST. Band detection was finally done using Western Chemiluminescent HRP Substrate in the Protein Simple detection machine.
2.11. Data analysis
Graph pad prism software was used for the analysis of data. The numerical values presentation was done as the means ± S.E.M. Mann-Whitney U test was used for analysis of data and results were significant were p < 0.05.
3. Results
3.1. miR-15a-5p is upregulated during osteogenic differentiation
Osteoporosis is a state of the bone characterized by reduction of the bone mass, degeneration of bone micro-architecture and quality, resulting in weakening and chances of fracture. MicroRNAs have a role in osteoclast behavior and osteoblast function during development, metastasis, and degeneration of bones. MiR-15a-5p is an important molecule which might contribute to osteogenic differentiation. The current investigation postulated that MicroRNA-15a-5p induce the osteogenic differentiation of MC3T3-E1 cells by targeting PDCD4 via Wnt/β-catenin dependent signaling pathway. Western blot, RT-qPCR and MTT assays were used to determine MicroRNA-15a-5p role in osteogenic differentiation of MC3T3-E1 cells.
The results showed that miR-15a-5p expression was considerably downregulated in the human OP samples (Figure 1(a)). To determine miR-15a-5p effect in osteogenic differentiation, cells were separately grown in a complete α-MEM media or in the presence of an osteogenic media (OM). Assessment of cell viability using MTT assay showed a significant increase of MC3T3-E1 cell growth when cultured in osteogenic media, as shown in Figure 1(b).
Figure 1.
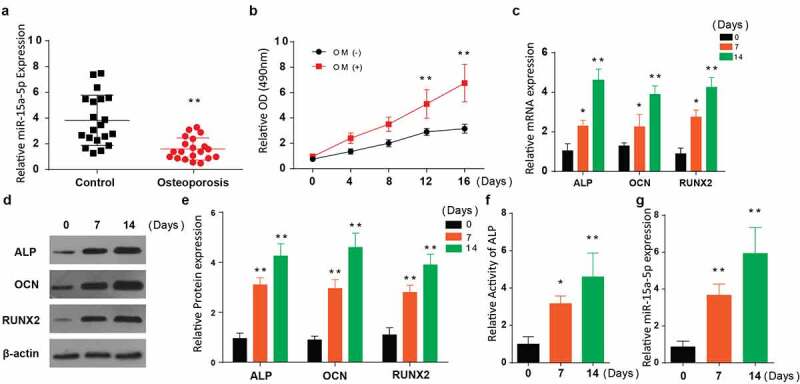
Expression of miR-15a-5p is upregulated during osteogenic differentiation in MC3T3-E1
a: Expression of miR-15a-5p in human osteoporosis specimens. b: Cell viability of MC3T3-E1 cells induced with osteogenic media. c-e: Relative mRNA (c), and protein (d) expression along with quantification (e) of ALP, OCN and RUNX2 genes detected in MC3T3-E1 induced with osteogenic media through RT-qPCR and Western blot. f: Relative activity of ALP in MC3T3-E1 induced with osteogenic media. G: Relative miR-15a-5p expression in MC3T3-E1 induced with osteogenic media. (Data presented as means ± SEM; *p < 0.05, **p < 0.01)
We then used RT-qPCR to assess the relative mRNA expression of osteogenesis-associated genes (ALP, RUNX2, and OCN in the cells post-osteogenic media treatment for 0, 7, and 14 days. According to the results, ALP expression, OCN, and RUNX2 were significantly increased 14 days after treatment with OM, as shown in Figure 1(c).
ALP, OCN, and RUNX2 protein expressions were further determined in the OM treated cells through western blotting. Similar to RT-qPCR results, these proteins’ expression was significantly elevated 14 days post-OM treatment (Figure 1(d,e)). Later, ALP activity was determined, and the results demonstrated a significantly elevated activity after 7 and 14 days of cell treatment with osteogenic media (Figure 1(f)). Finally, we determined the expression of miR-15a-5p in the cells post-OM treatment for 0, 7, and 14 days. Our observations indicated an increased miR-15a-5p expression at 7 and 14 days, as demonstrated in Figure 1(g). According to these results, miR-15a-5p expression is generally upregulated in osteogenic differentiation.
3.2. MiR-15a-5p promotes osteogenic differentiation of MC3T3-E1 cells
To assess miR-15a-5p role in osteogenic differentiation, MC3T3-E1 cells were transfected with mock, miR-15a-5p mimics, mimics-NC, miR-15a-5p inhibitors, and inhibitors-NC. Determination of miR-15a-5p relative expression was then done using RT-qPCR. According to our observations, miR-15a-5p relative expression was significantly elevated in cells transfected using miR-15a-5p-mimics but significantly lowered following transfection using miR-15a-5p-inhibitors, as shown in Figure 2(a). We also determined the levels of expression of osteogenesis-associated proteins in the transfected cells. The RT-qPCR results indicated a significant elevation of ALP, OCN, and RUNX2 in the cells transfected with miR-15a-5p mimics, but a significant reduction following transfection with miR-15a-5p-inhibitors, as shown in Figure 2(b). Determination of ALP activity demonstrated increased ALP activity in cells transfected with miR-15a-5p-Mimics, but a significant drop following transfection using miR-15a-5p-inhibitors, as shown in Figure 2(c). Finally, we used Alizarin Red staining to assess miR-15a-5p effects on mineralization. The results demonstrated a significantly increased and deep staining of miR-15a-5p mimics-transfected, than miR-15a-5p-inhibitors-transfected cells grown in osteogenic media, as compared to those cells grown in the general media and OM (Figure 2(d,e)). From the observations, we generally surmised that miR-15a-5p promotes osteogenic differentiation.
Figure 2.
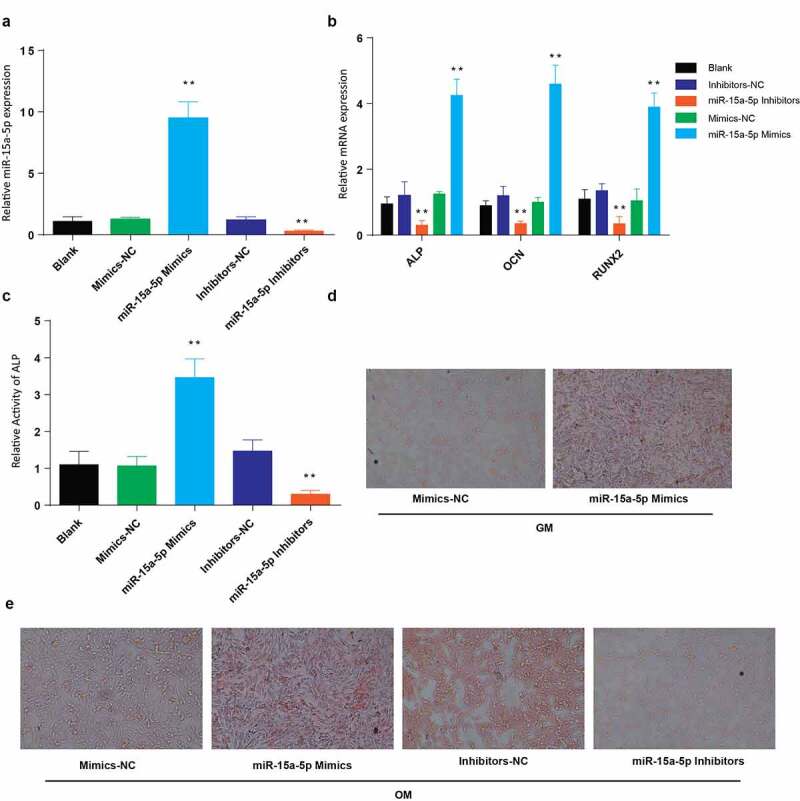
Overexpression of miR-15a-5p promotes osteogenic differentiation
a: Expression of miR-15a-5p in MC3T3-E1 after transfection of miR-15-a-5p mimics and inhibitors. b: Relative mRNA expression of ALP, OCN and RUNX2 genes detected in MC3T3-E1 cells after transfection of miR-15-a-5p mimics and inhibitors. c: Relative activity of ALP in MC3T3-E1 cells after transfection of miR-15-a-5p mimics and inhibitors. d: Alizarin Red staining of MC3T3-E1 cells after transfection of miR-15-a-5p mimics in presence of general media. e: Alizarin Red staining of MC3T3-E1 cells after transfection of miR-15-a-5p mimics and inhibitors in the presence of osteogenic media. (Data presented as means ± SEM; **p < 0.01)
3.3. PDCD4 is a target of miR-15a-5p and is involved in osteogenic differentiation of MC3T3-E1 cells
We then aimed at studying the mechanisms employed by miR-15a-5p promoting osteogenic differentiation. For confirmation, the activity of luciferase was determined by dual-luciferase reporter technique. Our findings indicated a significantly reduced luciferase activity in PDCD4-WT MC3T3-E1 cells treated using mimics of miR-15a-5p than in the PDCD4-MUT MC3T3-E1 cells treated using miR-15a-5p (Figure 3(a,b)). Next, we used RT-qPCR to determine the PDCD4 mRNA expression in MC3T3-E1 transfected using miR-15a-5p mimics, mimics-NC, miR-15a-5p inhibitors, and inhibitors-NC. Our observations indicated a significantly increased PDCD4 mRNA expression in miR-15a-5p-inhibitors, and decreased in the miR-15a-5p mimics as compared to their respective NC groups (Figure 3(c)). Western blot analysis also demonstrated a higher expression of PDCD4 in the miR-15a-5p inhibitors transfected cells and decrease in cells transfected with miR-15a-5p mimics as compared to NC, as shown in Figure 3(d,e).
Figure 3.
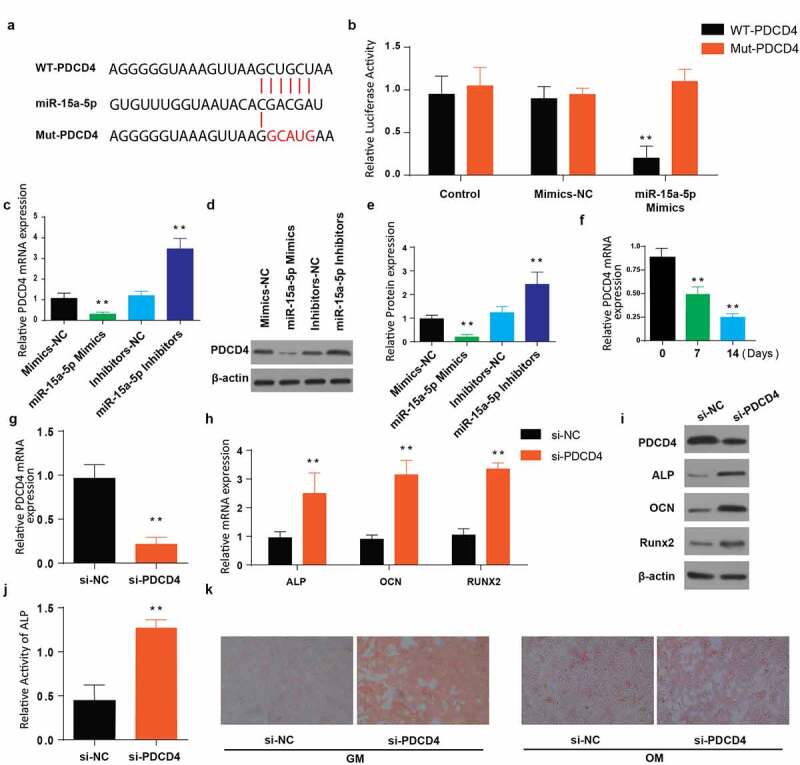
MiR-15a-5p targets 3ʹUTR of PDCD4 and regulate osteogenic differentiation
a: Binding site of mutant and wild type 3ʹUTR of PDCD4 compared to the miR-15a-5p. b: Relative luciferase activity to represent the binding of miR-15a-5p and 3ʹUTR of PDCD4. C: Relative mRNA expression of PDCD4 in MC3T3-E1 cells after transfection of miR-15-a-5p mimics and inhibitors. d-e: Relative protein expression of PDCD4 in MC3T3-E1 cells after transfection of miR-15-a-5p mimics and inhibitors. f: Relative PDCD4 mRNA expression in MC3T3-E1 induced with osteogenic media. g: Relative PDCD4 mRNA expression in MC3T3-E1 cells after transfection of si-PDCD4. h: Relative mRNA expression of ALP, OCN and RUNX2 genes detected in MC3T3-E1 cells after transfection of si-PDCD4. I: Western blot showing expression of PDCD4, ALP, OCN and RUNX2 proteins in MC3T3-E1 cells after transfection of si-PDCD4. j: Relative ALP activity in MC3T3-E1 cells after transfection of si-PDCD4. k: Alizarin Red staining of MC3T3-E1 cells after transfection of si-PDCD4. (Data presented as means ± SEM; **p < 0.01)
Further, expression of PDCD4 in the cells post-OM treatment for 0, 7, and 14 days was detected through RT-qPCR. Our findings showed a significantly reduced mRNA expression of PDCD4 after 14 days of treatment with the OM compared to the time point 0 (Figure 3(f)). To further elucidate the effect of PDCD4 on differentiation of osteogenic cells, MC3T3-E1 cells were transfected with si-PDCD4 and the corresponding si-NC. The PDCD4 mRNA expression was then determined using RT-qPCR. According to our observations, the expression of PDCD4 was significantly lowered in the cells transfected with si-PDCD4, as shown in Figure 3(g). We then determined the relative mRNA expression of osteogenic differentiation associated genes in the si-PDCD4 and si-NC transfected MC3T3-E1 cells. As indicated in Figure 3(h), mRNA expressions of ALP, OCN, and RUNX2 were significantly increased in the cells transfected with si-PDCD4 (Figure 3(h)). The western blot protein expression also indicated decreased PDCD4 expression, and increased ALP, OCN, and RUNX2 in the cells transfected with si-PDCD4, Figure 3(i). Finally, determination of the ALP activity and mineralization (Alizarin Red staining) indicated an increased ALP activity and increased mineralization in si-PDCD4 transfected cells (Figure 3(j,k)). These data summarized demonstrate that miR-15a-5p targets PDCD4 and the latter is involved in osteogenic differentiation.
3.4. Overexpression of miR-15a-5p attenuates the effects of PDCD4 overexpression in osteogenic differentiation of MC3T3-E1 cells
To understand PDCD4 overexpression effects on miR-15a-5p in osteogenic differentiation, the experimental MC3T3-E1 cells were transfected together with Mimics-NC+pcDNA3.1-NC, miR-15a-5p mimics+pcDNA3.1-NC, miR+ PDCD4 OE plasmid or miR-15a-5p+ PDCD4-OE. We then assessed the relative mRNA expression of PDCD4 post-transfection through RT-qPCR. According to our observations, PDCD4 mRNA was highly expressed after the overexpression of PDCD4 using pcDNA3.1-PDCD4 plasmid, but significantly reduced following the co-transfection with miR-15a-5p mimics+pcDNA3.1-PDCD4, as shown in Figure 4(a). We further determined the mRNA expressions of associated osteogenic genes in the same cells through RT-qPCR. The results also demonstrated lower ALP, OCN, and RUNX2 mRNA levels following transfection of PDCD4 overexpression plasmid, but a later significant upregulation of these genes following the co-transfection of miR-15a-5p+pcDNA3.1-PDCD4, as shown in Figure 4(b). The western blot assay confirmed the lower ALP OCN, and RUNX2 expression, whereas increased PDCD4 expression, following PDCD4 overexpression plasmid transfection. However, co-transfection of miR-15a-5p mimics and PDCD4-OE plasmid significantly reduced the expression of PDCD4 and improved the expression of ALP OCN, and RUNX2 than in PDCD4 overexpression cells, Figure 4(c,d).
Figure 4.
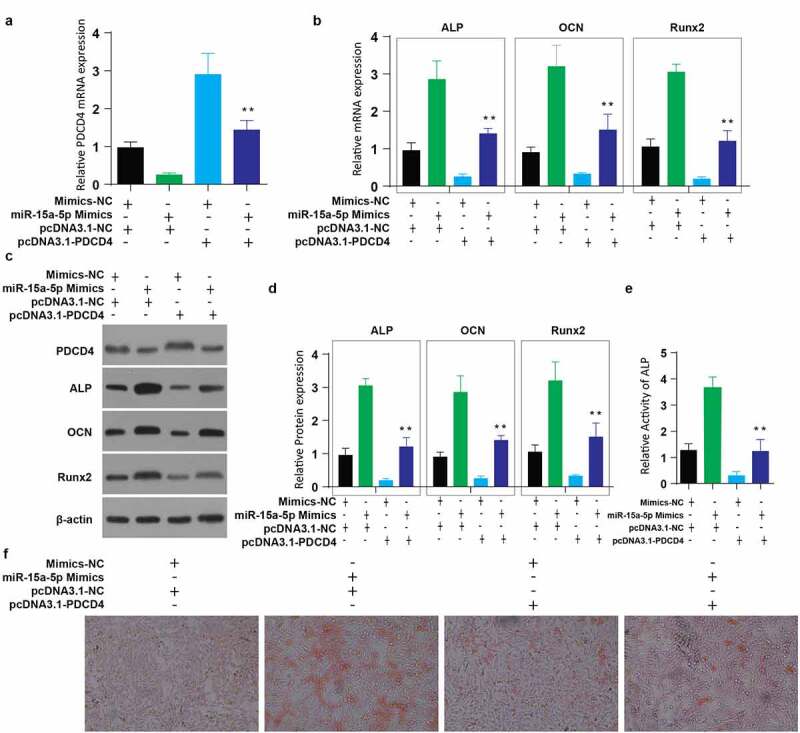
Overexpression of miR 15a 5p attenuates the effects of PDCD4 overexpression in osteogenic differentiation of MC3T3-E1 cells
a: Relative mRNA expression of PDCD4 in MC3T3-E1 cells after co-transfection of miR-15-a-5p mimics and PDCD4 overexpression plasmid. b: Relative mRNA expression of ALP, OCN and RUNX2 genes detected in MC3T3-E1 cells after co-transfection of miR-15-a-5p mimics and PDCD4 overexpression plasmid. c-d: Relative protein expression of ALP, OCN and RUNX2 genes detected in MC3T3-E1 cells after co-transfection of miR-15-a-5p mimics and PDCD4 overexpression plasmid. e: Relative activity of ALP in MC3T3-E1 cells after co-transfection of miR-15-a-5p mimics and PDCD4 overexpression plasmid. f: Alizarin Red staining of MC3T3-E1 cells after co-transfection of miR-15-a-5p mimics and PDCD4 overexpression plasmid. (Data presented as means ± SEM; **p < 0.01)
Similarly, the ALP activity significantly decreased after PDCD4 overexpression plasmid transfection, but co-transfection of miR-15a-5p+pcDNA3.1-PDCD4 plasmid significantly improved the level ALP activity (Figure 4(e)). Mineralization was then determined through Alizarin Red staining. According to our observations, the staining was considerably decreased after PDCD4 overexpression plasmid transfection, but co-transfection of miR-15a-5p+pcDNA3.1-PDCD4 plasmid significantly increased the staining intensity (Figure 4(f)). These data together show that miR-15a-5p overexpression attenuates the effects of PDCD4 overexpression in osteogenic differentiation.
3.5. Use of XAV939 (Wnt pathway inhibitor) downregulate osteogenic differentiation of MC3T3-E1 cells in miR-15a5p/PDCD4/Wnt-dependent signaling pathway
Finally, we aimed at investigating the possible effect of Wnt pathway inhibition on osteogenic differentiation in miR-15a5p/PDCD4/Wnt-dependent signaling pathway. MC3T3-E1 cells were transfected with 5, 10, 20 uM of XAV939 (Wnt-inhibitor), and relative miR-15a-5p and PDCD4 mRNA expressions were determined through RT-qPCR. We observed a reduction of miR-15a-5p mRNA proteins with an increasing XAV939 dose (Figure 5(a)), whereas increasing PDCD4 expression was observed with an increasing XAV939 dose (Figure 5(b)). We then determined the relative mRNA expressions of associated osteogenic proteins following transfection of cells with 20uM of XAV939. Our observations pointed to a significantly lowered ALP, OCN, and RUNX2, respectively, the expression following the treatment with 20uM of XAV939, Figure 5(c). Determination of protein expressions through western blot assay also significantly increased PDCD4, whereas decrease in ALP, OCN, and RUNX2 expression following the VAX939 treatment (20 µM), Figure 5(d). Lastly, Alizarin Red S staining following VAX939 treatment demonstrated a significant reduction of the intensity of staining (Figure 5(e)). Altogether, these data indicate that XAV939, as a Wnt pathway inhibitor, downregulates osteogenic differentiation in miR-15a5p/PDCD4/Wnt-dependent signaling pathway.
Figure 5.
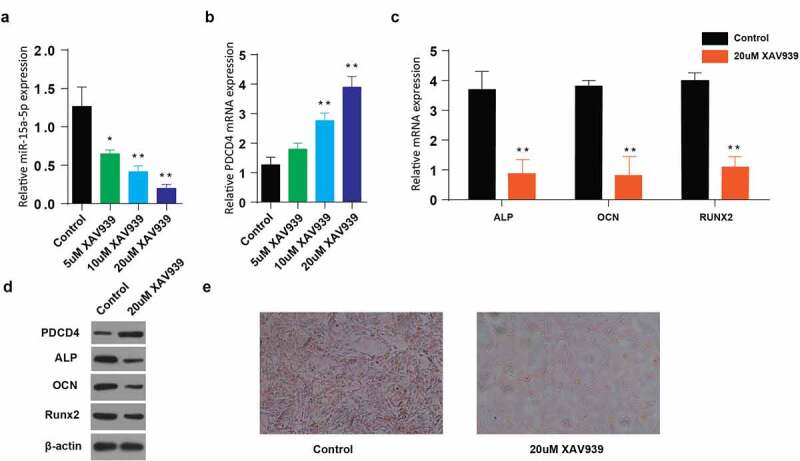
Use of Wnt pathway inhibitors (XAV939) decreases osteogenic differentiation of MC3T3-E1 cells in miR-15a5p/PDCD4/Wnt-dependent signaling pathway
a: Relative miR-15a-5p expression in MC3T3-E1 cells stimulated with different doses of XAV939. b: Relative mRNA expression of PDCD4 in MC3T3-E1 cells stimulated with different doses of XAV939. c: Relative mRNA expression of ALP, OCN and RUNX2 genes detected in MC3T3-E1 cells stimulated with XAV939. d: Western blot showing expression of PDCD4, ALP, OCN and RUNX2 proteins in MC3T3-E1 cells stimulated with XAV939. E: Alizarin Red staining of MC3T3-E1 cells after stimulation with XAV939. (Data presented as means ± SEM; **p < 0.01)
4. Discussion
Bone formation is multifaceted and involve osteoblast’s cell lineage proliferation, pre-osteoblast’s mobilization and differentiation, and osteoblast’s maturation. Recovery of bone tissue after bone damage, including migration, differentiation of cells, and mineralization of the bone matrix, also depends on osteoblasts. Consequently, osteoblasts regulate homeostasis of bones from birth to death, and osteoblast dysregulation leads to bone marrow diseases, such as osteoporosis. This study confirmed that miR-15a-5p upregulation promotes osteogenic differentiation and that miR-15a-5p induces differentiation by targeting PDCD4. We finally confirmed that Wnt pathway inhibition suppresses differentiation in miR-15a5p/PDCD4/Wnt-dependent signaling pathway.
Changes in osteogenic media have been reported to cause a significant increase in miRNA profiling, for instance, the elevated miRNA levels in the skeletal muscles [24] and miR-378 is significantly increased during the myogenic differentiation of C2C12 [25]. Osteogenic media also accelerates osteogenic differentiation and cell proliferation and osteogenic differentiation markers such as ALP [26].
According to recent reports, miRNAs have essential functions in osteogenic differentiation via targeting important transcription factors 3ʹ-UTR for osteogenic differentiation [27]. MiR-346, which is increased during hBMSCs osteoblastic differentiation, enhances osteogenic differentiation via GSK-3beta targeting and controlling the Wnt/β catenin differentiation pathway. Overexpression of miR-21 has been reported to significantly initiate the hUMSCs osteogenic differentiation by directly suppressing PTEN. MiR-15a-5p expression has been linked with progression of tumor, especially regarding cell proliferation and differentiation. According to some reports, miR-15a-5p expression was suppressed in human Osteoarthritis chondrocytes [28]. In our findings, MiR-15a-5p expression was gradually increased during MC3T3-E1 cells osteogenic differentiation, signifying the direct participation of miR-15a-5p in the regulation of this complex process.
MiR-15a-5p has also been linked with the regulation of the cartilage matrix degradation through ADAMTS5 targeting, in human chondrocytes [29]. MiR-15a-5p further alleviates response of inflammation and arterial injury in diabetes through FASN targeting [30]
Studies have revealed that microRNAs plays a regulatory role on osteogenic differentiation markers, for instance, miR-125b overexpression was reported to inhibit ALP activity [31]. However, microRNA overexpression has also been linked with increased expression of osteogenic differentiation markers as reported severally, for instance, miR-223 increased the ALP, OCN and RUNX2 activity [32]. In our findings, MiR-15a-5p was also associated with increased osteogenic differentiation, which was demonstrated by increased osteoblastic markers ALP, OCN, OPN, and RUNX2, and increased levels of matric mineralization and ALP activity.
MicroRNAs and Wnt/β-catenin pathway both form network important in regulating key biological processes [33]. The Wnt/β-catenin signaling also has an essential role in osteogenesis. The Wnt/β-catenin pathway regulates osteoblast genic proliferation, differentiation, and mineralization [34]. Wnt signaling can also increase osteogenesis through the regulation of RUNX2 expression [35]. Further, Wnt/β-catenin is also inactivated by PDCD4 in osteoporosis [36]. In the current report, miR-15a-5p initiates the differentiation of MC3T3-E1 cells by directly suppressing PDCD4. Our findings agree with the previous work that reported PDCD4 as a repressor of osteoclastogenesis gene and are directly suppressed by miR-21 [37]. Inhibition of PDCD4 is important for osteogenesis since its presence suppresses and hampers the function of c-Fos [38]. In our findings, Runx2 expression was increased to integrate pathways of PDCD4 and Wnt/β-catenin in regulating of osteoblast differentiation.
5. Conclusion
Consequently, miR-15a-5p impacts both differentiation of osteoblast and osteoclastogenesis via cooperative association of Wnt/β-catenin and PDCD4 pathways. In conclusion, miR-15a-5p induced differentiation of osteoblasts and mineralization by modulating osteoblast differentiation factors, mainly ALP, OCN, and RUNX2, by inhibiting PDCD4 in Wnt signaling pathways. Wnt/β-catenin pathway may stimulate the expression of miR-15a-5p, and upregulate osteoblast differentiation proteins, including ALP, and OPN, and RUNX2. This study provides a possible novel alternative of using miR-15a-5p in the treatment and prevention of osteoporosis.
Funding Statement
The authors have no funding to report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Kim MB, Song Y, Hwang JK.. Kirenol stimulates osteoblast differentiation through activation of the BMP and Wnt/β-catenin signaling pathways in MC3T3-E1 cells. Fitoterapia. 2014;98:59–65. [DOI] [PubMed] [Google Scholar]
- [2].Das S, Crockett JC.. Osteoporosis - a current view of pharmacological prevention and treatment. Drug Des Devel Ther. 2013;7:435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Man X, Yang L, Liu S, et al. Arbutin promotes MC3T3‑E1 mouse osteoblast precursor cell proliferation and differentiation via the Wnt/β‑catenin signaling pathway. Mol Med Rep. 2019;19:4637–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hernlund E, Svedbom A, Ivergård M, et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos. 2013;8(1–2):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lo YC, Chang YH, Wei BL, et al. Betulinic acid stimulates the differentiation and mineralization of osteoblastic MC3T3-E1 cells: involvement of BMP/Runx2 and beta-catenin signals. J Agric Food Chem. 2010;58(11):6643–6649. [DOI] [PubMed] [Google Scholar]
- [6].Yoon HJ, Seo C-R, Kim M, et al. Dichloromethane extracts of Sophora japonica L. stimulate osteoblast differentiation in mesenchymal stem cells. Nutr Res. 2013;33(12):1053–1062. [DOI] [PubMed] [Google Scholar]
- [7].Vicente R, Noël D, Pers YM, et al. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat Rev Rheumatol. 2016;12(4):211–220. [DOI] [PubMed] [Google Scholar]
- [8].Weilner S, Skalicky S, Salzer B, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. [DOI] [PubMed] [Google Scholar]
- [9].Heilmeier U, Hackl M, Skalicky S, et al. Serum miRNA signatures are indicative of skeletal fractures in postmenopausal women with and without type 2 diabetes and influence osteogenic and adipogenic differentiation of adipose tissue-derived mesenchymal stem cells in vitro. J Bone Miner Res. 2016;31(12):2173–2192. [DOI] [PubMed] [Google Scholar]
- [10].Li CJ, Cheng P, Liang M-K, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lian WS, Ko J-Y, Chen Y-S, et al. MicroRNA-29a represses osteoclast formation and protects against osteoporosis by regulating PCAF-mediated RANKL and CXCL12. Cell Death Dis. 2019;10(10):705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Alderman C, Yang Y. The anti-melanoma activity and oncogenic targets of hsa-miR-15a-5p. RNA & disease (Houston, Tex.). 2016;3. [PMC free article] [PubMed] [Google Scholar]
- [13].Long J, Jiang C, Liu B, et al. MicroRNA-15a-5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF. Tumour Biol. 2016;37(5):5821–5828. [DOI] [PubMed] [Google Scholar]
- [14].Shang J, He Q, Chen Y, et al. miR-15a-5p suppresses inflammation and fibrosis of peritoneal mesothelial cells induced by peritoneal dialysis via targeting VEGFA. J Cell Physiol. 2019;234(6):9746–9755. [DOI] [PubMed] [Google Scholar]
- [15].Duan L, Zhao H, Xiong Y, et al. miR-16-2* Interferes with WNT5A to regulate osteogenesis of mesenchymal stem cells. Cell Physiol Biochem. 2018;51(3):1087–1102. [DOI] [PubMed] [Google Scholar]
- [16].Chen H, Tian Y. MiR-15a-5p regulates viability and matrix degradation of human osteoarthritis chondrocytes via targeting VEGFA. Biosci Trends. 2017;10(6):482–488. [DOI] [PubMed] [Google Scholar]
- [17].Wei X, Wang W, Wang L, et al. Micro RNA-21 induces 5-fluorouracil resistance in human pancreatic cancer cells by regulating PTEN and PDCD 4. Cancer Med. 2016;5(4):693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhen Y, Li D, Li W, et al. Reduced PDCD4 expression promotes cell growth through PI3K/Akt signaling in non-small cell lung cancer. 2016;23:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walter BA, Gómez-Macias G, Valera VA, et al. miR-21 expression in pregnancy-associated breast cancer: a possible marker of poor prognosis. J Cancer. 2011;2:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Y, Su D, Song T. Programmed cell death 4 inhibits proliferation and differentiation and induces apoptosis of human mesenchymal stem cells through suppressing the Wnt/β-catenin pathway. RSC Adv. 2017;7(43):26566–26573. [Google Scholar]
- [21].Zhang R, Oyajobi BO, Harris SE, et al. Wnt/β-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone. 2013;52(1):145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wrobel E, Leszczynska J, Brzoska E. The characteristics of Human Bone-Derived Cells (HBDCS) during osteogenesis in vitro. Cell Mol Biol Lett. 2016;21(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu C, Bai H, Hu Q, et al. Enhanced proliferation and osteogenic differentiation of MC3T3-E1 pre-osteoblasts on graphene oxide-impregnated PLGA–gelatin nanocomposite fibrous membranes. RSC Adv. 2017;7:8886–8897. [Google Scholar]
- [24].Hou X, Tang Z, Liu H, et al. Discovery of MicroRNAs associated with myogenesis by deep sequencing of serial developmental skeletal muscles in pigs. PLoS One. 2012;7(12):e52123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hupkes M, Sotoca AM, Hendriks JM, et al. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells. BMC Mol Biol. 2014;15(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nishimura I, Hisanaga R, Sato T, et al. Effect of osteogenic differentiation medium on proliferation and differentiation of human mesenchymal stem cells in three-dimensional culture with radial flow bioreactor. Regen Ther. 2015;2:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang XM, Song YQ, Li L, et al. miR-1249-5p regulates the osteogenic differentiation of ADSCs by targeting PDX1. J Orthop Surg Res. 2021;16(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Duan Z-X, Huang P, Tu C, et al. MicroRNA-15a-5p regulates the development of osteoarthritis by targeting PTHrP in chondrocytes. Biomed Res Int. 2019;2019:3904923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lu X, Lin J, Jin J, et al. hsa-miR-15a exerts protective effects against osteoarthritis by targeting aggrecanase-2 (ADAMTS5) in human chondrocytes. Int J Mol Med. 2016;37(2):509–516. [DOI] [PubMed] [Google Scholar]
- [30].Liu Y, Liu LY, Jia Y, et al. Role of microRNA-15a-5p in the atherosclerotic inflammatory response and arterial injury improvement of diabetic by targeting FASN. Biosci Rep. 2019;39. DOI: 10.1042/bsr20181852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [31].Nakasa T, Yoshizuka M, Andry Usman M, et al. MicroRNAs and bone regeneration. Current Genomics. 2015;16:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen J, He G, Wang Y, et al. MicroRNA‑223 promotes osteoblast differentiation of MC3T3‑E1 cells by targeting histone deacetylase 2. Int J Mol Med. 2019;43:1513–1521. [DOI] [PubMed] [Google Scholar]
- [33].Onyido EK, Sweeney E, Nateri AS. Wnt-signalling pathways and microRNAs network in carcinogenesis: experimental and bioinformatics approaches. Mol Cancer. 2016;15(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Park KH, Kang JW, Lee E-M, et al. Melatonin promotes osteoblastic differentiation through the BMP/ERK/Wnt signaling pathways. J Pineal Res. 2011;51(2):187–194. [DOI] [PubMed] [Google Scholar]
- [35].Gaur T, Lengner CJ, Hovhannisyan H, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280(39):33132–33140. [DOI] [PubMed] [Google Scholar]
- [36].Liu Y, Su D, Song TJRA. Programmed cell death 4 inhibits proliferation and differentiation and induces apoptosis of human mesenchymal stem cells through suppressing the Wnt/β-catenin pathway. PLoS One. 2017;7:26566–26573. [Google Scholar]
- [37].Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117(13):3648–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ell B, Kang Y. MicroRNAs as regulators of bone homeostasis and bone metastasis. Bonekey Rep. 2014;3:549. [DOI] [PMC free article] [PubMed] [Google Scholar]


