Abstract
Background
The purpose of this study was to explore the reason for guanine nucleotide binding‐protein gamma subunit‐4 (GNG4) overexpression and the relationship between GNG4 overexpression and the poor prognosis of lung adenocarcinoma (LUAD) patients.
Methods
The genes and phenotypes related to GNG4 expression in patients with lung adenocarcinoma were analyzed by bioinformatics. The phenotype indicated by bioinformatic analysis was confirmed by experiments.
Results
GNG4 expression is elevated in lung adenocarcinoma, and overexpressed GNG4 is related to the poor prognosis of patients with lung adenocarcinoma. The hypoxic microenvironment of lung adenocarcinoma can promote GNG4 expression and GNG4 promotes the migration and proliferation of lung adenocarcinoma cells.
Conclusions
GNG4 expression in lung adenocarcinoma was significantly higher than in paired adjacent tissues. GNG4 overexpression is associated with a variety of malignant phenotypes of lung adenocarcinoma. Increased GNG4 expression is related to the hypoxic microenvironment in lung adenocarcinoma.
Keywords: GNG4, HIF‐1A, lung adenocarcinoma
The differences in the hypoxic microenvironment and immune cell infiltration of lung adenocarcinoma shape the heterogeneity among different patients. By analyzing the above conditions, we screened out genes that were differentially expressed in different modes of tumor microenvironment, and verified their influence on the malignant biological behavior of tumor cells.

INTRODUCTION
Lung adenocarcinoma (LUAD) is the most common cause of death from cancer, accounting for more than 40%. 1 In the past decade, advances in diagnosis and treatment technology have greatly improved the prognosis of lung cancer patients, especially targeted therapy. 2 However, LUAD has a strong ability to proliferate and metastasize. 3 , 4 , 5 , 6 Once it develops to an advanced stage, the effect of drug treatment is very poor. Behind this is the synergy of multiple genes and the mechanism has not yet been clearly explained. 5 Identification and discovery of corresponding genes for targeted therapy is an important strategy to improve LUAD patient outcome. 7
Guanine nucleotide binding‐protein gamma subunit‐4 (GNG4) is a member of the guanine nucleotide‐binding protein complex. 8 Previous studies have found that GNG4 expression is elevated in a variety of tumors and is related to the poor prognosis of these patients, including bladder, liver, breast and non‐small‐cell lung cancers. 7 , 9 , 10 , 11 , 12 Most studies confirmed the significance of high GNG4 expression by analyzing the correlation of sequencing data and clinical data and their related mechanisms. Maina et al. confirmed that GNG4 is the target gene of VHL and is related to the response pathways. The hypoxic microenvironment is one of the important microenvironmental characteristics of lung cancer, which is closely related to the poor prognosis of lung cancer patients. In‐depth analysis of the mechanism of the hypoxic microenvironment and targeted measures are of great significance in predicting the prognosis of lung cancer patients and improving the current status of patients' treatment. 13
The study by Zhao et al. considered GNG4 was essential for the tumor immune microenvironment in colorectal cancer (CRC). 10 The immune microenvironment of LUAD also has an important role in supporting the growth, invasion, and metastasis of cancer cells. 14 The hypoxic and immune microenvironments are linked to each other to further promote tumor progression. Therefore, we studied the role of GNG4 in hypoxia and the immune microenvironment in LUAD.
METHODS
Cells and tissues
Human LUAD cell lines A549 and H1299 were purchased from American Type Culture Collection (ATCC). Cells were cultured in an incubator at 37°C and 5% CO2 using RPMI1640 medium with 10% fetal bovine serum (FBS). The shRNA sequence of GNG4 is 5′‐ CCGGGCGGGAAGATCCTCTCATCATCTCGAGATGATGA GAGGATCTTCCCGCTTTTTG −3′.
Chromatin immunoprecipitation (ChIP)
We analyzed the binding site of HIF‐1A (ab243860, Abcam) in the promotor region of GNG4 by the JASPAR database and designed primers of the region,Forward primer: 5′‐CCTTTGTGATCTCCGCGTTC‐3′, reverse primer: 5′‐ATTTCGGGAAGATCTGGGCG‐3′. We then performed the ChIP experiment using a EZ ChIP kit (Merk 17–371) according to the manufacturer’s instructions.
RT‐qPCR
Cells and tissues were dissolved with TRIzol (Thermo Fisher),and mRNAs were then extracted and reverse transcripted to cDNA by cDNA synthesis SuperMix (Trans, AE301). Expression of GNG4 was detected by q‐PCR using Green qPCR SuperMix (Trans, AQ101) with CFX96 system according to the manufacturer's instructions. The forward primer was 5′‐ACAGCACCACTAGCATCTCC‐3′ and reverse was 5′‐GGCACTGGAATGATGAGAGG‐3′.
Western blot
Cells were lysed by RIPA lysis buffer with cocktail and the protein concentration was quantified with Pierce protein assay kit (Pierce). Then, 20 μg protein lysates were added to each lane of SDS PAGE. The antibodies used in this study were GNG4 polyclonal antibody(PA5‐103877; Thermo Fisher Scientific) at a dilution with 1:800 and β‐tubulin (MG7) mouse monoclonal antibody(RM2003,Ray Antibody Biotech) 1:5000.
Wound healing assay
Wound healing assay was performed according to the previously published protocol. 15 Cells were cultured by 1640 without FBS and the experiments were repeated independently at least three times.
Transwell assay
Transwell assay was performed as described by Zhao et al. 16 Inserts with 8.0 μM pore size were used in the study. Then, 1.0 × 105 cells in 200 μl medium were added to the upper chambers and cultured with RPMI 1640 medium without FBS and 500 μl RPMI 1640 with 10% FBS was added to each lower chamber. Cells were strained by three‐step‐strain set (Thermo Scientific).
Bioinformatic analysis
The String database (Version 11) was used in the study. 17 The packages used in this study were Consensus Cluster Plus, 18 survival package (version 3.2–10), ggplot2 package, DEaeq2 package, 19 ComplexHeatmap, 20 GSVA package. 21 Immune infiltration analysis was performed using CIBERSORT. 22 GSEA analysis was performed as previously described. 23 , 24
Statistical analysis
Mean values of paired data were compared by Student's t‐test and the Chi‐square method was used to analyze categorical data. All experiments were conducted independently at least three times. Analyses were performed using SPSS (version 19.0).
RESULTS
Group lung adenocarcinoma samples from TCGA database according to hypoxia gene expression status and identify differences between groups
We analyzed the RNA‐seq data and the corresponding clinical prognosis information in the TCGA‐LUAD data set. The clinicopathological information of the patients are shown in Table 1. The hypoxia‐related gene list is from the GSEA HALLMARK‐hypoxia gene set. We used the STRING online database and Cytoscape software to analyze the protein–protein interactions between the genes in the gene set and further constructed a protein interaction network (PPI) (Figure 1a) and screened out the 50 most adjacent nodes in the network. These genes are referred to as hub genes (Figure 1b). The TCGA‐LUAD samples were analyzed by cluster analysis according to the expression patterns of hub‐gene in each sample, and the corresponding cumulative distribution function curves (Figure S1a) when divided into 2–9 groups were calculated, and the area under the curve increased value (Figure S1b) and the consistency matrix were also shown (Figure 1C, Figure S1c). Based on the above results, we chose to divide all samples into two clusters, and combined the clinical information in order to calculate the overall survival period of the two groups. Among them, the OS of cluster 2 was shorter and statistically significant (Figure 1d), suggesting that cluster 2 and cluster 1 may be in a state of high and low hypoxia.
TABLE 1.
The clinicopathological information of TCGA‐LUAD patients
| Characteristic | Levels | Overall |
|---|---|---|
| n | 513 | |
| T stage, n (%) | T1 | 168 (32.9%) |
| T2 | 276 (54.1%) | |
| T3 | 47 (9.2%) | |
| T4 | 19 (3.7%) | |
| N stage, n (%) | N0 | 330 (65.9%) |
| N1 | 95 (19%) | |
| N2 | 74 (14.8%) | |
| N3 | 2 (0.4%) | |
| M stage, n (%) | M0 | 344 (93.2%) |
| M1 | 25 (6.8%) | |
| Pathological stage, n (%) | Stage I | 274 (54.3%) |
| Stage II | 121 (24%) | |
| Stage III | 84 (16.6%) | |
| Stage IV | 26 (5.1%) | |
| Gender, n (%) | Female | 276 (53.8%) |
| Male | 237 (46.2%) | |
| Age, n (%) | <=65 | 238 (48.2%) |
| >65 | 256 (51.8%) | |
| Age, median (IQR) | 66 (59, 72.75) |
FIGURE 1.
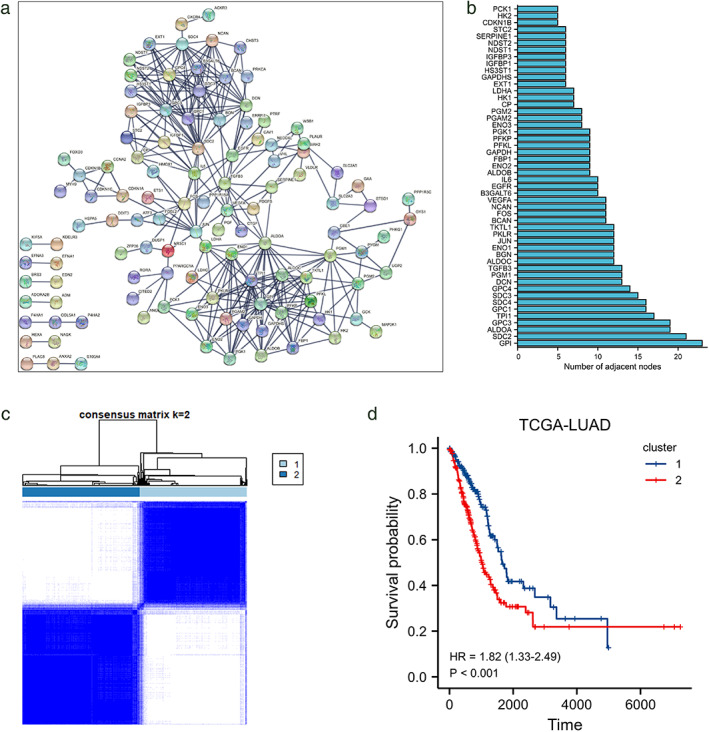
Classification based on expression patterns of hypoxic genes. (a) Protein–protein interaction analysis of genes contained in the HALLMARK_HYPOXIA pathway in the GSEA hallmark gene set (version v7.4) in the STRING database. (b) Screen the top 50 genes with the largest number of adjacent nodes in the network. (c) The LUAD samples in TCGA database were analyzed by cluster analysis and OS survival analysis (d)
We further analyzed the expression changes of the genes contained in the KEGG HIF‐1 signaling pathway in cluster 2 relative to cluster1. Among the 15 genes contained in the “increase oxygen delivery” gene set, nine were highly expressed in cluster 2, and 11 of the 13 genes contained in “reduce oxygen consumption” were more expressed in cluster 2 than in cluster 1 (Figure 2a). At the same time, single‐ and multifactor COX analysis was carried out on the influence of 50 hub‐genes on OS in TCGA‐LUAD samples (Figure 2b, c), and heat maps were drawn for genes with statistically significant results (Figure 2d). The genes with HR value greater than 1 were highly expressed in cluster 2 and the HR values of genes highly expressed in cluster 1 were all less than 1. Based on these results, cluster2 was defined as the hypoxia‐high group, and cluster1 was defined as the hypoxia‐low group. On this basis, we identified the differentially expressed genes of cluster 2 relative to cluster 1, of which 1288 genes were highly expressed in the hypoxia‐high group, with low expression in 1079 genes in the hypoxia‐high group (Figure 2e).
FIGURE 2.
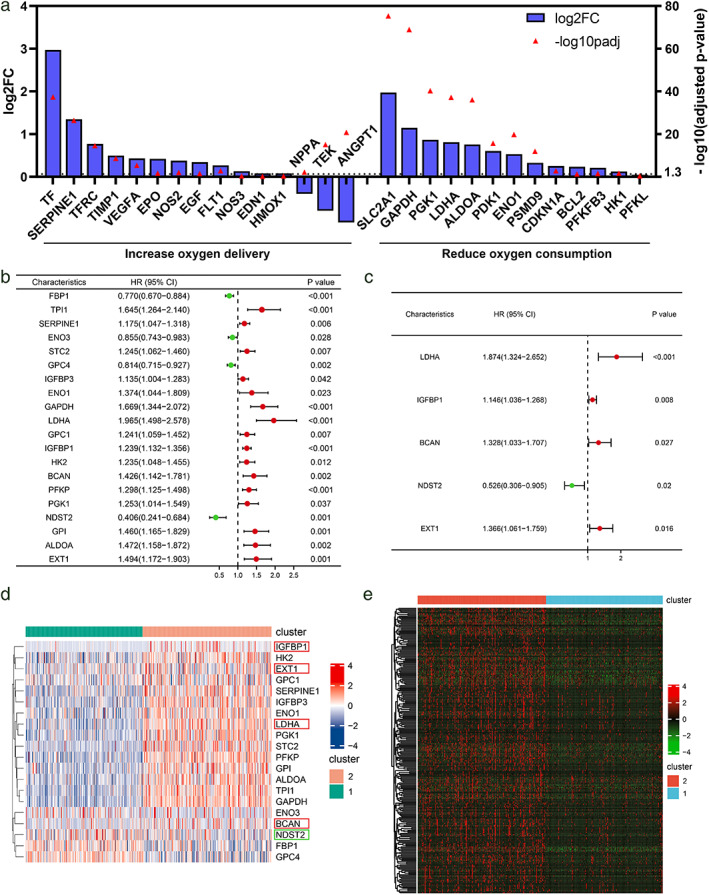
Different gene expression patterns in two clusters. (a) Fold change of 28 genes in the HIF1A signaling pathway between different clusters. (b) and (c) COX analysis of the hub‐gene combined with the clinical data of TCGA‐LUAD. (d) Heat map of the expression of statistically significant genes in cluster1 and cluster2 in univariate COX analysis. (e) Heat map of all genes upregulated in cluster2 relative to cluster1
Immune scoring analysis of lung adenocarcinoma samples from TCGA database
The immune score of TCGA‐LUAD samples was calculated using the ESTIMATE method to evaluate the degree of immune infiltration in each sample. In all samples, the immune score was distributed in the range of −1355.85 to 3286.67. On the basis of integrating the survival information of each sample, the best cutoff value of immune score was determined to be 1246.27 through the method of maximally selected rank statistics, and all samples were divided into immune‐high and immune‐low groups (Figure 3a). Further survival analysis showed that the survival of the immune‐high group was better than that of the immune‐low group (Figure 3b). A total of 684 genes were highly expressed in the immune‐high group, with low expression of 921 genes in the hypoxia‐high group (Figure 3C).
FIGURE 3.
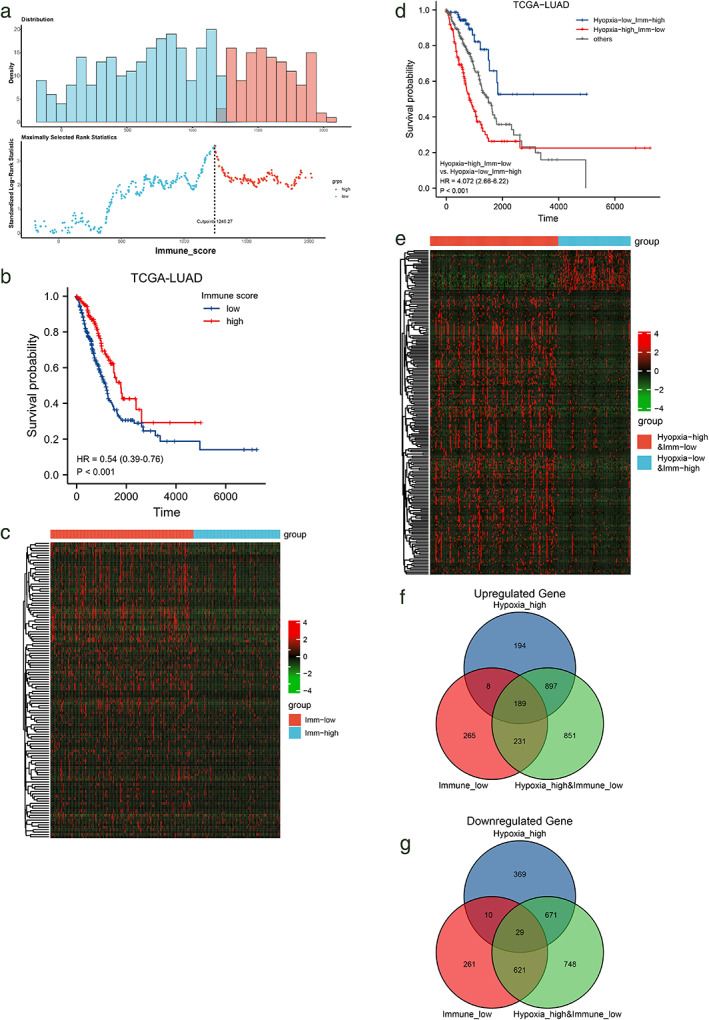
Immunological and hypoxic groups. (a) ESTIMATE immune scores. (b) Survival curve of the high/low immune score group. (c) Heat map of all upregulated genes in the low immune score group. (d) OS survival curve of hypoxia‐high&Imm‐low, hypoxia‐low&Imm‐high and others, (e) and (f) The intersection of up‐ and downregulated genes of the groups. (g) Heat map of expression of up‐ and downregulated genes in hypoxia‐high&Imm‐low, hypoxia‐low&Imm‐high group
At the same time, we used the CIBERSORT algorithm to calculate the proportion of various types of immune cell infiltration in each TCGA‐LUAD sample (Figure S2a) and evaluated the proportion of different immune cells and their relationship with ESTIMATE matrix score and ESTIMATE immune score. The ESTIMATE immune score was positively correlated with memory B cells, CD4+ T cells, CD8+ T cells, M1 macrophages, and Trag cells; it had a positive correlation with initial B and plasma cells, and activated dendritic cells etc. had a strong negative correlation (Figure S2b). In group comparison, CD8+ T cells, CD4+ T cells, monocytes and M1 macrophages were increased in the immune‐high group while B cells naïve, plasma and Th cells were decreased (Figure S2c). It is therefore suggested that the better prognosis of the immune‐high group may be related to the more active immune response mediated by T cells and macrophages.
Hypoxia‐immune related grouping and identification of differential genes
According to the above classification results of hypoxia and immune status, we further combined these two indicators and divided all cases into three groups, namely hypoxia‐low & immune‐high (80 cases), hypoxia‐high & immune‐low (142 cases) and others (224 cases) of the three groups. Survival analysis showed that the difference in overall survival was statistically significant. The overall survival of the hypoxia‐high & immune‐low group was significantly shorter than that of hypoxia‐low & immune‐high group (Figure 3d), suggesting hypoxia and immunity status may have the opposite effect on the prognosis of patients.
In order to identify hypoxia‐immune‐related differentially expressed genes, we performed differential analysis of the gene expression of hypoxia‐high & immune‐low relative to hypoxia‐low & immune‐high samples (Figure 3e), and combined hypoxia‐related differential genes and immune‐related differential genes to extract intersection, 189 upregulated intersection genes (Figure 3f) and 29 downregulated intersection genes (Figure 3g) were obtained. The results of single‐factor COX regression analysis of the above genes showed that the analysis results of 30 genes out of 189 upregulated genes were statistically significant. Among them, 28 genes had HR values greater than 1, and their high expression are risk factors that affects the prognosis of patients. (Figure S3a); The analysis results of 11 genes out of 29 downregulated genes are statistically significant, with HR values less than 1, and their low expression is a risk factor that affects the prognosis of patients (Figure S3b). Based on the above results, we defined the differential genes whose expression was upregulated as the “risk differential gene set” and the differential genes whose expression was downregulated as the “protective gene set”.
In order to further study the key genes that may exist in the “risk gene set”, we analyzed the protein–protein interaction between each gene and constructed a protein interaction network (Figure 4a) and counted the genes that constitute the network. The number of adjacent nodes (Figure 4b) shows that the GNG4 gene is at the core of the network.
FIGURE 4.
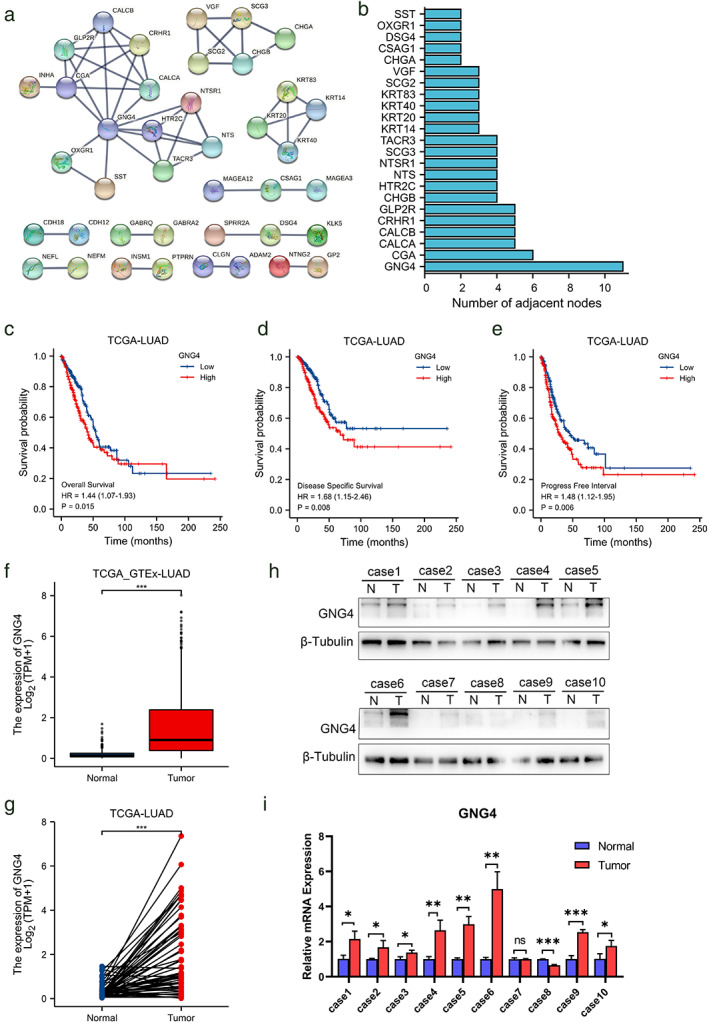
GNG4 gene was identified in LUAD. (a) Protein–protein interaction analysis of the genes in Figure 3. (b) Number of adjacent nodes of genes in the network. (c)–(e) The OS, DSS and PFI of different GNG4 expression in TCGA‐LUAD. (f) GNG4 expression of LUAD tumorous and nontumorous lung tissues in TCGA and GTEx database and in paired tissues in TCGA database (g). (h) Western blot analysis showed GNG4 protein expression levels in 10 total paired human LUAD tumorous and matched adjacent nontumorous tissues and mRNA levels (i)
Expression and main functional phenotype of GNG4 gene in LUAD
We first analyzed the influence of the GNG4 gene in the TCGA database on the prognosis of LUAD patients. The baseline data of the GNG4 high expression and low expression groups are shown in Table 2. Overall survival, disease specific survival and progression‐free survival of the GNG4 high expression group were all shorter (Figure 4c–e), suggesting that high GNG4 expression may be a risk factor affecting the prognosis. The analysis performed in the unpaired samples of cancer and adjacent cancers in the TCGA combined with GTEx database (Figure 4f) and the matched samples in the TCGA database (Figure 4g) showed that the expression level of GNG4 in cancerous tissues was significantly higher than that in adjacent tissues. The results have statistical significance. Our research group conducted Western blot (Figure 4h) and qPCR (Figure 4i) experiments on the cancer and paracancerous surgical specimens collected from 10 LUAD patients and confirmed that the mRNA and protein expression levels of the GNG4 gene in the cancer tissues were higher than those normal tissues adjacent to the cancer of most patients.
TABLE 2.
The baseline data of the GNG4 high expression and low expression groups from TCGA‐LUAD
| Characteristic | Levels | Low expression of GNG4 | High expression of GNG4 | p‐value |
|---|---|---|---|---|
| n | 256 | 257 | ||
| T stage, n (%) | T1 | 88 (17.3%) | 80 (15.7%) | 0.249 |
| T2 | 131 (25.7%) | 145 (28.4%) | ||
| T3 | 28 (5.5%) | 19 (3.7%) | ||
| T4 | 7 (1.4%) | 12 (2.4%) | ||
| N stage, n (%) | N0 | 185 (36.9%) | 145 (28.9%) | < 0.001 |
| N1 | 37 (7.4%) | 58 (11.6%) | ||
| N2 | 27 (5.4%) | 47 (9.4%) | ||
| N3 | 0 (0%) | 2 (0.4%) | ||
| M stage, n (%) | M0 | 179 (48.5%) | 165 (44.7%) | 0.084 |
| M1 | 8 (2.2%) | 17 (4.6%) | ||
| Pathological stage, n (%) | Stage I | 155 (30.7%) | 119 (23.6%) | 0.004 |
| Stage II | 57 (11.3%) | 64 (12.7%) | ||
| Stage III | 31 (6.1%) | 53 (10.5%) | ||
| Stage IV | 9 (1.8%) | 17 (3.4%) | ||
| Gender, n (%) | Female | 139 (27.1%) | 137 (26.7%) | 0.892 |
| Male | 117 (22.8%) | 120 (23.4%) | ||
| Age, n (%) | <=65 | 103 (20.9%) | 135 (27.3%) | 0.005 |
| >65 | 144 (29.1%) | 112 (22.7%) | ||
| Age, median (IQR) | 68 (60.5, 74) | 64 (57.5, 71.5) | < 0.001 |
We further conducted bioinformatic analysis on the relationship between GNG4 gene expression and various signaling pathways. Among them, the results of GSEA analysis showed that high expression of GNG4 is associated with EMT, G2M checkpoint, mitosis, hypoxia and other HALLMARK signaling pathways (Figure 5a), and cell cycle, DNA replication, P53 signaling pathway and mismatch repair and other KEGG pathways (Figure 5b) related to enrichment. ssGSEA analysis showed that GNG4 was positively correlated with the infiltration of Th2 T cells, Tgd cells, and CD56dim NK cells, and negatively correlated with the infiltration of DC cells, mast cells, and eosinophils (Figure S4a).
FIGURE 5.
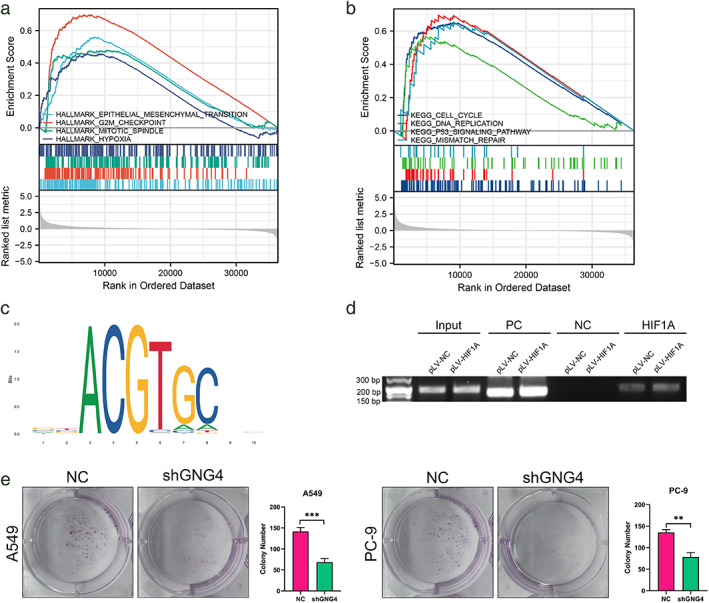
Influencing factors that regulate GNG4. (a) and (b) GSEA analysis of different GNG4 expression group in TCGA‐LUAD. (c) Predicted binding motif of HIF‐1 in the promoter region of GNG4 by JASPAR database. (d) Chromatin immunoprecipitation analysis showed HIF‐1 binding to GNG4 promoter in A549 cells. (e) Clone formation experiment showed reduced proliferation capacity in A549‐shGNG4 and PC‐9‐shGNG4 cells
Based on the above results, we first analyzed the regulation effect of the hypoxia‐inducible factor HIF‐1A on GNG4 under hypoxia and performed ChIP experiments (Figure 5D) for the DNA binding site of HIF‐1A (Figure 5c). The results show that HIF‐1A has a transcriptional regulatory effect on GNG4.
We studied the function of the gene by constructing a stable GNG4 downexpression line. The results showed that in the plate cloning experiment using A549 and PC‐9 cells, the proliferation ability of the GNG4‐reduced expression group was significantly reduced (Figure 5e). Analysis of GNG4 expression in patients with different T, N, and M stages showed that there was no significant difference in GNG4 expression in different T stages (Figure 6a), and N2, N3, and N4 stages were relative to N1 stages (Figure 6b) and the expression of GNG4 in M1 relative to M0 staging (Figure 6c) increased. Based on the above results and GSEA analysis conclusions, we used A549 and PC‐9 cells to perform scratch (Figure 6d, e) and cell migration experiments (Figure 6f, g). The results showed that the motility of cells and other cells was significantly weakened after GNG4 expression was reduced.
FIGURE 6.
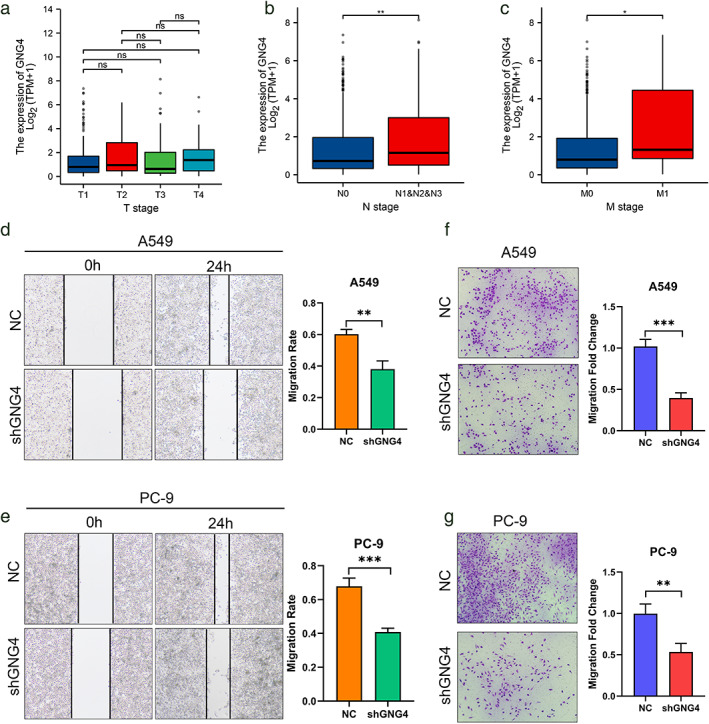
GNG4 affects the malignant phenotype of cancer cells. (a) Expression of GNG4 in different T stages of LUAD and different N stages (b) and different M stages (c). (d) Wound‐healing assays comparing the motility of A549/A549‐shGNG4 cells and PC‐9/PC‐9‐shGNG4 cells (e). (f) Comparison of migration potential of A549/A549‐shGNG4 cells and PC‐9/PC‐9‐shGNG4 cells (g)
DISCUSSION
In recent years, more studies have focused on the interaction mechanism and influence between cancer cells and the tumor microenvironment, especially immune cells, and related experimental techniques and bioinformatic algorithms have gradually matured. Our research shows that CD4 T cells, CD8 T cells and M1 Macrophages in Imm‐high group increased significantly compared to Imm‐low group in the calculation results, suggesting that the formation of high immune status may relate to the killing effect of immune cells to cancer cells, and so on. Therefore, the evaluation of tumor immune status may help patients receive more targeted immune therapy or targeted therapy.
It has been reported that GNG4 is overexpressed in multiple kinds of tumors as demonstrated here and that overexpressed GNG4 is associated with poor patient prognosis. In those studies and our results, GNG4 might therefore be an oncogene. However, according to the study by Maina et al., GNG4 might be a tumor suppressor gene. GNG4 has been approved to be a target gene of VHL. VHL inhibits GNG4 expression and renal cell carcinoma cell growth was suppressed when GNG4 and MLC2 were re‐expressed. 25 But in multiple other tumors, the role of GNG4 is different. In gall bladder cancer, PSMC2 promoted gall bladder cancer through the regulation of GNG4 and predicted poor patient prognosis. Growth of gall bladder cancer cells was inhibited when expression of GNG4 was downregulated, and was inhibited further when PSMC2 was knocked down at the same time. 9
In colorectal cancer, GNG4 overexpression is associated with poor prognosis and can be a biomarker. The overall survival rate of the igh GNG4 expression group was significantly lower than low GNG4 expression group. A similar result has been shown in liver cancer 12 and also other cancers. 11 , 26
As for lung cancer, GNG4 expression had also been demonstrated to be overexpressed in NSCLC. 27 However, why GNG4 is overexpressed in LUAD and the reason for this is still unclear. Our results have discussed the significance of GNG4 overexpression in LUAD. The increase in GNG4 expression may enhance the proliferation ability of cancer cells and may change the invasion and migration ability of cancer cells by affecting their epithelial mesenchymal transformation status, thus promoting tumor metastasis. These phenotypic changes may be part of the reason for the poor prognosis of patients with high GNG4 expression. However, the mechanism of GNG4 promoting tumor progression needs to be further explored.
CONFLICT OF INTEREST
No authors report any conflict of interest.
Supporting information
Figure S1 The computational process of unsupervised clustering, (a) CDF curve under different groups and the increase of the area of CDF curve compared to the previous grouping number (b) (c) Gene expression matrix when the number of groups is 3–9 respectively
Figure S2 Prediction of immune cell infiltration and its correlation, (a) The infiltration ratio in TCGA‐LUAD sample, (b) The infiltration ratio and ESTIMATE matrix score in TCGA‐LUAD samples and their correlation. (c) Differences in the proportion of different immune cell infiltration between Imm‐high and Imm‐low groups
Figure S3 Univariate COX regression analysis of differential genes, (a) Single‐factor COX regression analysis of the up‐ and downregulated genes in Figure3
Figure S4 Correlation between GNG4 immune cell infiltration, Correlation between GNG4 expression and immune cell infiltration analyzed by ssGSEA
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (grant nos. 82072657) and Natural Science Foundation of Tianjin (19JCYBJC26200).
Zhou B, Zhu W, Yuan S, Wang Y, Zhang Q, Zheng H, et al. High GNG4 expression is associated with poor prognosis in patients with lung adenocarcinoma. Thorac Cancer. 2022;13:369–379. 10.1111/1759-7714.14265
Bodong Zhou, Wenbo Zhu, and Shuai Yuan are these authors contributed equally to this work and should be considered co‐first authors.
Funding information National Natural Science Foundation of China, Grant/Award Number: 82072657; Natural Science Foundation of Tianjin City, Grant/Award Number: 19JCYBJC26200
Contributor Information
Lei Zhu, Email: zhulei0519@163.com.
Jie Xu, Email: xujie508@126.com.
REFERENCES
- 1. Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao JJ, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7(6):596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pakkala S, Ramalingam SS. Personalized therapy for lung cancer: striking a moving target. JCI Insight. 2018;3(15):e120858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Auperin A, Le Péchoux C, Rolland E, Curran WJ, Furuse K, Fournel P, et al. Meta‐analysis of concomitant versus sequential radiochemotherapy in locally advanced non‐small‐cell lung cancer. J Clin Oncol. 2010;28(13):2181–90. [DOI] [PubMed] [Google Scholar]
- 4. Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol. 2013;31(8): 992–1001. [DOI] [PubMed] [Google Scholar]
- 5. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei Y, et al. HIF‐1a‐regulated miR‐1275 maintains stem cell‐like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/beta‐catenin and Notch signaling. Theranostics. 2020;10(6):2553–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang L, Xu H, Ma C, Zhang J, Zhao Y, Yang X, et al. Upregulation of deubiquitinase PSMD14 in lung adenocarcinoma (LUAD) and its prognostic significance. J Cancer. 2020;11(10):2962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalyanaraman S, Copeland NG, Gilbert DG, Jenkins NA, Gautam N. Structure and chromosomal localization of mouse G protein subunit gamma 4 gene. Genomics. 1998;49(1):147–51. [DOI] [PubMed] [Google Scholar]
- 9. Zhu D, Gu X, Lin Z, Yu D, Wang J. High expression of PSMC2 promotes gallbladder cancer through regulation of GNG4 and predicts poor prognosis. Oncogenesis. 2021;10(5):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao H, Danli S, Qian Z, Ye S, Chen J, Tang Z. Identifying GNG4 might play an important role in colorectal cancer TMB. Cancer Biomark. 2021;32(4):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X, Kang X, Jin L, Bai J, Zhang H, Liu W, et al. ABCC9, NKAPL, and TMEM132C are potential diagnostic and prognostic markers in triple‐negative breast cancer. Cell Biol Int. 2020;44(10):2002–10. [DOI] [PubMed] [Google Scholar]
- 12. Tanaka H, Kanda M, Miwa T, Umeda S, Sawaki K, Tanaka C, et al. G‐protein subunit gamma‐4 expression has potential for detection, prediction and therapeutic targeting in liver metastasis of gastric cancer. Br J Cancer. 2021;125(2):220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ancel J, Perotin JM, Dewolf M, Launois C, Mulette P, Nawrocki‐Raby B, et al. Hypoxia in lung cancer management: a translational approach. Cancers (Basel). 2021;13(14):3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y, Li N, Wang TM, Di L. Natural products with activity against lung cancer: a review focusing on the tumor microenvironment. Int J Mol Sci. 2021;22(19):10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang S, Huang XY. Ca2+ influx through L‐type Ca2+ channels controls the trailing tail contraction in growth factor‐induced fibroblast cell migration. J Biol Chem. 2005;280(29):27130–7. [DOI] [PubMed] [Google Scholar]
- 16. Zhao T, Ren H, Li J, Chen J, Zhang H, Xin W, et al. LASP1 is a HIF1alpha target gene critical for metastasis of pancreatic cancer. Cancer Res. 2015;75(1):111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta‐Cepas J, et al. STRING v11: protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
- 21. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. [DOI] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maina EN, Morris MR, Zatyka M, Raval RR, Banks RE, Richards FM, et al. Identification of novel VHL target genes and relationship to hypoxic response pathways. Oncogene. 2005;24(28):4549–58. [DOI] [PubMed] [Google Scholar]
- 26. Kishibuchi R, Kondo K, Soejima S, Tsuboi M, Kajiura K, Kawakami Y, Kawakami Y et al., DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common epigenetic alteration in thymic carcinoma. Int J Oncol. 2020;56(1):315–326. [DOI] [PubMed] [Google Scholar]
- 27. Zhang JJ, Hong J, Ma YS, Shi Y, Zhang DD, Yang XL, et al. Identified GNGT1 and NMU as combined diagnosis biomarker of non‐small‐cell lung cancer utilizing bioinformatics and logistic regression. Dis Markers. 2021;2021:6696198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The computational process of unsupervised clustering, (a) CDF curve under different groups and the increase of the area of CDF curve compared to the previous grouping number (b) (c) Gene expression matrix when the number of groups is 3–9 respectively
Figure S2 Prediction of immune cell infiltration and its correlation, (a) The infiltration ratio in TCGA‐LUAD sample, (b) The infiltration ratio and ESTIMATE matrix score in TCGA‐LUAD samples and their correlation. (c) Differences in the proportion of different immune cell infiltration between Imm‐high and Imm‐low groups
Figure S3 Univariate COX regression analysis of differential genes, (a) Single‐factor COX regression analysis of the up‐ and downregulated genes in Figure3
Figure S4 Correlation between GNG4 immune cell infiltration, Correlation between GNG4 expression and immune cell infiltration analyzed by ssGSEA


