Abstract
Oligonucleotide primers specific for gene clusters involved in the biosynthesis of serotype-specific polysaccharide antigens were designed to identify Actinobacillus actinomycetemcomitans serotypes a to e using the multiplex PCR. This method may be useful for serotype-specific genotyping rapidly and directly from clinical samples containing various organisms.
Actinobacillus actinomycetemcomitans is a nonmotile, gram-negative, capnophilic, fermentative coccobacillus that has been implicated in the etiology and pathogenesis of localized juvenile periodontitis (2, 8), some forms of adult periodontitis (20), and severe nonoral human infections (6). A. actinomycetemcomitans strains are classified into five distinct serotypes: a, b, c, d, and e (4, 18, 30). The serologic specificity is defined by the polysaccharides on the surface of the organism (7), and the serotype-specific polysaccharide antigens (SPAs) are immunodominant antigens in the organism (1, 3, 12).
The serotypes of A. actinomycetemcomitans strains may have differences in virulence potential. We reported that the serotype b-specific polysaccharide antigen of A. actinomycetemcomitans plays an important role in resistance to phagocytosis and killing by human polymorphonuclear leukocytes (25). Takahashi et al. (23) reported that the ability of SPAs from serotypes a and c to induce the release of interleukin-1 by murine macrophages is lower than that of SPA from serotype b. Pajukanta et al. (15) demonstrated variation in the antimicrobial susceptibility patterns of different serotypes of A. actinomycetemcomitans.
Patients are usually infected by only one serotype, not multiple serotypes, and the serotypes are stable over time (2, 11, 18, 19). The frequency distribution of A. actinomycetemcomitans serotypes differs among populations. In the United States, serotype b is detected more frequently than serotypes a and c in patients with localized juvenile periodontitis (30). In Finland, serotype b is predominant in periodontitis patients and serotype c is frequently isolated in periodontally healthy individuals (2). In Japanese patients with periodontitis, serotypes a, c, and e are predominant (26). These findings suggest that the virulence and pathogenic role of the organism in the induction of periodontitis may differ among serotypes, and further work on the geographical distribution of serotypes of A. actinomycetemcomitans is required to understand the etiology of periodontitis and to control this disease.
PCR has become a powerful and increasingly popular tool in microbial identification (17). This technique requires the specific oligonucleotide primers designed from sequences of the target organism. The LKT2 and LKT3 primers specific to the lktA gene of A. actinomycetemcomitans are actually useful for identification of A. actinomycetemcomitans (5). Recently, we reported the gene clusters responsible for the synthesis of the SPAs of all serotypes of A. actinomycetemcomitans (9, 10, 22, 28, 29). In this study, we designed five pair of primers from specific DNA sequences for each serotype and then developed and evaluated a genetic method of identifying serotypes of A. actinomycetemcomitans strains, using a multiplex PCR assay with these primers.
All the bacterial strains used in this study are described in Table 1. The A. actinomycetemcomitans strains were cultured at 37°C in a CO2-enriched atmosphere for 48 h in THY broth (Todd-Hewitt broth supplemented with 1.0% yeast extract [Difco Laboratories, Detroit, Mich.]) or on THY agar plates. Escherichia coli DH5α was grown aerobically in 2× TY (1.6% Bacto Tryptone, 1.0% yeast extract, 0.5% NaCl) medium at 37°C. Streptococcus mutans Xc, Streptococcus sobrinus 6715, Streptococcus salivarius HT9R, Fusobacterium nucleatum ATCC 10953, Eikenella corrodens 1085, Haemophilus aphrophilus NCTC 5908, and Porphyromonas gingivalis ATCC 33277 were selected from the culture collection in the Department of Preventive Dentistry, Kyushu University Faculty of Dental Science, Fukuoka, Japan.
TABLE 1.
Bacterial strains and amplification of the serotype-specific fragments
| Strain | Serotype | Source or reference | Product size (bp) |
|---|---|---|---|
| A. actinomycetemcomitans | |||
| ATCC 29523 | a | ATCCa | 428 |
| TN-1 | a | Nishiharab | 428 |
| SUNYaB 75 | a | SUNYaBc | 428 |
| Y4 | b | Socranskyd | 298 |
| JP2 | b | 24 | 298 |
| ATCC 29522 | b | ATCC | 298 |
| ATCC 29524 | b | ATCC | 298 |
| NCTC 9710 | c | NCTCe | 559 |
| NCTC 9709 | c | NCTC | 559 |
| SUNYaB 67 | c | SUNYaB | 559 |
| IDH 781 | d | Asikainenf | 690 |
| 392 | d | Asikainen | 690 |
| 1344 | d | Asikainen | 690 |
| 3381 | d | Asikainen | 690 |
| IDH 1705 | e | Asikainen | 211 |
| OMZ 534 | e | Gmürg | 211 |
| OMZ 541 | e | Gmür | 211 |
| OMZ 546 | e | Gmür | 211 |
| H. aphrophilus NCTC 5908 | KUh | NDi | |
| F. nucleatum ATCC 10953 | KU | ND | |
| E. corrodens 1085 | KU | ND | |
| P. gingivalis ATCC 33277 | KU | ND | |
| S. mutans Xc | KU | ND | |
| S. sobrinus 6715 | KU | ND | |
| S. salivarius HT9R | KU | ND | |
| P. intermedia ATCC 25611 | ATCC | ND | |
| E. coli DH5α | ND |
ATCC, American Type Culture Collection, Rockville, Md.
T. Nishihara, Kyushu Dental College, Fukuoka, Japan.
SUNYaB, State University of New York, N.Y.
S. S. Socransky, Forsyth Dental Center, Mass.
NCTC, National Collection of Type Cultures, London, England.
S. Asikainen, University of Helsinki, Helsinki, Finland.
R. Gmür, University of Zurich, Zurich, Switzerland.
KU, the culture collection in Department of Preventive Dentistry, Kyushu University Faculty of Dental Science, Fukuoka, Japan.
ND, not detected.
Genomic bacterial strains DNA was isolated and purified as described below. Whole cells were collected from 50-ml cultures by centrifugation, suspended in 1 ml of a solution containing 25 mM Tris-HCl (pH 7.5), 50 mM glucose, and 10 mM EDTA, and incubated at 37°C for 10 min with 100 μg of lysozyme per ml. The suspension was then incubated with 500 U of proteinase K (Sigma Chemical Co., St. Louis, Mo.) per ml until it became viscous. After 10 μg of RNase per ml and 1% sodium dodecyl sulfate were added, the DNA was purified by repeated phenol-chloroform extraction. Genomic DNA precipitated by adding 100% ethanol was resuspended in TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and stored at 4°C.
Colonies of A. actinomycetemcomitans as PCR templates were picked up from THY agar plates, suspended in 50 μl of cell lysis buffer (1.0% Triton X-100, 20 mM Tris-HCl, 2 mM EDTA [pH 8.0]), and boiled at 100°C for 5 min (5). After the lysed cells were centrifuged, the supernatant containing the bacterial DNA was removed and frozen at −30°C until use.
For clinical evaluation, subgingival plaque samples from patients with periodontitis were obtained by inserting a sterile endodontic paperpoint into the subgingival site for 10 s. The paperpoint was transferred into 200 μl of phosphate-buffered saline (0.12 M NaCl, 0.01 M Na2HPO4, 5 mM KH2PO4 [pH 7.5]) and centrifuged at 15,000 × g for 5 min. The cells resuspended in 100 μl of cell lysis buffer were boiled at 100°C for 5 min, and the supernatant was used as a PCR template.
A 10-μl volume of PCR mixture consisted of 0.25 mM each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.6), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 5 U of Taq polymerase (Amersham Pharmacia Biotech UK Ltd., Little Chalfont, England), 2 μM each primer, and 1 μl of template DNA. The PCR assays were performed in a T3 thermocycler (Biometra, Göttingen, Germany). After denaturation at 96°C for 2 min, a total of 25 PCR cycles were performed; each cycle consisted of 15 s of denaturation at 94°C, 30 s of annealing at 54°C, and 60 s of extension at 72°C. Amplification products were loaded into 1.8% (wt/vol) agarose gels by electrophoresis, stained with ethidium bromide (0.5 μg/ml), and photographed under UV light.
The sequences of the DNA primers used in this study are listed in Table 2. Each primer was designed from the DNA sequence of the respective gene cluster involved in the biosynthesis of the SPA from A. actinomycetemcomitans serotype a, b, c, d, or e with the DNASIS sequence analysis program (Hitachi Software Engineering Co., Yokohama, Japan): the mannosyltransferase homologue gene of SUNYaB 75 (serotype a) (22), the gene encoding dTDP-4-keto-6-deoxy-d-glucose reductase of Y4 (serotype b) (27, 29), the putative acetyltransferase gene of NCTC 9710 (serotype c) (10), the mannosyltransferase homologue gene of IDH 781 (serotype d) (9), and an unknown gene of IDH 1705 (serotype e) (28). Each amplification product is a distinct size between 211 and 690 bp. Primers SA-F, SA-R, SB-F, SB-R, SC-F, SC-R, SD-F, SD-R, SE-F, and SE-R were each used at 2 μM (final concentration) in the reaction solution of the multiplex PCR. The LKT2 and LKT3 primers (5) were used as primers specific for A. actinomycetemcomitans.
TABLE 2.
Primer sequences used in this study
| Name | Sequence | Product size (bp) | Protein coded by the gene involving primers | Source or reference |
|---|---|---|---|---|
| SA-F | 5′-GCAATGATGTATTGTCTTCTTTTGGA-3′ | 428 | Putative mannosyltransferase | SUNYaB 75 |
| SA-R | 5′-CTTCAGTTGAATGGGGATTGACTAAAAC-3′ | |||
| SB-F | 5′-CGGAAATGGAATGCTTGC-3′ | 298 | dTDP-4-keto-6-deoxy-d-glucose reductase | Y4 |
| SB-R | 5′-CTGAGGAAGCCTAGCAAT-3′ | |||
| SC-F | 5′-AATGACTGCTGTCGGAGT-3′ | 559 | Putative acetyltransferase | NCTC 9710 |
| SC-R | 5′-CGCTGAAGGTAATGTCAG-3′ | |||
| SD-F | 5′-TTACCAGGTGTCTAGTCGGA-3′ | 690 | Putative mannosyltransferase | IDH 781 |
| SD-R | 5′-GGCTCCTGACAACATTGGAT-3′ | |||
| SE-F | 5′-CGTAAGCAGAAGAATAGTAAACGT-3′ | 211 | Unknown | IDH 1705 |
| SE-R | 5′-AATAACGATGGCACATCAGACTTT-3′ | |||
| LKT2 | 5′-CTAGGTATTGCGAAACAATTTG-3′ | 262 | LktA | 5 |
| LKT3 | 5′-CCTGAAATTAAGCTGGTAATC-3′ |
To determine whether the amplification product of each pair of primers is serotype specific among A. actinomycetemcomitans strains, Southern blotting was performed using each digoxigenin-labeled PCR product as a probe. The expected bands of the sizes listed in Table 2 were observed and were serotype specific (data not shown). Moreover, the PCR tests with multiplex primers using chromosomal DNA from various oral bacteria and E. coli as templates were performed. No bands were amplified from all the bacteria except for A. actinomycetemcomitans, tested in this study (Table 1). When the LKT primers were used, the expected fragments were amplified from all strains of A. actinomycetemcomitans but not from other bacteria (data not shown). These results demonstrate that each set of primers is specific for A. actinomycetemcomitans strains and serotypes.
To standardize and optimize PCR, 18 A. actinomycetemcomitans strains, including 3 serotype a strains (ATCC 29523, TN-1, and SUNYaB 75), 4 serotype b strains (Y4, JP2, ATCC 29522, and ATCC 29524), 3 serotype c strains (NCTC 9710, NCTC 9709, and SUNYaB 67), 4 serotype d strains (IDH 781, 392, 1344, and 3381), and 4 serotype e strains (IDH 1705, OMZ 534, OMZ 541, and OMZ 546) were used. The multiplex PCR assay was optimized using genomic DNA samples from these strains. The concentration of the reaction mixture was based on the conditions recommended by the manufacturer. Annealing temperatures from 48 to 57°C were tested, and the clearest bands were obtained at 54°C. PCR assays of 25, 30, and 35 cycles were tested, and all resultant bands were clear enough after 25 cycles, which was used for the multiplex PCR. In a 10-μl reaction mixture, 3 to 5 ng of genomic DNA was used as a template.
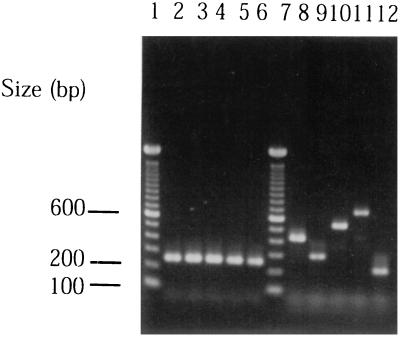
Under the above standard condition, colonies of A. actinomycetemcomitans ATCC 29523 (serotype a), JP2 (serotype b), NCTC 9710 (serotype c), IDH 781 (serotype d), and IDH 1705 (serotype e) were used as templates for the multiplex PCR. The resulting amplification products produced respective single bands of 428 bp (ATCC 29523), 298 bp (JP2), 559 bp (NCTC 9710), 690 bp (IDH 781), or 211 bp (IDH 1705) (Fig. 1, lanes 8 to 12), which corresponded to the size of the amplification band when the genomic DNA sample was used.
FIG. 1.
Agarose gel electrophoresis of PCR products amplified from whole cells of serotypes a through e. Lanes: 1 and 7, molecular size markers; 2 and 8, ATCC 29523 (serotype a); 3 and 9, JP2 (serotype b); 4 and 10, NCTC 9710 (serotype c); 5 and 11, IDH 781 (serotype d); 6 and 12, IDH 1705 (serotype e). PCR products in lanes 2 to 6 and lanes 8 to 12 were amplified with primers LKT2 and LKT3 and with multiplex primers, respectively.
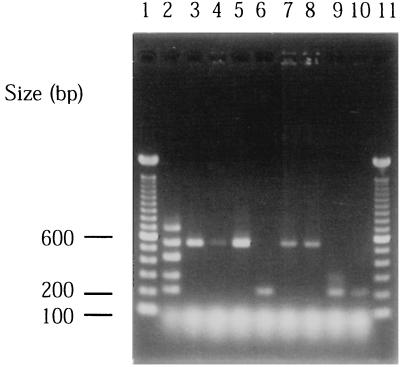
Subgingival plaque samples from subjects with periodontitis were analyzed by multiplex PCR. Eight plaque samples from five subjects, which produced a 262-bp DNA fragment after amplification with primers LKT2 and LKT3, were used as templates for the multiplex PCR assay. A total of 35 cycles were performed under the same conditions as described above. The amplification products of samples from three different sites of subject 1 and two different sites of subject 3 showed a single band of 559 bp, which corresponded to the size of serotype c (Fig. 2, lanes 3 to 5, 7, and 8). With the templates prepared from subjects 2, 4, and 5, the product size was 211 bp, which corresponded to serotype e (lanes 6, 9, and 10). Only a single serotype of A. actinomycetemcomitans was harbored in five subjects analyzed in this study.
FIG. 2.
PCR amplification using subgingival plaque samples as a template. Lanes: 1 and 11, molecular size markers; 2, reference marker mixed PCR products of five serotypes; 3 to 5, subject 1; 6, subject 2; 7 and 8, subject 3; 9, subject 4; 10, subject 5.
Serotypes of A. actinomycetemcomitans have been classified by an immunodiffusion assay with antisera directed against whole cells of A. actinomycetemcomitans strains (4, 18, 30). Although this assay is useful for identification, it can be time-consuming and ambiguous. To prepare antigens for the immunodiffusion assay, the organism should be cultured in growth medium. Culturing of clinical isolates is also important because it is valuable for some additional analyses including serotyping. However, culture-based analysis relies on the growth of organisms, and hence only viable organisms can be detected. On the other hand, PCR offers a practical alternative that does not need antigens and antibodies. Therefore, our multiplex PCR method for serotyping of A. actinomycetemcomitans is independent of the different growth patterns of the organism. Furthermore, the PCR test is more sensitive and specific than the immunodiffusion test, which often shows nonspecific reactions.
To detect genotypes, an arbitrarily primed PCR (AP-PCR) is usually used (21). AP-PCR is a comparative method based on variability in the genomes detected using oligonucleotide primers with random sequences. This method is used in studies on the genetic diversity of nonserotypeable or cross-reactive strains (13, 16). Using random primers, however, A. actinomycetemcomitans strains cannot be detected directly from clinical samples contaminated with various other bacteria. In this study, the successful application of the multiplex PCR assay to clinical samples provides an effective method for serotype-specific genotyping of A. actinomycetemcomitans. Our multiplex PCR may be useful for screening for and monitoring periodontal disease and planning treatment, because we can detect serotypes of A. actinomycetemcomitans strains directly from subgingival plaque samples harboring various oral bacteria.
Three to eight percent of isolates of A. actinomycetemcomitans remain nonserotypeable (4, 18). Paju et al. (14) reported that both serotypeable and nonserotypeable A. actinomycetemcomitans strains belong to identical AP-PCR genotypes. It is possible that some of these nonserotypeable strains are derived from serotypeable strains (14). Our multiplex PCR test may be useful for identifying the origin of nonserotypeable A. actinomycetemcomitans strains.
Acknowledgments
We thank Toshihiro Ansai, Kyushu Dental College, Fukuoka, Japan, for kindly providing Prevotella intermedia chromosomal DNA.
This study was supported in part by a Grant-in-Aid for Encouragement of Young Scientists 11771157 (Y.N.) and Grants-in-Aid for Scientific Research (A) 10307054 (T.K.) and (B) 11470452 (T.K.) from the Ministry of Education, Science, Sports, and Culture, Tokyo, Japan.
REFERENCES
- 1.Amano K, Nishihara T, Shibuya N, Noguchi T, Koga T. Immunochemical and structural characterization of a serotype-specific polysaccharide antigen from Actinobacillus actinomycetemcomitans Y4 (serotype b) Infect Immun. 1989;57:2942–2946. doi: 10.1128/iai.57.10.2942-2946.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asikainen S, Lai C-H, Alaluusua S, Slots J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991;6:115–118. doi: 10.1111/j.1399-302x.1991.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 3.Califano J V, Schenkein H A, Tew J G. Immunodominant antigens of Actinobacillus actinomycetemcomitans serotypes a and c in high-responder patients. Oral Microbiol Immunol. 1991;6:228–235. doi: 10.1111/j.1399-302x.1991.tb00482.x. [DOI] [PubMed] [Google Scholar]
- 4.Gmür R, McNabb H, van Steenbergen T J M, Baehni P, Mombelli A, van Winkelhoff A J, Guggenheim B. Seroclassification of hitherto nonserotypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993;8:116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 5.Goncharoff P, Figurski D H, Stevens R H, Fine D H. Identification of Actinobacillus actinomycetemcomitans: polymerase chain reaction amplification of lktA-specific sequences. Oral Microbiol Immunol. 1993;8:105–110. doi: 10.1111/j.1399-302x.1993.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan A H, Weber D J, Oddone E Z, Perfect J R. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11:46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- 7.Koga T, Nishihara T, Amano K, Takahashi T, Nakashima K, Ishihara Y, Shibuya N. Chemical and biological properties of cell-surface components of Actinobacillus actinomycetemcomitans. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens & host immune responses. Tokyo, Japan: Quintessence Publishing Co., Ltd.; 1991. pp. 117–127. [Google Scholar]
- 8.Meyer D H, Fives-Taylor P M. The role of Actinobacillus actinomycetemcomitans in the pathogenesis of periodontal disease. Trends Microbiol. 1997;5:224–228. doi: 10.1016/S0966-842X(97)01055-X. [DOI] [PubMed] [Google Scholar]
- 9.Nakano Y, Yoshida Y, Suzuki N, Yamashita Y, Koga T. A gene cluster for the synthesis of serotype d-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 2000;1493:259–263. doi: 10.1016/s0167-4781(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 10.Nakano Y, Yoshida Y, Yamashita Y, Koga T. A gene cluster for 6-deoxy-l-talan synthesis in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 1998;1442:409–414. doi: 10.1016/s0167-4781(98)00174-2. [DOI] [PubMed] [Google Scholar]
- 11.Olsen I, Shah H N, Gharbia S E. Taxonomy and biochemical characteristics of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontology 2000. 1999;7:69–82. doi: 10.1111/j.1600-0757.1999.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 12.Page R C, Sims T J, Engel L D, Moncla B J, Bainbridge B, Stray J, Darveau R P. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect Immun. 1991;59:3451–3462. doi: 10.1128/iai.59.10.3451-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paju S, Saarela M, Alaluusua S, Fives-Taylor P, Asikainen S. Characterization of serologically nonserotypeable Actinobacillus actinomycetemcomitans isolates. J Clin Microbiol. 1998;36:2019–2022. doi: 10.1128/jcm.36.7.2019-2022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paju S, Saarela M, Chen C, Jousimies-Somer H, Uitto V-J, Asikainen S. Altered antigenicity is seen in the lipopolysaccharide profile of non-serotypeable Actinobacillus actinomycetemcomitans strains. FEMS Immunol Med Microbiol. 2000;27:171–177. doi: 10.1111/j.1574-695X.2000.tb01428.x. [DOI] [PubMed] [Google Scholar]
- 15.Pajukanta R, Asikainen S, Saarela M, Alaluusua S, Jousimies-Somer H. In vitro antimicrobial susceptibility of different serotypes of Actinobacillus actinomycetemcomitans. Scand J Dent Res. 1993;101:299–303. doi: 10.1111/j.1600-0722.1993.tb01124.x. [DOI] [PubMed] [Google Scholar]
- 16.Preus H R, Haraszthy V I, Zambon J J, Genco R J. Differentiation of strains of Actinobacillus actinomycetemcomitans by arbitrarily primed polymerase chain reaction. J Clin Microbiol. 1993;31:2773–2776. doi: 10.1128/jcm.31.10.2773-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodu B. Molecular biology in medicine. The polymerase chain reaction: the revolution within. Am J Med Sci. 1999;299:210–216. doi: 10.1097/00000441-199003000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai C-H, Jousimies-Somer H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol. 1992;7:277–279. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 19.Saarela M, Asikainen S, Jousimies-Somer H, Asikainen T, von Troil-Linden B, Alaluusua S. Hybridization patterns of Actinobacillus actinomycetemcomitans serotypes a–e detected with an rRNA gene probe. Oral Microbiol Immunol. 1993;8:111–115. doi: 10.1111/j.1399-302x.1993.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 20.Slots J, Bragd L, Wikström M, Dahlén G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 21.Slots J, Liu Y B, DiRienzo J M, Chen C. Evaluating two methods for fingerprinting genomes of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1993;8:337–343. doi: 10.1111/j.1399-302x.1993.tb00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki N, Nakano Y, Yoshida Y, Nakao H, Yamashita Y, Koga T. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 2000;1517:135–138. doi: 10.1016/s0167-4781(00)00229-3. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi T, Nishihara T, Ishihara Y, Amano K, Shibuya N, Moro I, Koga T. Murine macrophage interleukin-1 release by capsularlike serotype-specific polysaccharaide antigens of Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:18–23. doi: 10.1128/iai.59.1.18-23.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai C-C, Shenker B J, DiRienzo J M, Malamud D, Taichman N S. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infect Immun. 1984;43:700–705. doi: 10.1128/iai.43.2.700-705.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi N, Kawasaki M, Yamashita Y, Nakashima K, Koga T. Role of the capsular polysaccharide-like serotype-specific antigen in resistance of Actinobacillus actinomycetemcomitans to phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1995;63:4589–4594. doi: 10.1128/iai.63.12.4589-4594.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto M, Nishihara T, Koseki T, He T, Yamato K, Zhang Y J, Nakashima K, Oda S, Ishikawa I. Prevalence of Actinobacillus actinomycetemcomitans serotypes in Japanese patients with periodontitis. J Periodontal Res. 1997;32:676–681. doi: 10.1111/j.1600-0765.1997.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida Y, Nakano Y, Nezu T, Yamashita Y, Koga T. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-d-fucose. J Biol Chem. 1999;274:16933–16939. doi: 10.1074/jbc.274.24.16933. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida Y, Nakano Y, Suzuki N, Nakao H, Yamashita Y, Koga T. Genetic analysis of the gene cluster responsible for synthesis of serotype e-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 1999;1489:457–461. doi: 10.1016/s0167-4781(99)00192-x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y, Nakano Y, Yamashita Y, Koga T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect Immun. 1998;66:107–114. doi: 10.1128/iai.66.1.107-114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]