Abstract
Fifty-three rotavirus-positive fecal specimens from children with diarrhea admitted to a Rio de Janeiro, Brazil, children's hospital between January 1997 and December 1998 were characterized for P and G types by using reverse transcription-PCR. Genotype P[4]G2 accounted for 21% of isolates, while uncommon genotypes P[8]G9, P[6]G9, and P[4]G9 accounted for 13% of the isolates.
Group A rotaviruses (RV) are the leading cause of severe gastroenteritis in infants and young children worldwide and are associated with 600,000 to 800,000 deaths each year, mostly in developing countries (9).
Belonging to the Reoviridae family, RV are characterized by the presence of 11 segments of double-stranded RNA (dsRNA) enclosed in a triple-shelled protein capsid. Serotypes of RV are defined by the two outer proteins: the protease-sensitive protein, VP4, and the major glycoprotein, VP7. The two outer proteins induce neutralizing antibodies and designate the G (VP7) and P (VP4) viral serotypes (8). Using genotyping methods to determine P and G genotypes, the most common combinations found in humans worldwide are P[8]G1, P[4]G2, P[8]G3, and P[8]G4 (9). Each serotype of RV induces serotype-specific protective immunity, which has been used as the basis for the development of an RV vaccine including the four most important human strains (13). However, uncommon genotypes and/or serotypes infecting humans in developed and developing countries throughout the world have been detected (9, 19). In Brazil, uncommon RV serotypes and/or genotypes, such as P[8]G5, P[6]G2, G8, G10, mixed infections, and a significant number of untypeable strains, have been previously reported (14, 20). Serotype G9 RV was first described in the United States (4) and later in different localities, including Japan (15), Thailand (22), India (18), Italy (1), Bangladesh (21), and Malawi (6). Griffin et al. (12) detected and characterized G9 RV strains in 10 of 12 cities in the United States and suggested that this strain could be emerging as significant, reaching epidemic proportions. Other recent studies support the increasing importance of serotype G9 as a cause of severe diarrhea in children around the world (2, 5, 16, 17). Therefore, serotype G9 must be taken into consideration for prospective RV vaccine studies, since this serotype is not constituted in any of the preceding RV vaccines.
Considering the large diversity of RV infecting Brazilian children, we report strain genotyping results from 53 positive RV fecal specimens collected from hospitalized children less than 3 years old with acute gastroenteritis at Hospital Municipal Jesus (Rio de Janeiro, RJ, Brazil), a reference center for children care. The samples were collected between January 1997 and December 1998 during the first 8 h of hospitalization, and the children took medical care for at least 2 days. Approximately 10% (wt/vol) stool suspensions were prepared in Tris-HCl Ca2+ (0.01 M) (pH 7.2), and an enzyme immunoassay for RV and adenovirus antigen detection (EIARA) was carried out. Concomitantly, stool suspensions were used for dsRNA extraction by the glass powder method (3), followed by a polyacrylamide gel electrophoresis (PAGE). Electrophoresed dsRNA segments were visualized by silver staining. The EIARA, PAGE, and silver staining procedures were performed as previously described (14).
The viral dsRNA, extracted from RV-positive clarified stool supernatants by the glass powder method (3), was first reverse transcribed (RT) and amplified by PCR (first amplification step) with a pair of consensus primers corresponding to a conserved nucleotide sequence of the VP7 (7, 11) or VP4 (10) genes. The DNA fragment obtained of 904 bp (VP7) or 876 bp (VP4) was then used as a template in a second PCR, carried out by using 1 μl of the first amplicon and a pool of genotype-specific primers complementary to variable regions of the VP7 (11) or VP4 (10) gene. Temperature and time conditions for PCR amplifications were performed as originally described (10, 11). Distilled Milli-Q water was used as a negative control in all techniques, and recommended manipulations for PCR procedures were carried out as a precaution to avoid false-positive results.
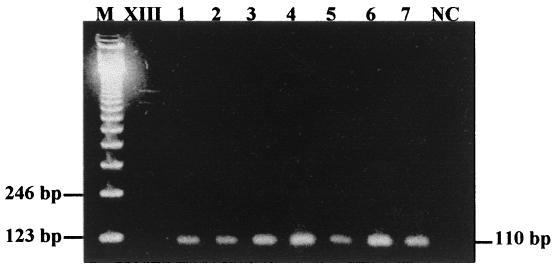
The P[4]G2 genotype was the most common strain detected in 21% of these isolates, followed by P[8]G2 (17%), P[8]G1 (11%), P[8]G3 (6%), P[8]G4 (6%), P[8]G10 (6%), P[6]G4 (4%), and one mixed infection by P[8+4]G3 (2%). In addition, we detected seven G9 strains: five P[8]G9 (9%), one P[6]G9 (2%), and one P[4]G9 (2%). As shown in Fig. 1, DNA bands corresponding to amplified segments of 110 bp for genotype G9 were visualized. The G9 strains were confirmed using Southern hybridization and chemiluminescent detection (data not shown), performed as previously described (14). The 5′-end-labeled digoxigenin-oligonucleotide probe for the G9 gene strain US1205 (5′ GC ATC AAC TCA AAT TGG AGA T; VP7 gene, nucleotides 313 to 333, plus-sense) was designed to be homologous to internal regions of the consensus G9 first-round RT-PCR product produced by primers 9con1 and 9con2. In addition, we sequenced the G9 genotype-specific PCR products with the Big Dye Terminator (PE Biosystem Inc.) on an automated sequencer, ABI 377. The sequenced Brazilian strains were very similar to United States and Malawi G9 strains (data not shown), suggesting that they belong to type G9. The nucleotide and amino acid homology between Brazilian strain 1527 and other G serotypes showed very high homology to prototype G9 strain US1205 (Table 1), confirming that these strains belong to serotype G9.
FIG. 1.
Nested PCR products for seven samples characterized as G9 genotype. Lane M, 123-bp ladder molecular size marker (Life Technologies, Inc.); lane XIII, molecular weight marker type XIII, digoxigenin labeled (Boehringer Mannheim Biochemicals); lanes 1 through 7, Brazilian strains of genotype G9; lane NC, negative control.
TABLE 1.
Nucleotide and amino acid homology between Brazilian strain 1527 (nucleotides 28 to 1034, amino acids 1 to 326) and the VP7 gene of other rotavirus G serotypes
| Strain | % Nucleotide homology | % Amino acid homology |
|---|---|---|
| G1 Wa | 72.3 | 81.0 |
| G2 S2 | 75.8 | 78.2 |
| G3 Y0 | 77.8 | 86.2 |
| G4 ST3 | 75.5 | 79.8 |
| G5 OSU | 76.9 | 82.5 |
| G6 UK | 75.0 | 85.3 |
| G7 TY-1 | 65.1 | 62.7 |
| G8 69M | 76.7 | 82.8 |
| G9-WI61 | 89.8 | 95.7 |
| G9-US1205 | 99.1 | 99.4 |
| G9-US1212 | 98.9 | 98.2 |
| G10 B223 | 75.5 | 83.1 |
| G11 YM | 78.2 | 85.3 |
| G12 L27 | 76.0 | 82.8 |
| G13 L338 | 76.2 | 79.1 |
| G14 F123 | 78.1 | 85.9 |
Analysis of dsRNA by PAGE and silver staining revealed that all P[8] and P[4] strains had long electropherotypes, while the P[6] strain had a short profile. Samples that could not be characterized for G type (4 samples), P type (1 sample), or G and P types (3 samples) represented 14% of the strains. Despite the methodologies used in this study, some strains remain untypeable, raising the possibility that additional genotypes may be present in the population. It is important to characterize these strains, since they may emerge as being predominant in the future, as type G9 did between March and May of 1998 in the present work. The present study reports for the first time G9 strains infecting humans in Brazil; thus, these findings have significant implications, since this serotype has recently emerged in Australia (17), France (2), England (5), Ireland (16), and the United States (12), raising the possibility that RV type G9 may represent a fifth globally important serotype. The identification of novel P and G combinations of RV in Rio de Janeiro emphasizes the ability of this segmented virus to form reassortants (21) which may result in the establishment of new important strains.
Nucleotide sequence accession number.
The VP7 gene sequence of Brazilian RV strain 1527 has been assigned the accession number AJ279082.
Acknowledgments
This work was supported by FIOCRUZ, COLAB/MS, and CNPq.
We thank members of the WHO/PAHO Rotavirus Collaborating Center in the Viral Gastroenteritis Section of the Centers for Disease Control and Prevention (Dixie Griffin and Jon Gentsch) for assistance with this investigation and for critical review of the manuscript (Jon Gentsch).
ADDENDUM IN PROOF
Since the submission of this report, N. Santos et al. (J. Clin. Microbiol. 39:1157–1160, 2001) reported rotavirus serotype G9 in the state of Rio de Janeiro from 1997 to 1999, and the present study reinforces the previous description with the identification of G9 strains infecting hospitalized children (inpatient).
REFERENCES
- 1.Arista S, Vizzi E, Ferraro D, Cascio A, Di Stefano R. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch Virol. 1997;142:2065–2071. doi: 10.1007/s007050050224. [DOI] [PubMed] [Google Scholar]
- 2.Bon F, Fromantin S, Aho S, Pothier P, Kohli E The Azay Group. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J Clin Microbiol. 2000;38:1681–1683. doi: 10.1128/jcm.38.4.1681-1683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark H F, Hoshino Y, Bell L M, Groff J, Hess G, Bachman P, Offit P A. Rotaviruses isolate W161 representing a presumptive new human serotype. J Clin Microbiol. 1987;25:1757–1762. doi: 10.1128/jcm.25.9.1757-1762.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cubitt W D, Steele A D, Iturriza M. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6] J Med Virol. 2000;61:150–154. doi: 10.1002/(sici)1096-9071(200005)61:1<150::aid-jmv24>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 6.Cunliffe N A, Gondwe J S, Broadhead R L, Molyneux M E, Woods P A, Bresee J S, Glass R I, Gentsch J R, Hart C A. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57:308–312. [PubMed] [Google Scholar]
- 7.Das B K, Gentsch J R, Cicirello H G, Woods P A, Gupta A, Ramachandran M, Kumar R, Bhan M K, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentsch J R, Woods P A, Ramachandran M, Das B K, Leite J P G, Alfieri A, Kumar R, Bhan M K, Glass R I. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174(Suppl. 1):S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 10.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea V, Glass R I, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin D D, Kirkwood C D, Parashar U D, Woods P A, Bresee J S, Glass R I, Gentsch J R. Surveillance of rotavirus strains in the United States: identification of unusual strains. J Clin Microbiol. 2000;38:2784–2787. doi: 10.1128/jcm.38.7.2784-2787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapikian A Z, Hoshino Y, Chanock R M, Pérez-Schael I. Efficacy of a quadrivalent rhesus rotavirus based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis. 1996;174(Suppl. 1):S65–S72. doi: 10.1093/infdis/174.supplement_1.s65. [DOI] [PubMed] [Google Scholar]
- 14.Leite J P G, Alfieri A A, Woods P A, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 15.Nakagomi T, Akatani K, Ikegami N, Katsushima N, Nakagomi O. Occurrence of changes in human rotavirus serotypes with concurrent changes in genomic RNA electropherotypes. J Clin Microbiol. 1988;26:2586–2592. doi: 10.1128/jcm.26.12.2586-2592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Halloran F, Lynch M, Cryan B, O'Shea H, Fanning S. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J Clin Microbiol. 2000;38:3370–3374. doi: 10.1128/jcm.38.9.3370-3374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palombo E A, Masendycz P J, Bogdanovic-Sakran N, Barnes G L, Bishop R F. Emergence of serotype G9 human rotaviruses in Australia. J Clin Microbiol. 2000;38:1305–1306. doi: 10.1128/jcm.38.3.1305-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P A, Holmes J L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Tanaguchi K, Bresee J S, Glass R I. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos N, Lima R C C, Pereira C F A, Gouvea V. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J Clin Microbiol. 1998;36:2727–2729. doi: 10.1128/jcm.36.9.2727-2729.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unicomb L E, Podder G, Gentsch J R, Woods P A, Hasan K Z, Faruque A S G, Albert M J, Glass R I. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J Clin Microbiol. 1999;37:1885–1891. doi: 10.1128/jcm.37.6.1885-1891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urasawa S, Hasegawa A, Urasawa T, Taniguchi K, Wakasugi F, Suzuki H, Inouye S, Pongprot B, Supawadee J, Suprasert S, Rangsiyanond P, Tonusin S, Yamazi Y. Antigenic and genetic analyses of human rotaviruses in Chiang Mai, Thailand: evidence for a close relationship between human and animal rotaviruses. J Infect Dis. 1992;166:227–234. doi: 10.1093/infdis/166.2.227. [DOI] [PubMed] [Google Scholar]