(See the Brief Report by Nussenblatt et al, on pages 1118–23.)
In the fall of 2020, the B.1.1.7 or Alpha variant emerged [1], launching the first of several, deadly, international waves of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants (Figure 1). The Alpha variant was notable for a sudden leap in the rate of accumulation of SARS-CoV-2 mutations, and for the eerie familiarity of its constituent mutations such as spike (S) N501Y and S:del69-70, which had been observed in people chronically infected with SARS-CoV-2 [2, 3]. The sequence of subsequent variants of concern elicited the same sense of déjà vu. For example, S:E484K (Beta and Gamma), S:T478K (Delta), and S:E484A (Omicron) had been detected in the case report of Choi et al [2]. Thus, people with chronic SARS-CoV-2 infection provided a window into the future of circulating variants.
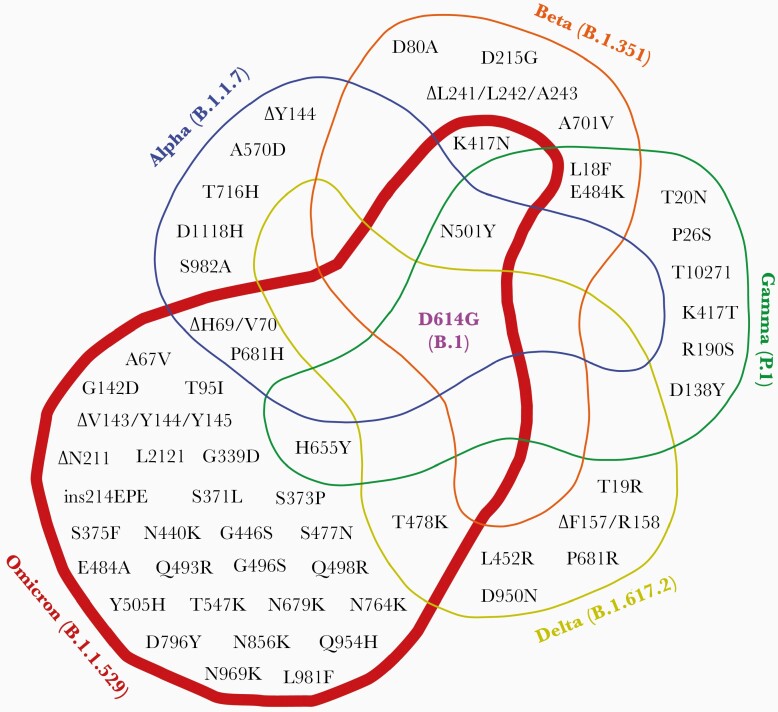
Figure 1.
Venn diagram showing the shared, nonsynonymous S mutations in B.1 (S-D614G), Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) variants.
Why have chronic SARS-CoV-2 infections seemingly conferred such clairvoyance? At a minimum, chronic infections provide a fertile environment for evolution, permitting the virus ample time to explore potential genetic space without requirement for transmission to another susceptible host. If selection for resistance to antiviral immunity is a matter of “time and titer” [4], chronic infections provide both in abundance. And while chronic virus infection may select for the ability to escape neutralization by monoclonal or polyclonal antibodies (either endogenous or exogenous), the intrahost environment differs from the Petri dish in that the virus must also remain viable for replication within native human tissues. As a result, sequencing of viruses after chronic infection has revealed mutations that confer both escape from neutralizing antibodies and increase in infectivity, with the apparent ability to balance the structural requirements for these 2 properties. Chronic infections have also highlighted complex clonal dynamics, epistatic effects of individual mutations, and emphasized how much we don’t yet understand about within-host evolution of SARS-CoV-2, including the possibility that multiple compartments of viral replication are present across different host tissues.
Perhaps, though, persistent infection is not simply a curiosity of SARS-CoV-2 evolution, but the actual breeding ground for variants of concern, which then intermittently spill over into the general population. While there are other highly plausible hypotheses for the genesis of variants of concern, including reverse zoonosis [5] and SARS-CoV-2 circulation in regions with minimal genomic surveillance [6], many scientists point the finger at chronic infection, swayed by the repeated emergence of extraordinary clinical cases like the one described in this issue of The Journal of Infectious Diseases [7]. Nussenblatt et al report a nearly year-long infection in a patient who had B-cell aplasia and hypogammaglobulinemia secondary to anti-CD19 chimeric antigen receptor T-cell therapy administered for diffuse B-cell lymphoma. The patient presented with fever and cough in May 2020, and though nasopharyngeal swabs were initially negative for SARS-CoV-2, bronchial lavage tested positive. Over the course of the SARS-CoV-2 infection the patient received 2 treatments with convalescent plasma, as well as immunoglobulin, prednisone, and remdesivir. Ultimately, a year later, the patient tested negative for SARS-CoV-2 but, unfortunately, had no detectable anti–SARS-CoV-2 antibodies. This case is notable for being the longest duration SARS-CoV-2 infection on record.
The SARS-CoV-2 sequences from this patient all mapped to a common branch of the phylogenetic tree and the sequences corresponded to B.1.332, a Pango lineage that was circulating at the time of initial presentation but disappeared from the community soon thereafter. This sequence analysis convincingly demonstrates that this was a case of a single prolonged infection, rather than a reinfection. Over the course of the infection, SARS-CoV-2 acquired 28 mutations with respect to the sequence from the earliest time point. Nineteen of the mutations were nonsynonymous, with 4 in the S gene. Two deletions were observed in the viral genome that are informative regarding the SARS-CoV-2–host interaction. One was a unique deletion in the N-terminal domain of the spike protein (S:del244-246). This domain is a common target of neutralizing antibodies [8], similar to an adjacent deletion in the Beta variant, which may have been selected by treatment with convalescent plasma. The authors also describe a much larger deletion of ORF7/8. This is among the largest SARS-CoV-2 deletions to date and provides compelling evidence that this region is not essential for within-host replication of SARS-CoV-2.
The article by Nussenblatt et al adds to the growing collection of case reports of prolonged SARS-CoV-2 infection, which include people treated with immunosuppressive drugs for leukemia and lymphoma or for autoimmune disease [2, 3], and people with immunosuppression due to untreated human immunodeficiency virus type 1 infection [9]. Interestingly, in the latter case report, SARS-CoV-2 infection resolved within a few weeks of starting effective antiretroviral therapy. Few cohort-based studies and no randomized treatment trials have been conducted in these populations, and critical knowledge gaps remain about them. What are the risk factors for persistent infection and what are the best management strategies to control viral replication in patients with chronic SARS-CoV-2 infection? Answering these questions is urgent, not only for the individual patients, but also for the public health response to the pandemic because of the potential role these infections play in generating new variants of concern.
The report by Nussenblatt et al appears just as the Omicron variant is sweeping across the globe with lightning speed. In its evolutionary leap, Omicron resembles Alpha and the other variants of concern, but it is much more extreme, with an unprecedented number of mutations that distinguish it from its nearest known neighbor. It occurs on a long branch of the SARS-CoV-2 phylogeny, leaving little trace of precursor strains bearing intermediate numbers of mutations. Yet, like other variants before it, the mutational changes that comprise this extreme evolutionary jump are familiar, similar, or identical to those that have arisen from situations in which large viral populations have been propagated in the face of antibody pressure, both in experimental settings [10] and in infected hosts [2].
If Alpha, Omicron, and other variants of concern resulted from an extreme long-tail of infectivity in a small number of patients, it would be a reminder of the outsized consequence of rare events. The COVID-19 pandemic itself—in which millions of deaths resulted from a coronavirus outbreak that was first noted in a single market—has been a case study of this phenomenon. Yet even if variants evolve by some other mechanism, or their origins remain obscure, the insight about the pandemic that comes from studying mutations in SARS-CoV-2 sampled from chronic infections in immunocompromised hosts remains as valuable as ever, serving an oracular function in the pandemic. Thus, in this time of increasing turbulence wrought by SARS-CoV-2 variants, and with the availability of new classes of therapeutic monoclonal antibodies and small-molecule antiviral drugs on the horizon, wise scientists would do well to heed an ancient practice: Consult frequently the oracle and consider carefully her predictions.
Notes
Acknowledgments. The authors thank Dr Caterina Strambio De Castillia for generation of the Venn diagram.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant number R37AI147868) and by the Massachusetts Consortium on Pathogen Readiness and the China Evergrande Group.
Potential conflicts of interest. The authors: No potential conflicts of interest.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Volz E, Mishra S, Chand M, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021; 593:266–9. [DOI] [PubMed] [Google Scholar]
- 2. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021; 592:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drew WL. Ganciclovir resistance: a matter of time and titre. Lancet 2000; 356:609–10. [DOI] [PubMed] [Google Scholar]
- 5. Hammer AS, Quaade ML, Rasmussen TB, et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis 2021; 27:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mandolo J, Msefula J, Henrion MYR, et al. SARS-CoV-2 exposure in Malawian blood donors: an analysis of seroprevalence and variant dynamics between January 2020 and July 2021. BMC Med 2021; 19:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nussenblatt V, Roder AE, Das S, et al. . Year-long COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B cell depletion. medRxiv [Preprint]. Posted online 5 October 2021. doi: 10.1101/2021.10.02.21264267. [DOI] [PMC free article] [PubMed]
- 8. McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021; 371:1139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karim F, Moosa MYS, Gosnell BI, et al. . Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv [Preprint]. Posted online 4 June 2021. doi: 10.1101/2021.06.03.21258228. [DOI]
- 10. Weisblum Y, Schmidt F, Zhang F, et al. . Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020; 9:e61312. [DOI] [PMC free article] [PubMed]