Abstract
Background
A protective antibody response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is crucial to decrease morbidity and mortality from severe coronavirus disease 2019 (COVID-19) disease. The effects of preexisting anti-human coronavirus (HCoV) antibodies on the SARS-CoV-2–specific immunoglobulin G (IgG) responses and severity of disease are currently unclear.
Methods
We profiled anti-spike (S), S1, S2, and receptor-binding domain IgG antibodies against SARS-CoV-2 and 6 HCoVs using a multiplex assay (mPLEX-CoV) with serum samples from SARS-CoV-2 infected (n = 155) and pre–COVID-19 (n = 188) cohorts.
Results
COVID-19 subjects showed significantly increased anti-S SARS-CoV-2 IgG levels that were highly correlated with IgG antibodies against OC43 and HKU1 S proteins. However, OC43 and HKU1 anti-S antibodies in pre–COVID-19 era sera did not cross-react with SARS-CoV-2. Unidirectional cross-reactive antibodies elicited by SARS-CoV-2 infection were distinct from the bidirectional cross-reactive antibodies recognizing homologous strains RaTG13 and SARS-CoV-1. High anti-OC43 and anti-S2 antibody levels were associated with both a rapid anti–SARS-CoV-2 antibody response and increased disease severity. Subjects with increased sequential organ failure assessment (SOFA) scores developed a higher ratio of S2- to S1-reactive antibodies.
Conclusions
Early and rapid emergence of OC43 S- and S2-reactive IgG after SARS-CoV-2 infection correlates with COVID-19 disease severity.
Keywords: SARS-CoV-2, common cold human coronaviruses (HCoVs), cross-reactive antibody response, immune imprinting
A rapid and robust increase in SARS-COV-2 anti-S2 subunit IgG antibodies after infection is highly associated with disease severity. This is likely due to preexisting cross-reactive humoral immunity against human betacoronaviruses OC43 and HKU1, leading to early, nonneutralizing IgG production.
Infection with the human coronavirus (HCoV) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause coronavirus disease (COVID-19). Most infected individuals have only mild symptoms, but some quickly develop severe pneumonia associated with high mortality [1, 2]. At the time of this study, COVID-19 has caused over 240 million known infections and over 5.7 million deaths worldwide [3]. Thus, understanding immunity to HCoVs is critical to developing vaccine strategies and therapeutic approaches.
SARS-CoV-2 belongs to the zoonotic Coronaviridae family [4]. The HCoVs include SARS- CoV-1 and Middle Eastern respiratory syndrome (MERS) virus, which are both beta-HCoVs and can cause severe acute respiratory syndrome (SARS). Four common HCoVs in 2 classes, beta (OC43, HKU1) and alpha (229E, NL63) [5], circulate in the human population, causing 30% of mild upper respiratory infections [6, 7]. Although HCoV infections occur throughout life, antibody seroconversion rates vary greatly by age and geography [6, 8–10].
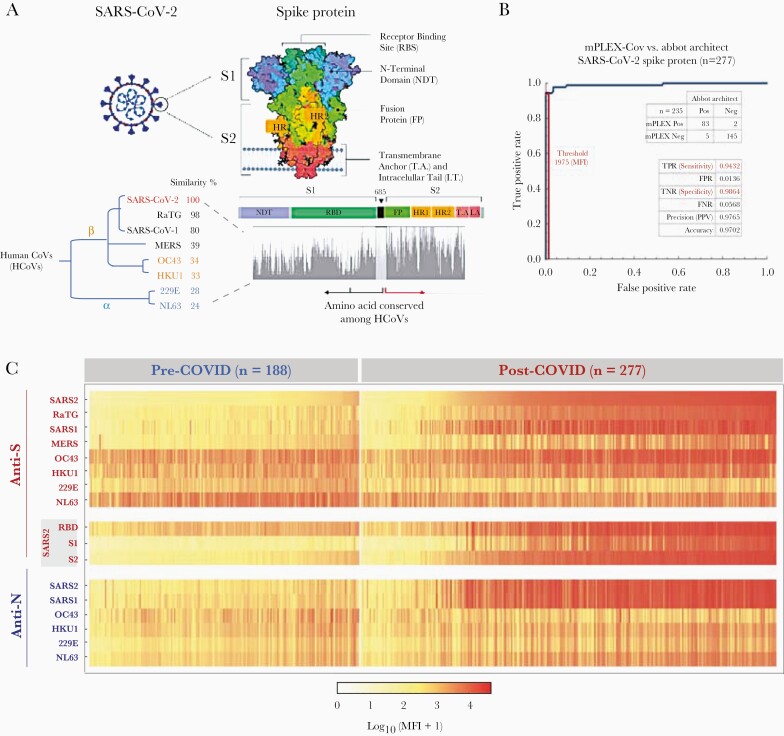
The immunodominant antigens for HCoVs include the nucleocapsid (N) and the homotrimeric glycoprotein spike (S) viral protein antigens [4]. HCoVs share S- and N-protein sequence homologies and antibodies against HCoV S and N proteins are found in varying frequencies across all age groups [6, 7]. The SARS-CoV-2 S protein has a receptor-binding domain (RBD) on the N-terminal S1 subunit, which mediates viral binding via high-affinity interaction with the host cell surface angiotensin-converting enzyme 2 (ACE2). In contrast, the S2 subunit is responsible for virus-cell membrane fusion [11] and displays more sequence homology between HCoV strains than the S1 subunit (Figure 1A) [12, 13].
Figure 1.
Structure of SARS-CoV-2 spike protein (S) and protein homology analysis from common human coronaviruses (HCoVs). A, The trimerized S (spike) protein structure and the phylogenetic groups of HCoVs used in this study. The homologous protein analysis and the phylogenic tree were generated with Unipro UGENE V33.0. B, The sensitivity and specificity of multiplex assay for the SARS-CoV-2 anti-spike antibody detection receiver operating characteristic curve with the Abbot ARCHITECT test for SARS-CoV-2 N (nucleocapsid) protein antibodies as the reference standard to account for acute infection. C, The anti-S and -N IgG repertoires against HCoVs in the pre–COVID era and COVID-19 cohorts. Each column represents a serum sample; each row is the IgG reactivity against an HCoV S- or N- protein. The heat-map shows the mean of duplicate median fluorescence intensity (MFI) measurements from the mPLEX-CoV multiplex assay after background subtraction. Abbreviations: SARS2, SARS-CoV-2; SARS1, SARS-CoV-1; RBD, Receptor binding domain.
Several studies suggest an association between anti–SARS-CoV-2 S immunoglobulin G (IgG) levels and COVID-19 disease severity [1, 2, 14, 15], raising concerns that some patterns of humoral immunity may be associated with increased morbidity and mortality during infection [16]. Less clear is the contribution of cross-reactive antibody responses to common HCoVs in the time course and severity of initial COVID-19 disease. OC43 anti-S IgG levels correlate with an increase in both SARS-CoV-2–specific IgG levels and disease severity in acutely ill COVID-19 patients [17]. In contrast, others report that immune imprinting to conserved HCoV epitopes is negatively correlated with the induction of IgG and IgM against the SARS-CoV-2 S protein [18], and that primary SARS-CoV-2–specific anti-S antibody responses in acutely ill patients target homologous epitopes on the S2 segment [14, 19, 20].
The goal of this study was to better understand the effect of preexisting HCoV-reactive IgG antibodies on the development of humoral immune responses to, and the pathogenesis of, SARS-CoV-2 infection. Specifically, we tested the hypothesis that preexisting anti-OC43 S protein and anti–SARS-CoV-2 S2 segment IgG correlate with the severity of illness in hospitalized patients, as measured by sequential organ failure assessment (SOFA) scores [21]. To this end, we profiled anti-S and anti-N IgG levels against multiple HCoVs in sera from pre–COVID-19 and acute COVID-19 cohorts using a Luminex-based multiplex assay [22]. We describe high seroconversion rates of β-HCoVs OC43 and HKU1 anti-S antibodies in the pre–COVID-19 era cohort and evidence to support a potential role for immune imprinting in de novo SARS-CoV-2 antibody responses.
METHODS
Human Subjects Protection
The collection of pre–COVID-19 and COVID-19 infected subjects’ serum samples, and secondary use of infant serum samples, were approved by the University of Rochester Research Subjects Review Board (RSRB protocols 00004836 and 00058437).
Pre–COVID-19 Era Cohort
Serum samples (n = 188) from before December 2019 were obtained. In addition to healthy subjects (n = 55), these included sera from subjects positive for bacterial and viral infections (Lyme disease, syphilis, cytomegalovirus, Epstein-Barr virus, respiratory syncytial virus, and influenza) and autoimmune disease markers (antinuclear antigen and rheumatoid factor) that might create false-positives for anti–SARS-CoV-2 S reactivity (Table 1). The SARS-CoV-2, SARS-CoV-1 anti-S, and anti-N cutoff values were set at 6 standard deviations above the negative sample mean, using serum samples from infants younger than 12 months (n = 16; Supplementary Table 2).
Table 1.
The Study Cohorts
| Cohort Characteristics | No. Total (Male/Female) | Age, y, Mean/Median |
|---|---|---|
| Pre–COVID-19 | ||
| Cytomegalovirus | 10 (4/6) | 52/55 |
| Epstein-Barr virus | 10 (4/6) | 55/51 |
| Respiratory syncytial virus | 10 (6/4) | 51/55 |
| Lyme disease | 10 (2/8) | 49/52 |
| Syphilis | 10 (4/6) | 43/55 |
| Autoimmune disease, ANA | 19 (9/10) | 45/52 |
| Autoimmune disease, RF | 9 (5/4) | 45/43 |
| Acute viral respiratory illness | 55 (20/25) | 48/62 |
| Othera | 55 (22/23) | 55/58 |
| Total | 188 (80/108) | 49/52 |
| COVID-19 | ||
| Outpatients | 42 (19/23) | 68/70 |
| Inpatients | 33 (19/14) | 68/66 |
| ICU | 57 (25/32) | 67/67 |
| Death | 23 (11/12) | 74/67 |
| Total | 155 (88/73) | 68/67 |
One sample was obtained from each pre–COVID-19 cohort subject.
Abbreviations: ANA, antinuclear antigen-positive sera; COVID-19, coronavirus disease 2019; ICU, intensive care unit; RF, rheumatoid factor-positive sera.
Samples with no serological diagnosis.
COVID-19 Cohort
Serum samples were collected from 155 symptomatic subjects positive for SARS-CoV-2 nucleic acid (Cobas SARS-CoV-2; Roche Molecular Systems) and SARS-CoV-2 N protein antibody (ARCHITECT test i2000SR platform; Abbot Laboratories [23]) from April to June 2020. A total of 155 acute COVID-19 patients with 277 longitudinal serum samples (days from symptom onset [DFSO], ≤ 42) were divided into 4 cohorts by illness severity and mortality: (1) outpatients that were never hospitalized (n = 42); (2) inpatients with moderate disease (n = 33); (3) an intensive care unit (ICU) group (n = 57); and (4) a severe COVID-19 group that died in hospital (n = 23). We quantified disease severity using the maximum daily SOFA score for all hospitalized subjects. Subject demographics were abstracted from the electronic medical record (Table 2). Serum samples were stored at 4°C or −80°C until analysis.
Table 2.
Demographics of COVID-19 Cohort and Subgroups of Acute COVID-19 Patients
| Characteristic | Outpatient | Inpatient | ICU Survivor | Death, Nonsurvivor | Total |
|---|---|---|---|---|---|
| Subjects | |||||
| No. | 42 | 33 | 57 | 23 | 155 |
| Age, y, mean/median | 69/70 | 69/70 | 67/67 | 70/68 | |
| Male/female | 23/19 | 13/20 | 28/29 | 10/13 | |
| Maximum SOFA scorea | 0 | 2.12 + 3.12 | 5.48 + 3.90 | 6.78 + 3.84 | |
| Median SOFA score | 0 | 1 | 5 | 7 | |
| Serum samples | |||||
| No. | 47 | 45 | 162 | 23 | 277 |
| DFSO = 0–7 | 15 | 14 | 36 | 9 | 74 |
| DFSO = 8–14 | 12 | 7 | 42 | 2 | 63 |
| DFSO > 14 | 17 | 13 | 72 | 12 | 114 |
| UNK | 3 | 11 | 12 | 0 | 26 |
Some COVID-19 cohort subjects had longitude samples. Total No. serum samples = 277.
Abbreviations: COVID-19, coronavirus disease 2019; DFSO, days from symptom onset; ICU, intensive care unit; SOFA, sequential organ failure assessment; UNK, samples without DFSO information.
Mean + standard deviation.
Anti-HCoV IgG Multiplex Assay
We estimated the levels of IgG antibodies against S and N proteins of SARS-CoV-2 and other HCoVs using the multiplex assay (mPLEX-CoV) [22]. In brief, we cloned and expressed S proteins of SARS-CoV-2, SARS-CoV-1, the homologous RaTG CoV (positive control), MERS, and 4 seasonal HCoVs (OC43, HKU1, NL63, and 229E) using a baculovirus insect expression system (Supplementary Table 1), and commercial proteins for SARS-CoV-2 S1, S2, and RBD domains (Sino Biological). All proteins were coupled to Luminex beads (xMAP Antibody Coupling kit; Luminex) at 40 mole/million beads. Protein-coupled beads were incubated with 1:1000 diluted duplicate serum samples. Bound IgG was detected with phycoerythrin-conjugated anti-human IgG (Southern Biotech), and the median fluorescence intensity (MFI) was read on a Luminex MagPix (Figure 1B).
To determine the sensitivity and specificity of the mPlex-CoV assay we generated receiver operating characteristic (ROC) curves for the SARS-CoV-2 anti-S antibody with SARS-CoV-2 N protein antibody Abbott ARCHITECT test as the gold standard. ROC curves were generated using the pre–COVID-19 and polymerase chain reaction (PCR)-positive COVID-19 era cohort samples (Mathematica version 12.0.0; Wolfram Research).
Statistical Methods
We estimated correlations between anti-HCoV and anti–SARS-CoV-2 IgG levels using Pearson correlation coefficients. For subjects with more than 2 longitudinal samples, we used the mean IgG antibody MFI level of each HCoV strain. Immune repertoires of the pre–COVID-19 era and COVID-19 cohorts were visualized using multidimensional scaling analyses of anti-S and anti-N reactive anti-HCoV IgG levels [24]. Density plots of anti-S and anti-N IgG HCoV repertoires of the pre–COVID-19 era and COVID-19 cohorts were compared using the bootstrap method. Generalized estimating equation models were used to compare anti-S IgG levels among COVID-19 severity groups (outpatients, inpatients, severe/ICU, death) grouped by sample days from symptom onset (0–7, 8–14, 15+ days). Generalized linear mixed-effects models were used to evaluate the association of maximum SOFA score with anti-S IgG levels. Statistical analyses were conducted using SAS version 9.4 (SAS Institute) and R (R Core Team, 2017), and hypotheses tests were 2-sided with P < .05.
RESULTS
Pre–COVID-19 and COVID-19 Cohorts
Pre–COVID-19 era cohort sera were collected from 188 subjects (mean age 49 years). To assess for false-positive SARS-CoV-2 anti-S results, sera were selected from pre–COVID-19 era subjects with a history of other bacterial and viral infections (n = 50), with autoantibodies (antinuclear antigen, rheumatoid factor; n = 28), and healthy subjects with no serological diagnosis (n = 55) (Table 1). A positive result for SARS-CoV-2 IgG was set at 6 standard deviations above the mean anti–SARS-CoV-2 (1975 MFI) or N (1128 MFI) protein levels in the pre–COVID-19 era cohort. Consistent with our prior study, we found no false-positive SARS-CoV-2 anti-S or anti-N results [23]. The COVID-19 cohort included 277 serum samples from 155 subjects (mean ± standard deviation of age 62 ± 7 years) with PCR-confirmed acute SARS-CoV-2 infections diagnosed between April and June 2020 (Table 1 and Table 2) [23]. Given the limitations of sample collection, subjects in the outpatient group did not have longitudinal samples collected. The cohorts were not fully matched, including uneven disease distribution across age and risk factors, as well as limitations in pre–COVID-19 era sera availability.
Pre–COVID-19 Era Sera Contain High Anti-S IgG Levels Against Common Human Coronaviruses
To determine if HCoV antibody levels changed after acute SARS-CoV-2 infection, we measured anti-S and anti-N IgG levels in cohort sera against SARS-CoV-2, SARS-CoV-1, MERS, OC43, HKU1, 229E, NL63, and RaTG bat coronavirus (Figure 1) [22, 25, 26]. Prior studies have shown that > 75% of children > 2.5 years old are HCoV seropositive [8, 9, 27]; thus, we used sera from 2 to 3-year-old subjects (n = 11) as negative controls to compare the prevalence of HCoVs in the pre–COVID-19 era cohort (Supplementary Table 2). The adult pre–COVID-19 era sera demonstrated a high antibody prevalence to the 4 common HCoVs (Figure 2C). Over 90% of pre–COVID-19 era subjects had anti-S IgG against OC43 (92.7%) and 229E (93.6%), but fewer had anti-N IgG against OC43 (53.2%) and 229E (16.8%) (Supplementary Table 2), consistent with previous reports [6, 8–10]. The mismatch between anti-S and anti-N IgG to HCoVs suggests that most anti-S IgG developed from distant exposure. The above results suggest that abundant anti-HCoV, but not SARS-CoV-2, antibodies existed in the population prior to the pandemic.
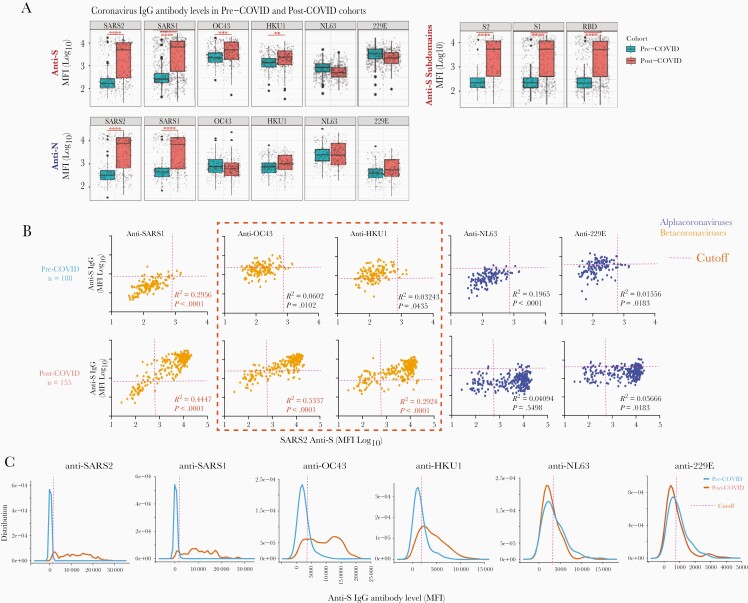
Figure 2.
“Uni-directional” IgG cross-reactivity against spike protein of β-HCoVs after SARS-CoV-2 infection. The mean antibody level for each subject with multiple longitude samples was used to avoid sampling bias for both pre-COVID (n = 188) and COVID-19 (n = 155) cohorts. A, Comparison of SARS-COV-2 and HCoV IgG levels between the pre-COVID-19 and COVID-19 cohorts. Generalized estimating equation models were used for the statistical analysis; ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, and ∗P < .05. B, The correlation of mean anti-S and N IgG levels against HCoVs and SARS-CoV-2 for each individual with multiple samples. Significant correlations (P < .05) are shown in red. The threshold cutoffs for SARS-CoV-2 and SARS-CoV-1 were defined as 5 standard deviations above the mean of all negative samples. The threshold cutoffs of OC43, HKU1, 229E, and NL63 were determined with infant serum samples (<12 month-old, n = 16; Supplementary Table 2). C, Distribution of SARS-COV-2 and HCoV antibody levels between pre-COVID-19 era and COVID-19 cohorts, with the threshold of each HCoV strain. The cutoff value for a positive test for each strain was set at 5 times the standard deviation of the mean measure in negative control infant sera.
β-HCoVs Anti-S IgG Antibody Levels Associated With SARS-CoV-2 Anti-S in COVID-19 Patients
In the acute SARS-CoV-2–infected cohort, the profile of IgG levels against multiple HCoV strains showed that the infection elicited anti-S IgG against SARS-CoV-2, antigenically similar CoV strains (RaTG, SARS-CoV-1), and cross-reactive IgG against the β-HCoV OC43 and HKU1 S-proteins. In contrast, SARS-CoV-2 infection did not increase anti-S IgG against genetically dissimilar α-HCoV (229E, NL63; Figure 2A and 2B). These results were visualized using multidimensional scaling (Supplementary Figure 1B) [22, 25, 26, 28]. Pre–COVID-19 and COVID-19 cohorts could be graphically distinguished by anti-S, but not by anti-N, IgG immune repertoire cartography. As expected, SARS-CoV-2 infection did not change anti-N IgG levels against α- and β-HCoVs.
Pre–COVID-19 Era Unidirectional Cross-Reactivity of OC43 and HKU1 Anti-S IgG Antibodies
SARS-CoV-2 infection significantly increased anti-S IgG levels against β-HCoVs OC43 and HKU1, but not against the α-HCoVs (229E, NL63; Figure 2B). Moreover, high levels of RaTG and SARS-CoV-1–reactive anti-S antibodies were still detected in both a small number of pre–COVID-19 era, and a much larger number of post–COVID-19, subjects. These results suggest that before SARS-CoV-2 infection, OC43 and HKU1 anti-S antibodies did not cross-react with SARS-CoV-2, RaTG, or SARS-CoV-1 at detectable levels, despite their sequence homologies. However, SARS-CoV-2 infection resulted in high levels of anti-S IgG against SARS-CoV-2, SARS-CoV-1, RaTG, and β-HCOVs, but not α-HCoVs. These results suggest that either SARS-CoV-2 infection elicits anti-β-HCoV IgG with low specificity and broad cross-reactivity, or high-affinity IgG against shared epitopes, which are immunodominant in SARS-CoV-2 but not OC43 or HKU1, and are shared by RaTG and SARS-CoV-1.
Elevated OC43 and HKU1 anti-S IgG levels were highly correlated with SARS-CoV-2 anti-S IgG levels in serum samples of COVID-19 patients (Supplementary Figure 1A), demonstrating the same conclusions (Figure 2C). If IgG antibodies cross-reacted with common epitopes across HCoVs, we would expect bidirectional binding to both S proteins. In contrast, we found that while OC43 and HKU1 anti-S IgG antibodies were present in sera from both cohorts, pre–COVID-19 samples lacked reactivity against the SARS-CoV-2 S protein (Figure 2A and 2B). We refer to this as unidirectional reactivity, which is distinct from the cross-reactivity of the sera from COVID-19 patients that bind both SARS-CoV-2 and OC43 [17, 29].
OC43 and HKU1 Anti-S IgG Levels Correlate With the Rapid Increase in SARS-CoV-2 Anti-S IgG in De Novo Infection
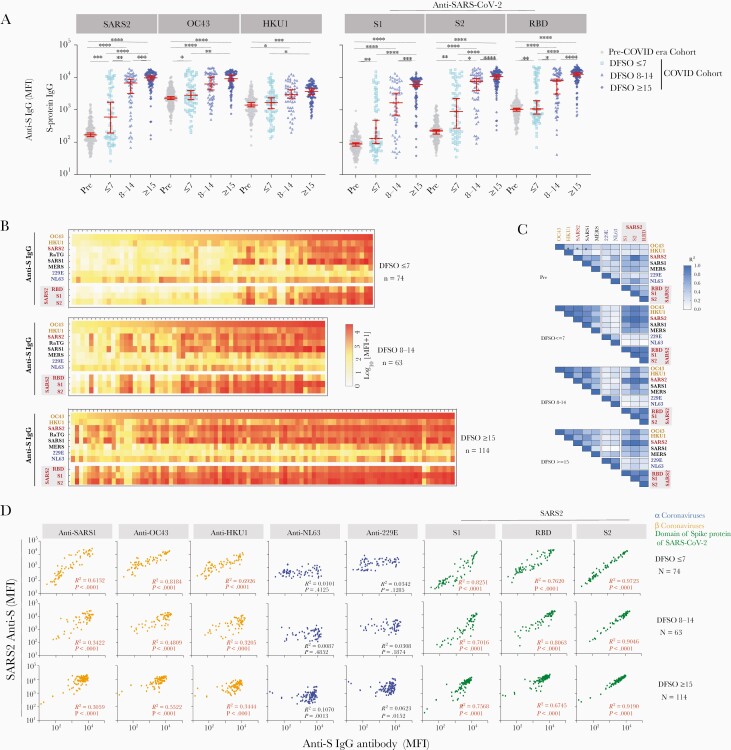
To understand the kinetics of SARS-CoV-2, OC43, and HKU1 anti-S antibodies in acute COVID- 19 subjects, we measured the anti-S IgG repertoire profile against all HCoVs and SARS-CoV-2 in samples binned by DFSO (0–31 DFSO; n = 54 subjects with > 2 longitudinal samples). SARS-CoV-2 anti-S IgG antibodies appeared in 42% (29/69) of patients’ sera within 7 DFSO. Among them, 62% (18/29) of subjects’ SARS-CoV-2 anti-S IgG levels peaked 8–14 DFSO (Figure 3A). Notably, serum samples from subjects with high SARS-CoV-2 IgG levels also showed higher levels of OC43 anti-S antibody (Figure 3B). SARS-CoV-2 anti-S IgG seroconversion occurred within 1– 2 weeks of infection for most subjects. OC43 and HKU1 anti-S antibodies showed a similar pattern but less statistical significance (Figure 3A).
Figure 3.
Kinetics of HCoV-OC43 and HKU1 anti-spike (S) IgG levels correlate with anti-SARS-CoV-2 IgG during acute infection. The profile of S-reactive IgG antibodies against SARS-CoV-2 and common HCoVs in the acute COVID-19 cohort, binned by days from symptoms onset (DFSO). 251 serum samples with known DFSO from 155 subjects in the acute COVID-19 cohort, including 54 subjects who had samples from more than two longitudinal time points ranging from post symptom onset day 0 to day 31. A, The changes in anti-S IgG levels versus SARS-CoV-2, OC43 and HKU1, as well SARS-CoV-2 S protein segments from the S1, S2, and RBD regions. ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, and ∗P < .05). B, Heatmaps of weekly anti-S IgG antibody repertoire against HCoVs, and S protein domains of SARS-CoV-2, sorted by OC43 IgG antibody levels. Each column represents a serum sample from a subject, and each cell represents the average median fluorescence intensity (MFI) of duplicate samples after background subtraction. C, The correlation matrix of anti-S IgG antibodies against OC43 with SARS-CoV-2 and other viruses, binned by the weeks from symptom onset in COVID-19 cohort. The ∗ represents statistically significant correlation (P < .01). D, The correlation of anti-S IgG against SARS-CoV-2 with that against other HCoVs binned by weeks from symptom onset in acute COVID-19 patients. The individual antibody level against each HCoV virus (x-axis) is plotted against the SARS-CoV-2 IgG level (y-axis) from the same subjects. Pearson correlation analysis R-squared and P values are shown, with statistically significant correlations (P < .001) shown in red.
To analyze the correlations between the HCoV and anti-S IgG levels in the rapid-response subjects, we binned serum samples by DFSO and visualized subject antibody profiles (Figure 3B). Within-subject analysis showed a high correlation between anti-β-HCoV (OC43, HKU1) and anti- SARS-CoV-2 spike IgG levels in all 3 time periods, which was not seen with the α-HCoVs (Figure 3C). These results demonstrate that increased OC43 (R2 = 0.8184) and HKU1(R2 = 0.6926) anti-S IgG levels are highly correlated with the rapid appearance of SARS-CoV-2 anti-S IgG during early infection (DFSO ≤ 7). These data support the hypothesis that higher OC43 and HKU1 anti-S antibodies early in COVID-19 disease may accelerate the anti–SARS-CoV-2 S IgG response. After 2 weeks (DFSO ≥ 15), 95% of COVID-19 subjects developed high IgG levels against SARS-CoV-2, OC43, and HKU1 S-proteins. Thus, preexisting IgG targeting OC43 and HKU1 may also influence the de novo antibody response to SARS-CoV-2 infection.
Anti-OC43 S IgG Levels Correlate With COVID-19 Severity
Accumulating data demonstrate that rapid and robust increases in SARS-CoV-2 S-reactive IgG correlate with disease severity and plasmablast expansion [2, 11, 15]. Broadly cross-reactive anti- SARS-CoV-2 antibody responses in patients with more severe COVID-19 [29], and higher OC43 anti-S antibody levels, are also associated with disease severity [17]. To our knowledge, there are no published data demonstrating this correlation between SARS-CoV-2 and HCoV anti-S IgG levels and SOFA scores, a standard measure of disease severity [21].
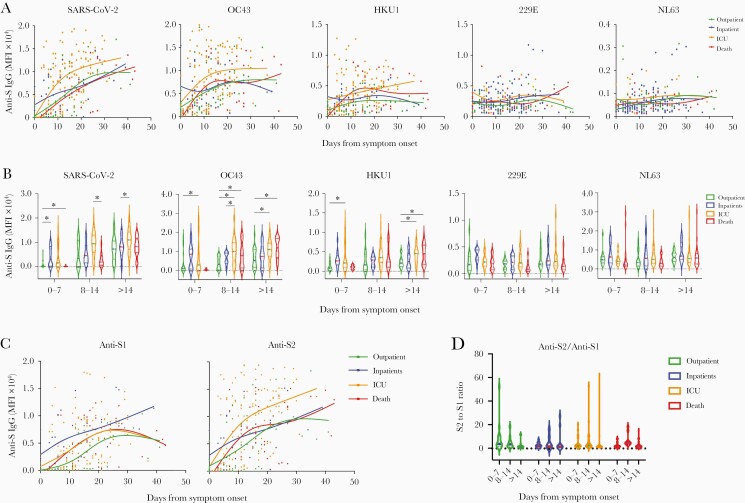
We first profiled the anti-S IgG antibodies against SARS-CoV-2 and other HCoVs across moderate to severe illness within the COVID-19 cohorts: outpatients (n = 42), inpatients (n = 33), ICU (survivors, n = 57), and those who died (n = 23). Each subject had 1–5 serum samples available within 42 days after infection, and all hospitalized subjects (inpatient, ICU, or death) had maximum SOFA scores for the sampling date (Table 2). Figure 4A shows the rapid and robust rise of anti-S SARS-CoV-2 IgG levels in the severe (ICU) versus moderate (in- and outpatient) disease cohorts. Mean maximum SOFA score changes were associated with a 1-unit increase in subject IgG levels against HCoV spike proteins using linear mixed-effects models (Table 3). Notably, changes in SARS-CoV-2 (P = .0112) and OC43 (P = .0047) anti-S IgG were highly associated with the mean change in maximum SOFA scores on the day of serum sampling. We also observed a delayed increase of anti-S SARS-CoV-2 IgG levels in the nonsurvivor group compared with severe disease survivors [15]. Notably, the dynamics of OC43 anti-S IgG over the clinical course of the disease were very similar to S-reactive SARS-CoV-2 IgG, with a rapid rise in antibody levels in the surviving severe patients. The weekly anti-S antibody level comparison results also showed that anti-S antibodies against OC43 and HKU1 were significantly higher in the severe survivors than moderate disease patients (Figure 4B).
Figure 4.
The anti-S IgG repertoire against SARS2 and common HCoVs after SARS2 infection by disease severity. The anti-S IgG antibody response profile to multiple HCoV strains in SARS-CoV-2-infected subjects. Data are stratified by illness severity over days from symptom onset (DFSO). A, Kinetics of HCoV anti-S IgG in acute COVID-19. Each point shows the mean flourescent intensity (MFI) values of a subject’s measurements of S-reactive IgG against each HCoV strain plotted versus DFSO (277 samples; 155 subjects). Plots were smoothed using non-parametric regression (LOWESS) splines with 4 knots, and grouped by disease severity (Out-patients: not-hospitalized; On-patient: hospitalized, ICU: hospitalization in the intensive care unit; Death: non-survivors). B, Correlations between anti-S IgG against multiple HCoVs in serum samples from SARS-CoV-2-infected patients by DFSO, binned by week. Lines indicate means. Differences between groups were calculated by the trend test within the generalized estimating equation model framework: ∗∗P < .01, ∗P < .05 . C, Kinetics of IgG responses against S1 and S2 domains of the SARS-CoV-2 S protein by clinical courses in acute COVID-19 subjects. D, The ratio of IgG antibody level against S2 over S1 domain of SARS-CoV-2 S protein showed a predominant anti-S2 response in the group with higher Sequential Organ Failure Assessment scores, requiring intensive care unit hospitalization.
Table 3.
The Correlation Coefficients (r, Spearman Correlation) and Mean Maximum SOFA Score Changes Associated With 1-Unit Increase in HCoV anti-S IgG Within the COVID-19 Cohort
| Spike Strain or Subunit | Spearman Correlation | Maximum SOFA Change | ||
|---|---|---|---|---|
| r | P Value | Mean | P Value | |
| SARS-CoV-2 S | 0.2249 | .0041 | 0.26 | .0112 |
| OC43 | 0.2326 | .0030 | 0.40 | .0465 |
| HKU1 | 0.2453 | .0017 | 0.33 | .1312 |
| 229E | −0.0136 | .8644 | −0.48 | .1268 |
| MNL63 | 0.0663 | .4036 | −0.07 | .7989 |
| SARS-CoV-2 S1 | 0.2679 | .0006 | 0.18 | .0504 |
| SARS-CoV-2 RBD | 0.2415 | .0020 | 0.23 | .0777 |
| SARS-CoV-2 S2 | 0.2301 | .0033 | 0.27 | .0125 |
Abbreviations: COVID-19, coronavirus disease 2019; HCoV, human coronavirus; IgG, immunoglobulin G; RBD, receptor-binding domain; S, spike; SARS-CoV-2, severe acute respiratory syndrome 2; SOFA, sequential organ failure assessment. Values in bold are statistically significant with P < .05.
Rapidly Elevated Anti-Spike S2-Reactive IgG Correlates With COVID-19 Severity
To further characterize cross-reactive anti–SARS-CoV-2 S IgG, we evaluated binding to the S1, S2, and RBD subunits. As expected, S1, S2, and RBD-reactive IgG levels were highly correlated with the development of anti–SARS-CoV-2 IgG during acute SARS-CoV-2 infection (Figure 2 and Figure 3). Interestingly, we found a higher linear correlation between anti-S compared with anti-S2 IgG levels (R2 = 0.9723) than with anti-S1 (R2 = 0.825). In addition, the dynamics of IgG levels against the S1 and S2 subunits were very different. The trajectory of the S1 antibody was significantly higher in the severe disease (ICU) group with higher SOFA scores compared to the mild disease inpatient group (P < .05, trend testing within the generalized estimating equation framework). However, the S2 antibody increased rapidly in the severe group, leading to an increased S2/S1 IgG ratio (Figure 4C). The S2/S1 ratio decreased over time in outpatients with mild symptoms but increased in the hospitalized inpatients, including high SOFA score ICU patients (Figure 4D). Notably, a higher level of anti-S IgG antibody against the S1 domain of SARS-CoV-2 and other common cold HCoVs did not correlate with illness severity. We could not detect a higher trajectory of anti-S1 antibody in the outpatient cohort in this study. This is an intriguing finding, which should be further investigated by a future analysis with more longitudinal data.
DISCUSSION
We report a multidimensional analysis of pre–COVID-19 and COVID-19 era sera using multiplex analysis to profile comprehensively S- and N-reactive IgG against SARS-CoV-2 and common HCoVs. We found a high prevalence of anti-S and anti-N IgG antibodies against seasonal HCoVs in the pre–COVID-19 era, without cross-reactive binding to SARS-CoV-2 S and N proteins. This occurred despite moderate S-protein sequence homologies between SARS-CoV-2 and the β-HCoVs OC43 (74.9%) and HKU1 (75.7%) [30]. Consistent with recent reports, we also found that SARS-CoV-2 infection was associated with elevated anti–SARS-CoV-2 S IgG and correspondingly high IgG levels against OC43 and HKU1 S proteins [15, 17, 18, 31]. This association was especially prominent in the first week after symptom onset, suggesting that preexisting β-HCoV immunity may contribute to the initial SARS-CoV-2 antibody response. This unidirectional IgG binding was distinct from the bidirectional cross-reactivity after SARS-CoV-2 infection. Post-COVID, there was strong serum reactivity against RaTG13 and SARS-CoV-1, which have 96.1% [4] and 81.2% homology, respectively, with SARS-CoV-2 [32]. Of note, during the first week of infection, increased β-HCoV and S2-reactive IgG highly correlated with SOFA scores, illness severity, and mortality [33–35].
Our analysis found a significant correlation between COVID-19 severity and the kinetics of anti-S1 and anti-S2 IgG levels during acute infection. Early, elevated anti-S2 IgG levels were associated with higher SOFA scores and mortality. Several reports have shown that preexisting S2-specific memory B cells [14] and CD4+ T cells [36–38] contribute to an early immune response strongly biased towards the S2 domain. The high prevalence of anti-S IgG against the similar OC43 and HKU1 β-HCoVs, described here and by others [39], suggests that imprinting may bias an early, nonneutralizing immune response towards S2 epitopes. In contrast, the slower emergence of anti-S1 and anti-RBD IgG in subjects with higher SOFA scores is consistent with the hypothesis that anti-S1 kinetics reflect a primary antibody response. It also suggests that the lag in protective anti-RBD IgG may contribute to increased COVID-19 disease severity. Whether imprinting against S1 and RBD epitopes after vaccination reverses this trend is yet to be determined. We also found unidirectional anti-S IgG binding to HCoV S proteins from OC43 and HKU1 in pre–COVID-19 era sera.
In contrast, COVID-19 sera had strong bidirectional reactivity against OC43, HKU1, and SARS-CoV-2 S proteins. The precise epitopes involved in high cross-reactivity with OC43 and HKU1 β-HCoVs in the COVID-19 sera remain unclear. One possibility is the presence of unique immunodominant epitopes on OC43 and HKU1 S proteins. Alternatively, low-prevalence memory B cells recognizing conserved subimmunodominant epitopes during infection may produce HCoV cross-reactive antibodies, consistent with immune imprinting [18], as described with influenza infection and vaccination [2, 11, 15, 40]. Infected and vaccinated individuals acquire the ability to produce high-affinity specific antibodies against novel epitopes from new virus strains [41]. Such cross-reactive anti-HCoV antibodies have also been isolated from convalesced COVID-19 patients [29].
Relevant to both disease severity and population immunity, the above findings suggest a role for immune imprinting in SARS-CoV-2 anti-S protein IgG responses. Elevated IgG antibodies against OC43 S and SARS-CoV-2 S2 early (<7 days) in acute COVID-19 subjects with high SOFA scores may be due to prior β-HCoV immune imprinting of memory B cells. Broadly cross-reactive antibodies recognizing both SARS-CoV-2 and OC43 most likely developed from rapidly expanding broadly memory B cells that recognize conserved epitopes. Further study with both linear and spatial B-cell epitopes with cloned IgG from B cells during acute infection is needed to better characterize the molecular basis of the switch from uni- to bidirectional IgG binding.
Finally, this study provides direct evidence that preexisting β-HCoV S-reactive antibodies are associated with the SARS-CoV-2–specific antibody response, and the OC43 anti-S and SARS-CoV-2 anti-S2 antibodies highly correlate with the COVID-19 severity. One possible mechanism for this finding is that anti-S2 IgG does not prevent receptor binding and viral fusion, enabling higher viral loads. Alternatively, complement activation by anti-spike IgG1 and IgG3 could potentiate thrombosis via complement-dependent cytotoxicity of vascular endothelial cells [2, 42–45]. However, several caveats apply. First, while anti-RBD antibodies have been shown to correlate with viral neutralization, we did not directly measure neutralizing activity. In addition, sera were obtained for secondary analysis from clinical samples and not a longitudinal series spanning from pre- to postinfection in single individuals. This may introduce a selection bias, where subjects with the most longitudinal samples tended to be those with severe COVID-19, limiting our ability to detect the significant impact of preexisting OC43 antibodies in mild illness. Similarly, while we observed a trend of delayed development of SARS-CoV-2 S-reactive IgG in the nonsurvivor group [15, 31], we could not isolate the effects of OC43 and HKU1 antibodies on specific antibody development. Future, prospective, longitudinal studies would help address some of these questions.
CONCLUSION
Our study suggests that a rapid and robust increase in anti-S2 subunit IgG antibodies after SARS-CoV-2 infection may be a marker of disease severity. This quick de novo response may be due to immune imprinting of preexisting β-HCoVs cross-reactive memory B cells or plasmablasts, which may, in turn, lead to nonneutralizing IgG production. These findings may impact future vaccine development strategies targeting new emerging SARS-CoV-2 variants.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank the anonymous reviewers for their valuable suggestions; and Dr Shannon Hilchey, Samantha King, and Eric Mendleson for their critical reading of the manuscript.
Author contributions. J. W., N. D. P., and M. S. Z. conceived the investigation plan designed the experiments and analytic methods. J. W., Q. Z., A. W., and A. C. carried out the experiments. J. W., D. L., and M. S. Z. analyzed the data. J. W., M. S. Z., Q. Z., D. L., A. W., J. N., and A. C. wrote and edited the manuscript.
Disclaimer . The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. None of the funders had any role in study design, data collection, analysis, the decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the National Institutes of Health (NIH) Institute of Allergy, Immunology and Infectious Diseases (grant numbers R21 AI138500 and R01 AI129518 to M. Z. and J. W.); and the NIH National Center for Advancing Translational Sciences University of Rochester Clinical and Translational Science Award (grant number UL1 TR002001 to D. L. and M. Z.) and Rochester Vaccine Fellowship (to A. C.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Jiong Wang, Department of Medicine, Division of Nephrology, University of Rochester, Rochester, New York, USA.
Dongmei Li, Clinical and Translational Science Institute, University of Rochester, Rochester, New York, USA.
Andrew Cameron, Clinical Microbiology, Department of Pathology and Laboratory Medicine, University of Rochester, Rochester, New York, USA.
Qian Zhou, Department of Medicine, Division of Nephrology, University of Rochester, Rochester, New York, USA.
Alexander Wiltse, Department of Medicine, Division of Nephrology, University of Rochester, Rochester, New York, USA.
Jennifer Nayak, Department of Pediatrics, Division of Infectious Diseases, University of Rochester, Rochester, New York, USA.
Nicole D Pecora, Clinical Microbiology, Department of Pathology and Laboratory Medicine, University of Rochester, Rochester, New York, USA.
Martin S Zand, Department of Medicine, Division of Nephrology, University of Rochester, Rochester, New York, USA; Clinical and Translational Science Institute, University of Rochester, Rochester, New York, USA.
References
- 1. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zohar T, Alter G.. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol 2020; 20:392–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World. Health Organization. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/. Accessed February 13, 2022.
- 4. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF.. The proximal origin of SARS-CoV-2. Nat Med 2020; 26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui J, Li F, Shi ZL.. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019; 17:181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lim YX, Ng YL, Tam JP, Liu DX.. Human coronaviruses: a review of virus-host interactions. Diseases 2016; 4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tyrrell DA, Bynoe ML.. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J 1965; 1:1467–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dijkman R, Jebbink MF, El Idrissi NB, et al. Human coronavirus NL63 and 229E seroconversion in children. J Clin Microbiol 2008; 46:2368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng ZQ, Chen DH, Tan WP, et al. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur J Clin Microbiol Infect Dis 2018; 37:363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobano C, Santano R, Jimenez A, et al. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: utility and limitations in seroprevalence and immunity studies. Transl Res 2021; 232:60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020; 584:443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect 2020; 81:e28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020; 581:215–20. [DOI] [PubMed] [Google Scholar]
- 14. Ng KW, Faulkner N, Cornish GH, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020; 370:1339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zohar T, Loos C, Fischinger S, et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 2020; 183:1508–19.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beretta A, Cranage M, Zipeto D.. Is cross-reactive immunity triggering COVID-19 immunopathogenesis? Front Immunol 2020; 11:567710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo L, Wang Y, Kang L, et al. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study. Emerg Microbes Infect 2021; 10:664–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aydillo T, Rombauts A, Stadlbauer D, et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat Commun 2021; 12:3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hicks J, Klumpp-Thomas C, Kalish H, et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal Betacoronaviruses. J Clin Immunol 2021; 41:906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ladner JT, Henson SN, Boyle AS, et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep Med 2021; 2:100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Angus DC, Seymour CW, Coopersmith CM, et al. A framework for the development and interpretation of different sepsis definitions and clinical criteria. Crit Care Med 2016; 44:e113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J, Li D, Zhou Q, Wiltse A, Zand MS.. Antibody mediated immunity to SARS-CoV-2 and human coronaviruses: multiplex beads assay and volumetric absorptive microsampling to generate immune repertoire cartography. Front Immunol 2021; 12:2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cameron A, Porterfield CA, Byron LD, et al. A multiplex microsphere IgG assay for SARS-CoV-2 using ACE2-mediated inhibition as a surrogate for neutralization. J Clin Microbiol 2021; 59:e02489-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zand MS, Wang J, Hilchey S.. Graphical representation of proximity measures for multidimensional data: classical and metric multidimensional scaling. Math J 2015; 17:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J, Hilchey SP, Hyrien O, et al. Multi-dimensional measurement of antibody-mediated heterosubtypic immunity to influenza. PLoS One 2015; 10:e0129858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Hilchey SP, DeDiego M, et al. Broad cross-reactive IgG responses elicited by adjuvanted vaccination with recombinant influenza hemagglutinin (rHA) in ferrets and mice. PLoS One 2018; 13:e0193680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dijkman R, Jebbink MF, Gaunt E, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol 2012; 53:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J, Li D, Perry S, et al. Broadly reactive IgG responses to heterologous H5 prime-boost influenza vaccination are shaped by antigenic relatedness to priming strains. mBio 2021; 12:e0044921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shrock E, Fujimura E, Kula T, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020; 370:eabd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cueno ME, Imai K.. Structural comparison of the SARS CoV 2 spike protein relative to other human-infecting coronaviruses. Front Med (Lausanne) 2021; 7:594439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaplonek P, Wang C, Bartsch Y, et al. Early cross-coronavirus reactive signatures of humoral immunity against COVID-19. Sci Immunol 2021; 6:eabj2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu Y, Yu D, Han Y, et al. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci Adv 2020; 6:eabc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hejazi ME, Malek Mahdavi A, Navarbaf Z, et al. Relationship between chest CT scan findings with SOFA score, CRP, comorbidity, and mortality in ICU patients with COVID-19. Int J Clin Pract 2021; 75:e14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Z, Hu Q, Huang F, Xiong S, Sun Y.. The prognostic value of the SOFA score in patients with COVID-19: a retrospective, observational study. Medicine (Baltimore) 2021; 100:e26900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su C, Xu Z, Hoffman K, et al. Identifying organ dysfunction trajectory-based subphenotypes in critically ill patients with COVID-19. Sci Rep 2021; 11:15872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao J, Zhao J, Mangalam AK, et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 2016; 44:1379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mateus J, Grifoni A, Tarke A, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020; 370:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020; 5:eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen-Contant P, Embong AK, Kanagaiah P, et al. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio 2020; 11:e01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020; 183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turner JS, Zhou JQ, Han J, et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 2020; 586:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu F, Han K, Blair R, et al. SARS-CoV-2 infects endothelial cells in vivo and in vitro. Front Cell Infect Microbiol 2021; 11:701278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rambaldi A, Gritti G, Mico MC, et al. Endothelial injury and thrombotic microangiopathy in COVID-19: treatment with the lectin-pathway inhibitor narsoplimab. Immunobiology 2020; 225:152001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manolis AS, Manolis TA, Manolis AA, Papatheou D, Melita H.. COVID-19 infection: viral macro- and micro-vascular coagulopathy and thromboembolism/prophylactic and therapeutic management. J Cardiovasc Pharmacol Ther 2021; 26:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colling ME, Kanthi Y.. COVID-19-associated coagulopathy: an exploration of mechanisms. Vasc Med 2020; 25:471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.