Abstract
We describe rapid detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant using targeted spike single-nucleotide polymorphism polymerase chain reaction and viral genome sequencing. This case occurred in a fully vaccinated and boosted returning traveler with mild symptoms who was identified through community surveillance rather than clinical care.
Keywords: genomic surveillance, multiplex PCR, Omicron, SARS-CoV-2, vaccine
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant was identified in November 2021 and classified as a variant of concern (VOC) by the World Health Organization on 26 November 2021 [1]. The Omicron variant has been rapidly detected throughout the world, has demonstrated a concerning rise in frequency in some locations, and contains an unusually large number of mutations in the spike gene [2], some of which have been associated with increased transmissibility and partial immune evasion in other SARS-CoV-2 lineages. Critical work is ongoing to understand the epidemiological, clinical, virological, and immunological implications of this variant.
CASE DESCRIPTION
We identified a previously healthy woman in her 30s who presented with upper respiratory symptoms following a trip to South Africa and was diagnosed with coronavirus disease 2019 (COVID-19) on return to Fulton County, Georgia, in November 2021, 4 days after identification of Omicron as a VOC. She was fully vaccinated with the Pfizer-BioNTech BNT162b2 mRNA vaccine as of February 2021 and had received a booster of the same vaccine in early October 2021. She had a negative polymerase chain reaction (PCR) test prior to departing the United States and wore a medical-grade procedure mask and glasses on all flights. She stayed in a hotel in Cape Town for 6 days and attended an indoor, unmasked event with 9 family members and friends, 4 of whom subsequently also tested positive for SARS-CoV-2. She had a negative PCR test 48 hours prior to return travel while asymptomatic but then developed mild congestion on her last day in Cape Town, followed by a sore throat during travel home the next day. After arrival in Georgia, she also began experiencing nausea, fatigue, cough, and myalgias. She therefore had nasopharyngeal PCR testing performed at a community testing site on day 3 after symptom onset, which was positive. This sample was unable to be located during a contact tracing investigation. She never developed fevers, shortness of breath, or chest discomfort and did not require medical attention. All symptoms had resolved by day 9, with the exception of ongoing myalgias and significant fatigue.
MOLECULAR TESTING
The patient was enrolled in a research study for emerging pathogens (Emory Institutional Review Board STUDY00022371). A mid-turbinate nasal swab was obtained on day 4 after symptom onset. Nucleic acids were extracted from 400 µL of swab sample and tested using real-time reverse-transcription (rRT) PCR for the SARS-CoV-2 N2 target as previously described [3], yielding a cycle threshold value of 22.2. The sample was also tested for specific single-nucleotide polymorphisms (SNPs) in the spike receptor binding motif using the previously described multiplex rRT-PCR Spike SNP assay [4, 5]. Briefly, the Spike SNP assay uses a single primer pair to amplify a 348bp region and tiled hydrolysis probes to differentiate mutations that are associated with VOCs and variants of interest (VOIs).
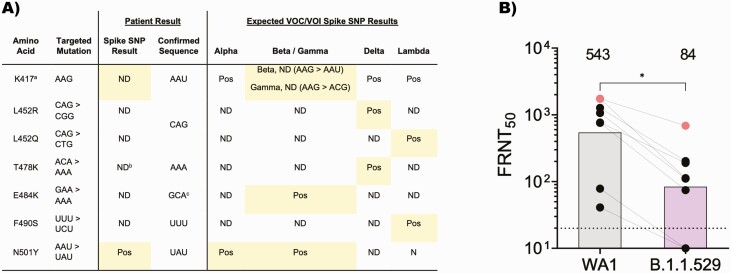
Within 24 hours after sample collection, Spike SNP results from this sample detected a variant sequence at amino acid position 417 and the mutation conferring N501Y (Figure 1A). It did not detect the mutations conferring L452R/L452Q, E484K, or F490S. This pattern of SNPs is consistent with the sequence expected for the Omicron variant and different from results observed with prior VOCs/VOIs (Figure 1A) [6–8]. Interestingly, the T478K mutation was not detected in this sample, though 90% of Omicron sequences bear this mutation.
Figure 1.
A, Mutations identified in the Spike SNP assay. Results from Spike SNP and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome sequencing are shown for this patient’s sample, in comparison with the pattern of mutations detected by Spike SNP for previous VOCs/VOIs. aProbe for 417 is designed to detect the ancestral sequence and is negative in the presence of mutations conferring K417N/T. bPredicted false-negative result due to mutation at position 477, which lies within this probe sequence. cOmicron variant contains E484A and yields an expected negative result for E484K. B, Neutralization responses against WA1/2020 and B.1.1.529 SARS-CoV-2 variants. The FRNT50 geometric mean titer (GMT) for sera from the patient (gray dots) and 7 fully vaccinated control patients (black dots) are shown against WA1/2020 and B.1.1.529. The connecting lines represent matched serum samples. The horizontal dashed line indicates the limit of detection (FRNT50 GMT = 10). Vertical bars and numbers above them indicate the mean FRNT50 GMT across all samples. Nonparametric tests were performed using a Kruskal-Wallis test with Dunn’s multiple comparisons test. ∗P < .05. Abbreviations: FRNT50, focus reduction neutralization test; ND, not detected; Pos, positive; SNP, single-nucleotide polymorphism; VOC, variant of concern; VOI, variant of interest.
SARS-CoV-2 sequencing libraries were generated using the SuperScript IV First Strand Synthesis kit (Thermo Fisher) followed by the Swift Amplicon SARS-CoV-2 Research Panel (Swift Biosciences) [9]. Sequencing was performed with paired-end 150bp reads on an Illumina MiSeq, and 1 034 420 reads were generated for this sample. The consensus SARS-CoV-2 genome was assembled using the viralrecon analysis pipeline [10] with reference sequence MN908947.3.
Within 72 hours after sample collection, the SARS-CoV-2 lineage from this sample’s sequence was classified as B.1.1.529, and the variant was classified as Omicron using cov-lineages.org [11]. The sequence of the spike protein confirmed results from the multiplex Spike SNP PCR assay (Figure 1A) and additionally detected both T478K and the adjacent mutation S477N. The negative result for T478K by the Spike SNP assay is most likely because the mutation conferring S477N is located within the probe for T478K, preventing its binding. Overall, the spike sequence from this sample contained all mutations originally described for the Omicron variant, including amino acid substitutions at 30 positions; deletions at positions 69–70, 143–145, and 212; and insertion EPE after residue R214. Across the entire SARS-CoV-2 genome, the sequence from this sample did not harbor any unique mutations compared with available Omicron reference genomes (N = 538, downloaded from Global Initiative on Sharing All Influenza Data [GISAID] 2021-12-05).
NEUTRALIZATION SUSCEPTIBILITY
Virus from the patient’s nasal swab was successfully isolated in Vero-TMPSS2 cells (Supplementary Methods). Its neutralization susceptibility was tested to serum collected from the patient on day 12, as well as sera from 7 individuals who had undergone a similar vaccination regimen as the patient, with 2 doses plus a booster of the Pfizer-BioNTech BNT162b2 mRNA vaccine (Supplementary Table 1). The patient’s post-infection serum was able to neutralize autologous virus with a focus reduction neutralization test (FRNT50) geometric mean titer (GMT) of 687, substantially higher than neutralization of this virus by sera from the other 7 individuals (Figure 1B). In all cases, neutralization of WA1 was 2.5- to 10-fold higher than the Omicron variant. For all sera, FRNT50 values for neutralization of B.1.617.2 and B.1.351 variants were lower than WA1 and higher than B.1.1.529 (Supplementary Figure 1).
DISCUSSION
In summary, we rapidly detected the SARS-CoV-2 Omicron variant in a patient with compatible epidemiological exposure using a combination of multiplex SARS-CoV-2 Spike SNP PCR and viral genome sequencing. This was the first case detected by SARS-CoV-2 genomic surveillance in the state of Georgia. Identifying this case required eliciting an appropriate travel history and being able to identify and perform sequencing for COVID-19 patients in the community, since the patient had mild symptoms and did not seek clinical care. This underscores the importance of strong public health surveillance systems across a variety of settings.
This case also emphasizes the importance of ongoing research to answer key clinical questions about the virulence of the Omicron variant and the efficacy of existing vaccines. The patient had symptomatic infection despite having completed vaccination with the Pfizer-BioNTech mRNA vaccine, including a booster approximately 6 weeks prior to symptom onset. She experienced only mild illness, consistent with reports that the SARS-CoV-2 Omicron variant is associated with fewer presentations to care and hospital admissions [12]. However, there is currently too little data to draw conclusions about the clinical severity of the SARS-CoV-2 Omicron variant in vaccinated, unvaccinated, and previously infected individuals. Consequently, further study remains crucial. Reassuringly, our patient mounted a robust antibody response against autologous virus, and her post-infection serum was also able to neutralize other VOCs.
Our work highlights the strength of existing systems for SARS-CoV-2 genomic surveillance and also illustrates mechanisms for overcoming potential logistical barriers. For example, despite improvements in turnaround time, viral genome sequencing generally takes several days. In this case, the SARS-CoV-2 genome sequence and lineage classification were available approximately 72 hours after sample collection. However, the Spike SNP PCR assay takes only 2 hours and 12 minutes to run; in our laboratory, this is typically performed alongside testing for the N2 target. This protocol allows the preliminary lineage classification to be completed within 24 hours after sample collection and at the same time as SARS-CoV-2 detection. One remarkable feature of the Omicron variant is the large number of mutations in the spike gene, and this allowed us to leverage our existing Spike SNP assay to clearly distinguish Omicron from Alpha, Delta, and other variant samples. Importantly, the Spike SNP assay can be modified to include new mutations as needed. For example, a new T478K probe has been designed to account for the single base pair mutation in the codon for S477N, and this is predicted to detect both Omicron and Delta variants with equal efficiency. The Spike SNP assay can also be readily redesigned to use a previously developed probe for the ancestral sequence at position 484. Similar to the current 417 probe, this probe would result in signal dropout in the presence of mutations conferring E484K/Q/A, which have been identified among different variants in 2021. Thus, targeted molecular assays can serve as critical tools for flexible, rapid, and high-throughput screening of samples prior to SARS-CoV-2 genome sequencing.
Finally, while the scope and throughput of SARS-CoV-2 genomic surveillance in the United States have made incredible strides, these efforts fundamentally rely on the availability of samples and corresponding metadata. Not all positive samples will enter the pipelines for SARS-CoV-2 genomic surveillance due to the appropriate abundance of SARS-CoV-2 testing options, including the soon-to-be increased availability of home testing. As illustrated by our case, redundant systems for sample collection are essential, including the rapid recruitment of patients into research studies.
Overall, our work illustrates the strengths of a flexible, multimodal approach in the detection and characterization of emerging SARS-CoV-2 variants, which will inform ongoing and future public health surveillance. In addition to their clinical and public health benefits, surveillance efforts also serve as a critical foundation for studies that characterize the capacity of the SARS-CoV-2 Omicron variant to escape natural immunity to prior strains and acquire immunity to vaccines, including boosters.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are very grateful to the patient for participating in this study and granting permission to present this information. We thank staff of the Emory Vaccine Center Hope Clinic for assistance with sample processing.
Financial support. This work was supported by Centers for Disease Control and Prevention (CDC) contract 75D30121C10084 under BAA ERR 20-15-2997; by National Institutes of Health (NIH) grants U54 EB027690 02S1, U54 EB027690 03S1, U54EB027690 03S2, and UL1TR002378; by NIH P51 OD011132, 3U19AI057266-17S1, 1U54CA260563; NIH/National Institute of Allergy and Infectious Diseases (NIAID) Centers of Excellence for Influenza Research and Response (CEIRR)\\Center of Excellence for Influenza Research and Surveillance (CEIRS) contracts HHSN272201400004C and 75N93021C00017 from the NIAID NIH; by intramural funding from the NIAID (Daniel Douek); Emory Executive Vice President for Health Affairs Synergy Fund award; by the Pediatric Research Alliance Center for Childhood Infections and Vaccines and Children’s Healthcare of Atlanta; by the Emory-University of Georgia Center of Excellence for Influenza Research and Surveillance (Atlanta, Georgia); by COVID (coronavirus disease 2019)-Catalyst-I3 Funds from the Woodruff Health Sciences Center and Emory School of Medicine; and by the Emory Woodruff Health Sciences Center COVID-19 Urgent Research Engagement Center, which is supported by generous donations from the O. Wayne Rollins Foundation and the William Randolph Hearst Foundation.
Potential conflicts of interest. M. S. S serves on the advisory boards for Moderna and Ocugen (payments made to self). N. R. reports grants or contracts made to their institution from NIH, CDC, Quidel, Lilly, Pfizer, Sanofi Pasteur, and Merck outside of the submitted work and participation on a data safety monitoring board or advisory board for EMMES, LLC and ICON (payments made to self), and ARLG (unpaid). M. E. S. reports consulting fees for providing COVID infection prevention advice to State Farm Arena, the Atlanta Braves, and Tyler Perry Studios in 2020 and early 2021 (payments were made to Emory Healthcare, not to self). All remaining authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. Severe acute respiratory syndrome coronavirus 2 sequence data generated in this study are available at GISAID under accession EPI_ISL_7171744 and in GenBank under BioProject PRJNA634356.
Contributor Information
Mary Elizabeth Sexton, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Jesse J Waggoner, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Ludy R Carmola, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Phuong Vi Nguyen, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Ethan Wang, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Dara Khosravi, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Azmain Taz, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
Robert A Arthur, Emory Integrated Computational Core, Emory Integrated Core Facilities, Emory University, Atlanta, Georgia, USA.
Mit Patel, Center for Childhood Infections and Vaccines of Children’s Healthcare of Atlanta, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
Venkata Viswanadh Edara, Center for Childhood Infections and Vaccines of Children’s Healthcare of Atlanta, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA; Emory Vaccine Center, Emory University, Atlanta, Georgia, USA; Yerkes National Primate Research Center, Atlanta, Georgia, USA.
Stephanie L Foster, Center for Childhood Infections and Vaccines of Children’s Healthcare of Atlanta, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA; Emory Vaccine Center, Emory University, Atlanta, Georgia, USA; Yerkes National Primate Research Center, Atlanta, Georgia, USA.
Kathryn M Moore, Center for Childhood Infections and Vaccines of Children’s Healthcare of Atlanta, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA; Emory Vaccine Center, Emory University, Atlanta, Georgia, USA; Yerkes National Primate Research Center, Atlanta, Georgia, USA.
Matthew Gagne, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Jesmine Roberts-Torres, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Amy R Henry, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Sucheta Godbole, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Daniel C Douek, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Nadine Rouphael, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Hope Clinic of the Emory Vaccine Center, Emory University School of Medicine, Decatur, Georgia, USA.
Mehul S Suthar, Center for Childhood Infections and Vaccines of Children’s Healthcare of Atlanta, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA; Emory Vaccine Center, Emory University, Atlanta, Georgia, USA; Yerkes National Primate Research Center, Atlanta, Georgia, USA; Department of Microbiology and Immunology, Emory University, Atlanta, Georgia, USA.
Anne Piantadosi, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia, USA.
References
- 1. World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. Accessed 6 December 2021.
- 2. Latif AA, Mullen JL, Alkuzweny M, et al. B.1.1.529 lineage report. https://outbreak.info/situation-reports?pango=B.1.1.529. Accessed 7 December 2021.
- 3. Waggoner JJ, Stittleburg V, Pond R, et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020; 26:1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martinez M, Nguyen P-V, Su M, et al. SARS-CoV-2 variants in Paraguay: detection and surveillance with a readily modifiable, multiplex real-time RT-PCR. medRxiv 2021. doi: 10.1101/2021.09.15.21263618 [DOI] [Google Scholar]
- 5. Babiker A, Immergluck K, Stampfer SD, et al. Single-amplicon multiplex real-time reverse transcription-PCR with tiled probes to detect SARS-CoV-2 spike mutations associated with variants of concern. J Clin Microbiol 2021; 59:e0144621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep 2021; 70:95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438–43. [DOI] [PubMed] [Google Scholar]
- 8. Wilhelm A, Toptan T, Pallas C, et al. Antibody-mediated neutralization of authentic SARS-CoV-2 B.1.617 variants harboring L452R and T478K/E484Q. Viruses 2021; 13:1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Addetia A, Lin MJ, Peddu V, Roychoudhury P, Jerome KR, Greninger AL.. Sensitive recovery of complete SARS-CoV-2 genomes from clinical samples by use of Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. J Clin Microbiol 2020; 59:e02226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patel H, Varona S, Monzón S, et al. nf-core/viralrecon: nf-core/viralrecon v2.2 - Tin Turtle. 2021. [Google Scholar]
- 11. O’Toole A, Hill V, Pybus OG, et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res 2021; 6:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 33. 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1043680/technical-briefing-33.pdf. Accessed 27 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.