ABSTRACT
Ovarian cancer (OC) brings about serious physical and psychological burden for female patients. LncRNA CASC9 has been reported to be intimately linked with the occurrence and development of several tumors. However, the biological role of lncRNA CASC9 in OC still lacks sufficient evidence. The expressions of CASC9 and miR-488-3p in OC cell lines and xenograft mice were detected by qRT-PCR assay. Cell Counting Kit-8 (CCK-8) assay was used to assess cell inhibition rate and cell proliferation in OVCAR-3 and OVCAR-3/DDP cells. Wound healing assay and transwell assay were performed to evaluate the capacity of migration and invasion, respectively. In addition, cell apoptosis was measured by TUNEL assay and cell cycle was assessed by flow cytometric analysis. Moreover, western blotting was carried out to detect the cyclinG1 (CCNG1)/TP53/MMP7 signaling and apoptosis-related proteins. Furthermore, luciferase reporter assay was performed to verify the combination of CASC9 with CCNG1 and miR-488-3p. The results of our study revealed that CASC9 expression was upregulated while miR-488-3p and CCNG1 expression was downregulated in OC cells with significant higher TP53 and MMP7 protein levels compared with normal ovarian surface epithelial cells. Additionally, luciferase reporter assay confirmed CASC9 bond to miR-488-3p/CCNG1. CASC9 silencing inhibited cell proliferation, migration, and invasion whereas promoted cell inhibition rate and apoptosis in vitro and in vivo. However, CASC9 overexpression showed the opposite effects. In summary, LncRNA CASC9 played a regulative role in ovarian carcinoma by cyclinG1/TP53/MMP7 signaling via binding to miR-488-3p in vivo and in vitro.
KEYWORDS: Ovarian cancer, lncrna casc9, cyclinG1/tp53/mmp7 signaling, miR-488-3p
Introduction
Ovarian cancer (OC) is the commonest gynecological cancer that ranks the eighth-leading cause of female cancer death around the world [1]. The disease usually appears in postmenopausal women, along with 3–4 months of abdominal pain and distension [2]. The symptoms of ovarian carcinoma is unconspicuous and asymptomatic until advanced stages, and the lack of screening methods and early diagnostics of this disease, leading to little chance of a cure [3]. Thus, this cancer often is identified as the ‘silent killer’ and survival rates for women with advanced stage ovarian carcinoma range from 20% to 30% [4]. So far, advanced equipment and technology in surgery and chemotherapy have been applied for treating this disease, which promote the high incidence of complete remissions, but the recurrence rate is also high [5,6]. Therefore, it is vital to further explore the carcinogenesis and underlying mechanism of OC for female patients.
Long noncoding RNAs (lncRNAs) are a kind of transcripts with over 200 nucleotides in length, and they play crucial roles in a variety of biological activities in organism [7,8]. Numerous studies have shown that lncRNAs take part in the cellular progresses, such as gene transcription, epigenetic regulation, and post-transcription regulation in many cancers [9,10]. LncRNAs can regulate tumor cell proliferation, invasive, migration, cell cycle, and epithelial–mesenchymal transition [11–13]. The long noncoding RNA cancer susceptibility candidate 9 (CASC9), located in human chromosome 8q21.11, was first identified in esophageal squamous cell carcinoma. Recent studies have reported that lncRNA CASC9 was aberrantly expressed in several cancers. For example, CASC9 was upregulated in esophageal squamous cell carcinoma and accelerated cell metastasis via regulating LAMC2/CREB-binding protein [14]. Luo et al. reported that CASC9 was overexpressed in colorectal cancer and higher CASC9 level was implicated in poor patient outcomes. CASC9 promoted colorectal cancer cell proliferation in vitro and in vivo by binding to CPSF3 via regulating TGF-β signaling [15]. However, the biological role of CASC9 and potential mechanism in OC is elusive.
In this study, we are aimed to explore the functional role of CASC9 in OC. Given its high expression in OC cells, we hypothesized that CASC9 may play a tumor-promoting role in OC. Then we investigated the expression of CASC9 in OC cell lines and detected the influences of CASC9 overexpression or silencing on cell proliferation, invasion, migration, apoptosis, and cell cycling in OC or DPP-resistant OC cells and in mouse model. In addition, the oncogenic mechanism of CASC9 in OC was also demonstrated.
Materials and methods
Cell culture
Human immortalized ovarian surface epithelial cells IOSE-80 and OC cell lines SKOV3, OVCAR-3, TOV-21 G and CoC1 were purchased from China Center for Type Culture Collection. The cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C in a humid wet atmosphere containing 5% CO2 at 37°C.
Cell transfection
The specific shRNA targeting CASC9 (shRNA-CASC9-1/2), CASC9-specific pcDNA overexpression vector (OverExp-CASC9-1/2) and corresponding control vectors were synthesized by Gene Pharma (Shanghai, China). The sequences are shown in Supplementary Table.S1. The overexpression of miR-488-3p was achieved by transfection of miR-488-3p mimic (GeneCopoeia, Guangzhou, Guangdong, China). The recombinants were transfected into OVCAR or OVCAR-3/DDP cells using Lipofectamine 2000 reagent (Invitrogen-Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. After 48 h transfection, transfected OVCAR or OVCAR-3/DDP cells were storage for following experiments [16].
Establishment of DDP-resistant OC cells
DDP-resistant OVCAR cells (OVCAR-3/DDP) were induced by increasing doses of DDP (Sigma-Aldrich Co., USA). Firstly, OVCAR cells were cultured in the medium containing 0.02 mg/L DDP every day and the DDP concentration was gradually increased until the cells were stable in the medium with 0.2 mg/L DDP. Then the DDP-resistant cell culture lasted until the experiments were executed.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed to measure the mRNA levels of CASC9 and miR-488-3p in OC cells transfected with or without different vectors. Briefly, total RNA from treated OC cells or control cells was extracted by using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Then the RNA was converted to cDNA by PrimeScript RT Master Mix (Takara, Japan). The qRT-PCR assay was conducted using the SYBR Green 2X Real-Time PCR Master Mix in an ABI PRISM 7900 Real-Time system (Applied Biosystems, Foster City, CA, USA). The primer sequences for PCR are presented as below: CASC9: 5′-AGATGAAGCCGGTACCTCAGAT-3′ (forward), and 5′-TCACTTTAAAGAGGGAGAGGAG-3′ (reverse); miR-488-3p: 5′-CGGGGCAGCUCAGUACAG-3 (forward), 5′-CAGTGCGTGTCGTGGAGT-3′ (reverse); U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward), 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). GAPDH: 5′-GGGAAACTGTGGCGTGAT-3′ (forward) and 5′-GAGTGGGTGTCGCTGTTGA-3′ (reverse). U6 was used to normalize the expression of miR-30a-5p, while GAPDH was chosen as the internal control for CASC9. The results were measured by using the 2−ΔΔCt method [17].
Cell Counting Kit-8 (CCK-8) assay
OVCAR-3 and OVCAR-3/DDP cells were seeded into 96-well plates at a density of 8 × 103 cells/well until reaching 80–90% confluence. After the cell attachment, 10 µL DDP (25 µg/mL) was added to each well in treated group. The control group was set by adding medium only and the negative control group was set by adding OVCAR-3 or OVCAR-3/DDP cells with medium. Each group was repeated six times. After the exposure period of 72 h, 10 μL of WST-8 was added to each well d for another 3 h incubation. Finally, the absorbance was detected by a microplate reader (BioTek Instruments, VT) at the wavelength of 490 nm. The cell inhibition rate = 1 − (Aexperimental group −Acontrol group)/(ANC group − Acontrol group) × 100%. All of the experiments were performed in triplicate.
To assess the cell proliferative ability, OVCAR-3 and OVCAR-3/DDPs were placed in 96-well plates for 48 h and treated with 10 μL CCK-8 reagent for 2 h in the dark. The OD value at 450 nm was measured with the microplate reader [18]. Six parallel wells were set for this assay and all assays were performed in triplicate.
Wound healing assay
OVCAR-3 and transfected OVCAR-3/DDP cells were seeded into a six-well plate and were allowed to grow to 90% confluency. The cell monolayers were then wounded with white pipette tips to make a straight scratch. After washing three times with phosphate-buffered saline (PBS) to remove cell debris, the cells were incubated for 24 h. The number of migrated cells in the scratch was counted with an inverted microscopy [19].
Transwell assay
A transwell assay was employed to measure tumor invasion ability after transfection with shRNA-CASC9 or OverExp-CASC9. OVCAR-3 and transfected OVCAR-3/DDP cells were placed into the upper wells of the transwell chambers with 0.1 mL of matrigel (Becton Dickinson, MA). At the same time the medium with 10% fetal bovine serum was added in the lower chamber. The cells on the upper wells of the transwell chambers were removed with a cotton swab. After 48 h, the invaded cells on the bottom of the chamber were stained with H&E and counted with a microscope. The invasion assay were repeated at least three times [20].
TdT-mediated dUTP nick-end labeling (TUNEL) assay
TUNEL assay was carried out to evaluate effects of CASC9 expression on the apoptosis of OVCAR-3/DDP cells or animal models [21]. Briefly, the treated cells and the sections from mouse model were fixed with 4% paraformaldehyde and then incubated with proteinase K for 15 min in 37°C. After washing three times, cells and sections were inactivated by 3% H2O2 at room temperature for 15 min. Then, cells and tissues were stained by 3, 3′-diaminobenzidine (DAB) and hematoxylin for staining the cell nuclei. TUNEL-positive cells were quantified by counting positively stained cells. Five randomly chosen fields were counted for each group.
Flow cytometric analysis
OVCAR-3 and OVCAR-3/DDP cells were transfected with shRNA-CASC9 or OverExp-CASC9 for 48 h and were harvested by trypsinization. Then, the adhered cells were fixed with 70% ethanol and stained with 50 μg/ml propidium iodide and 100 μg/ml RNase I at 37°C for 30 min. Finally, the cell cycle distribution of OVCAR-3 and DDP-resistant OVCAR-3 cells was detected using the FACSCalibur Flow Cytometer (BD Bioscience, USA) and FlowJo software (TreeStar Inc) [22].
Luciferase reporter assay
The wild type of cyclinG1 (wt-CCNG1) or CASC9 (wt-CASC9) and mutant type of CCNG1 (mut-CCNG1) or CASC9 (mut-CASC9) containing miR-488-3p binding sites were subcloned into a pGL3 vector (Promega, USA) to create a luciferase plasmid. For luciferase reporter analysis, OVCAR-3 cells were co-transfected with these luciferase reporter vectors and miR-488-3p mimic/mimic NC using lipofectamine 2000 (Invitrogen) for 48 h. The relative luciferase activities were detected by using a Dual-Luciferase Reporter Assay (Promega) in accordance with manufacturer’s guidance [23].
Xenograft experiments
All procedures using animals were approved by Animal Care and Use Committee of Tangdu Hospital, Air Force Medical University and performed in accordance with Chinese legislation regarding experiment animals. Four-week-old sixty female nude mice (weight 14–16 g) were raised in pathogen-free rooms at 25 ± 2°C with 12 h light/dark cycles, 22 ± 2°C, and fed with standard laboratory food and water. The mice were injected subcutaneously with 1 × 105 OVCAR-3 and OVCAR-3/DDP cells transfected with shRNA-CASC9 or OverExp-CASC9 or empty vectors on flanks (12 mice per group). When the tumors appeared, six mice in each group received intraperitoneal injections of 8 mg/kg DDP and another six mice were treated with PBS per day for 21 days. The tumor volumes in each group were measured every 3 days. After DDP treatment for 21 days, all mice were sacrificed and the tumors were excised. Tumor volumes and weight were measured, and tumor tissues were stored for further research. Tumor volume was measured with a caliper and calculated using the formula: volume = (length × width2)/2 [23].
Western blotting assay
OVCAR-3, OVCAR-3/DDP cells, and the tissue from mice with transfection of shRNA-CASC9 or OverExp-CASC9 were lysed using cold RIPA lysis buffer (Beyotime Institute of Biotechnology, China). The protein samples were then separated by 10% SDS-PAGE (Bio-Rad, Hercules, CA) and transferred onto PVDF membranes (Amersham, Germany). After blockage with 5% nonfat milk for 1 h at room temperature, the membranes were incubated at 4°C overnight with the following primary antibodies: CCNG1 (1:200; Abcam, Cambridge, MA, USA; ab49274), p53 (1:1000; Dako Products, Santa Clara, CA; ABCA0332729), MMP7 (2 µg/ml; ab5706), CCND1 (1:200; ab16663), Bax (1:2000; ab182733), Bcl-2 (1:1000; ab32124), caspase-3 (1:500; ab13847), cleaved caspase-3 (1:500; ab2302) and GAPDH (1:1000; ab8245). After that, membranes were incubated with horseradish peroxidase-labeled secondary antibody (Cell Signaling Technology) for 1 h. GAPDH served as the internal control. The bands were visualized by an Imagequant LAS 4000 (GE Healthcare) system with the Western Lighting Plus – chemiluminescence (PerkinElmer, 203–17,201) [24].
Statistical analysis
SPSS 19.0 statistical software (IBM, Armonk, NY, USA) were performed to analyze the data. All results were presented by the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used for multiple comparisons and two group comparisons were performed with a Student’s t-test. Statistical significance was defined as P values < 0.05.
Results
In this study, we explored the biological roles and the potential molecular mechanism of LncRNA CASC9 in OC. The data showed the upregulation of CASC9 and downregulation of miR-488-3p in vivo and in vitro. CASC9 silencing inhibited cell proliferation, invasion and migration but promoted apoptosis and blocked cell cycle. CASC9 overexpression revealed the opposite results. CASC9 upregulated CCNG1/TP53/MMP7 expression by targeting miR-488-3p. Overall, our study demonstrated a functional role of the CASC9/miR-488-3p/CCNG1 axis in OC, providing a potential novel diagnostic and therapeutic target in OC treatment.
Expressions of CCNG1/TP53/MMP7 and LncRNA CASC9/miR-488-3p were aberrant in ovarian carcinoma
To explore the roles of CASC9 and potential molecular mechanism in OC, we first detected the expressions of CASC9 and its downstream gene in several OC cell lines by western blot assay and qRT-PCR. As shown in Figure 1a, the CCNG1 level was decreased while protein levels of TP53 and MMP7 were increased in OC cells compared with human ovarian surface epithelial cell line IOSE-80. In addition, CASC9 expression was significantly elevated and miR-488-3p expression was downregulated in OC cell lines compared with human ovarian surface epithelial cell line IOSE-80 (Figures 1B and 1 C). Given the results above, OVCAR-3 cell line was used for further experiments.
Figure 1.
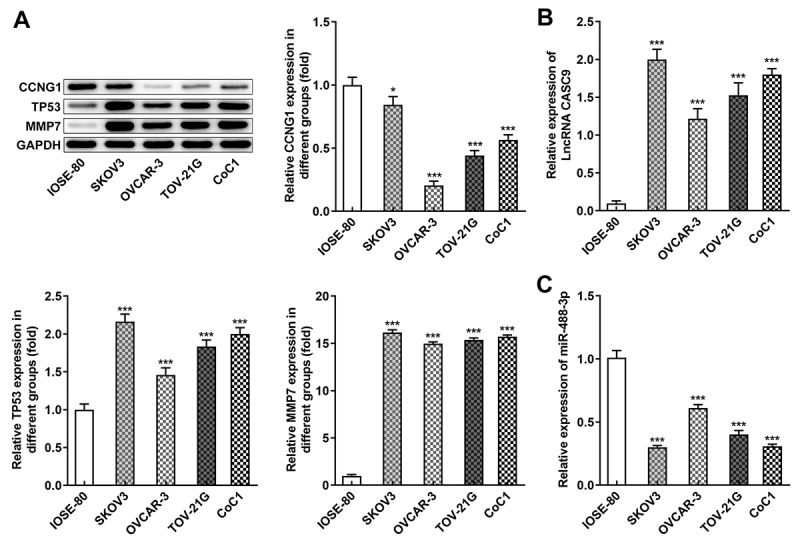
Expressions of CCNG1/TP53/MMP7 and LncRNA CASC9/miR-488-3p were abnormal in OC cells. A, Protein levels of CCNG1, TP53 and MMP7 in several OC cells were detected by western blot analysis. B-C, mRNA levels of CASC9 and miR-488-3p in several OC cells were measured by qRT-PCR. Data are expressed as mean ± SD. *P < 0.05, ***P < 0.001 versus IOSE-80 cells
CASC9 was involved in cell proliferation, migration, and invasion of OVCAR-3 and OVCAR-3/DDP cells
To investigate the biological role of CASC9 in OC cells and DDP-resistant OC cells, DDP-resistant OVCAR-3 cells were cultured and the cell inhibition rate was evaluated (Figure 2a). Then, we transfected shRNA-CASC9-1/2 and OverExp-CASC9-1/2 into OVCAR-3 cells and the transfection efficiency was detected (Figure 2b). According to Figure 2c, the proliferation ability of OVCAR-3/DDP cells was remarkably inhibited by shRNA-CASC9 whereas CASC9 overexpression enhanced the proliferation compared with the negative control. Additionally, CASC9 silencing improved cell inhibitory rate obviously but CASC9 overexpression repressed the inhibitory rate in OVCAR-3/DDP cells (Figure 2d). Consistently, cell migration and invasion capacity of OVCAR-3/DDP cells were suppressed by transfection with shRNA-CASC9 while CASC9 overexpression had an opposite effects on migration and invasion (Figures 2E and 2 F).
Figure 2.
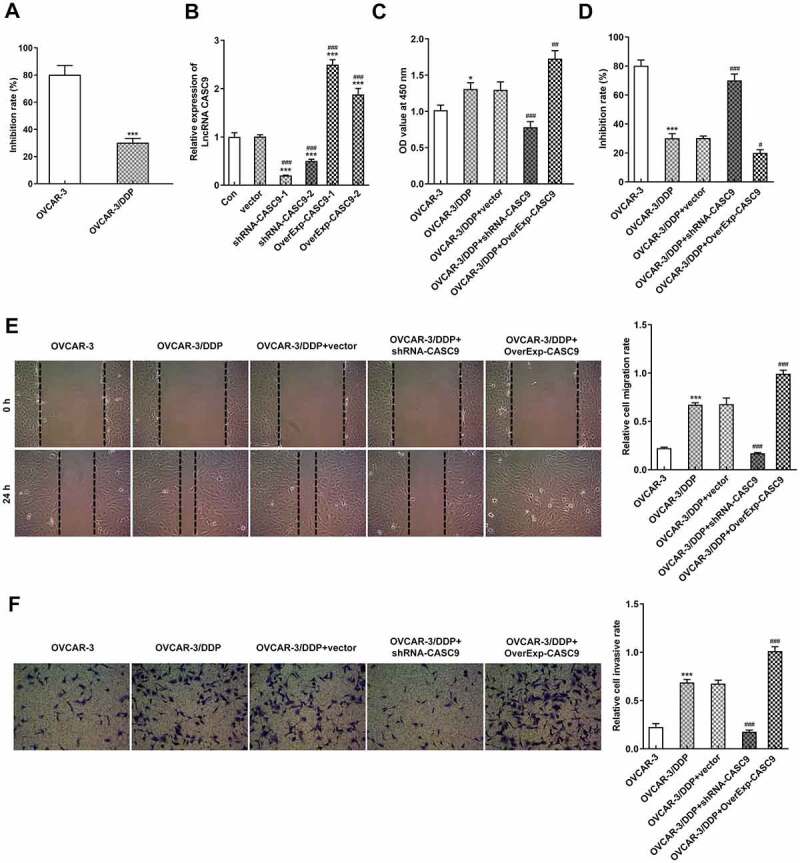
CASC9 promoted cell proliferation, migration and invasion in OVCAR-3 and OVCAR-3/DDP cells. A, Cell inhibitory rate in OVCAR-3 and OVCAR-3/DDP cells was evaluated by CCK-8 assay. *P < 0.05. B, CASC9 mRNA level was detected after transfection with shRNA-CASC9 or OverExp-CASC9. ***P < 0.001 versus control; ###P < 0.001 versus vector. C, Cell proliferation were identified by CCK-8 assay after transfection with shRNA-CASC9 or OverExp-CASC9. D, Cell inhibitory rate was identified by CCK-8 assay after transfection with shRNA-CASC9 or OverExp-CASC9. E, Cell migration was evaluated by wound healing assay after transfection with shRNA-CASC9 or OverExp-CASC9. F, Cell invasion was examined by transwell assay after transfection with shRNA-CASC9 or OverExp-CASC9. Data are expressed as mean ± SD. *P < 0.05, ***P < 0.001 versus OVCAR-3 cells; #P < 0.05, ##P < 0.01, ###P < 0.001 versus OVCAR-3/DDP+vector
CASC9 inhibited the apoptosis and cell cycle of OVCAR-3 and OVCAR-3/DDP cells
Next, we explored the effects of CASC9 expression on cell apoptosis and cell cycle in OVCAR-3 and OVCAR-3/DDP cells. As presented in Figure 3a, Tunel assay showed that cell apoptosis was extremely induced by shRNA-CASC9 but undermined by OverExp-CASC9 in OVCAR-3/DDP cells when compared with the negative control cells. Furthermore, western blot analysis results showed that Bcl-2 level were markedly inhibited whereas levels of Bax and cleaved caspase 3 was increased in CASC9 silenced cells. However, CASC9 overexpression reduced levels of Bax and cleaved caspase 3 but accelerated Bcl-2 level (Figure 3b). In addition, shRNA-CASC9 induced a G1 cell cycle arrest and reduced the percentage of cells in G2 phase. Nevertheless, CASC9 overexpression decreased the G1 phase population, and the same time increased the G2 phase population in OVCAR-3/DDP cells (Figure 3c).
Figure 3.
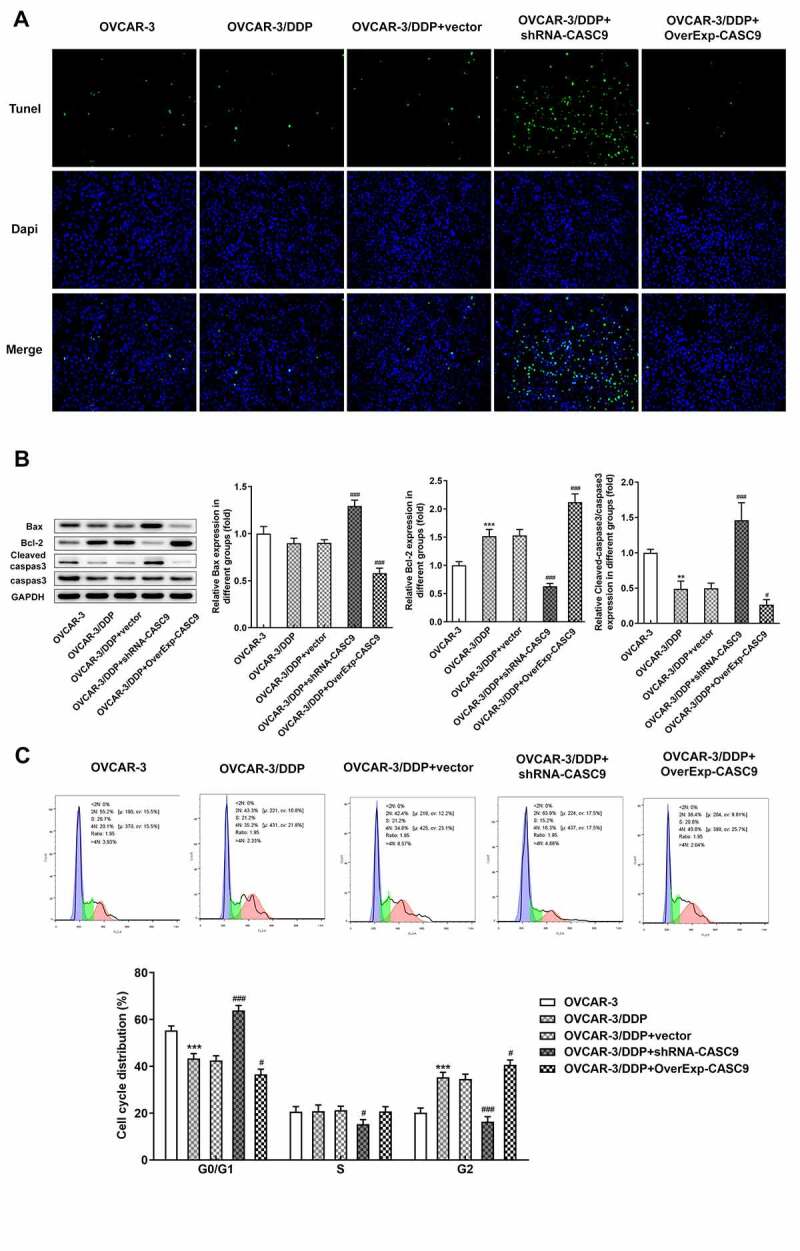
CASC9 suppressed cell apoptosis and cell cycle in OVCAR-3 and OVCAR-3/DDP cells. A, Tunel assay was performed to assess cell apoptosis after transfection with shRNA-CASC9 or OverExp-CASC9. B, Protein levels of Bax, Bcl-2, caspase 3 and cleaved caspase 3 were measured by western blot analysis after transfection with shRNA-CASC9 or OverExp-CASC9. C, Flow cytometric analysis was carried out to detect cell apoptosis after transfection with shRNA-CASC9 or OverExp-CASC9. Data are expressed as mean ± SD. **P < 0.01, ***P < 0.001 versus OVCAR-3 cells; #P < 0.05, ###P < 0.001 versus OVCAR-3/DDP+vector
CCNG1/TP53/MMP7 was regulated by aberrant expression of CASC9
To study the potential mechanism by which CASC9 expression affects OC cells, the expressions of CCNG1/TP53/MMP7 pathway were investigated. As shown in Figure 4, the protein levels of CCNG1, TP53, and MMP7 were noticeably improved in OVCAR-3/DDP cells compared with that in OVCAR-3 cells. Inhibition of CASC9 repressed the contents of CCNG1, TP53, and MMP7 while upregulation of CASC9 promoted the protein expression levels of CCNG1, TP53, and MMP7 in OVCAR-3/DDP cells as compared with the negative control.
Figure 4.
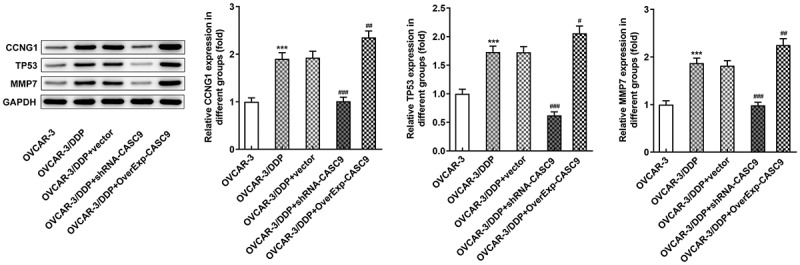
Expressions of CCNG1/TP53/MMP7 were suppressed by CASC9 silencing. Western blot analysis was performed to identify the protein levels of CCNG1, TP53 and MMP7 in OVCAR-3 and OVCAR-3/DDP cells transfected with shRNA-CASC9 or OverExp-CASC9. Data are expressed as mean ± SD. ***P < 0.001 versus OVCAR-3 cells; #P < 0.05, ##P < 0.01, ###P < 0.001 versus OVCAR-3/DDP+vector
CASC9/miR-488-3p targets CCNG1
Afterward, we used bioinformatics analysis to predict the target of CASC9 in OC. miR-488-3p was chosen for its potential to interact with CCNG1 and its documented role in OC from previous studies. Then, the qRT-PCR result revealed that downregulated CASC9 restrained CASC9 expression while promoting miR-488-3p expression in OVCAR-3/DDP cells. However, CASC9 overexpression elicited the opposite results (Figures 5A and 5B). Additionally, the binding sequences of CASC9 and miR-488-3p were shown in Figure 5c. Luciferase reporter assay results showed that miR-488-3p overexpression markedly inhibited the luciferase activity in OVCAR-3/DDP cells cotransfected with a reporter vector containing wild-type CASC9 segment (Figure 5d). In addition, CCNG1 was chosen as the target of miR-488-3p by bioinformatics analysis and the miR-488-3p binding sequence was shown in Figure 5e. The luciferase activity of wild-type CCNG1 reporter was found to be abated and no effects on the luciferase activity of cells cotransfected with a reporter vector containing mutated CCNG1 segment (figure 5f).
Figure 5.
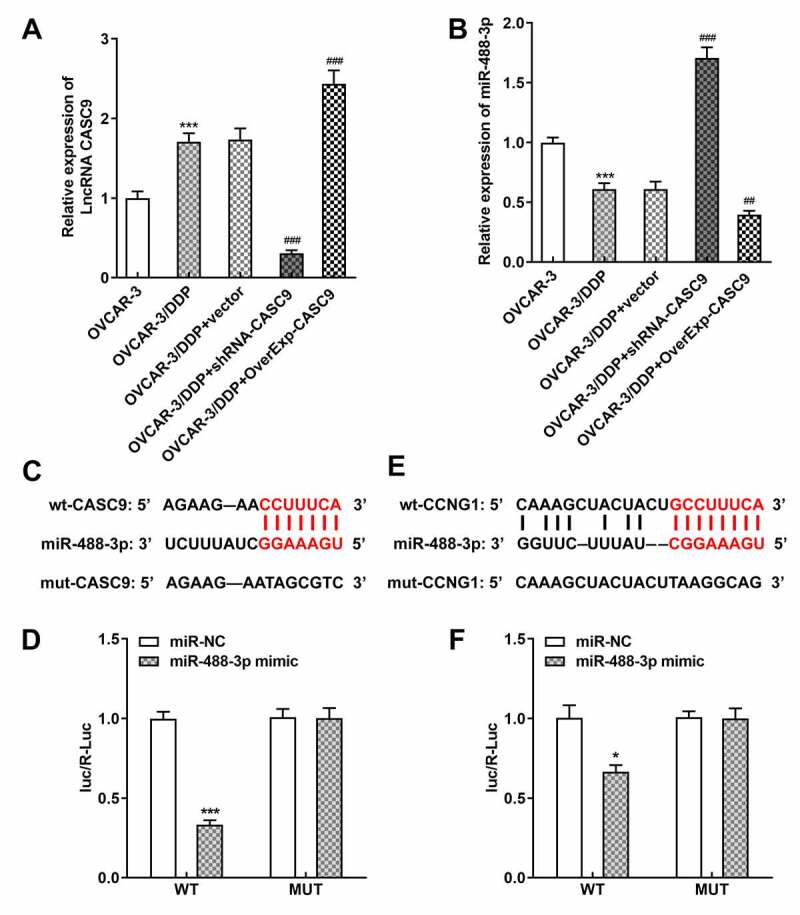
CASC9/miR-488-3p directly targets CCNG1. A-B, mRNA levels of CASC9 and miR-488-3p were identified by qRT-PCR after transfection with shRNA-CASC9 or OverExp-CASC9. C, The binding sequence of CASC9 and miR-488-3p. D, Luciferase reporter assay was performed to verify the combination between CASC9 and miR-488-3p. E, The binding sequence of miR-488-3p and CCNG1. F, Luciferase reporter assay was performed to verify the combination between miR-488-3p and CCNG1. Data are expressed as mean ± SD. *P < 0.05, ***P < 0.001 versus control; ##P < 0.01, ###P < 0.001 versus OVCAR-3/DDP+vector
Upregulation of miR-488-3p repressed cell resistance and anti-apoptosis activity of OVCAR-3/DDP cells
To study whether miR-488-3p expression is responsible for OC cells, we overexpressed miR-488-3p in OVCAR-3/DDP cells. As shown in Figure 6a, overexpressed CASC9 greatly diminished the inhibitory rate of OVCAR-3/DDP cells, while upregulation of miR-488-3p reversed the suppressive effects of CASC9 overexpression. Moreover, cell apoptosis was abolished by transfection with OverExp CASC9. However, miR-488-3p overexpression induced the CASC9-mediated apoptosis of OVCAR-3/DDP cells (Figure 6b).
Figure 6.
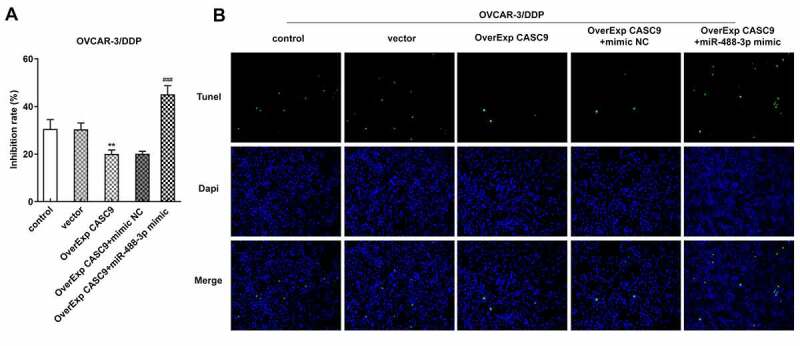
Effects of miR-488-3p overexpression on cell resistance and apoptosis in OVCAR-3/DDP cells. A, Cell inhibitory rate was detected by CCK-8 assay after transfection with OverExp-CASC9 in the presence and absence of miR-488-3p mimic. B, Tunel assay was carried out to assess cell apoptosis after transfection with OverExp-CASC9 in the presence and absence of miR-488-3p mimic. Data are expressed as mean ± SD. ***P < 0.01 versus vector; ##P < 0.01, ###P < 0.001 versus OverExp-CASC9+ mimic NC
Interference of CASC9 inhibited tumor growth and accelerated apoptosis in vivo
To further determine the effects of CASC9 on OC, we structured an OC xenograft mouse model (Figure 7a). As shown in Figures 7B and 7 C, the weight and volume of tumor in mice were significantly increased after injection with cells transfected with OverExp CASC9 compared with the negative control, but CASC9 silencing suppressed the elevated weight and volume of tumor. In addition, the results from tunel assay and western bolt assay revealed that CASC9 overexpression specifically prevented cell apoptosis in model mice while CASC9 knockdown exhibited the opposite results (Figures 7D and 8B). Moreover, we also measured the expressions of CCNG1/TP53/MMP7 pathway in tumor tissues. The results showed that CASC9 overexpression increased the levels of CCNG1, TP53 and MMP7, but silencing of CASC9 blocked the expressions of CCNG1/TP53/MMP7 pathway in tumor tissues of mice (Figure 8a). Also, qRT-PCR results showed that upregulation of CASC9 increased CASC9 expression while reduced miR-488-3p expression in tissues. Nevertheless, CASC9 silencing have the inverse effects on expressions of CASC9 and miR-488-3p (Figures 8C and8 D).
Figure 7.
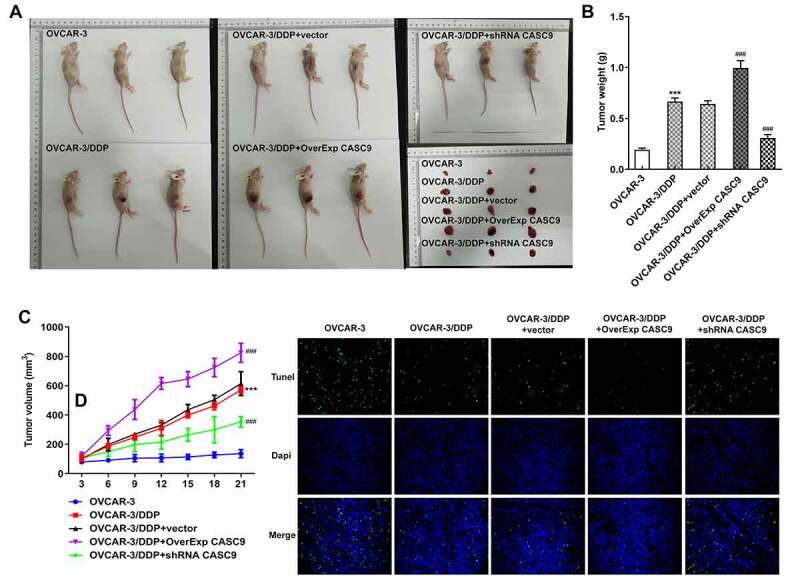
Effects of CASC9 silencing or overexpression on tumor growth and apoptosis in vivo. A, Ovarian cancer xenograft mouse model was established. B, Tumor weight of mice model was measured after transfection with shRNA-CASC9 or OverExp-CASC9. C, Tumor volume of mice model was evaluated after transfection with shRNA-CASC9 or OverExp-CASC9. D, Cell apoptosis was assessed in tumor tissues of mice by tunel assay after transfection with shRNA-CASC9 or OverExp-CASC9. Data are expressed as mean ± SD. ***P < 0.001 versus OVCAR-3 cells; ###P < 0.001 versus OVCAR-3/DDP+vector
Figure 8.
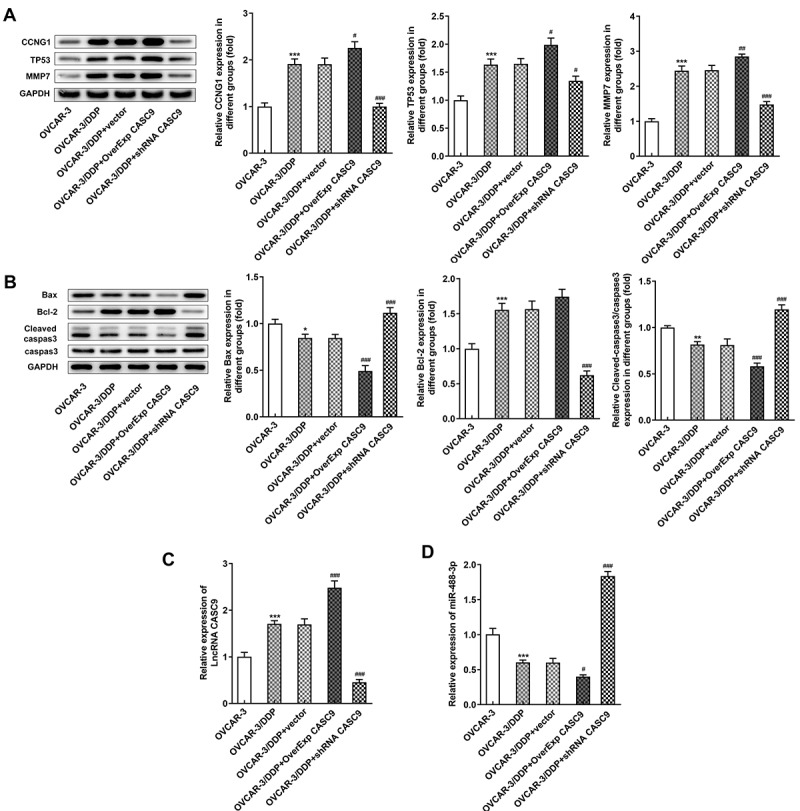
Effects of CASC9 on protein expressions involved in CCNG1/TP53/MMP7 and apoptosis. A, Protein levels of CCNG1, TP53 and MMP7 in tumor tissues of mice was evaluated by western blot assay transfected with shRNA-CASC9 or OverExp-CASC9. B, Protein levels of Bax, Bcl-2, caspase 3 and cleaved caspase 3 in tumor tissues of mice were identified by western blot analysis after transfection with shRNA-CASC9 or OverExp-CASC9. C-D, Expression of CASC9 and miR-488-3p in tumor tissues of mice was determined with qRT-PCR after transfection with shRNA-CASC9 or OverExp-CASC9. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01, ***P < 0.001 versus OVCAR-3 cells; #P < 0.05, ##P < 0.01, ###P < 0.001 versus OVCAR-3/DDP+vector
Discussion
Ovarian cancer is the most common gynecological malignancies and there is a lack of effective treatments for ovarian cancer to date [25]. Accumulating studies have shown that special lncRNAs play important roles in Pathology and Pathophysiology of ovarian cancer [26–28]. In the present study, we demonstrated that CASC9 was upregulated in several OC cells compared with ovarian surface epithelial cells. In addition, CASC9 silencing inhibited cell proliferation, migration and invasion, facilitated cell apoptosis and blocked cell cycle; CASC9 overexpression showed the opposite functions. Further mechanistic investigations showed that CASC9 regulated OV progress through cyclinG1/TP53/MMP7 pathway via targeting miR-488-3p.
On the basis of the previous researches, CASC9 has played oncogenic roles in several types of tumors [29,30]. A previous study revealed that CASC9 was significantly overexpressed in esophageal squamous cell carcinoma and CASC9 silencing markedly suppressed cell migration and invasion [31]. Shao et al. reported that CASC9 contributed to breast cancer cell proliferation and cell cycle progression, and repressed cell apoptosis by sponging the miR-195/497 cluster [32]. Recent studies also showed that dysregulation of CASC9 were involved in lung cancer, bladder cancer, and nasopharyngeal carcinoma [33–35]. In addition, Hu et al. also revealed that CASC9 was upregulated in ovarian cancer tissues and cell lines. CASC9 was associated with cell proliferation and metastasis, and promoted tumor growth by facilitating LIN7A expression via targeting miR-758-3p in ovarian cancer [36]. In our study, upregulated CASC9 expression was observed in several OC cells and in vivo. Knockdown of CASC9 notably inhibited OVCAR-3/DDP cell proliferation, migration and invasion, enhanced cell inhibitory rate and apoptosis, and blocked cell cycle progression, as well as inhibition of tumor growth in Xenograft mice. However, the opposite results were observed after transfection with overexpressed CASC9 and in mouse models, which was in line with previous results.
It is widely accepted that lncRNAs can serve as ceRNA for microRNAs to modulate expression of specific mRNA [37]. Based on bioinformatics analysis, miR-488-3p was identified as a potential target of CASC9. miR-488-3p has been reported to express variously in many different tissues and diseases, especially in cancers [38]. For example, miR-488 was downregulated in gastric cancer tissues and cells. miR-488 displayed anti – tumor activity by inhibition of cell proliferation, cell cycle, colony information, and migration in gastric cancer [39]. Moreover, miR-488-3p was found to be implicated in chemoresistance of ovarian cancer [40]. Another study revealed that higher CASC9 expression was related with malignant phenotypes of papillary thyroid cancer (PTC) and CASC9 expression was negatively related to miR-488-3p. lncRNA CASC9 accelerate PTC progress by inhibition of miR-488-3p /ADAM9 pathway [36]. In the study, we verified the combination of CASC9 and miR-488-3p by luciferase reporter assay and revealed that overexpression of miR-488-3p rescues the decreased cell inhibitory rate and apoptosis rate induced by CASC9 overexpression, which indicates CASC9 may play roles in OC by regulating miR-488-3p expression. LncRNA CASC9 is one of long noncoding RNAs and it cannot be transcribed into proteins, thus we can’t detect CASC9 expression by immunohistochemistry or western blot. To ensure the consistency of the results, we used the same method (qRT-PCR assay) to analysis the expression of CASC9 and miR-488-3p in OC cell lines and xenograft mice. Other technologies such as immunohistochemistry or western blot to further explore the effects of CASC9 and miR-488-3p in OC cell lines and xenograft mice are needed. In addition, most experiments in this study were done in OC cells and xenograft mice. We will verify our results in clinical and further animal experiments in next study.
Several cyclin proteins were expressed abnormally in various human cancer, such as Cyclin A, B, D1, D3, and E [41]. CCNG1 is a subtype of Cyclin G of the cyclin family and it induces failure of mitosis, polyploid formation, and blockage of M phase arrest [42]. It was found that Cyclin G1 was upregulated in epithelial ovarian cancer tissues was and was associated with poor survival [43]. CCNG1 could facilitate the degradation of TP53 by the MDM2 pathway and the inactivation of TP53 can induce the high expression of CCNG1 [44,45]. Besides, MMP7 is generally regulated by TP53 and functions as an oncogene for OC [46]. Thus, we hypothesized that cyclinG1/TP53/MMP7 was associated with the regulation of CASC9/miR-488-3p in OC. In our study, we used luciferase reporter assay to confirm the binding of miR-488-3p and CCNG1. The expressions of TP53 and MMP7 were significantly upregulated in OC cell lines. Furthermore, CASC9 silencing or overexpression regulated the protein levels of cyclinG1/TP53/MMP7 in vitro and in vivo. Thus, the data demonstrated that cyclinG1/TP53/MMP7 was mediated by CASC9 in OC cells.
Conclusion
In summary, our findings for the first time showed that CASC9 exerts oncogenic roles in OC cells and OC mice models. Furthermore, a cyclinG1/TP53/MMP7 pathway is involved in the regulation of OC progression by CCNG1/miR-488-3p, providing novel underlying mechanisms and therapeutic targets for patients with OC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
The authors declare that they have no competing interests.
Highlights
1. CASC9 is significantly upregulated in ovarian cancer cells.
2. CASC9 silencing inhibits ovarian cancer metastasis, facilitates cell apoptosis.
3. CASC9 overexpression promotes ovarian cancer metastasis, suppresses cell apoptosis.
4. CASC9 regulates ovarian cancer through cyclinG1/TP53/MMP7 pathway via miR-488-3p.
Authors’ contributions
MS and YC designed the research, drafted and revised the manuscript. MS, YC, XL and YC performed the experiments. YC and XL searched the literature and analyzed the data. YC guided the experiments. All authors read and approved the final manuscript.
Ethics approval
All procedures using animals were approved by Animal Care and Use Committee of Tangdu Hospital, Air Force Medical University and performed in accordance with Chinese legislation regarding experimental animals.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. [DOI] [PubMed] [Google Scholar]
- [2].Goff BA, Mandel LS, Melancon CH, et al. Frequency of symptoms of ovarian cancer in women presenting to primary care clinics. Jama. 2004;291(22):2705–2712. [DOI] [PubMed] [Google Scholar]
- [3].Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495–2505. [DOI] [PubMed] [Google Scholar]
- [4].Kommoss S, Gilks CB, du Bois A., Kommoss F.. Ovarian carcinoma diagnosis: the clinical impact of 15 years of change. British. J Cancer. 2016;115(8):993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reid BM, Permuth JB, Sellers TA.. Epidemiology of ovarian cancer: a review. Cancer Biol Med. 2017;14(1):9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cortez AJ, Tudrej P, Kujawa KA, et al. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol. 2018;81(1):17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ip JY, Nakagawa S.. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54(1):44–54. [DOI] [PubMed] [Google Scholar]
- [8].Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. [DOI] [PubMed] [Google Scholar]
- [10].Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu G, Hu X, Zhou G. Long non-coding RNA OR3A4 promotes proliferation and migration in breast cancer. Biomed Pharmacother. 2017;96:426–433. [DOI] [PubMed] [Google Scholar]
- [12].Shi D, Wu F, Mu S, et al. LncRNA AFAP1-AS1 promotes tumorigenesis and epithelial-mesenchymal transition of osteosarcoma through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J Exp Clin Cancer Res. 2019;38(1):375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cheng Y. FEZF1-AS1 is a key regulator of cell cycle, epithelial-mesenchymal transition and Wnt/β-catenin signaling in nasopharyngeal carcinoma cells. Biosci Rep. 2019;39(1). DOI: 10.1042/BSR20180906 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25(11):1980–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Luo K, Geng J, Zhang Q, et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang F, Qin Y, Lv J, et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis. 2018;9(10):1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morimoto A, Kannari M, Tsuchida Y, et al. An HNF4α-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem. 2017;292(25):10574–10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu K, Zhao D, Wang D. LINC00528 regulates myocardial infarction by targeting the miR-143-3p/COX-2 axis. Bioengineered. 2020;11(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cheng Q, Zhang M, Zhang M, et al. Long non-coding RNA LOC285194 regulates vascular smooth muscle cell apoptosis in atherosclerosis. Bioengineered. 2020;11(1):53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tong L, Tang C, Cai C, et al. Upregulation of the microRNA rno-miR-146b-5p may be involved in the development of intestinal injury through inhibition of Kruppel-like factor 4 in intestinal sepsis. Bioengineered. 2020;11(1):1334–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vanzyl EJ, Rick KRC, Blackmore AB, et al. Flow cytometric analysis identifies changes in S and M phases as novel cell cycle alterations induced by the splicing inhibitor isoginkgetin. PLoS One. 2018;13(1):e0191178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hua Q, Jin M, Mi B, et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol. 2019;12(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang JY, Lu AQ, Chen LJ. LncRNAs in ovarian cancer. Clin Chim Acta. 2019;490:17–27. [DOI] [PubMed] [Google Scholar]
- [27].Fu LL, Li CJ, Xu Y, et al. Role of lncRNAs as novel biomarkers and therapeutic targets in ovarian cancer. Crit Rev Eukaryot Gene Expr. 2017;27(2):183–195. [DOI] [PubMed] [Google Scholar]
- [28].Tripathi MK, Doxtater K, Keramatnia F, et al. Role of lncRNAs in ovarian cancer: defining new biomarkers for therapeutic purposes. Drug Discov Today. 2018;23(9):1635–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu H, Li C, Yang J, et al. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med. 2018;22(12):6338–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li X, Chen B, Chi D, et al. lncRNA CASC9 regulates cell migration and invasion in hemangioma endothelial cells by targeting miR-125a-3p/Nrg1. Onco Targets Ther. 2019;12:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pan Z, Mao W, Bao Y, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5(9):2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shao G, Wang M, Fan X, et al. Ji, lncRNA CASC9 positively regulates CHK1 to promote breast cancer cell proliferation and survival through sponging the miR‑195/497 cluster. Int J Oncol. 2019;54:1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou J, Xiao H, Yang X, et al. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 2018;8(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhan Y, Zhang L, Yu S, et al. Long non-coding RNA CASC9 promotes tumor growth and metastasis via modulating FZD6/Wnt/β-catenin signaling pathway in bladder cancer. J Exp Clin Cancer Res. 2020;39(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu L, Zhang Y, Wang J, et al. Long non-coding RNA CASC9 knockdown inhibits the progression of nasopharyngeal carcinoma by regulating miR-145. Int J Clin Exp Pathol. 2019;12:4024–4033. [PMC free article] [PubMed] [Google Scholar]
- [36].Hu X, Li Y, Kong D, et al. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234(7):10800–10808. [DOI] [PubMed] [Google Scholar]
- [37].Liu D, Yu X, Wang S, et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7(35):57228–57238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li Y, Li S, Luo Y, et al. LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. DNA Cell Biol. 2017;36(7):571–580. [DOI] [PubMed] [Google Scholar]
- [39].Zhao Y, Lu G, Ke X, et al. miR-488 acts as a tumor suppressor gene in gastric cancer. Tumour Biol. 2016;37(7):8691–8698. [DOI] [PubMed] [Google Scholar]
- [40].Yang Z, Feng Z, Gu J, et al. microRNA-488 inhibits chemoresistance of ovarian cancer cells by targeting Six1 and mitochondrial function. Oncotarget. 2017;8(46):80981–80993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ye XX, Liu CB, Chen JY, et al. The expression of cyclin G in nasopharyngeal carcinoma and its significance. Clin Exp Med. 2012;12(1):21–24. [DOI] [PubMed] [Google Scholar]
- [42].Russell P, Hennessy BT, Li J, et al. Cyclin G1 regulates the outcome of taxane-induced mitotic checkpoint arrest. Oncogene. 2012;31(19):2450–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jiang L, Liu R, Wang Y, et al. The role of Cyclin G1 in cellular proliferation and apoptosis of human epithelial ovarian cancer. J Mol Histol. 2015;46(3):291–302. [DOI] [PubMed] [Google Scholar]
- [44].Xu Y, Zhang Q, Miao C, et al. CCNG1 (Cyclin G1) regulation by mutant-P53 via induction of Notch3 expression promotes high-grade serous ovarian cancer (HGSOC) tumorigenesis and progression. Cancer Med. 2019;8(1):351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ohtsuka T, Jensen MR, Kim HG, et al. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene. 2004;23(31):5405–5408. [DOI] [PubMed] [Google Scholar]
- [46].Al-Alem LF, McCord LA, Southard RC, et al. Activation of the PKC pathway stimulates ovarian cancer cell proliferation, migration, and expression of MMP7 and MMP10. Biol Reprod. 2013;89(3):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


