Abstract
Extracellular vesicles (EVs) are important mediators of intercellular communication. However, EV biogenesis remains poorly understood. We previously defined a role for Arrdc4 (Arrestin domain containing protein 4), an adaptor for Nedd4 family ubiquitin ligases, in the biogenesis of EVs. Here we report that ubiquitination of Arrdc4 is critical for its role in EV secretion. We identified five potential ubiquitinated lysine residues in Arrdc4 using mass spectrometry. By analysing Arrdc4 lysine mutants we discovered that lysine 270 (K270) is critical for Arrdc4 function in EV biogenesis. Arrdc4K270R mutation caused a decrease in the number of EVs released by cells compared to Arrdc4WT, and a reduction in trafficking of divalent metal transporter (DMT1) into EVs. Furthermore, we also observed a decrease in DMT1 activity and an increase in its intracellular degradation in the presence of Arrdc4K270R. K270 was found to be ubiquitinated with K‐29 polyubiquitin chains by the ubiquitin ligase Nedd4‐2. Thus, our results uncover a novel role of K‐29 polyubiquitin chains in Arrdc4‐mediated EV biogenesis and protein trafficking.
Keywords: DMT1, extracellular vesicles, Nedd4‐2, Ubiquitin K‐29 chains, ubiquitination
1. INTRODUCTION
Extracellular vesicles (EVs) are membrane bound vesicles secreted by most cells (Mathivana et al., 2010; Zappulli et al., 2016). Based on the mode of biogenesis, EVs can be classified as exosomes or microvesicles (ectosomes). Exosomes are small EVs (30–150 nm) that are released into the extracellular microenvironment upon the fusion of multivesicular bodies with the plasma membrane, whereas microvesicles (100–1000 nm) bud directly from the plasma membrane (Thery et al., 2018). EVs contain a cargo comprising proteins, nucleic acids and lipids from the cell of origin which can be transferred to a recipient cell, thus altering the signalling and physiology of the target cells (Simpson et al., 2009; Valadi et al., 2007; Vidal et al., 1989). Despite their role in intercellular communication, little is known about EV biogenesis. Most well studied are exosomes, which require ESCRT (endosomal sorting complex required for transport) dependent biogenesis or a lesser‐known mechanism involving the ceramide biosynthesis pathway (Castro et al., 2014; Henne William et al., 2011; Hurley, 2015). The biogenesis of microvesicles is distinct from that of exosomes and involves Ca2+ dependent cytoskeletal reformation resulting in the budding of the outer leaflet of plasma membrane (Whitlock & Hartzell, 2017). RhoA and ARF6 have been proposed to be regulators of microvesicle budding (Ishizaki et al., 1996; Muralidharan‐Chari et al., 2009).
It is well established that protein modification via ubiquitination regulates endocytosis and protein trafficking (Foot et al., 2017). In addition, ubiquitination is critical for regulating membrane budding, including egress of many viruses, and hence can have a key role in the biogenesis of EVs. However, very little is known as to how ubiquitination regulates EV biogenesis. The ubiquitin protein ligases (E3s) constitute the substrate recognition part of the ubiquitin machinery. Among the various types of E3s, the Nedd4 family of E3s are key regulators of cellular signalling, often by controlling the abundance, trafficking and functions of membrane proteins (Foot et al., 2017). These E3s, including Nedd4, Nedd4‐2, WWP1/2, and Smurf1/2 through their WW domains bind PPxY (PY) or similar motifs in their substrates and adaptors (Rotin & Kumar, 2009; Scheffner & Kumar, 2014). The Nedd4 family adaptor proteins play important role in recruiting substrates or promoting the activity of the E3s. These adaptors include Ndfip1 and Ndfip2, transmembrane proteins containing PY motifs that bind and recruit Nedd4 E3s to intracellular membranes (Shah & Kumar, 2021). Our previous studies have shown that Ndfip1 is a substrate for ubiquitination by Nedd4 E3s (Harvey et al., 2001). It is also required for ubiquitination‐dependent regulation of divalent metal ion transporter (DMT1), the apical iron importer expressed in many tissues, including on the surface of duodenal enterocytes, through recruitment of Nedd4 E3 ligases (Foot et al., 2008, 2011; Howitt et al., 2009). As stated above, Ndfip1 and Nedd4 are also involved in the exosomal transport of ubiquitinated cargo (Putz et al., 2008).
Another family of Nedd4 E3 adaptors are the α‐arrestins, evolutionarily conserved arrestin domain‐containing (Arrdc) proteins with important functions in the regulation of signalling and trafficking (Alvarez, 2008; Kahlhofer et al, 2021). The six α‐arrestins include Arrdc1–5 and TXNIP, and except for Arrdc5, all contain two PY motifs near their C terminus. Arrdc1 was shown to be involved in EV biogenesis requiring Tsg101 and Vps4, and the Nedd4 family member WWP2 (Nabhan et al., 2012). The analysis of embryonic fibroblasts isolated from Arrdc1‐deficient (Arrdc1–/– ) mice also show reduced EV production and altered protein cargo (Anand et al., 2018). Our earlier work showed that Arrdc1 and Arrdc4 act as the Nedd4 E3 ligase adaptors to regulate DMT1 release in EVs (Mackenzie et al., 2016). Cells with Arrdc4 knockdown and embryonic fibroblasts from Arrdc4–/– mice show significantly reduced EV production. However, the Arrdc4‐dependent EV release may involve a different mechanism than Arrdc1 (Mackenzie et al., 2016). Using the Arrdc4–/– mice, Arrdc4 was also found to be important for maturation of sperm through the control of EV biogenesis (Foot et al., 2021). Although Arrdc1 and Arrdc4 are E3 adaptors and Nedd4 family members appear to be involved in Arrdc1 and Arrdc4 dependent EV biogenesis, the precise role of ubiquitination of these adaptors in the regulation of EV biogenesis by these proteins remains unknown.
Here, we investigated the possible role of Arrdc4 ubiquitination in EV biogenesis. We identified five potential ubiquitinated lysine (K) residues in Arrdc4 by mass spectrometry and established that the ubiquitination of the highly conserved K270 is crucial for Arrdc4 function in EV biogenesis. The Arrdc4 K270R mutation resulted in mislocalisation of the protein and an increase in cellular levels leading to an increase in intracellular degradation. We show that Nedd4‐2‐dependent ubiquitination involving K‐29‐linked ubiquitin chains at K270 results in an increase in EV biogenesis. Mutation at K270 also altered the trafficking of an Arrdc4 target protein, the metal ion transporter DMT1. Overall, our data suggest that K‐29‐linked polyubiquitination of Arrdc4 at K270 by Nedd4‐2 is a putative novel signal for Arrdc4‐mediated EV release.
2. MATERIAL AND METHODS
2.1. Antibodies and reagents
Source of antibodies used in this study are as follows: rabbit polyclonal anti‐GFP, Annexin A1 and CD9 from Abcam (Cambridge, MA,USA); rabbit polyclonal for Nedd4‐2 generated and purified in‐house (Konstas et al., 2002); rat monoclonal anti‐HA (3F10) and mouse monoclonal anti‐c‐Myc (9E10) from Roche Diagnostics (Indianapolis, IN, USA); mouse monoclonal anti flag (M2) and goat polyclonal anti‐GFP from Rockland Immuno‐chemicals (Limerick, PA, USA); rabbit polyclonal anti‐Rab7 from Cell Signalling Technology (MA, USA). Secondary antibodies used for this study are as follows: donkey anti‐rabbit horseradish peroxidase and ECL Plex goat anti‐mouse Cy5 from GE Healthcare (Buckinghamshire, UK); goat anti‐mouse Alkaline Phosphatase, goat anti‐rabbit Alkaline Phosphatase and goat anti‐rat Alkaline Phosphatase from Merck Millipore (Billerica, MA, USA). Alexa‐Fluor donkey anti‐goat 488, donkey anti‐mouse 568 and donkey anti‐rabbit 647 were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
2.2. Expression vectors
Wild type Arrdc4 and PY mutant cDNA were cloned into pEGFPN1 and pcDNA3.1 (with an N‐terminal HA/6X His tag) as described previously (Mackenzie et al., 2016). Arrdc4 K‐R mutants (K110R, K131R, K226R, K267R and K270R) were generated using site‐directed mutagenesis. Flag‐tagged wild type ubiquitin and KallR mutant (ubiquitin in which all Lysines are mutated to Arginine) constructs were purchased from the MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee. Specific polyubiquitin chain mutants were then generated by reverting specific Arginine residues back to Lysine in the KallR construct (R6K, R11K, R27K, R29K, R33K, R48K, R63K). The HA‐ubiquitin expression plasmid was provided by Dr. Dirk Bohmann (University of Rochester, Rochester, NY). Nedd4, Nedd4CM (Cysteine mutant), Nedd4‐2, Nedd4‐2CM constructs have been described previously (Fotia et al., 2003; Harvey et al., 2002; Kumar et al., 1997). Smurf1 and Smurf1CM constructs were from Prof. Takeshi Imamura (University of Tokyo, Japan). DMT1 constructs were from Prof. Philippe Gros (McGill University, Montreal, Canada).
2.3. Cell culture and plasmid transfection
Cells were grown in either Dulbecco's modified Eagle's medium (DMEM; CHO) or RPMI‐1640 (HEK293T) supplemented with 10% foetal calf serum, 2 mM L‐glutamine, 0.5 M HEPES and 100 units/ml penicillin‐streptomycin at 37°C with 5% CO2. Cells stably transfected with myc‐tagged DMT1 (isoform 2/+IRE) were provided by Prof Philippe Gros (McGill University, Montreal, Canada). Fugene HD (Promega, Madison, WI, USA) was used for transfection of plasmids according to manufacturer's instructions. The siRNA‐mediated knockdown of Arrdc4 was performed as described previously (MacKenzie et al., 2016). Cells were harvested 24 h post‐transfection and used for re‐expression of WT or mutant Arrdc4 proteins.
2.4. Quantitative real‐time PCR
Total RNA isolated from cells using TRIzol (Thermo Fisher Scientific) was reverse‐transcribed using a High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, MA, USA) according to manufacturer's instructions. Quantitative PCR (qPCR) was performed with the KAPA SYBR FAST Universal Kit (Sigma Aldrich, MA, USA) on a Rotor‐Gene Q Real‐time PCR cycler (Qiagen, MD, USA) with the following conditions: 95°C for 3 min, 40 cycles of 95°C for 3 s and 60°C for 25 s. Results were analysed using the Rotor‐Gene Q Series software and data was normalised to β‐actin. Primer sequences are as follows (5′–3′): β‐actin F: GATCATTGCTCCTCCTGAGC, β‐actin R: AGTCCGCCTAGAAGCACTTG, Arrdc4 F: GGCTGCTATTCCAGCGGCGA and Arrdc4 R: CAGGCTGAGGCGCACGTTCA.
2.5. Proteomic analysis
Protein samples were resuspended in 6 M Urea, 100 mM DTT and 100 mM Tris‐HCl pH7.0 and subjected to protein digestion using FASP (filter aided sample preparation) before lyophilisation to dryness using a SpeedVac AES 1010 (Savant, Thermo Fisher). Peptides were resuspended in 2% acetonitrile, 1% formic acid and injected and separated by reversed‐phase liquid chromatography on a M‐class UPLC system (Waters) using a 250 mm × 75 μm column (1.6 μm C18, packed emitter tip; IonOpticks, Australia) with a linear 90 min gradient at a flow rate of 400 nl/min from 98% solvent A (0.1% Formic acid in Milli‐Q water) to 34% solvent B (0.1% Formic acid, 99.9% acetonitrile). The UPLC was coupled on‐line to a freshly cleaned Q‐Exactive HF‐X mass spectrometer (Thermo Fisher, MA, USA). The Q‐Exactive HF‐X was operated in a data‐dependent mode, switching automatically between one full‐scan and subsequent MS/MS scans of the 15 most abundant peaks. Full‐scans (m/z 350–1600) were acquired with a resolution of 60,000 at 200 m/z. The 15 most intense ions were sequentially isolated with a target value of 100,000 ions and an isolation width of 1.4 m/z and fragmented using HCD with normalized collision energy of 27. Maximum ion accumulation times were set to 50 ms for full MS scan and 40 ms for MS/MS. Dynamic exclusion was enabled and set to 20 s.
Raw files were analysed using MaxQuant software (version 1.5.8.3) and the database search was performed using mouse sequences obtained from Uniprot including isoforms with strict trypsin specificity allowing up to two missed cleavages. The minimum required peptide length was set to seven amino acids. Carbamidomethylation of cysteine was set as a fixed modification while N‐acetylation of proteins N‐termini and oxidation of methionine were set as variable modifications. During the MaxQuant main search, precursor ion mass error tolerance was set to 4.5 ppm and fragment ions were allowed a mass deviation of 20 ppm. PSM and protein identifications were filtered using a target‐decoy approach at a false discovery rate (FDR) of 1%. Further analysis was performed using a custom pipeline developed in R, which utilizes the LFQ intensity values in the MaxQuant output file proteinGroups.txt. Proteins not found in at least 50% of the replicates in one group were removed. Missing values were imputed using a random normal distribution of values with the mean set at mean of the real distribution of values minus 1.8 s.d., and a s.d. of 0.3 times the s.d. of the distribution of the measured intensities. The probability of differential protein expression between groups was calculated using the Limma R package. Probability values were corrected for multiple testing using Benjamini–Hochberg method.
2.6. Isolation of extracellular vesicles
Isolation of EVs was carried out as described previously with some modifications (Mackenzie et al., 2016). Briefly, 24 h after transfection, cells were incubated in serum free media for a further 24 h to prevent contamination of samples with serum derived EVs. The cell culture supernatant was then sequentially centrifuged at 800 × g for 10 min and 2600 × g for 15 min at 4°C to remove cell debris and aggregates. Ultracentrifugation of the supernatant in ultra‐clear centrifuge tubes (Beckman Coulter, Brea, CA, USA) was carried out using the Beckman TLA100.4 rotor in an Optima TLX centrifuge (Beckman Coulter) at 100,000 × g at 4°C for 2 h. The pellet was washed twice with ice‐cold phosphate buffer saline (PBS) and resuspended in RIPA (150 mm NaCl, 5 mM EDTA, 50 mM Tris‐HCl pH 7.4, 0.1% SDS, 1% NP40, 0.5% sodium deoxycholate) buffer and sonicated twice or pellets were resuspended in PBS to perform nanoparticle tracking analysis (NTA). NTA was performed using a NanoSight NS300 (Malvern Panalytical, UK). The data was normalised for equal number of cells.
2.7. Immunoprecipitation and immunoblotting
Following transfection, cells were treated with 50 μM MG132 (Boston Biochemical, Boston, MA, USA) and 400 μM chloroquine (Sigma‐Aldrich) for 4 h before lysis in Verhagan lysis buffer (1% triton X‐100, 10% glycerol, 150 mM NaCl, 20 mM Tris HCl pH 7.4 and 2 mM EDTA). For ubiquitination assays, 5 mM N‐ethylemaleimide (Sigma Aldrich) was added to the lysis buffer to inhibit protein deubiquitination. Immunoprecipitation was performed on protein G Sephrose beads with goat polyclonal anti‐GFP antibody. Samples were loaded onto 4%–20% precast polyacrylamide gels (Bio‐Rad Laboratories, Hercules, CA, USA) for separation by electrophoresis. Proteins were then transferred to polyvinylidene difluoride (PVDF) membrane using a Trans‐blot turbo instrument (Bio‐Rad). Membranes were then blocked with 5% skim milk and immunoblotted with primary antibody diluted in 5% skim milk in TBS‐T (Tris‐buffered saline with 0.05% Tween 20) incubated overnight at 4°C, followed by incubation with an alkaline phosphatase (AP), horseradish peroxidase (HRP) or Cy5‐conjugated secondary antibody. Visualization of HRP signals was using an ImageQuant LAS4000 (GE Healthcare) whereas a Typhoon FLA biomolecular imager (GE healthcare) was used to visualize signals from alkaline phosphatase and Cy5.
2.8. Immunocytochemistry
Cells were seeded onto cover slips at a density of 1 × 105 and then transfected with respective plasmids. Twenty‐four h following transfection, cells were fixed in 4% paraformaldehyde for 15 min at room temperature and then permeabilized in 0.1% Triton X‐100 in PBS for 2 min. After permeabilization cells were blocked in 1% donkey serum for 30 min at room temperature and then incubated overnight with primary antibody at 4°C. Next day cells were washed 3X with 1% PBS and incubated with secondary antibody for 2 h at room temperature. Primary and secondary antibodies were diluted in 1% donkey serum in PBS. All confocal images were obtained on a Zeiss LSM 700 confocal microscope (Zeiss, Oberkochen, Germany). The fluorescence was sequentially developed for multiple channels to avoid emission spectra bleed‐through. The relative expression or fluorescence intensities were quantified using ImageJ (1.52a) software. The degree of colocalisation of DMT1 with Rab7 was measured by Pearson's correlation coefficient using ImageJ software. The scale lay between −1 and 1, where 1 represents colocalisation, −1 represents negative colocalisation and 0 means no colocalisation.
2.9. Protein stability assays
HEK293T cells were seeded in six‐well plates (2 × 105 cells per well) and transfected with appropriate plasmids. Cells were then treated with 100 μg/ml cycloheximide (CHX) in RPMI 1640 media and incubated for different time intervals (0, 2, 6, and 24 h). The cells were then lysed in RIPA lysis buffer with HALT protease and phosphatase inhibitor cocktail, incubated at 4°C for 30 min. Lysates were cleared by centrifugation and then 25 μg protein used for immunoblotting as described above.
2.10. Fluorescence quenching assays
To measure the relative transport activity of DMT1 in CHO cells we performed a fluorescence quenching assay as described previously (Mackenzie et al., 2016). Briefly, transfected cells were loaded with 0.25 μM calcein‐AM (Thermo Fisher Scientific) at 37°C for 20 min. Cells were then washed to remove excess calcein‐AM from the media, then suspended in transport buffer containing 150 mM NaCl, 20 mM 2‐(N‐morpholino) ethanesulfonic acid and plated in triplicate with 2 × 105 cell density in a 96‐well flat bottom transparent plate (Nalgene Nunc International, Rochester, NY, USA). Fluorescence readings were recorded using a BMG Labtech FLUOStar Optima microplate reader (Melbourne, VIC, Australia) with excitation at 485 nm and emission at 520 nm. Baseline fluorescence was measured for 20 s. CoCl2 (final concentration 100 μM; pH 6.7) was then injected and readings were taken every 0.5 s for 150 s. The degree of calcein quenching is an indication of the amount of DMT1 present on the cell surface. DMT1 transport activity of each sample was measured as the rate of fluorescence quenching (slope) observed in the 1 min period after CoCl2 injection.
2.11. Three‐dimensional structure prediction and protein sequence alignment
The amino acid sequence of mouse Arrdc4 (Accession No. A0A0B4J1F4) was retrieved from the UniProt database. I‐TASSER (Yang & Zhang, 2015) online tool was used to generate the three‐dimensional structure of Arrdc4 and Arrdc4K270R proteins. The template used for this purpose was 4ll1‐A (structure of the TRX and TXNIP complex). After construction of five models, the one having the best stereochemistry was selected following evaluation by PROCHECK (Laskowski et al., 1993). The Arrdc4 sequence from different species were obtained from NCBI database (The UniProt Consortium, 2019) and multiple sequence alignment was performed using CLUSTAL W tool (Thompson et al., 1994).
2.12. Quantification and statistical analysis
For immunoblots Image J software (version 1.52a, National Institute of Health, USA) was utilised to calculate the band intensity and data normalised to levels of β‐Actin. Statistical Analysis were performed using GraphPad Prism software (v8.0). One‐phase decay equation was used to assess protein half‐life, while t‐tests were used for all other analyses. All results presented as mean ± standard deviation (SD). Statistical significance was determined as being P < 0.05. + indicates significance when data compared with the control or empty vector, whereas * indicates significance when compared with Arrdc4WT. + P < 0.05, ++/** P < 0.01, +++/*** P < 0.001, ns = not significant. Unless stated otherwise, the data were derived from at least three independent experiments.
3. RESULTS
3.1. Identification of potential ubiquitination sites in Arrdc4
The adaptors of Nedd4 family of E3s also act as E3 substrates themselves but it is not established whether ubiquitination is required for their physiological function. The disruption of the interaction between Arrdc4 and the E3 Nedd4‐2 has been shown to alter Arrdc4 localisation and it has been postulated that Arrdc4 ubiquitination may be important for its function in EV release (Mackenzie et al., 2016).
To explore Arrdc4 ubiquitination and the role of Arrdc4 ubiquitination in EV biogenesis, we performed mass spectrometry to identify the potential ubiquitination sites in Arrdc4, which can be recognized by MS/MS spectra through detection of peptide adducts derived from the ubiquitin molecule. From this analysis, we identified K110, K131, K226, K267, and K270 as potential ubiquitinated sites in Arrdc4 (Figures 1a and S1). K110 and K131 are in the Arrestin‐N domain, whereas K226, K267, and K270 are in the Arrestin‐C domain (Figure 1b). The crystal structure of Arrdc4 is not available so we generated a 3D model using bioinformatic tools. As shown in Figure 1c, the model shows the two arrestin domains (N and C) and highlights the potential ubiquitinated K residues identified from the MS analysis. Multiple sequence alignment using ClustalW showed that Arrdc4K270 is the most highly conserved site among different species (Figure 1d).
FIGURE 1.
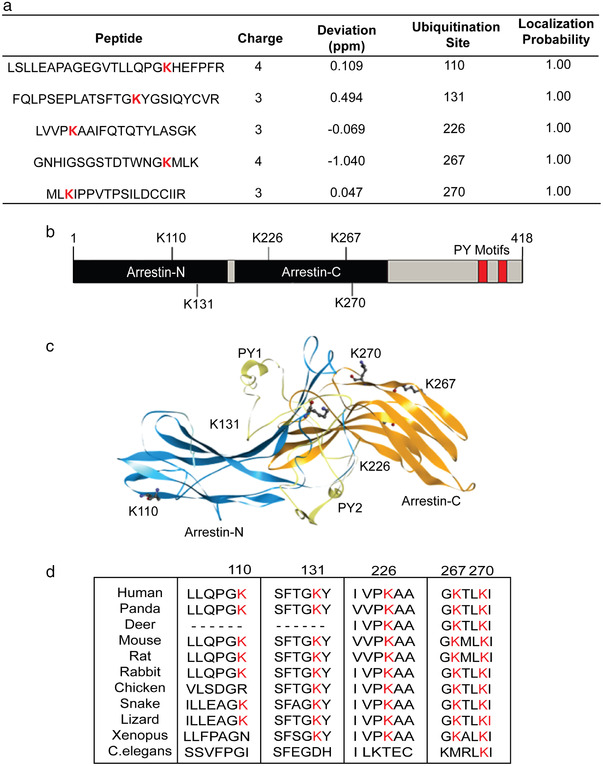
Identification of potential ubiquitinated lysine residues in Arrdc4. (a) Mass spectrometric analysis identifies five potential ubiquitination sites. (b) Schematic representation of Arrdc4 protein showing position of lysines within the Arrestin N and C domains and the relative positions of the PY motifs. (c) The structure of Arrdc4 predicted using the I‐TASSER online tool. Highlighted are the ubiquitinated K residues K110, K131, K226, K267, and K270, along with the PY motifs necessary for binding to Nedd4 family of ubiquitin ligases. (d) Multiple sequence alignment of putative Arrdc4‐like proteins from various species, showing high evolutionary conservation of K270
3.2. Arrdc4K270 is important for protein turnover, localisation and EV biogenesis
To address whether the K residues identified by MS contribute to protein ubiquitination, we constructed mutants of Lys to Arg (R). The stability of the mutant proteins was measured using cycloheximide chase analysis of protein degradation. Arrdc4K226R and Arrdc4K270R were found to be more stable than Arrdc4WT and the other mutants over a 24 h period (Figures 2a and S2). We further determined the half‐life of the proteins using one phase decay analysis that confirmed that Arrdc4K226R and Arrdc4K270R are more stable than Arrdc4WT (Figure 2b). We then examined whether these K‐R mutations affect protein localisation as Arrdc4WT is normally localised to the plasma membrane and early endosomes. We found that most mutants showed predominantly plasma membrane localisation similar to Arrdc4WT, whereas Arrdc4K270R was more cytoplasmic (Figure 2c,d)
FIGURE 2.
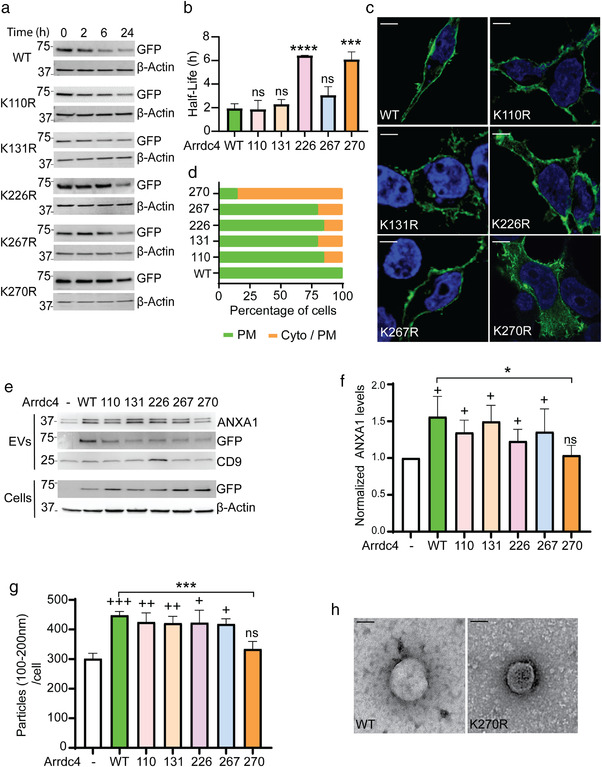
Lysine 270 is important for protein turnover, localisation and EV biogenesis. (a) HEK293T cells were transfected with GFP‐tagged Arrdc4 WT or K‐R mutants, treated with cycloheximide (CHX; 100 μg/ml) for 0, 2, 6, and 24 h, and stability of the protein measured using immunoblotting. Arrdc4K270R and Arrdc4K226R were found to be more stable than the other mutants. (b) The half‐life of K‐R mutant proteins were determined using one phase decay equation in GraphPad. (c, d) Cellular localisation of Arrdc4WT and K‐R mutants. Scale bar (for all panels) 10 μm. The localisation of Arrdc4WT and its mutant was quantified for at least 20 cells (n = 20). (e, f) Representative immunoblots (25 μg protein/lane) showing expression of Arrdc4WT and other K‐R mutants. Arrdc4K270R shows decreased expression of ANXA1 in EVs and increased level of Arrdc4K270R in the cells when compared with Arrdc4WT. The data show a similar pattern that is, significant decrease in expression of ANXA1 in Arrdc4K270R expressing cells when quantified using three independent blots while comparing with Arrdc4WT. +, denotes that data were compared with untransfected control, whereas * indicates significance when compared with Arrdc4WT transfected cells. +/*P < 0.05, ++ P < 0.01, ns = not significant. (g) Nanoparticle tracking analysis (NTA) showing significantly decreased number of particles of 100–200 nm size (normalised to equal number of cells) released by Arrdc4K270R compared to Arrdc4WT and other K‐R mutants. */+ P < 0.05, ++ P < 0.01, ***/+++ P < 0.001, ns = not significant. (h) Transmission electron microscopy of EVs released from Arrdc4WT compared to Arrdc4K270R mutant. Scale bar, 100 nm
We have previously shown a role for Arrdc4 in EV release via a direct plasma membrane budding mechanism (Mackenzie et al., 2016). To explore the effect of K mutations on the release of EVs, we collected EVs from the culture media of HEK293T cells transfected with Arrdc4WT or mutants. Annexin A1 (ANXA1) is a marker for EVs budding directly from plasma membrane (Jeppesen et al., 2019). Immunoblotting showed a decreased level of ANXA1 in EVs released by Arrdc4K270R in EVs as compared to Arrdc4WT and other mutants (Figure 2e). As shown in Figure 2f quantification of the ANXA1 levels for each mutant showed a significant decrease in number of EVs released by Arrdc4K270R. We then quantified the number of EVs released by cells expressing the various K mutant proteins (Figure S3A) using Nanoparticle Tracking Analysis (NTA) (Figure S3B). We observed a significant increase in the number of EVs (100–200 nm size) released by Arrdc4WT compared with vector control cells as expected (Figure S3B). As Arrdc4 mostly regulates a specific subset of EVs in the >100 nm size (Foot et al., 2021) we used this range (EV100‐ 200nm) for all further analyses. Arrdc4K270R was the only mutant to show a significant decrease in the number of EV100‐200 nm released from the cells compared to Arrdc4WT (Figures 2g and S3B). Transmission electron microscopy (TEM) showed the presence of intact vesicles in the supernatant harvested from both Arrdc4WT and Arrdc4K270R transfected cells (Figure 2h).
FIGURE 3.
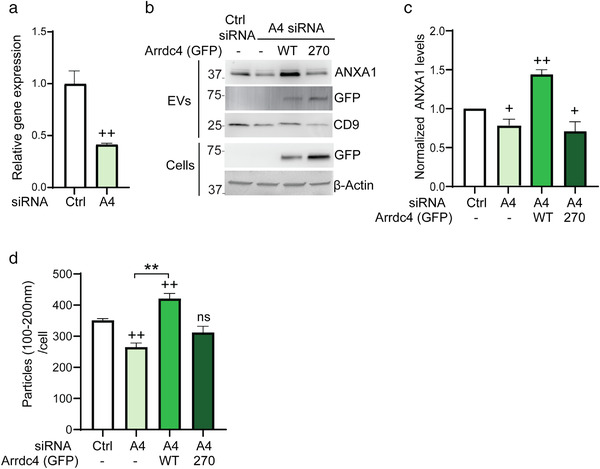
Knockdown of Arrdc4 affects EV biogenesis. (a) Arrdc4 mRNA expression was examined by real time qPCR in HEK293Ts cells using β‐Actin as an internal control. +, comparing data with control; ++ P < 0.01. (b, c) ANXA1 expression was examined by immunoblotting following Arrdc4 knockdown (using A4 siRNA) in HEK293Ts cells and re‐expressing Arrdc4WT or Arrdc4K270R as indicated. +, data compared with control siRNA; + P < 0.05, ++ P < 0.01. (d) Nanoparticle analysis (NTA) showing EV100‐200nm release normalised to equal number of cells. The Arrdc4 knockdown reduces EV production, whereas the re‐expression of Arrdc4WT, but not Arrdc4K270R significantly increases EV100‐200nm release. +, data compared with untransfected control, whereas * indicates significance when compared with Arrdc4WT. ++/**P < 0.01, ns = not significant
3.3. Knockdown of Arrdc4 affects EVs release
To further understand the role of Arrdc4 and its mutant K270 in relation to EV biogenesis, we used siRNA to knockdown Arrdc4, followed by reexpression Arrdc4WT or Arrdc4K270R in the Arrdc4‐depleted cells. Real‐time qPCR analysis indicated that siRNA transfection reduced the expression of endogenous Arrdc4 by approximately 60% as compared to control siRNA (Figure 3a). We then assessed EV release using immunoblotting and NTA analysis (Figures 3b–d and S4A). As expected, Arrdc4 knockdown significantly reduced EV release (Figure 3b,c) and NTA size profile suggested that Arrdc4 mostly affects EV100‐200nm (Figures 3d and S4A). The re‐expression of Arrdc4WT significantly increased the release of EV100‐200nm whereas Arrdc4K270R failed to show any significant effect on EV100‐200nm release (Figure 3d). These data suggest that K270R mutation abrogates the activity of Arrdc4 in promoting EEV100‐200nm release.
3.4. Arrdc4 K270 is ubiquitinated by specific ubiquitin ligases
Earlier studies have shown that Arrdc4 recruits Nedd4 family of ubiquitin ligases for the ubiquitination and trafficking of proteins (Mackenzie et al., 2016). To determine the importance of K270 in Arrdc4 ubiquitination we performed a ubiquitination assay using Nedd4, Nedd4‐2 and Smurf1 (WT and catalytically inactive cysteine mutants; CM) (Figure 4a). We found that ubiquitination of Arrdc4 by Nedd4, Nedd4‐2 and Smurf1 was decreased when K270 was mutated (Figure 4b). To further investigate the effect of ubiquitination of Arrdc4 on the release of EV100‐200nm we harvested EVs from cells transfected with Arrdc4WT or Arrdc4K270R with Nedd4, Nedd4‐2 and Smurf1 E3 ligases. On their own, Nedd4, Nedd4‐2, and Smurf1 did not show notable increase in EV100‐200nm release when compared with control. However, when Arrdc4WT was co‐expressed with Nedd4‐2 or Smurf1 an increase in EV release was observed. Strikingly co‐expression of Nedd4 had no significant effect on Arrdc4 dependent promotion of EV100‐200nm production. When Arrdc4 K270 mutant was used the E3 ligases had no effect on EV release (Figures 4c–e, S4B, and S5A,B). Taken together these findings suggest that K270 is important for Arrdc4 ubiquitination and EV release mediated by both Nedd4‐2 and Smurf1.
FIGURE 4.
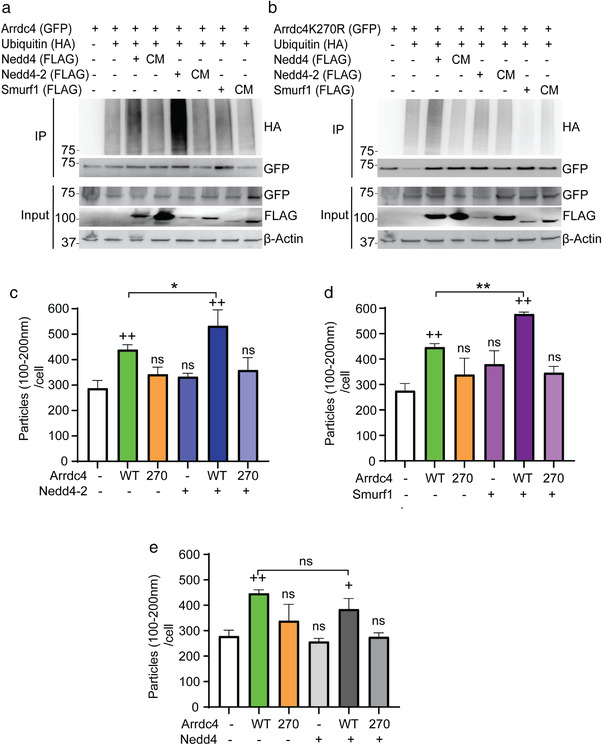
K270 mutation prevents ubiquitination of Arrdc4 and inhibits Arrdc4‐mediated EV biogenesis. (a, b) Representative immunoblots of ubiquitination assay performed in HEK293T cells by expressing GFP‐tagged Arrdc4 (WT/K270R), Flag‐tagged Nedd4, Nedd4‐2 and Smurf1 (WT and catalytically inactive mutant, CM) and HA‐tagged ubiquitin. Arrdc4 ubiquitination by Nedd4, Nedd4‐2 and Smurf1 is decreased by mutation at K270. (c–e) Release of extracellular vesicles (EV100‐200nm normalised to equal number of cells) by Arrdc4WT is significantly increased in presence of Nedd4‐2 and Smurf1, but not Nedd4, and this is inhibited by the Arrdc4K270R mutant. +, data compared with control whereas * indicates significance when compared with Arrdc4WT. +/*P < 0.05, ++/**P < 0.01, ns = not significant
3.5. Arrdc4K270 is ubiquitinated by K‐29 polyubiquitin chains
Next, we analysed the type of ubiquitination occurring on Arrdc4 K270. We performed ubiquitination assays using ubiquitin mutants that allow only single ubiquitin chain types. We identified ubiquitination of Arrdc4WT most prominently by K29 and K63 ubiquitin chains (Figure 5a), whereas Arrdc4K270R shows a significant reduction in ubiquitination by K29 chains (Figure 5b). We therefore hypothesised that Ub K‐29 chains may be a signal for EV release. To further investigate the relationship between Arrdc4 and Ub K‐29 chains in EV biogenesis we harvested EVs from cells transfected with Ub K‐29 alone and with Arrdc4WT or Arrdc4K270R. Our data indicated that Ub K‐29 alone does not trigger EV100‐200nm release but when in combination with Arrdc4 there was a large increase in EV100‐200nm release (Figures 5c and S6A). When K270 was mutated, the effect of Ub K‐29 on EV100‐200nm release was abrogated (Figure 5c). To further validate these results, we analysed protein levels in EVs released from cells transfected with Ub K‐29 and Arrdc4WT or Arrdc4K270R (Figure 5d). For immunoblots we loaded the entire EV pellet, while our NTA analysis reflects the size profiles of EV released from the cells. The data in Figure S6A suggest that expression of Ub K‐29 alone resulted in an increased release of EV0‐100nm and we observed a significant increase in EV100‐200nm release from cells co‐expressing Arrdc4WT and Ub K‐29, which was not seen with the Arrdc4K270R mutant (Figure 5d). We also observed an increase in ANXA1 levels in EVs from Arrdc4WT/Ub K‐29 expressing cells compared to Arrdc4K270R /Ub K‐29 cells. These results demonstrate a novel role for Ub K‐29 chains on K270 in Arrdc4‐mediated EV biogenesis.
FIGURE 5.
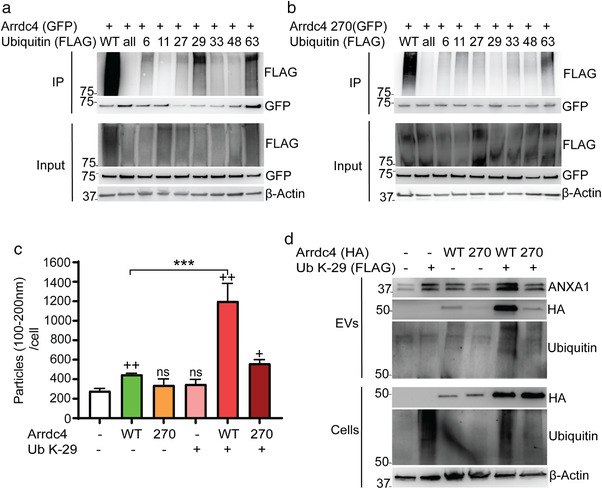
Arrdc4 is ubiquitinated by K‐29 ubiquitin chains at K270. (a) Immunoblot analysis of ubiquitination using Flag‐tagged ubiquitin and various lysine ubiquitin mutants (KallR, R6K, R11K, R27K, R29K, R33K, R48K, R63K) with Arrdc4 (WT or K270R). The results show that Arrdc4WT is mostly ubiquitinated by K‐29 and K‐63 ubiquitin chains. (b) Ubiquitination by Ub K‐29 is absent in Arrdc4K270R mutant. (c) EV100‐200nm biogenesis (normalised to equal number of cells) by Arrdc4WT is significantly enhanced in the presence of ubiquitin K29 and this is inhibited by Arrdc4 K270 mutation. +, data compared with untransfected control, whereas * indicates significance when compared with Arrdc4WT. + P < 0.05, ++/**P < 0.01, ns = not significant. (d) Expression of ANXA1 is increased in EVs by Ub K‐29 and this is inhibited by K270 mutation
3.6. Nedd4‐2 promotes K‐29‐linked polyubiquitination of Arrdc4
To determine the role of E3 ligases in the ubiquitination of Arrdc4 by Ub K‐29 chains we performed a ubiquitination assay using Ub K‐29 together with Nedd4‐2 or Smurf1. We found that Nedd4‐2 is involved in ubiquitinating Arrdc4 via Ub K‐29 chains, whereas Smurf1 is less able to do so (Figure 6a). We further demonstrated that mutation of K270 reduces K29‐linked ubiquitination of Arrdc4 by Nedd4‐2 (Figure 6b). To gain insight into the mechanism of Arrdc4‐mediated EV released by Nedd4‐2 and K‐29‐linked ubiquitination, we measured the EVs released by transfected cells both by NTA and immunoblotting (Figures 6c,d and S6B). Interestingly, there was a significant increase in EV100‐200nm release by cells expressing Nedd4‐2, Arrdc4WT and Ub‐K29, and this was inhibited when K270 was mutated (Figure 6c,d). This did not occur when Smurf1 was included as the ubiquitin ligase, demonstrating that this effect is specific to Nedd4‐2.
FIGURE 6.
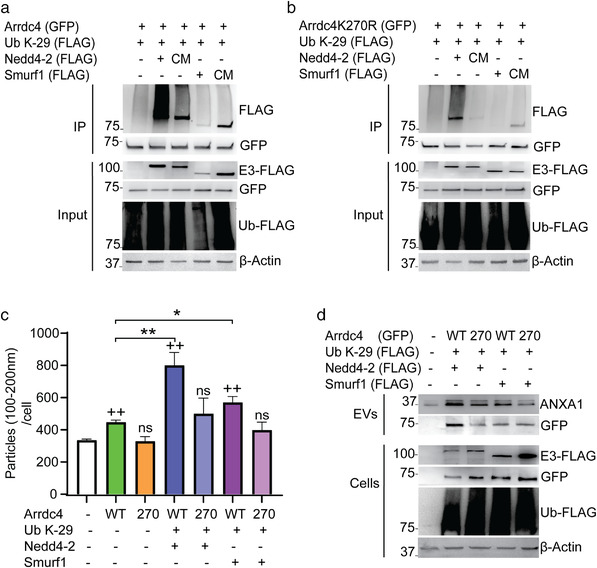
Nedd4‐2 promotes K29‐linked polyubiquitination of Arrdc4. (a, b) Ubiquitination of Arrdc4WT by K29‐linked polyubiquitination is strongly enhanced by Nedd4‐2 but not Smurf1, whereas K29‐linked polyubiquitination of Arrdc4 by Nedd4‐2 is reduced when K270 is mutated. HEK293T cells were transfected using GFP‐tagged Arrdc4 (WT/K270R), FLAG‐tagged Nedd4‐2 or Smurf1 (WT and catalytically inactive Cys mutant, CM) and FLAG‐tagged Ub K‐29. (c) EV100‐200nm (normalised to equal number of cells) released by Arrdc4WT is significantly enhanced in the presence of Nedd4‐2 and Ub K‐29, and this is inhibited when K270 is mutated. +/*P < 0.05, ++/**P < 0.01, ns = not significant. (d) Immunoblot analysis follows a similar pattern of increased expression of ANXA1 when cells were co‐transfected with Arrdc4WT, Nedd4‐2 and Ub K‐29
3.7. Arrdc4K270R inhibits DMT1 activity and its release in EVs
Given that Arrdc4 is involved in the regulation of DMT1 and plays a critical role in its release in EVs (Mackenzie et al., 2016), we investigated the potential role of Arrdc4K270 in DMT1 regulation. First, we measured DMT1 transport activity in CHO cells stably expressing Myc‐tagged DMT1 isoform II (CHO‐DMT1 cells) using a fluorescence quenching assay and found that there was a significant decrease in DMT1 activity in the presence of Arrdc4K270R, but not the other K mutants (Figure 7a). The Arrdc4 PY mutant that lacks the activity to bind Nedd4 family of E3s (Mackenzie et al., 2016), was included as a negative control. Secondly, we found that overexpression of Arrdc4K270R reduced DMT1 levels in cells and EVs compared with Arrdc4WT (Figure 7b). We therefore hypothesised that mutation of K270 results in altered trafficking of Arrdc4 targets such as DMT1. To confirm this, we carried out immunocytochemistry using CHO‐DMT1 cells co‐expressing Arrdc4WT or Arrdc4K270R. Earlier studies have shown that in the presence of Arrdc4, the plasma membrane localisation of DMT1 is reduced and it is trafficked to EVs utilising an alternative pathway (independent of ESCRT and Vps4A components) (Mackenzie et al., 2016). Notably, this depends upon the normal localisation of Arrdc4 at plasma membrane. Here we observed more intracellular DMT1 in the cytoplasm of Arrdc4K270R expressing cells when compared to Arrdc4WT (Figure 7c,d) because mutation at K270 affects the plasma membrane localisation of Arrdc4 itself influencing protein function. The punctate localisation corresponding to Rab7 positive vesicles, a marker for late endosomes, suggested that DMT1 is being targeted to an endosomal compartment (Figure 7c). Interestingly Rab7 levels were somewhat increased in Arrdc4K270R expressing cells compared to Arrdc4WT (Figure 7e). Importantly, the analysis of ImageJ data using Pearson's correlation confirmed increased colocalisation of DMT1 with Rab7 in cells expressing Arrdc4K270R compared to those expressing Arrdc4WT (Figure 7f). To understand how Arrdc4K270R may modulate DMT1 trafficking, we investigated whether Arrdc4K270R was targeting DMT1 to intracellular degradation pathways. In cells expressing Arrdc4K270R, we observed an increase in DMT1 levels when both the proteasomal and lysosomal degradation pathways were inhibited using MG132 and chloroquine, respectively (Figure 7g), whereas this was not observed in cells expressing Arrdc4WT, suggesting that Arrdc4K270R was alternatively targeting DMT1 to intracellular degradation rather than extracellular release in EVs. Increased accumulation of Arrdc4K270R compared to Arrdc4WT also confirms that mutation at K270 increases Arrdc4 stability. Taken together, these results suggest that Arrdc4K270R mutation causes mislocalisation of DMT1, resulting in a decrease in its activity and reduced targeting to EVs.
FIGURE 7.
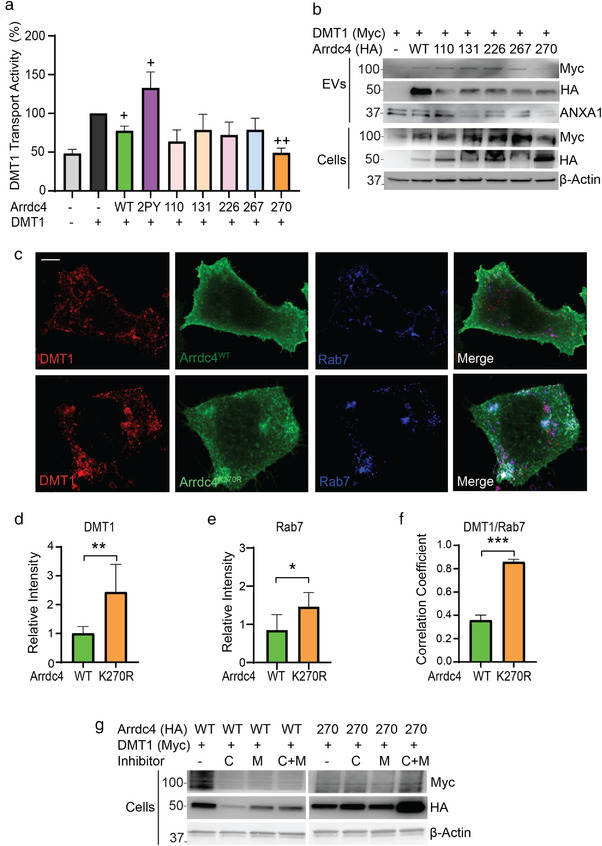
Arrdc4K270R targets DMT1 for intracellular degradation rather than trafficking to EVs. (a) Arrdc4K270R significantly decreases DMT1 transport activity as measured by the fluorescence quenching assay (three independent experiments). + P < 0.05, ++ P < 0.01. (b) Expression of DMT1 is decreased in both EVs and cells expressing Arrdc4K270R compared to Arrdc4WT and other lysine mutants. (c) Confocal images of CHO‐DMT1 cells shows that expression of Arrdc4K270R causes uneven distribution of DMT1 in the cytoplasm and it colocalises with Rab7. Scale bar 10 μm. (d, e) Quantification of immunofluorescence intensity (n = 15 cells) demonstrate significant increase in intracellular DMT1 and Rab7 in cells expressing Arrdc4K270R. *P < 0.05, **P < 0.01 (f) Pearson's correlation analysis showing increased colocalisation of DMT1 with Rab7 in cells expressing Arrdc4K270R. ***P < 0.001. (g) Arrdc4K270R targets DMT1 for both lysosomal and proteasomal degradation in comparison with Arrdc4WT which causes DMT1 release in EVs. Inhibitors used are Chloroquine (C) and MG132 (M)
4. DISCUSSION
Ubiquitination is one of the most common post‐translational modifications found in eukaryotic cells, however its role in EV biogenesis remains unclear, with some evidence suggesting it can assist in the sorting of proteins to EVs (Smith et al., 2015), whereas other studies suggest that ubiquitination is not a prerequisite for the release of a protein in EVs (Gauvreau et al., 2009; Liu et al., 2009). As Arrdc1 and Arrdc4 are adaptors for the Nedd4 family of ubiquitin ligases and play a role in EV biogenesis (Mackenzie et al., 2016; Nabhan et al., 2012; Puca & Brou, 2014), the current study was designed to understand the role of ubiquitination in EV biogenesis. We provide evidence here that links specific Arrdc4 ubiquitination with its function in EV release. Our data indicate that: (i) Arrdc4 is ubiquitinated, possibly at five K residues, (ii) ubiquitination at K270 affects Arrdc4 stability and alters its subcellular localisation, (iii) Arrdc4 is ubiquitinated at K270 via K‐29 linked chains, (iv) the K‐29‐linked K270 ubiquitination regulates Arrdc4 function in EV biogenesis, and (v) Nedd4‐2 is involved in ubiquitin K‐29 chain formation on Arrdc4 K270. As K270 is highly conserved in Arrdc4 proteins across species, we propose that ubiquitination at K270 may be an evolutionarily conserved mechanism to regulate the function of Arrdc4.
Arrdc4WT is primarily localised on the plasma membrane and its localisation plays an essential role in Arrdc4‐mediated EV biogenesis (Mackenzie et al., 2016). The increase in stability and mislocalisation of Arrdc4K270R to the cytoplasm inform the importance of ubiquitination at this residue. The study involving Arrdc4 knockdown, followed by re‐expression of WT and K270R Arrdc4 highlights that K270 is vital for release of EV100‐200nm and mutation at K270 eliminates its activity.
We show that Arrdc4 is ubiquitinated by K‐29 and K‐63 polyubiquitin chains, with K270 being targeted specifically by K‐29 chains. It is well known that K‐48 chains are a signal for proteasomal degradation (Grice & Nathan, 2016, Schreiner et al., 2008) and K‐63 chains play a role in intracellular signalling, DNA repair and endosomal‐lysosomal degradation (Ikeda & Dikic, 2008; Ohtake et al., 2018). However, the biological role of atypical ubiquitin chains such as K‐6, K‐11, K‐27, K‐29 and K‐33 are less well established (Swatek & Komander, 2016). Previous work suggests that K‐29 and K‐63 ubiquitin chains adopt an open conformation whereas K‐6, K‐11 and K‐48 adopt a compact conformation (Datta et al., 2009; Komander et al., 2009; Kristariyanto et al., 2015). UBR5, UBE3C, SMURF1 and ITCH E3 ligases are reported to be involved in Ub K‐29 linkages (Chastagner et al., 2006; Fei et al., 2014; Hay‐Koren et al., 2011). SMURF1‐mediated non‐proteolytic Ub K‐29 linked polyubiquitination of AXIN results in negative regulation of Wnt/β signalling pathway (Fei et al., 2014). Our data now provide the first evidence for a vital role of K‐29 chains as a specific signal involved in EV biogenesis via Arrdc4. Moreover, we also provide the evidence that this interaction occurs at a highly conserved K270 of Arrdc4 protein. We also identify a novel function of Nedd4‐2 ubiquitin ligase in the K‐29‐linked polyubiquitination of Arrdc4. Ubiquitination mediated by Nedd4‐2 regulates a number of membrane proteins, including many ion channels and transporters (Goel et al., 2015; Manning & Kumar, 2018). Both in Saccharomyces cerevisiae and in mammalian cells the α‐arrestin proteins (ARTs/ Arrdcs) function as adaptors for Rsp5 (yeast) and Nedd4 family (mammals) of E3s to facilitate the ubiquitin‐dependent endocytosis and trafficking of several transporters (Kahlhofer et al., 2021). We speculate that a similar machinery involving Arrdc4 and Nedd4‐2 is also involved in ubiquitination‐dependent release of EVs from the plasma membrane.
By assessing the fate of DMT1, a membrane transporter that is known to be regulated by Arrdc4‐dependent release in EVs, we provide functional significance of K‐29 ubiquitinated K270 of Arrdc4. Increased expression of both Arrdc4 and Nedd4‐2 allows DMT1 release in EVs originating from the plasma membrane (Mackenzie et al., 2016), whereas expression of the Arrdc4K270R mutant significant decreases DMT1 release in EVs. These results confirm the critical role of Arrdc4 K‐29 ubiquitination by Nedd4‐2 in EV secretion. In conclusion, our study provides novel insight into the role of atypical K‐29 linked ubiquitin chains in the Arrdc4‐dependent biogenesis of plasma membrane EVs.
AUTHOR CONTRIBUTIONS
A.F., N.J.F., S.M., and S.K. conceptualized the project and designed experiments; A.F. and K.G. performed experimental work; A.F., N.J.F., and S.K. analysed data and wrote the manuscript; J.A.M. and S.S.S. assisted with the revision of the manuscript, acquisition and analysis of the new data and writing of revised manuscript. J.J.S. generated and J.J.S., A.W., and S.M. analysed proteomics data; S.K. and S.M. acquired funding. N.J.F. and S.K. co‐supervised this study. All authors edited text and provided feedback.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
This work was supported by the National Health and Medical Research Council (NHMRC) project grant (GNT1122437) to S.K. and S.M., as well as a NHMRC project grant (GNT1099307), a NHMRC Senior Principal Research Fellowship (GNT1103006), a NHMRC Investigator Grant (GNT2007739) and a University of South Australia support package to S.K. A.F. was supported by an Australian Postgraduate Award. We thank the School of Pharmacy and Medical Sciences Instrument & Facilities for NanoSight and members of our laboratory for helpful suggestions.
Farooq, A. U. , Gembus, K. , Sandow, J. J. , Webb, A. , Mathivanan, S. , Manning, J. A. , Shah, S. S. , Foot, N. J. , & Kumar, S. (2022). K‐29 linked ubiquitination of Arrdc4 regulates its function in extracellular vesicle biogenesis. Journal of Extracellular Vesicles, 11, e12188. 10.1002/jev2.12188
Natalie J. Foot and Sharad Kumar co‐supervised the project.
REFERENCES
- Alvarez, C. E. (2008). On the origins of arrestin and rhodopsin. BMC Evolutionary Biology, 8, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, B. M. , Prieto, M. , & Silva, L. C. (2014). Ceramide: A simple sphingolipid with unique biophysical properties. Progress in Lipid Research, 54, 53–67. [DOI] [PubMed] [Google Scholar]
- Anand, S. , Foot, N. , Ang, C. ‐S. , Gembus, K. M. , Keerthikumar, S. , Adda, C. G. , Mathivanan, S. , & Kumar, S. (2018). Arrestin‐domain containing protein 1 (Arrdc1) regulates the protein cargo and release of extracellular vesicles. Proteomics, 18, 1800266. [DOI] [PubMed] [Google Scholar]
- Chastagner, P. , Israel, A. , & Brou, C. (2006). Itch/AIP4 mediates Deltex degradation through the formation of K29‐linked polyubiquitin chains. EMBO Reports, 7, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A. B. , Hura, G. L. , & Wolberger, C. (2009). The structure and conformation of Lys63‐linked tetraubiquitin. Journal of Molecular Biology, 392, 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, C. , He, X. , Xie, S. , Miao, H. , Zhou, Z. , & Li, L. (2014). Smurf1‐mediated axin ubiquitination requires Smurf1 C2 domain and is cell cycle‐dependent. Journal of Biological Chemistry, 289, 14170–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foot, N. J. , Dalton, H. E. , Shearwin‐Whyatt, L. M. , Dorstyn, L. , Tan, S. S. , Yang, B. , & Kumar, S. (2008). Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin‐dependent mechanism involving Ndfips and WWP2. Blood, 112, 4268–4275. [DOI] [PubMed] [Google Scholar]
- Foot, N. J. , Gonzalez, M. , Gembus, K. , Fonseka, P. , Sandow, J. J. , Nguyen, T. T. , Tran, D. , Webb, A. I. , Mathivanan, S. , Robker, R. L. , & Kumar, S. (2021). Arrdc4‐dependent extracellular vesicle biogenesis is required for sperm maturation. Journal of Extracellular Vesicles, 10, e12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foot, N. , Henshall, T. , & Kumar, S. (2017). Ubiquitination and the regulation of membrane proteins. Physiological Reviews, 97, 253–281. [DOI] [PubMed] [Google Scholar]
- Foot, N. J. , Leong, Y. A. , Dorstyn, L. E. , Dalton, H. E. , Ho, K. , Zhao, L. , Garrick, M. D. , Yang, B. , Hiwase, D. , & Kumar, S. (2011). Ndfip1 deficient mice have impaired DMT1 regulation and iron homeostasis. Blood, 117, 638–646. [DOI] [PubMed] [Google Scholar]
- Fotia, A. B. , Dinudom, A. , Shearwin, K. E. , Koch, J. P. , Korbmacher, C. , Cook, D. I. , & Kumar, S. (2003). The role of individual Nedd4‐2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. FASEB Journal, 17, 70–72. [DOI] [PubMed] [Google Scholar]
- Gauvreau, M. E. , Cote, M. H. , Bourgeois‐daigneault, M. C. , Rivard, L. D. , Xiu, F. , Brunet, A. , Shaw, A. , Steimle, V. , & Thibodeau, J. (2009). Sorting of MHC class II molecules into exosomes through a ubiquitin‐independent pathway. Traffic (Copenhagen, Denmark), 10, 1518–1527. [DOI] [PubMed] [Google Scholar]
- Goel, P. , Manning, J. , & Kumar, S. (2015). NEDD4‐2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene, 557, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, G. L. , & Nathan, J. A. (2016). The recognition of ubiquitinated proteins by the proteasome. Cellular and Molecular Life Sciences, 73, 3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, K. F. , Shearwin‐whyatt, L. M. , Fotia, A. , Parton, R. G. , & Kumar, S. (2002). N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi‐associated protein. Journal of Biological Chemistry, 277, 9307–9317. [DOI] [PubMed] [Google Scholar]
- Hay‐Koren, A. , Caspi, M. , & Zilberberg, A. , & Rosin‐arbesfeld, R. (2011). The EDD E3 ubiquitin ligase ubiquitinates and up‐regulates beta‐catenin. Molecular Biology of the Cell, 22, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne William, M. , Buchkovich Nicholas, J. , & Emr Scott, D. (2011). The ESCRT pathway. Developmental Cell, 21, 77–91. [DOI] [PubMed] [Google Scholar]
- Howitt, J. , Putz, U. , Lackovic, J. , Doan, A. , Dorstyn, L. , Cheng, H. , Yang, B. , Chan‐Ling, T. , Silke, J. , Kumar, S. , & Tan, S.‐S. (2009). Divalent metal transporter 1 (DMT1) regulation by Ndfip1 prevents metal toxicity in human neurons. Proceedings National Academy of Science USA, 106, 15489–15494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J. H. (2015). ESCRTs are everywhere. The EMBO Journal, 34, 2398–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, F. , & Dikic, I. (2008). Atypical ubiquitin chains: New molecular signals. EMBO Reports, 9, 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki, T. , Maekawa, M. , Fujisawa, K. , Okawa, K. , Iwamatsu, A. , Fujita, A. , Watanabe, N. , Saito, Y. , Kakizuka, A. , Morii, N. , & Narumiya, S. (1996). The small GTP‐binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. The EMBO Journal, 15, 1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlhofer, J. , Leon, S. , Teis, D. , & Schmidt, O. (2021). The α‐arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis. Biology of the Cell, 113, 183–219. [DOI] [PubMed] [Google Scholar]
- Komander, D. , Reyes‐turcu, F. , Licchesi, J. D. , Odenwaelder, P. , Wilkinson, K. D. , & Barford, D. (2009). Molecular discrimination of structurally equivalent Lys 63‐linked and linear polyubiquitin chains. EMBO Reports, 10, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstas, A.‐A. , Shearwin‐Whyatt, L. M. , Fotia, A. , Degger, B. , Riccardi, D. , Cook, D. I. , Korbmacher, C. , & Kumar, S. (2002). Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4‐2‐interacting protein. Journal of Biological Chemistry, 277, 29406–29416. [DOI] [PubMed] [Google Scholar]
- Kristariyanto, Y. A. , Abdul Rehman, S. A. , Campbell, D. G. , Morrice, N. A. , Johnson, C. , Toth, R. , & Kulathu, Y. (2015). K29‐selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Molecular Cell, 58, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Harvey, K. F. , Kinoshita, M. , Copeland, N. G. , Noda, M. , & Jenkins, N. A. (1997). cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics, 40, 435–443. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A. , Macarthur, M. W. , Moss, D. S. , & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283–291. [Google Scholar]
- Liu, Y. , Shah, S. V. , Xiang, X. , Wang, J. , Deng, Z. B. , Liu, C. , Zhang, L. , Wu, J. , Edmonds, T. , Jambor, C. , Kappes, J. C. , & Zhang, H. G. (2009). COP9‐associated CSN5 regulates exosomal protein deubiquitination and sorting. American Journal of Pathology, 174, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, K. , Foot, N. J. , Anand, S. , Dalton, H. E. , Chaudhary, N. , Collins, B. M. , Mathivanan, S. , & Kumar, S. (2016). Regulation of the divalent metal ion transporter via membrane budding. Cell Discovery, 2, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, J. A. , & Kumar, S. (2018). Physiological functions of Nedd4‐2: Lessons from knockout mouse models. Trends in Biochemical Sciences, 43, 635–647. [DOI] [PubMed] [Google Scholar]
- Mathivana, S. , Ji, H. , & Simpson, R. J. (2010). Exosomes: Extracellular organelles important in intercellular communication. Journal of Proteomics, 73, 1907–1920. [DOI] [PubMed] [Google Scholar]
- Muralidharan‐Chari, V. , Clancy, J. , Plou, C. , Romao, M. , Chavrier, P. , Raposo, G. , & Dsouza‐Schorey, C. (2009). ARF6‐regulated shedding of tumor cell‐derived plasma membrane microvesicles. Current Biology, 19, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan, J. F. , Hu, R. , Oh, R. S. , Cohen, S. N. , & Lu, Q. (2012). Formation and release of arrestin domain‐containing protein 1‐mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proceedings of the National Academy of Sciences, 109, 4146–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake, F. , Tsuchiya, H. , Saeki, Y. , & Tanaka, K. (2018). K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proceedings of the National Academy of Sciences, 115, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca, L. , & Brou, C. (2014). Alpha‐arrestins – New players in Notch and GPCR signaling pathways in mammals. Journal of Cell Science, 127, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Putz, U. , Howitt, J. , Lackovic, J. , Foot, N. , Kumar, S. , Silke, J. , & Tan, S. S. (2008). Nedd4 family‐interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. Journal of Biological Chemistry, 283, 32621–32627. [DOI] [PubMed] [Google Scholar]
- Rotin, D. , & Kumar, S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nature Reviews. Molecular Cell Biology, 10, 398–409. [DOI] [PubMed] [Google Scholar]
- Scheffner, M. , & Kumar, S. (2014). Mammalian HECT ubiquitin‐protein ligases: Biological and pathophysiological aspects. BBA‐Molecular Cell Research, 1843, 61–74. [DOI] [PubMed] [Google Scholar]
- Schreiner, P. , Chen, X. , Husnjak, K. , Randles, L. , Zhang, N. , Elsasser, S. , Finley, D. , Dikic, I. , Walters, K. J. , & Groll, M. (2008). Ubiquitin docking at the proteasome through a novel pleckstrin‐homology domain interaction. Nature, 453, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, R. J. , Lim, J. W. E. , Moritz, R. L. , & Mathivanan, S. (2009). Exosomes: Proteomic insights and diagnostic potential. Expert Review of Proteomics, 6, 267–283. [DOI] [PubMed] [Google Scholar]
- Smith, V. L. , Jackson, L. , & Schorey, J. S. (2015). Ubiquitination as a mechanism to transport soluble mycobacterial and eukaryotic proteins to exosomes. Journal of Immunology, 195, 2722–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S. S. , & Kumar, S. (2021). Adaptors as the regulators of HECT ubiquitin ligases. Cell Death and Differentiation, 28, 455–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek, K. N. , & Komander, D. (2016). Ubiquitin modifications. Cell Research, 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research, 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. , & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J. J. , & Lotvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vidal, M. , Sainte‐marie, J. , Philippot, J. R. , & Bienvenue, A. (1989). Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: Evidence precluding a role for “aminophospholipid translocase”. Journal of Cellular Physiology, 140, 455–462. [DOI] [PubMed] [Google Scholar]
- Whitlock, J. M. , & Hartzell, H. C. (2017). Anoctamins/TMEM16 proteins: Chloride channels flirting with lipids and extracellular vesicles. Annual Review of Physiology, 79, 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , & Zhang, Y. (2015). I‐TASSER server: New development for protein structure and function predictions. Nucleic Acids Research, 43, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulli, V. , Friis, K. P. , Fitzpatrick, Z. , Maguire, C. A. , & Breakefield, X. O. (2016). Extracellular vesicles and intercellular communication within the nervous system. Journal of Clinical Investigation, 126, 1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.


