Abstract
Translation elongation is a key step of protein synthesis, during which the nascent polypeptide chain extends by one amino acid residue during one elongation cycle. More and more data revealed that the elongation is a key regulatory node for translational control in health and disease. During elongation, elongation factor Tu (EF-Tu, eEF1A in eukaryotes) is used to deliver aminoacyl-tRNA (aa-tRNA) to the A-site of the ribosome, and elongation factor G (EF-G, EF2 in eukaryotes and archaea) is used to facilitate the translocation of the tRNA2-mRNA complex on the ribosome. Other elongation factors, such as EF-Ts/eEF1B, EF-P/eIF5A, EF4, eEF3, SelB/EFsec, TetO/Tet(M), RelA and BipA, have been found to affect the overall rate of elongation. Here, we made a systematic review on the canonical and non-canonical functions and regulation of these elongation factors. In particular, we discussed the close link between translational factors and human diseases, and clarified how post-translational modifications control the activity of translational factors in tumors.
Keywords: translation, elongation factors, regulation, expression, tumorigenesis
Introduction
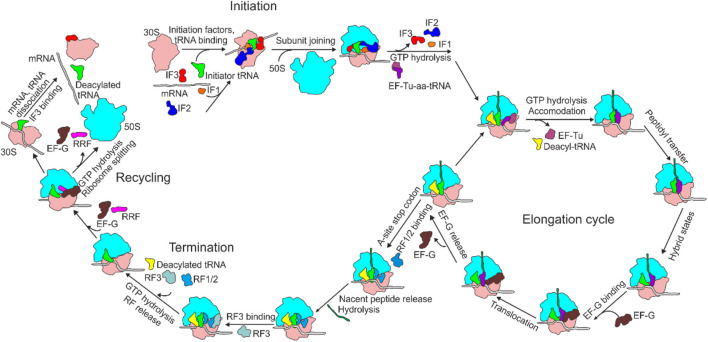
With the development of structural biology, especially the rapid development of cryo-electron microscopy (cryo-EM) (Benjin and Ling, 2019), the mechanism of intracellular protein translation and its regulation have been gradually clarified. As described by the central dogma, translation is the final stage of gene expression, during which the genetic information carried by an mRNA is transformed into the amino acid sequence of a protein catalyzed by a ribosome (Voorhees and Ramakrishnan, 2013). Translation is a highly dynamic and cyclic process, which is composed of four steps: initiation, elongation, termination, and ribosome recycling (Figure 1). During translation initiation, the ribosome, with an initiator fMet-tRNAfMet in the P-site, is assembled on mRNA with the assistance of the three initiation factors (IF1-3). The ribosome complex is then ready to accept the first elongator tRNA and form the first peptide bond, which marks the beginning of the next stage, elongation. During elongation, an aa-tRNA enters the ribosome A-site with the help of EF-Tu. If proper base-pairing between the three bases of the mRNA codon and those of the aa-tRNA anticodon is established, the aa-tRNA is cognate, a peptide bond is formed with the peptide attached to the tRNA in the P-site. The peptidyl-tRNA is then moved from the A-to the P-site, and the deacylated tRNA in the P-site is moved to the E-site. The mRNA is coordinately translocated by one codon. Termination occurs when the ribosome reaches a terminator codon in an mRNA. The nascent peptide is hydrolyzed by the release factors (RF1/2) and dissociated from the ribosome. In the end, the ribosome is split into two subunits by the concerted action of EF-G and RRF, releasing the deacylated tRNA and mRNA, and preparing for a new round of translation initiation. This step is called ribosome recycling.
FIGURE 1.
Overview of bacterial translation cycle. aa-tRNA, aminoacyl-tRNA; EF, elongation factor; IF, initiation factor; RF, release factor; RRF, ribosome recycling factor.
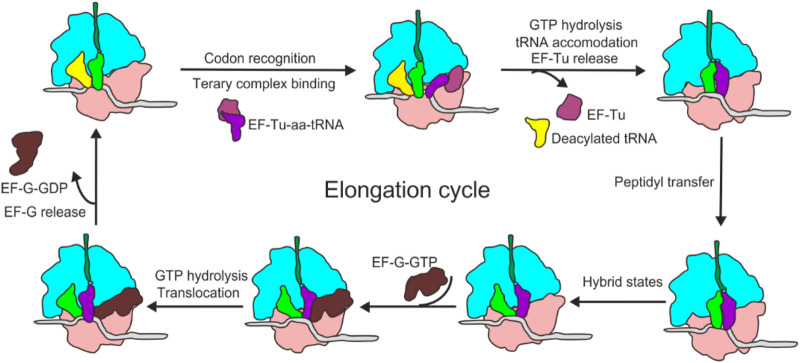
Translation elongation is a process of repeated decoding, peptidyl transfer and tRNA2-mRNA translocation. It starts with the binding of the second aminoacyl-tRNA at the A-site. During elongation, an aa-tRNA is delivered to the ribosome as a ternary complex (TC) with elongation factor Tu (EF-Tu) and GTP (EF-Tu∙GTP∙aa-tRNA). Binding of cognate aa-tRNA to mRNA in the A site of the ribosome induces the crucial and generally conserved bases A1493, A1492 and G530 to flip out and interact with the minor groove of the mRNA-tRNA duplex (Schmeing and Ramakrishnan, 2009), which further induces a domain closure in the 30S subunit (Ogle et al., 2002). 30S domain closure makes the 30S shoulder towards the ternary complex (Ogle and Ramakrishnan, 2005), leading to the stimulation of the GTP hydrolysis by EF-Tu and acceleration of tRNA selection. After decoding, EF-Tu dissociates from ribosome in the form of EF-Tu∙GDP, followed by the complete accommodation of aa-tRNA into the A-site. EF-Tu∙GDP is recycled to EF-Tu∙GTP by EF-Ts, a guanosine nucleotide-exchange factor, so as to participate in multiple rounds of peptide chain elongation. The next step is peptidyl transfer and peptide bond formation, which is catalyzed by the peptidyl transfer center (PTC) of the ribosome. During this stage, the nucleophilic α-amino group of the aa-tRNA in the A-site attacks the carbonyl carbon of the peptidyl-tRNA in the P-site, yielding a pre-translocation (PRE) ribosome complex with a deacylated tRNA in the P-site and a new, one residue longer peptidyl-tRNA in the A-site (Beringer and Rodnina, 2007). The third stage is EF-G∙GTP (eEF2∙GTP in eukaryotes) catalyzed translocation. Upon the addition of EF-G into the pre-translocational ribosome (PRE) system, EF-G in complexed with GTP facilitates movements of peptidyl-tRNA on the 50S subunit, and shifts the classical pre-translocation state to the hybrid state (Holtkamp et al., 2014a; Xie, 2016). And GTP hydrolysis induces a strong conformational change of 30S subunit within the 70S ribosome, allowing the movement of tRNA2-mRNA by one codon length inside the ribosome, shifting the tRNAs from A- and P-sites to P- and E-sites, respectively, resulting in a post-translocational ribosome (POST) (Figure 2).
FIGURE 2.
Schematic of the bacterial elongation cycle. EF-Tu delivers aa-tRNA to the A-site of the ribosome, where the ribosome decodes for the cognate tRNA. After aa-tRNA is fully accommodated and peptide bond formation, EF-G facilitates translocation of the tRNA2-mRNA duplex, then the next round of elongation begins.
There is a complex set of protein factors including EF-Tu/eEF1A, EF-G/eEF2, EF-P/eIF5A, and SelB/EFsec (Table 1), involved in translation elongation. Although the fundamental mechanism of the elongation cycle is very similar among three domains of life, the molecular mechanism of the elongation factors varies in different species. In the following, we will introduce in more detail the functions and regulation of the most important and well-studied canonical and non-canonical translation elongation factors that function in different stages of the elongation cycles of bacteria, archaea, and eukaryotes as well as that of organelles, including mitochondria.
TABLE1.
Translation elongation factors among bacteria, archaea, eukaryotes and mitochondria.
| Species | aa-tRNA incorporation | tRNA translocation | Other translation elongation factors | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | EF-Tu | EF-Ts | SelB | EF-G | EF4 | EF-P | — | Tet(O)/(M) | RelA | BipA |
| Archaea | aEF1A | — | — | aEF2 | — | aIF5A | — | — | — | — |
| Eukaryotes | eEF1A | eEF1B | EFsec | eEF2 | mtEF4 | eIF5A | eEF3 | — | — | — |
| Mitochondria | mtEF-Tu | mtEF-Ts | — | mtEF-G1 | mtEF4 | — | — | — | — | — |
None.
Factors Involved in AA-tRNA Incorporation
EF-Tu and EF-Ts
EF-Tu is encoded by two genes tufA and tufB in E. coli (Jaskunas et al., 1975), T. thermophilus (Satoh et al., 1991) and S. typhimurium (Hughes, 1990). It is a universally conserved GTPase in all species (Caldon and March, 2003). During translation elongation, EF-Tu∙GTP transports aa-tRNA to the ribosome A-site in the form of the ternary complex (Ramakrishnan, 2002). Upon binding of the ternary complex to the ribosome, proper base pairing between the anticodon of aa-tRNA and the mRNA codon within the 30S subunit decoding region stimulates EF-Tu to hydrolyze GTP. After the hydrolysis of GTP, the conformational change following the GTP hydrolysis to GDP and a leaving phosphate group (Pi) leads to the dissociation of EF-Tu from the ribosome and accommodation of the aa-tRNA on the A-site for a peptidyl transfer (Rodnina et al., 2005). The growing peptide chain extends by one amino acid under the catalysis of the ribosome. Recycling of EF-Tu∙GDP to EF-Tu∙GTP depends on EF-Ts, another elongation factor encoded by the tsf gene (Wang et al., 1997). EF-Tu can be reversibly phosphorylated on its serine and threonine residues, and this modification has been founded in multiple organisms including E. coli (Lippmann et al., 1993), Listeria monocytogenes (Archambaud et al., 2005), Thermus thermophilus (Lippmann et al., 1993), Streptococcus pneumoniae (Sun et al., 2010), Bacillus subtilis (Lévine et al., 2006), Corynebacterium glutamicum (Bendt et al., 2003), Mycoplasma pneumoniae (Schmidl et al., 2010), Mycobacterium tuberculosis (Sajid et al., 2011), and Streptomyces collinus (Mikulík and Zhulanova, 1995). Phosphorylation inhibits the GTPase activity of EF-Tu and prevents its dissociation from the ribosome (Pereira et al., 2015). Phosphorylated EF-Tu could not bind with aa-tRNA or kirromycin (Hughes, 2013). It has been reported that phosphorylation of EF-Tu plays a vital role in bacterial dormancy, sporogenesis, virulence, and stress tolerance (Archambaud et al., 2005; Holub et al., 2007; Misra et al., 2011; Pereira et al., 2015). The exact physiological significance of EF-Tu phosphorylation is still need to be clarified while phosphorylation of EF1A in eukaryotes has been shown to be involved in maintaining a proper elongation rate under various conditions (Hughes, 2013).
eEF1A and eEF1B
The GTPase eEF1A, the homolog of EF-Tu in bacteria, is one of the most widely expressed factors in eukaryotes (Schuller and Green, 2018). Human eEF1A has 33% sequence identity with bacterial EF-Tu (Cavallius et al., 1993). Similar to EF-Tu, another guanine nucleotide-exchange factor eEF1B is required to regenerate active eEF1A∙GTP (Gromadski et al., 2007). In lower eukaryotes, eEF1B contains a guanine nucleotide exchange subunit eEF1Bα and a structural subunit eEF1Bγ, while higher eukaryotes have another guanine nucleotide exchange subunit eEF1Bδ (plants) or eIF1Bβ (mammals). The mechanism of guanine-nucleotide exchange employed by eEF1B is very different from that of EF-Ts (Rodnina and Wintermeyer, 2009). Upon binding of EF-Ts to EF-Tu, the switch I region of EF-Tu is displaced by the C-terminal helix of EF-Ts. The switch II region is moved upon binding due to pushing by subdomain N. Altogether, these changes disrupt the coordination of the Mg2+ ion, leading to the dissociation of GDP. In contrast, eEF1Bα interacts with domains 1 and 2 of eEF1A, disrupting the binding pocket for Mg2+ and preventing the binding of the GDP to eEF1A.
In humans, there are two eEF1A homologues, named eEF1A1 and eEF1A2 (coded by two genes: EEF1A1 and EEF1A2). The sequences of eEF1A1 and eEF1A2 have 98% similarity and 92% identity, but the expression patterns of the two proteins are different (Tomlinson et al., 2005). eEF1A1 is ubiquitously expressed, whereas the expression of eEF1A2 is switched-on in adult life in specialized tissues such as skeletal muscle, cardio-myocytes and neurons (Lee and Surh, 2009). Overexpression of eEF1A2 has been reported to be linked with a variety of tumors (Lee and Surh, 2009), and mutations in EEF1A2 are related to a new type of epilepsy syndrome and intellectual disability (Inui et al., 2016; Lam et al., 2016). In addition to its canonical functions in transporting aa-tRNA to the ribosome, eEF1A is found to be involved in cellular activities such as regulation of cytoskeleton organization (Mateyak and Kinzy, 2010), protein degradation mediated by the proteasome (Mateyak and Kinzy, 2010), viral replication and propagation (Li et al., 2009), nuclear protein export (Khacho et al., 2008), signaling transduction pathway concerning apoptosis and oncogenesis (Schulz et al., 2014; Abbas et al., 2015) (Figure 3). eEF1A1 also plays an important role in the process of heat shock stress response (Vera et al., 2014). Single-cell transcriptomic analysis revealed that the expression level of eEF1A1 in neurons was low and changed with age in glial cells (Ximerakis et al., 2019). Therefore, eEF1A may represent a potential candidate for lifespan modulation (Skariah and Todd, 2020). In addition, eEF1A, along with eEF2, has been shown to be related to neurodegenerative disorders including Alzheimer disease (AD) and Parkinson disease (PD) with an unknown mechanism. The low expression levels of these factors in the brains of AD and PD patients indicating defects in the efficiency or fidelity of translation (Li et al., 2005; Vera et al., 2014; Garcia-Esparcia et al., 2015; Beckelman et al., 2016; Skariah and Todd, 2020). eEF1A has chaperone-like activity (Lukash et al., 2004) and may also be involved in antiviral response by interaction with Sgt1, a multifunctional protein (Novosylna et al., 2015).
FIGURE 3.
Canonical and non-canonical functions of eEF1A. In addition to its canonical functions in transporting aa-tRNA, eEF1A is also involved in cytoskeleton organization, protein degradation, viral replication and propagation, nuclear protein export, heat shock stress response, and signaling transduction pathway concerning apoptosis and oncogenesis.
Humans and yeast eEF1A are subjected to extensive methylation modifications at multiple conserved lysine residues (Hamey and Wilkins, 2018; Jakobsson et al., 2018; Robichaud et al., 2019). Methylation modification is the addition of 1∼3 (me1-me3) methyl groups to the side chains of lysine residues. Up to now, several methylation sites and corresponding methyltransferases of eEF1A have been identified in humans and yeasts. For example, human N6AMT2 (eEF1A-KMT1), METTL10 (eEF1A-KMT2), METTL21B (eEF1A-KMT3), and eEF1A-KMT4 (gene name EEF1AKMT4), which methylate eEF1A at K79 (me3), K318 (me3), K165 (me1/me2/me3), and K36 (me3), respectively (Shimazu et al., 2014; Hamey et al., 2016; Jakobsson et al., 2017; Malecki et al., 2017); In addition, two other methylation sites (N terminus and K55) have been reported in human cells, but the enzymes responsible for N-terminal trimethylation and K55 dimethylation in human eEF1A still need to be identified (Hamey et al., 2017). Saccharomyces cerevisiae Efm1, Efm4, Efm5, Efm6, and Efm7, which methylate eEF1A at K30 (me1), K316 (me2), K79 (me3), K390 (me1), and N-terminal (me3) and K3 (partially me1 and me2) respectively (Lipson et al., 2010; Dzialo et al., 2014; Jakobsson et al., 2015; Hamey et al., 2016). The abundant lysine methylations of eEF1A and the existence of multiple corresponding methyltransferases in various eukaryotes made it clear that the lysine methylation of eEF1A has important physiological significance. In METTL21B knockout (KO) human cells, the expression of proteins involved in cytoskeleton organization was downregulated, while the expression of proteins related to large ribosomal subunit biogenesis, mRNA turnover and rRNA processing were upregulated (Hamey et al., 2017). Moreover, mammalian METTL21B was found to be partially localized in the centrosome, which may reveal a non-canonical function for this protein (Malecki et al., 2017). In EEF1AKMT4 KO cells, the global translation is changed and the translation speed of codons for histidine (H), tryptophan (W), and asparagine (N) was altered compared with wild-type cells (Jakobsson et al., 2017). In EEF1AKMT1 KO cells, the expression of proteins related to tRNA aminoacylation and nuclear exosome were downregulated, while the expression of proteins related to ubiquitination regulation and small-subunit processome were upregulated (Hamey et al., 2017). Even though the physiological function of K318 methylation of eEF1A has not been clarified in mammalian, perhaps it can be speculated that K318 methylation may affect the replication of RNA virus and the migration of neural crest based on the highly conservative of this site between human and yeast (Shimazu et al., 2014). In yeast, Efm4 is involved in vesicle transport processes including secretory protein production, transfection and endocytosis (Martín-Granados et al., 2008). Efm4 is also play a vital role in Tombusvirus replication (Li et al., 2014). Similarly, the gene of EFM5 was shown to be crucial for virus replication in yeast (Kushner et al., 2003). In S. cerevisiae, EFM7 KO leads to decreased replicative lifespan (Anderson et al., 2003), which results from altered translation rate (Pan et al., 2007). Methylation of eEF1A by Efm6 occurs in its domain III, which is involved in protein translation and cytoskeleton organization (Gross and Kinzy, 2005; Liu et al., 2006). Finally, even though the five methyltransferases of eEF1A in S. cerevisiae are not absolutely necessary to its viability, the precise regulation of eEF1A function by distinct methyltransferases optimizes the cell physiology (White et al., 2019). Besides methylation, the lysine residues of eEF1A are also modified by acetylation, sumoylation and ubiquitination, as well as phosphorylation of tyrosine, threonine and serine residues (Hornbeck et al., 2012).
mtEF-Tu and mtEF-Ts
Eukaryotic cells, including those in animals and fungi, contain two translation systems, one in the cytosol and the other in the mitochondria. Mitochondria use their own translational system to synthesize proteins for respiratory chain complexes. mtEFs (mitochondrial translation elongation factors) are coded by the nuclear genome, synthesized and transported into mitochondria. These factors are more similar to their counterparts in bacteria than those from the cytoplasm of eukaryotes. Genes encoding mtEFs such as TUFM (mtEF-Tu), TFSM (mtEF-Ts), and GFM1(mtEF-G1), have been reported with mutations in cases causing down regulation of mitochondrial translation and early fatality (Hughes, 2013).
mtEF-Tu consists of 409 amino acids, and is 55∼60% identical to the homologous protein from bacteria. It was found that mtEF-Tu folded into three main domains similar to EF-Tu. One of the main differences is that the C-terminal of mtEF-Tu has an 11 amino acids extension, which may interact with aa-tRNA (Jeppesen et al., 2005). Early studies showed that, mtEF-Tu was compatible with E. coli aa-tRNAs, whereas E. coli EF-Tu was unable to catalyze polypeptide chain elongation when supplied with mitochondrial aa-tRNAs (Kumazawa et al., 1991). This is probably due to the incorrect positioning of the shorter mitochondrial aa-tRNAs on the ribosome by bacterial EF-Tu, which leads to the ineffective stimulation of the GTPase activity of EF-Tu (Christian and Spremulli, 2012). Unlike EF-Tu, the bacterial and mitochondrial EF-Ts show low (25∼30%) sequence conservation. The most widely studied form of mtEF-Ts comes from B. taurus. It consists of 283 amino acids, with a mitochondrial import signal about 55 residues reside in the N-terminal of the mature protein (Xin et al., 1995).
SelB
Selenocysteine (Sec) is a cysteine (Cys) residue analogue with a selenium-containing selenol group in place of the sulfur-containing thiol group in Cys. The selenium atom gives Sec quite different properties from Cys. Sec utilization is scattered across archaea (Mariotti et al., 2016) and bacteria (Zhang et al., 2006). In eukaryotes, selenoproteins exist in some algae and protozoa (Lobanov et al., 2009; Mariotti et al., 2015), and most metazoans (Mariotti et al., 2012). Recently, Mariotti et al. (2019) provided evidence for Sec usage in early-branching fungal phyla. In mammals, Sec exists in enzymes associated with ROS detoxification and hormone biosynthesis. It plays a vital role in many biological processes including development, reproduction, immune response, tumorigenesis, viral infections and cardiovascular diseases (Carlson et al., 2006). The terminator codon UGA behaves as the codon of Sec when the downstream of UGA possesses a selenocysteine insertion sequence (Ose et al., 2007). During the translation of selenoproteins such as glutathione peroxidase and bacterial formate dehydrogenase, SelB, a unique protein factor, is needed to deliver selenocysteinyl-tRNASec containing a UCA anticodon to the ribosome A-site to recognize UGA codon in proper position of an mRNA (Böck et al., 1991). Incorporation of Sec is governed by a unique mRNA hairpin that located 3′ near the Sec codon (Yoshizawa et al., 2005). This hairpin structure associates with the corresponding ternary complex that is composed of SelB, Sec-specific aa-tRNA and GTP (Yoshizawa et al., 2005). It is assumed that by this mechanism selenocysteinyl-tRNASec is delivered to the ribosome.
Factors Involved in Ribosome Translocation
EF-G
EF-G, which is the third most conserved trGTPase among all domains of life (Caldon and March, 2003), catalyzes the translocation of A-site peptidyl-tRNA and P-site deacylated tRNA to the P- and E-site, respectively (Ramakrishnan, 2002). Translocation of tRNA2-mRNA during translation elongation is associated with EF-G triggered GTP hydrolysis and a series of conformational changes of the ribosomes (Savelsbergh et al., 2005). Recently, Holtkamp et al. (2014b) concluded that EF-G integrates the energy regimes of a motor protein and a GTPase, and promotes tRNA motion through the combined use of power stroke and Brownian ratchet mechanisms. After GTP hydrolysis, EF-G departs from the ribosome in the form of EF-G∙GDP. EF-G is the only canonical trGTPase that functions at two distinct phases in bacterial translation (Ero et al., 2016). In addition to facilitating translocation in elongation, it also plays an essential role in ribosome recycling, during which EF-G∙GTP works together with RRF to split the post-termination complex (PoTC) into two subunits (Song et al., 2020).
The GTPase activity of EF-G involves two hydrophobic amino acids Ile61 and Ile19 (E. coli numbers), which facilitate approaching of His92 to GTP by forming an opened hydrophobic gate. The water molecule promoted by His92 attacks the γ phosphate of GTP, resulting in an active state of the GTPase center (Yamamoto et al., 2014). EF-G is extensively modified by reversible phosphorylation. An early study reported that E. coli EF-G can be phosphorylated by a serine/threonine specific protein kinase (gp 0.7 PK) encoded by the T7 early gene 0.7 (Robertson et al., 1994). This gene was expressed early following T7 infection, leading to rapid shutdown of host RNAP at 4 min after infection, and soon thereafter, most protein synthesis began to turn off gradually. Which established favorable conditions for T7 phage growth (McAllister and Barrett, 1977). And this modification may help to increase the translation elongation rate of T7 late genes that specify T7 virion assembly and structural proteins (Robertson et al., 1994). Another in vivo and in vitro study in B. subtilis showed that EF-G can be phosphorylated on at least one threonine residue by a membrane Ser/Thr kinase PrkC and dephosphorylated by phosphatase PrpC, and the dynamic control of EF-G phosphorylation may play a regulatory role in stationary-phase B. subtilis (Gaidenko et al., 2002). Later, Shah and Dworkin (2010) proved that phosphorylation of EF-G by PrkC in B. subtilis is in response to cell wall-derived muropeptides. E. coli EF-G can also be modified at the lysine residue essential for GTP binding by pyridoxal phosphate (PLP), a selective, site-specific lysine reagent, leading to progressive loss of the EF-G activity, and destruction of its interaction with 30S subunits as well as a conformational change required for GTP hydrolysis (Giovane et al., 1982). Although these effects have been more widely studied, the physiological significance of phosphorylated EF-G still needs to be elucidated (Hughes, 2013).
eEF2
eEF2 is the eukaryotic homolog of EF-G. When cells are starved of nutrients, eEF2 is phosphorylated by the Ca2+-activated kinase eEF2K, resulting in a lower binding affinity to the ribosome (Carlberg et al., 1990). The activity of eEF2K is regulated by nutrients through mTORC1 and AMPK (Kenney et al., 2014; Proud, 2019). During translation elongation, the active eEF2K can reduce termination read-through errors and codon-anticodon mismatches, and promote more accurate recognition of the start codon by reducing initiation at the near-AUG codons (Xie et al., 2019). Interestingly, despite the major role of eEF2’s phosphorylation in blocking whole protein translation, its phosphorylation in neurons is associated with elevated translation of Arc/Arg3.1 which plays a key role in postsynaptic endocytosis (Park et al., 2008).
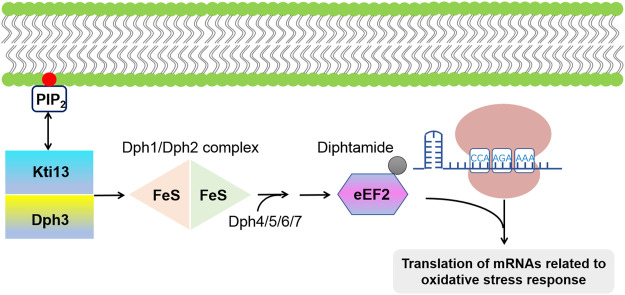
Diphthamide is another conserved modification in archaeal and eukaryal eEF2, where a conserved histidine (H715 in mammals; H699 in S. cerevisiae) at the eEF2 domain IV is modified with diphthamide (Su et al., 2013; Schaffrath et al., 2014). In eukaryotes, this modification event occurs through a four-step pathway involving multiple proteins including Dph1-Dph7 (Versées, 2015). Dph1/Dph2 is a [4Fe-4S] cluster-containing heterodimeric protein complex, which is responsible for catalyzing the first step of this modification pathway. In S. cerevisiae, the highly conserved proteins Dph3 (or Kti11) and Kti13 form a heterodimer, which is involved in eEF2 modification by acting as an electron donor for the Dph1/Dph2 complex (Dong et al., 2014; Glatt et al., 2015). With the help of Dph4/5/6/7, the diphthamide group was finally added to eEF2. Moreover, Kti13 was reported to specifically binding with PIP2 (Di Paolo and De Camilli, 2006), which might contribute to the regulation of the downstream eEF2 modification pathway (Figure 4). Lack of the diphthamide modification is fatal to mice due to severe developmental defects (Yu et al., 2014). It is worth noting that yeast lacking Dph1, an enzyme needed for diphthamide synthesis, grew normally, indicating that diphthamide likely functions in translation fidelity but not a basic mechanism of translation (Dever et al., 2018). Besides, some mammalian cells can survive in the absence of diphthamide (Stahl et al., 2015). However, related studies (Liu et al., 2012) had shown that lacking diphthamide modification results in an increase in the level of programmed -1 ribosomal frameshifting. Considering its location in eEF2, it is reasonable to speculate that diphthamide may enhance the eEF2 function by contacting with RNA in the ribosomal decoding center, thus facilitating the ribosomal translocation fidelity (Dever and Green, 2012).
FIGURE 4.
Schematic of the diphtamide modification of eEF2. The first step in this modification pathway is catalyzed by [4Fe-4S] cluster-containing protein complex Dph1/Dph2; with the help of Dph4/5/6/7. Dph3/Kti13 heterodimer act as electron donor for Dph1/Dph2 complex. It has been reported that diphtamide group of EF2 can further be ADP-ribosylated by the bacterial diphtheria toxin, leading to a global inhibition of protein synthesis as well as an upregulated translation of mRNAs associated with oxidative stress response.
Diphthamide of eEF2 can be further ADP-ribosylated by diphtheria and cholera toxins, which catalyze the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to the diphthamide imidazole ring to yield ADP ribosyl diphthamide (Argüelles et al., 2014). ADP-ribosylation inactivates eEF2, hinders protein translation and damages cell growth (Mateyak and Kinzy, 2013). Recently, it has been reported that cells possess intrinsic abilities to modify the diphthamide group by ADP-ribosylation, and this ability will improve under certain stress conditions, causing the overall down-regulation of protein synthesis at the cost of an increased translation of IRES-containing mRNA that involved in the response of oxidative stresses (Argüelles et al., 2014). However, the molecular mechanism of how ADP-ribosylation impairs the function of eEF2 has not been fully elucidated (Mateyak and Kinzy, 2013).
mtEF-G1 and mtEF-G2
In human mitochondria, the dual function of bacterial EF-G is fulfilled by mtEF-G1 and mtEF-G2 (Tsuboi et al., 2009; Christian and Spremulli, 2012). During mitochondrial translation elongation, ribosome translocation was catalyzed by mtEF-G1. Previous studies revealed that mtEF-G1 has a strong tolerance to fusidic acid, an antibiotic that inhibits EF-G release from the ribosome without influencing on GTP hydrolysis and translocation (Gao et al., 2009; Savelsbergh et al., 2009), while the tolerance mechanism of mtEF-G1 to fusidic acid remains to be understood (Christian and Spremulli, 2012). mtEF-G1 is active not only with 55S mammalian mitochondrial ribosome but also with 70S bacterial ribosome. In contrast, E. coli EF-G can’t work with mitochondrial ribosomes (Chung and Spremulli, 1990). mtEF-G2 mediates ribosome recycling in concert with human mitochondrial RRF after termination (Achenbach and Nierhaus, 2015). However, it should be noted that overexpression of mtEF-G2 can improve the translation of respiratory chain complexes slightly in cells with mtEF-G1 mutation, indicating that mtEF-G2 perhaps plays a part in the translation elongation (Coenen et al., 2004). Unlike ribosome recycling in bacteria, mtEF-G2 catalyzed GTP hydrolysis is not essential for ribosome dissociation. Rather, it seems to be necessary for the dissociation of mtRRF and mtEF-G2 from ribosomes. Since mtEF-G2 represents a type of trGTPase participating in ribosome recycling, it has been proposed to rename this factor as mitochondrial ribosome recycling factor 2 (mtRRF2) (Tsuboi et al., 2009).
Other Elongation Factors
EF4
EF4 (LepA) was originally identified in E. coli in 1985 (March and Inouye, 1985). The high conservation of EF4 in bacteria suggests its functional importance (Margus et al., 2007). Nierhaus and co-workers reported that cell membranes behave as EF4 reservoir pool, releasing it to the cytoplasm under certain conditions such as elevated intracellular Mg2+ concentrations or low temperature, leading to an increased rate of translation and efficient folding of newly synthesized peptides (Pech et al., 2011; Yamamoto et al., 2014). Functional studies showed that EF4 knockout affects bacterial growth under conditions of high Mg2+ concentration (Pech et al., 2011) or low pH (Yang et al., 2014). In S. cerevisiae, lacking EF4 (Guf1) results in growth defects under conditions of starvation and low temperature, and decreased expression of cytochrome oxidase (Bauerschmitt et al., 2008). Besides, EF4 knockout E. coli cells showed a decreased translation rate and slow ribosome maturation at unfavorable conditions (Yang et al., 2014). Overexpression of EF4 in E. coli seriously affects the growth of cells (Qin et al., 2006), while EF4 knockout cells showed no obvious phenotype under culture conditions of rich LB medium (Shoji et al., 2010). Therefore, EF4 may contribute to the cell survival under adverse conditions, but the physiological role of the protein remains unclear.
A previous study proposed that EF4 acts as a ‘back-translocase’ that has a unique property of recognizing ribosomes with mistranslocated tRNAs and back-translocating them via GTP hydrolysis during elongation cycle (Qin et al., 2006). However, several subsequent studies could not confirm that EF4 did have this biochemical activity (Liu et al., 2010; Balakrishnan et al., 2014; Gibbs and Fredrick, 2018). Cooperman and coworkers have done the most detailed biochemical characterization of EF4, studying its effect on both PRE and POST state complexes (Liu et al., 2010; Liu et al., 2011). They found that the addition of EF4 to POST can promote movement of tRNA with respect to the 50S subunit but does not catalyze back-translocation. Notably, Cooperman also showed that the EF4 preferentially engages the PRE complex, and competes with EF-G rather than EF-Tu ternary complex to influence elongation. In vitro kinetic measurements showed that EF4-dependent back translocation proceeds through a four-stage kinetic route (POST→I1→I2→I3→PRE), not just a reversal of translocation but exist three intermediate states, and the rate of reverse codon-anticodon movement observed in the presence of EF4 is virtually identical to that seen in its absence (Liu et al., 2010), in line with the independent work led by Fredrick and coworkers, whose ribosome profiling results revealed that EF4 contributes mainly to the initiation phase of translation in E. coli (Balakrishnan et al., 2014). This is consistent with a recent physiological study suggesting EF4 contributes to biogenesis of the 30S subunit, immature 30S particles will accumulate in cells lacking EF4 (Gibbs et al., 2017).
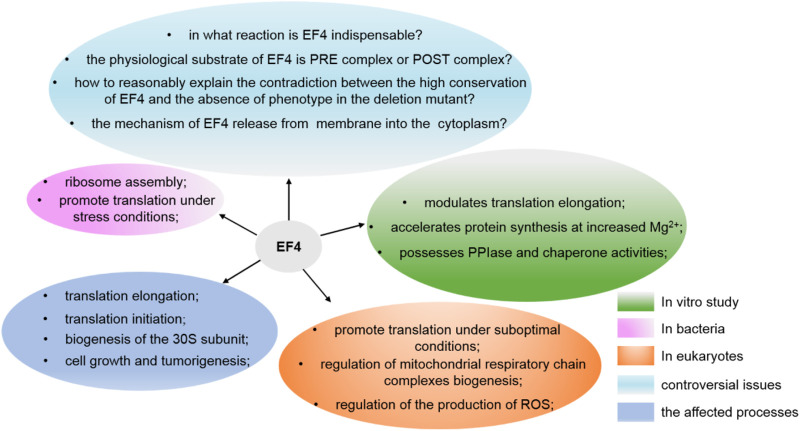
In summary, although the major role of EF4 is to facilitate bacteria in response to some stresses and affect protein synthesis in general, still some important issues concerning EF4 need to be resolved: What is the mechanism of EF4 release from the membrane into the cytoplasm? What is the real physiological substrate of EF4? How to reasonably explain the contradiction between the high conservation of EF4 and the obvious lack of phenotype in its deletion mutants (Figure 5)?
FIGURE 5.
Various functions of EF4 and the issues still need to be resolved.
mtEF4
mtEF4 is the homologue of bacterial EF4 in eukaryotic mitochondria. It consists of 651 residues with a mitochondrial-targeting signal in their N termini (Gao et al., 2016). mtEF4 is located in the mitochondrial matrix, close to the inner membrane (Bauerschmitt et al., 2008). mtEF4 binds to the ribosome in a GTP-dependent manner which is similar to that of bacterial EF4. It promotes the translation of mitochondrial proteins under non optimal conditions (Bauerschmitt et al., 2008). mtEF4 also plays a quality control role in the biogenesis of mitochondrial respiratory chain complexes. Previous studies showed that mtEF4 knockout induces respiratory chain defects as well as apoptosis, whereas overexpression of the protein stimulates cancer development (Zhu et al., 2018). mtEF4 ablation in mice results in testis-specific disorder of oxidative phosphorylation, and its deletion facilitated mitochondrial protein translation in the expense of synthesis unstable proteins (Gao et al., 2016). Increased expression of mtEF4 in multiple cancers suggested that mtEF4 probably facilitates tumorigenesis through an unbalanced regulation of mitochondrial activities and cellular redox (Zhu et al., 2018). Therefore, the proper level of mtEF4 in cell is requisite for the assembly of functional respiratory chain complexes as well as mitochondrial protein synthesis.
EF-P and eIF5A
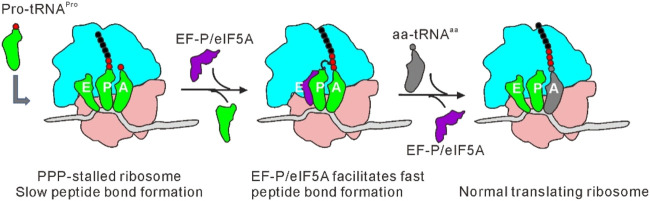
Bacterial EF-P (elongation factor P) is a homolog of eukaryotic and archaeal initiation factor 5A(e/aIF5A) (Kyrpides and Woese, 1998). It binds to the interface of the ribosome subunits and facilitates peptide bond formation through interactions with the P-site tRNA (Glick and Ganoza, 1975; Blaha et al., 2009). efp, the coding gene of EF-P, has been found throughout the bacteria (Hughes, 2013). It was reported that EF-P participates in the regulation of cell viability, growth, motility, virulence (Ude, 2013), and tolerance to multiple stresses including several classes of antibiotics, detergents, nutrient-limiting conditions and diverse growth inhibitors (Navarre et al., 2010; Zou et al., 2012). The ribosome needs the help of EF-P when successive prolines are incorporated into the nascent peptide chain (Rodnina, 2018). Without EF-P, the ribosome would stagnate at polyproline stretches, while the addition of EF-P could alleviate the translation stalling (Doerfel et al., 2013). EF-P is often subject to post-translational modification by PoxA catalyzed adding of a (R)-β-lysine to Lys34 (termed lysinylation) in bacteria like E. coli (Peil et al., 2012), S. enterica (Hersch et al., 2013) and S. typhimurium (Navarre et al., 2010; Zou et al., 2011), which could improve the catalytic activity of EF-P both in vitro and in vivo (Navarre et al., 2010; Park et al., 2012). β-lysylated EF-P also undergoes further hydroxylation, a second post-translational modification of this factor, but the function of hydroxylation does not seem to be critical for EF-P (Peil et al., 2012; Bullwinkle et al., 2013). In addition to the post-translational modifications mentioned above, other modifications of EF-P had been identified in other bacteria. For example, rhamnosylation of Arg32 in S. oneidensis, P. aeruginosa, N. gonorrhoeae, B. pertussis and N. meningitidis, which is closely related to bacterial fitness, pathogenicity and viability (Lassak et al., 2015; Rajkovic et al., 2015; Yanagisawa et al., 2016; Hummels and Kearns, 2020); 5-aminopentanolylation of Lys32 in B. subtilis, which can regulate the synthesis of diprolyl motifs of specific proteins needed for swarming motility (Rajkovic et al., 2016). There are hundreds of polyproline-containing peptides and proteins with different functions in all organisms, indicating EF-P (e/aIF5A) is necessary for the regulation of expression levels in various pathways.
eIF5A was firstly reported to promote the first peptide bond formation and was denoted as an initiation factor (Kemper et al., 1976). In archaea and eukaryotes, a conserved lysine located at the eIF5A domain I (Kim et al., 1998) is modified to hypusine post-translationally. Synthesis of hypusine is catalyzed by two consecutive enzymatic reactions involving deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH) (Park et al., 2010). Hydroxyl radical probing experiments revealed that the binding site of eIF5A on the ribosome is adjacent to the E-site, and the hypusine group is in proximity to the acceptor arm of the P-site tRNA (Gutierrez, 2013). An early study showed that unmodified eIF5A is unable to catalyze the formation of methionyl-puromycin (Park et al., 1991). Depletion of eIF5A leads to dysregulated translation initiation (Manjunath et al., 2019), elongation/termination (Schuller et al., 2017), and an increase in ribosomal transit times (Henderson and Hershey, 2011). In the absence of cycloheximide, inhibition of an eIF5A mutant that is sensitive to temperature leads to polysome accumulation (Saini et al., 2009), indicating eIF5A plays a vital role in the elongation phase. Moreover, if eIF5A is present, ribosome stalling will be restored when multiple prolines are to be incorporated in vitro. eIF5A depletion leads to defects in the synthesis of polyproline-containing proteins (Gutierrez, 2013), and ribosome stalling at tripeptides such as RDK, DVG, DDG, DDP, PDP and DNP (Schuller et al., 2017). Therefore, like EF-P in bacteria, eIF5A is considered to promote peptide transfer and improve the translation efficiency of poor substrates like proline (Gutierrez, 2013) (Figure 6). eIF5A was also found to play a role in the start codon selection during translation initiation. Depletion of eIF5A enhances upstream translation within 5′ UTRs across yeast and human transcriptomes (Manjunath et al., 2019).
FIGURE 6.
Mechanism of EF-P/eIF5A in alleviating ribosome stalling at polyproline stretches. Accomodation of the third consecutive Pro-tRNAPro into the A-site of translating ribosome leads to stalling. EF-P/eIF5A binds to a location between E- and P-sites of the stalled ribosome after E-site tRNA is dissociated, stimulating rapid proline-proline peptide bond formation. Translation can resume following dissociation of EF-P/eIF5A and binding of the next aa-tRNA.
eEF3-Fungal Specific Elongation Factor
eEF3, a third elongation factor which was reported to be important for the protein translation and viability in higher fungi including yeast and P. carinii (Triana-Alonso et al., 1995). eEF3 from S. cerevisiae is composed of 1,044 amino acids and the coding gene of this factor is YEF-3 (Qin et al., 1990). The crystal structure of S. cerevisiae eEF3 revealed that it is composed of a HEAT domain, two ATPase domains, a four-helix bundle, and a chromodomain (Andersen et al., 2006). It has a ribosome-dependent ATPase and GTPase (Dasmahapatra and Chakraburtty, 1981). eEF3 is mainly associated with cytosolic polysomes, and is needed for peptide bond formation (Kapp and Lorsch, 2004). The existence of eEF3 exclusively in fungi may be the most striking exception to the highly conserved translation elongation. It was reported that eEF2 and eEF1 from yeast can work with mammalian ribosomes to promote translation. In contrast, eEF2 and eEF1 from mammalian could function with yeast ribosomes only when eEF3 is present (Skogerson and Engelhardt, 1977), indicating eEF3 is required for yeast ribosome translation. Cryo-EM structures revealed that eEF3 has a new binding site close to the ribosome E-site, with the chromodomain stabilizing L1 stalk to facilitate tRNA release, which is consistent with the model that eEF3 may aid E-site tRNA release following translocation (Andersen et al., 2006).
The question that still needs to be answered is why do fungal ribosomes require eEF3 to promote E-site tRNA release and A-site occupancy while other ribosomes do not? Rodnina et al. (1994) reported that 80S from several higher eukaryotes can hydrolyse ATP and GTP without protein factors. This inherent ATPase activity functions as eEF3, promoting the dissociation of deacylated tRNA at the ribosome E-site (El’skaya et al., 1997), and cognate tRNA binding at the ribosome A-site seems to stimulate this activity in turn (Rodnina et al., 1994). Nevertheless, no homologues of fungal eEF3 have been found in mammalian cells. It is worth noting that a crystal structure of S. cerevisiae 80S contains an additional non-ribosomal protein, Stm1, which bound to the 40S subunit and precluded mRNA entry by placing a α-helix to the mRNA tunnel (Ben-Shem et al., 2011). In yeast, deletion of stm1 leads to increased binding affinity of eEF3 to ribosomes, whereas up-regulation of eEF3 expression in cells lacking Stm1 leads to growth defect and elevated anisomycin (a translation inhibitor) sensitivity. Moreover, a high level of Stm1 in ribosomes displays reduced eEF3 binding. Therefore, it can be concluded that Stm1 and eEF3 may jointly promote the elongation cycle (Van Dyke et al., 2009). Even though the characteristics of bacterial ribosome structures do not suggest the need for another elongation factor (Dever and Green, 2012), E. coli has an ATPase RbbA, which can bind to the ribosome and stimulates protein translation in vitro, and displays multiple biochemical properties similar to that of eEF3 (Kiel and Ganoza, 2001). More studies concerning biochemistry, structural biology and high resolution fluorescence imaging are required to fully reveal the function of eEF3 and to resolve its specific needs in fungi translation elongation (Dever and Green, 2012).
Tet(O) and Tet(M)
The tetracycline resistance proteins (Tet), which protect the bacterial ribosome from binding the antibiotic tetracycline, are another class of ribosome-associated GTPases (Margus et al., 2007). They are also called ribosomal protection proteins (RPPs). They are cytoplasmic proteins that display homology with the elongation factors EF-Tu and EF-G (Hughes, 2013). They bind to the ribosome, hydrolyze GTP and cause the release of tetracycline from the ribosome (Connell et al., 2003). RPPs are paralogs of elongation factors, and the best characterized and widely distributed RPPs are Tet(M) and Tet(O) (Hughes, 2013). Both Tet(M) and Tet(O) have ribosome-dependent GTPase activity, the hydrolysis of GTP providing the energy for the ribosomal conformational changes (Connell et al., 2003). Direct competition experiments showed that the ribosome binding site of Tet(M) is overlapping with that of EF-G (Dantley et al., 1998). To clarify the mechanism of the tetracycline resistance, Burdett purified Tet(M) protein (Burdett, 1991) and investigated its effects on several reactions that occur during protein translation (Burdett, 1996). The author found that Tet(M) could alleviate the inhibition of tetracycline on factor-dependent tRNA binding, and significantly reduce the affinity of ribosomes for tetracycline in the presence of GTP. Adding Tet(M) to the ribosome-tetracycline complex will replace the bound tetracycline. Therefore, the dissociation of tetracycline from the ribosome promoted by Tet(M) is GTP dependent. Structures of 70S∙Tet(O) complex revealed that Tet(O) really looks like EF-G and binds to the ribosome with an identical site (Spahn et al., 2001). In 2013, Li et al. identified the Tet(O) binding site on the ribosome, which involves three unique loops in Tet(O) domain IV (Li et al., 2013). The single glycine substitution of the residues in these loops leads to loss of tetracycline resistance.
RelA
In order to adapt to the changes in environment conditions, pathogens have evolved a variety of mechanisms to respond to various stresses, the most important of which is called stringent response (SR) (Winther et al., 2018). SR is a nearly universal mechanism mediated by pppGpp and ppGpp (Steinchen and Bange, 2016). Seeing that SR is related to metabolic regulation, biofilm formation, virulence gene expression (Yang et al., 2020), stress, antibiotic resistance, as well as the formation ability of bacteria retention (Hauryliuk et al., 2015), therefore, the signal pathway becomes a target for designing effective antibacterial agents (Kushwaha et al., 2019). The intracellular levels of pppGpp and ppGpp (collectively referred to as (p)ppGpp) are controlled by RelA/SpoT Homologue (RSH) proteins, which synthetize (p)ppGpp by transferring the pyrophosphate group of ATPs onto the 3′ of GDP or GTP, and degrade (p)ppGpp by removing the 3′ pyrophosphate moiety. It interacts with the ribosome to sense environmental pressure and leads to an adaptive response of pathogens. When pathogens encounter stresses like nutrition deficiency, deacylated tRNA binds to the empty ribosome A-site to form the so-called “starving” ribosome, triggering the (p)ppGpp synthetic activity of RelA (Haseltine and Block, 1973). At this time, RelA catalyzes the production of AMP and ppGpp or pppGpp from ATP and GDP or GTP, respectively (Wendrich et al., 2002), and the SR reaction is triggered.
Under stressed conditions, ppGpp, as a “global transcriptional regulator”, can up regulate the expression of many genes at the transcriptional level, and the concentration of intracellular ppGpp is largely determined by relA gene. Therefore, the function of relA gene in cell physiology is mainly reflected in controlling the concentration of intracellular ppGpp. Studies have shown that (P)ppGpp can change the physiology of bacteria from rapid growth to slow growth, so as to allow them to survive under harsh conditions (Hauryliuk et al., 2015). In E. coli, (P)ppGpp combines with RNA polymerase, up-regulating the expression of metabolic enzyme genes, especially amino acid biosynthesis genes, while reducing the transcription of tRNA, rRNA, ribosomal protein, translation factor and synthase genes (Barker et al., 2001; Lemke et al., 2011; Ross et al., 2016). Other main effects include activating the stress factor δE and inhibiting cell wall synthesis (Costanzo et al., 2008). It was found that more than 30% of the genes in E. coli were differentially expressed by (P)ppGpp, including the up regulation of SR related genes and the down regulation of macromolecular structure related genes under isoleucine starvation conditions (Traxler et al., 2008). Several models have been proposed for the molecular mechanism of SR (Sanchez-Vazquez et al., 2019). It is reported that (P)ppGpp directly binds to RNAP with RNAP binding protein dksA, which destroys the stability of its open complex. On the other hand, (P)ppGpp indirectly regulates gene expression by δ competition (Sanchez-Vazquez et al., 2019). It is worth noting that almost all bacterial pathogens need SR, otherwise they cannot survive and infect the host during stress conditions.
BPI-Inducible Protein A
BPI-inducible protein A (BipA) is a highly conserved ribosome-dependent trGTPase that regulates multiple cellular processes including bacterial attachment and effacement, motility, virulence gene expression, avoidance of host defenses. In addition, BipA is also associated with temperature, symbiosis, antimicrobial resistance and biofilm formation (Overhage et al., 2007; deLivron and Robinson, 2008; Neidig et al., 2013). BipA is widespread in most of the bacteria and plants (Margus et al., 2007) and is similar to EF4 and EF-G except for a unique C-terminal domain (Neidig et al., 2013; Kumar et al., 2015). Despite its conservation in bacteria, BipA was regarded as an essential factor only in adverse growth conditions such as low pH and temperature, nutrient deprivation, and other stresses (Starosta et al., 2014). Mutations in BipA leads to multiple phenotypes including cold sensitivity (Pfennig and Flower, 2001), hypermotility (Farris et al., 1998), decreased capsule synthesis (Rowe et al., 2000), increased sensitivity to chloramphenicol (Duo et al., 2008), and reduced pathogenicity (Grant et al., 2003). Moreover, it also participated in regulating some mRNAs translation under stresses (Ero et al., 2016). Interestingly, deLivron reported that BipA has two different binding modes to the ribosome. It is associated with 70S in the form of GTP-bound under normal cellular conditions, whereas it interacts with 30S subunit under stress conditions (deLivron and Robinson, 2008). Therefore, they speculated that there exist two ribosome∙BipA complexes that affect the response of bacteria to environmental conditions (deLivron and Robinson, 2008). Moreover, there is a growing body of evidence that BipA also functions in the 50S ribosomal subunit biogenesis (Choudhury and Flower, 2015; Del Peso Santos et al., 2021). Choi et al. (2019) reported that BipA is vital for 50S biogenesis at a low temperature, whose expression is involved in the incorporation of L6 protein. The exogenous expression of the L20 coding gene rplT can partially repair the defects in rRNA processing and ribosomal assembly, and then restore the growth of bipA-deficient strains at low temperatures. So, the authors speculated that BipA and L20 may play a coordinating role in proper ribosomal assembly under cold-shock conditions. Another study led by Fredrick (Gibbs et al., 2020) demonstrated the flexible nature of the 50S assembly process. They also found GTP hydrolysis was crucial to the function of BipA.
Elongation Factors and Tumorigenesis
Recently, more and more elongation factors have been reported to function as oncoproteins or tumor promoters in cancer cells (Table 2). Elevated levels of protein synthesis are a critical feature of tumor cells. Alterations in protein synthesis can increase the overall translation rate and stimulate the translation of certain mRNAs to facilitate tumorigenesis, oncological progress, and survival.
TABLE 2.
Alterations of translation elongation factors in human cancer.
| Elongation factors | Observed modification | Association with cancer |
|---|---|---|
| eIF5A1 | Increased expression | eIF5A is highly overexpressed in patients with glioblastoma (Preukschas et al., 2012). eIF5A1 is a diagnostic marker of vulvar intraepithelial neoplasia (Cracchiolo et al., 2004). eIF5A1 is upregulated in colorectal adenoma (Lam et al., 2010) |
| eIF5A2 | Increased expression | eIF5A2 is amplified in ovarian carcinoma (Guan et al., 2001) and is associated with metastatic progression in colorectal cancers (Xie et al., 2008) |
| Gain of EIF5A2 results in recurrence of hepatocellular carcinoma (Chen et al., 2000) | ||
| Ectopic expression of EIF5A2 causes tumorigenesis in naked mice (Guan et al., 2004) | ||
| Overexpression of eIF5A2 results in local invasion of non-small-cell lung cancer (He et al., 2011) | ||
| eEF1A1 | Increased expression | eEF1A1 was upregulated in the infiltrating edge of invasive breast cancers (Zhu et al., 2003) |
| Dimethylation of eEF1A is upregulated and can serve as a diagnostic marker for poor outcomes in lung and pancreatic cancer (Liu et al., 2019) | ||
| eEF1A2 | Increased expression | eEF1A2 is distributed in 30% of ovarian cancers, and 26% of cancers with amplifications near the EEF1A2 |
| eEF1A2 acts as an oncoprotein which is upregulated in 67% of breast cancers (Tomlinson et al., 2005) | ||
| eEF2 | Increased expression | eEF2 is overexpressed in most of colorectal and gastric tumors and promotes cancer growth in vivo and in vitro (Nakamura et al., 2009) |
eEF1A and Tumorigenesis
eEF1A is overexpressed in malignancies such as ovarian tumors. It can be inactivated by cytotoxic and anti-adipogenic ternatin and its derivatives (Carelli et al., 2015), with unclear mechanisms. Mutation in domain III of eEF1A hinders the binding of ternatin and confers resistance to its cytotoxic effects (Carelli et al., 2015). Cancers driven by the activation of PI3K-AKT axis are sensitive to the inhibitors of eEF1A (Lee and Surh, 2009). eEF1A1 was reported to be increased in the periphery of mammary cancer compared with the central region (Zhu et al., 2003), and the levels of eEF1A1 in neoplastic are relatively higher than in normal tissue (Lee and Surh, 2009). eEF1A2, an isoform of eEF1A, functions as cancer protein via a variety of mechanisms, such as facilitating cell invasion and migration by up-regulating MMP-9 and stimulating AKT in pancreatic tumors (Xu et al., 2013). It can accelerate proliferation and block apoptosis via down-regulating caspase 3 in prostate cancer tissues (Sun et al., 2014). Previous studies showed that the expression levels of eEF1A2 are elevated (Anand et al., 2002; Lee and Surh, 2009) and their genes (genomic region: 20q13) are amplified in a high proportion of solid tumors, such as ovarian (Iwabuchi et al., 1995) and breast cancers (Kallioniemi et al., 1994). For malignancy in the blood system, eEF1A2 is also up-regulated in cells of multiple myeloma (Losada et al., 2016). Therefore, eEF1A2 can be used as a diagnostic marker and target for some breast tumours (Tomlinson et al., 2005) and hematological malignancies (Shan et al., 2020).
Besides phosphorylation, other post-translational modifications on elongation factors have been found to be essential for tumorigenesis. Recently, it was reported that lysine 55 of eEF1A is dimethylated (eEF1AK55me2) by METTL13 (Methyltransferase-like 13), resulting in an increase of the inherent GTPase activity of eEF1A (Liu et al., 2019). This modification of eEF1A was utilized by RAS signal cascade to promote translational output and facilitate carcinogenesis (Figure 7). Therefore, METTL13-eEF1AK55me2 signal pathway is vital for tumors to cope with increased translational demand, and METTL13 inhibition might be an effective way to do targeted intervention for RAS driven cancers.
FIGURE 7.
Methylation of eEF1A and the role of eEF1AK55me2 mediated translational control in tumorigenesis. Expression of METTL13 and eEF1AK55me2 are upregulated in cancer, and negatively correlate with the survival of pancreatic and lung cancer patient. Increased METTL13 and eEF1AK55me2 promotes Ras-driven tumorigenesis in vivo.
eEF2 and Tumorigenesis
eEF2 plays an essential role in many biological processes including cell cycle (Hizli et al., 2013) and genotoxic stress (Kruiswijk et al., 2012). Previous studies have shown that eEF2 is overexpressed in various tumors. Translation of eEF2 was up-regulated in most of colorectal (91.7%) and gastric (92.9%) tumors, leading to the elevation of in vivo tumorigenicity (Nakamura et al., 2009). Therefore, eEF2 is a potential target for tumor immunotherapy in multiple cancers (Oji et al., 2014).
Under conditions of energy depletion or nutrient deprivation, tumor cells redistribute energy resources through weakening overall translation, while translating specific mRNAs to cope with stresses and fight for survival. AMPK, a sensor of energy, exists in a variety of tumors. When AMPK is activated, it can stimulate eEF2K, which further inhibits eEF2 activity by phosphorylating the Thr56 of eEF2. Inhibition of eEF2 leads to slowing down of protein translation (Proud, 2015), and prevents tumor cells from growing under nutritional deficiency (Leprivier et al., 2013) (Figure 8). Therefore, eEF2K is a negative regulator of protein synthesis (Wang et al., 2017), and inhibition of eEF2K activity may have therapeutic significance in preventing the survival of tumor cells and recovering protein translation.
FIGURE 8.

Regulation mechanism of eEF2 phosphorylation modification in cancer cells.
eEF2K is an atypical kinase, and plays an important role in the migration and survival of tumor cells. It is overexpressed in glioblastoma (Leprivier et al., 2013), medulloblastoma, mammary cancer (Liu et al., 2014) and pancreatic tumor (Ashour et al., 2014). eEF2K is activated by stresses such as nutrition deficiency, hypoxia (Moore et al., 2015), acidosis (Xie et al., 2015) and cellular energy depletion (Browne et al., 2004). Inhibiting eEF2K decreases the invasion and migration of tumor cells, while deletion of eEF2 or eEF2K overexpression promotes wound healing and invasion. These results indicated that eEF2K has a protective function in tumor cells and therefore can be used as a molecular target to prevent cancer metastasis (Xie et al., 2018). Nevertheless, a recent study showed that eEF2K can protect cells under stress conditions and make tumor cells adapt to stresses (Leprivier et al., 2013). Therefore, stimulating eEF2K-eEF2 axis to suppress tumor requires serious assessment, and more in-depth studies are required to comprehensively evaluate its effectiveness from a treatment perspective (Knight et al., 2020).
eIF5A and Tumorigenesis
eIF5A is involved in the invasive and/or metastatic processes of several types of human cancer (Mathews and Hershey, 2015). There are two subtypes of eIF5A in mammals, both of which were modified with hypusine on the same lysine (Clement et al., 2003). eIF5A1 is widely distributed in rapidly proliferating cells while eIF5A2 is expressed in a tissue-specific manner and is almost undetectable in most cases (Clement et al., 2006). It was found that both eIF5A1 and eIF5A2 are associated with several malignancies (Mathews and Hershey, 2015). Interestingly, the expression of eIF5A1 and eIF5A2, along with DOHH and DHPS, are increased in multiple tumors (Nakanishi and Cleveland, 2016). Recently, it was found that eIF5A2 promotes doxorubicin resistance of colon cancer cells by regulating EMT, suggesting that inhibition of eIF5A2 can be used as a way to reverse the drug resistance of colorectal cancer (Figure 9) (Bao et al., 2015). An early study suggested that eIF5A2 may aid tumorigenesis via promoting the translation of some mRNAs that boost DNA replication and provoke excessive proliferation of tumor cells (Hanauske-Abel et al., 1995). Nevertheless, more in-depth studies on the function and molecular mechanism of eIF5A and its hypusination modification in tumors are needed.
FIGURE 9.

The role of eIF5A mediated translational control in tumorigenesis.
Concluding Remarks
The elongation phase of translation is an important regulatory node in health and disease. Dysregulation of this process is often related to various disorders including tumors, neurodegenerative diseases and cardiovascular diseases. The close link between translational factors and human diseases are well coincident with the concept that gene expression is accurately regulated at the translational level (Dever et al., 2018). There are multiple levels at which protein translation can be regulated to hinder disease progression. Clarifying how post-translational modifications control the activity of translational factors in tumors is helpful to reveal the regulation mechanism in tumorigenesis, and provides a rationale for the new interventional treatment (Xu and Ruggero, 2020). There are still many questions about translation elongation that need to be resolved: 1) The exact physiological function of phosphorylated EF-G and EF-Tu; 2) The molecular mechanism of how ADP-ribosylation impairs the function of eEF2; 3) The molecular mechanism of EF4 in protein translation under stress conditions; 4) The unique requirement of eEF3 in fungal and its action mechanism in translation elongation; 5) The feasibility of eEF2K as a tumor therapeutic target; 6) The molecular mechanism of eIF5A and its hypusine modification in protein translation and tumorigenesis.
Author Contributions
BX and LL: Prepared the figures and tables and wrote the original draft; GS: Revised the manuscript.
Funding
This work was supported by the Key R&D Projects of Introducing High-Level Scientific and Technological Talents in Lvliang City (2021RC-1-4), Starting Fund for Talent Introduction of Fenyang College of Shanxi Medical University (2020A01), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (2020L0749), National College Students’ Innovation and Entrepreneurship Training Program (202117114001), Key Projects of Innovation and Entrepreneurship Training Program for College Students in Shanxi Province (S202117114008), Project of Lvliang City Science and Technology Program (2020SHFZ29), and National Natural Science Foundation of China (31771015).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbas W., Kumar A., Herbein G. (2015). The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 5, 75. 10.3389/fonc.2015.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achenbach J., Nierhaus K. H. (2015). The Mechanics of Ribosomal Translocation. Biochimie 114, 80–89. 10.1016/j.biochi.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Anand N., Murthy S., Amann G., Wernick M., Porter L. A., Cukier I. H., et al. (2002). Protein Elongation Factor EEF1A2 Is a Putative Oncogene in Ovarian Cancer. Nat. Genet. 31, 301–305. 10.1038/ng904 [DOI] [PubMed] [Google Scholar]
- Andersen C. B. F., Becker T., Blau M., Anand M., Halic M., Balar B., et al. (2006). Structure of eEF3 and the Mechanism of Transfer RNA Release from the E-Site. Nature 443, 663–668. 10.1038/nature05126 [DOI] [PubMed] [Google Scholar]
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003). Nicotinamide and PNC1 Govern Lifespan Extension by Calorie Restriction in Saccharomyces cerevisiae . Nature 423, 181–185. 10.1038/nature01578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. (2005). Translation Elongation Factor EF-Tu Is a Target for Stp, a Serine-Threonine Phosphatase Involved in Virulence of Listeria Monocytogenes. Mol. Microbiol. 56, 383–396. 10.1111/j.1365-2958.2005.04551.x [DOI] [PubMed] [Google Scholar]
- Argüelles S., Camandola S., Cutler R. G., Ayala A., Mattson M. P. (2014). Elongation Factor 2 Diphthamide Is Critical for Translation of Two IRES-dependent Protein Targets, XIAP and FGF2, under Oxidative Stress Conditions. Free Radic. Biol. Med. 67, 131–138. 10.1016/j.freeradbiomed.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour A. A., Gurbuz N., Alpay S. N., Abdel‐Aziz A. A. H., Mansour A. M., Huo L., et al. (2014). Elongation Factor‐2 Kinase Regulates TG 2/β1 integrin/Src/u PAR Pathway and Epithelial-Mesenchymal Transition Mediating Pancreatic Cancer Cells Invasion. J. Cel. Mol. Med. 18, 2235–2251. 10.1111/jcmm.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R., Oman K., Shoji S., Bundschuh R., Fredrick K. (2014). The Conserved GTPase LepA Contributes Mainly to Translation Initiation in Escherichia coli . Nucleic Acids Res. 42, 13370–13383. 10.1093/nar/gku1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Lu Y., Wang X., Feng W., Sun X., Guo H., et al. (2015). Eukaryotic Translation Initiation Factor 5A2 (eIF5A2) Regulates Chemoresistance in Colorectal Cancer through Epithelial Mesenchymal Transition. Cancer Cell Int 15, 109. 10.1186/s12935-015-0250-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. M., Gaal T., Gourse R. L. (2001). Mechanism of Regulation of Transcription Initiation by ppGpp. II. Models for Positive Control Based on Properties of RNAP Mutants and Competition for RNAP. J. Mol. Biol. 305, 689–702. 10.1006/jmbi.2000.4328 [DOI] [PubMed] [Google Scholar]
- Bauerschmitt H., Funes S., Herrmann J. M. (2008). The Membrane-Bound GTPase Guf1 Promotes Mitochondrial Protein Synthesis under Suboptimal Conditions. J. Biol. Chem. 283, 17139–17146. 10.1074/jbc.m710037200 [DOI] [PubMed] [Google Scholar]
- Beckelman B. C., Zhou X., Keene C. D., Ma T. (2016). Impaired Eukaryotic Elongation Factor 1A Expression in Alzheimer's Disease. Neurodegener Dis. 16, 39–43. 10.1159/000438925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011). The Structure of the Eukaryotic Ribosome at 3.0 Å Resolution. Science 334, 1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bendt A. K., Burkovski A., Schaffer S., Bott M., Farwick M., Hermann T. (2003). Towards a Phosphoproteome Map ofCorynebacterium Glutamicum. Proteomics 3, 1637–1646. 10.1002/pmic.200300494 [DOI] [PubMed] [Google Scholar]
- Benjin X., Ling L. (2019). Developments, Applications, and Prospects of Cryo-Electron Microscopy. Protein Sci. 29 (4), 872–882. 10.1002/pro.3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M., Rodnina M. V. (2007). The Ribosomal Peptidyl Transferase. Mol. Cell 26, 311–321. 10.1016/j.molcel.2007.03.015 [DOI] [PubMed] [Google Scholar]
- Blaha G., Stanley R. E., Steitz T. A. (2009). Formation of the First Peptide Bond: The Structure of EF-P Bound to the 70 S Ribosome. Science 325, 966–970. 10.1126/science.1175800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., et al. (1991). Selenocysteine: the 21st Amino Acid. Mol. Microbiol. 5, 515–520. 10.1111/j.1365-2958.1991.tb00722.x [DOI] [PubMed] [Google Scholar]
- Browne G. J., Finn S. G., Proud C. G. (2004). Stimulation of the AMP-Activated Protein Kinase Leads to Activation of Eukaryotic Elongation Factor 2 Kinase and to its Phosphorylation at a Novel Site, Serine 398. J. Biol. Chem. 279, 12220–12231. 10.1074/jbc.m309773200 [DOI] [PubMed] [Google Scholar]
- Bullwinkle T. J., Zou S. B., Rajkovic A., Hersch S. J., Elgamal S., Robinson N., et al. (2013). (R)-β-Lysine-modified Elongation Factor P Functions in Translation Elongation. J. Biol. Chem. 288, 4416–4423. 10.1074/jbc.m112.438879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. (1991). Purification and Characterization of Tet(M), a Protein that Renders Ribosomes Resistant to Tetracycline. J. Biol. Chem. 266, 2872–2877. 10.1016/s0021-9258(18)49928-0 [DOI] [PubMed] [Google Scholar]
- Burdett V. (1996). Tet(M)-promoted Release of Tetracycline from Ribosomes Is GTP Dependent. J. Bacteriol. 178, 3246–3251. 10.1128/jb.178.11.3246-3251.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon C. E., March P. E. (2003). Function of the Universally Conserved Bacterial GTPases. Curr. Opin. Microbiol. 6, 135–139. 10.1016/s1369-5274(03)00037-7 [DOI] [PubMed] [Google Scholar]
- Carelli J. D., Sethofer S. G., Smith G. A., Miller H. R., Simard J. L., Merrick W. C., et al. (2015). Ternatin and Improved Synthetic Variants Kill Cancer Cells by Targeting the Elongation factor-1A Ternary Complex. Elife 4. 10.7554/eLife.10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg U., Nilsson A., Nygård O. (1990). Functional Properties of Phosphorylated Elongation Factor 2. Eur. J. Biochem. 191, 639–645. 10.1111/j.1432-1033.1990.tb19169.x [DOI] [PubMed] [Google Scholar]
- Carlson B. A., Francisco Javier M.-R., Kumaraswamy E., Moustafa M. E., Zhi H., Hatfield D. L., et al. (2006). Selenium : its Molecular Biology and Role in Human Health. Free Radic. Res. 10.1080/10715760290006394 [DOI] [Google Scholar]
- Cavallius J., Zoll W., Chakraburtty K., Merrick W. C. (1993). Characterization of Yeast EF-1α: Non-conservation of post-translational Modifications. Biochim. Biophys. Acta (Bba) - Protein Struct. Mol. Enzymol. 1163, 75–80. 10.1016/0167-4838(93)90281-u [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Yeh S.-H., Chen J.-T., Wu C.-C., Hsu M.-T., Tsai S.-F., et al. (2000). Chromosomal Changes and Clonality Relationship between Primary and Recurrent Hepatocellular Carcinoma. Gastroenterology 119, 431–440. 10.1053/gast.2000.9373 [DOI] [PubMed] [Google Scholar]
- Choi E., Jeon H., Oh J. I., Hwang J. (2019). Overexpressed L20 Rescues 50S Ribosomal Subunit Assembly Defects of bipA-Deletion in Escherichia coli . Front. Microbiol. 10, 2982. 10.3389/fmicb.2019.02982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P., Flower A. M. (2015). Efficient Assembly of Ribosomes Is Inhibited by Deletion of bipA in Escherichia coli . J. Bacteriol. 197, 1819–1827. 10.1128/jb.00023-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian B. E., Spremulli L. L. (2012). Mechanism of Protein Biosynthesis in Mammalian Mitochondria. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1819, 1035–1054. 10.1016/j.bbagrm.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. K., Spremulli L. L. (1990). Purification and Characterization of Elongation Factor G from Bovine Liver Mitochondria. J. Biol. Chem. 265, 21000–21004. 10.1016/s0021-9258(17)45317-8 [DOI] [PubMed] [Google Scholar]
- Clement P. M. J., Henderson C. A., Jenkins Z. A., Smit-McBride Z., Wolff E. C., Hershey J. W. B., et al. (2003). Identification and Characterization of Eukaryotic Initiation Factor 5A-2. Eur. J. Biochem. 270, 4254–4263. 10.1046/j.1432-1033.2003.03806.x [DOI] [PubMed] [Google Scholar]
- Clement P. M. J., Johansson H. E., Wolff E. C., Park M. H. (2006). Differential Expression of eIF5A-1 and eIF5A-2 in Human Cancer Cells. FEBS J. 273, 1102–1114. 10.1111/j.1742-4658.2006.05135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenen M. J. H., Antonicka H., Ugalde C., Sasarman F., Rossi R., Heister J. G. A. M. A., et al. (2004). Mutant Mitochondrial Elongation Factor G1 and Combined Oxidative Phosphorylation Deficiency. N. Engl. J. Med. 351, 2080–2086. 10.1056/nejmoa041878 [DOI] [PubMed] [Google Scholar]
- Connell S. R., Trieber C. A., Dinos G. P., Einfeldt E., Taylor D. E., Nierhaus K. H. (2003). Mechanism of Tet(O)-mediated Tetracycline Resistance. Embo j 22, 945–953. 10.1093/emboj/cdg093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A., Nicoloff H., Barchinger S. E., Banta A. B., Gourse R. L., Ades S. E. (2008). ppGpp and DksA Likely Regulate the Activity of the Extracytoplasmic Stress Factor σEinEscherichia Coliby Both Direct and Indirect Mechanisms. Mol. Microbiol. 67, 619–632. 10.1111/j.1365-2958.2007.06072.x [DOI] [PubMed] [Google Scholar]
- Cracchiolo B. M., Heller D. S., Clement P. M. J., Wolff E. C., Park M.-H., Hanauske-Abel H. M. (2004). Eukaryotic Initiation Factor 5A-1 (eIF5A-1) as a Diagnostic Marker for Aberrant Proliferation in Intraepithelial Neoplasia of the Vulva. Gynecol. Oncol. 94, 217–222. 10.1016/j.ygyno.2004.03.018 [DOI] [PubMed] [Google Scholar]
- Dantley K. A., Dannelly H. K., Burdett V. (1998). Binding Interaction between Tet(M) and the Ribosome: Requirements for Binding. J. Bacteriol. 180, 4089–4092. 10.1128/jb.180.16.4089-4092.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra B., Chakraburtty K. (1981). Protein Synthesis in Yeast. I. Purification and Properties of Elongation Factor 3 from Saccharomyces cerevisiae . J. Biol. Chem. 256, 9999–10004. 10.1016/s0021-9258(19)68730-2 [DOI] [PubMed] [Google Scholar]
- Del Peso Santos T., Alvarez L., Sit B., Irazoki O., Blake J., Warner B. R., et al. (2021). BipA Exerts Temperature-dependent Translational Control of Biofilm-Associated colony Morphology in Vibrio cholerae . Elife 10. 10.7554/eLife.60607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLivron M. A., Robinson V. L. (2008). Salmonella enterica Serovar Typhimurium BipA Exhibits Two Distinct Ribosome Binding Modes. J. Bacteriol. 190, 5944–5952. 10.1128/jb.00763-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Dinman J. D., Green R. (2018). Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb Perspect. Biol. 10, a032649. 10.1101/cshperspect.a032649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Green R. (2012). The Elongation, Termination, and Recycling Phases of Translation in Eukaryotes. Cold Spring Harbor Perspect. Biol. 4, a013706. 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in Cell Regulation and Membrane Dynamics. Nature 443, 651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M. V. (2013). EF-P Is Essential for Rapid Synthesis of Proteins Containing Consecutive Proline Residues. Science 339, 85–88. 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- Dong M., Su X., Dzikovski B., Dando E. E., Zhu X., Du J., et al. (2014). Dph3 Is an Electron Donor for Dph1-Dph2 in the First Step of Eukaryotic Diphthamide Biosynthesis. J. Am. Chem. Soc. 136, 1754–1757. 10.1021/ja4118957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duo M., Hou S., Ren D. (2008). Identifying Escherichia coli Genes Involved in Intrinsic Multidrug Resistance. Appl. Microbiol. Biotechnol. 81, 731–741. 10.1007/s00253-008-1709-6 [DOI] [PubMed] [Google Scholar]
- Dzialo M. C., Travaglini K. J., Shen S., Loo J. A., Clarke S. G. (2014). A New Type of Protein Lysine Methyltransferase Trimethylates Lys-79 of Elongation Factor 1A. Biochem. Biophysical Res. Commun. 455, 382–389. 10.1016/j.bbrc.2014.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El'skaya A. V., Ovcharenko G. V., Palchevskii S. S., Petrushenko Z. M., Triana-Alonso F. J., Nierhaus K. H. (1997). Three tRNA Binding Sites in Rabbit Liver Ribosomes and Role of the Intrinsic ATPase in 80S Ribosomes from Higher Eukaryotes. Biochemistry 36, 10492–10497. 10.1021/bi970631e [DOI] [PubMed] [Google Scholar]
- Ero R., Kumar V., Chen Y., Gao Y.-G. (2016). Similarity and Diversity of Translational GTPase Factors EF-G, EF4, and BipA: From Structure to Function. RNA Biol. 13, 1258–1273. 10.1080/15476286.2016.1201627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris M., Grant A., Richardson T. B., O'Connor C. D. (1998). BipA: a Tyrosine-Phosphorylated GTPase that Mediates Interactions between Enteropathogenic Escherichia coli (EPEC) and Epithelial Cells. Mol. Microbiol. 28, 265–279. 10.1046/j.1365-2958.1998.00793.x [DOI] [PubMed] [Google Scholar]
- Gaidenko T. A., Kim T.-J., Price C. W. (2002). The PrpC Serine-Threonine Phosphatase and PrkC Kinase Have Opposing Physiological Roles in Stationary-phase Bacillus Subtilis Cells. J. Bacteriol. 184, 6109–6114. 10.1128/jb.184.22.6109-6114.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.-G., Selmer M., Dunham C. M., Weixlbaumer A., Kelley A. C., Ramakrishnan V. (2009). The Structure of the Ribosome with Elongation Factor G Trapped in the Posttranslocational State. Science 326, 694–699. 10.1126/science.1179709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Bai X., Zhang D., Han C., Yuan J., Liu W., et al. (2016). Mammalian Elongation Factor 4 Regulates Mitochondrial Translation Essential for Spermatogenesis. Nat. Struct. Mol. Biol. 23, 441–449. 10.1038/nsmb.3206 [DOI] [PubMed] [Google Scholar]
- Garcia-Esparcia P., Hernández-Ortega K., Koneti A., Gil L., Delgado-Morales R., Castaño E., et al. (2015). Altered Machinery of Protein Synthesis Is Region- and Stage-dependent and Is Associated with α-synuclein Oligomers in Parkinson's Disease. Acta Neuropathol. Commun. 3, 76. 10.1186/s40478-015-0257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M. R., Fredrick K. (2018). Roles of Elusive Translational GTPases Come to Light and Inform on the Process of Ribosome Biogenesis in Bacteria. Mol. Microbiol. 107, 445–454. 10.1111/mmi.13895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M. R., Moon K.-M., Chen M., Balakrishnan R., Foster L. J., Fredrick K. (2017). Conserved GTPase LepA (Elongation Factor 4) Functions in Biogenesis of the 30S Subunit of the 70S Ribosome. Proc. Natl. Acad. Sci. USA 114, 980–985. 10.1073/pnas.1613665114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M. R., Moon K.-M., Warner B. R., Chen M., Bundschuh R., Foster L. J., et al. (2020). Functional Analysis of BipA in E. coli Reveals the Natural Plasticity of 50S Subunit Assembly. J. Mol. Biol. 432, 5259–5272. 10.1016/j.jmb.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A., Balestrieri C., Gualerzi C. (1982). Structure-function Relationships in Escherichia coli Translational Elongation Factor G: Modification of Lysine Residues by the Site-specific Reagent Pyridoxal Phosphate. Biochemistry 21, 5224–5230. 10.1021/bi00264a018 [DOI] [PubMed] [Google Scholar]
- Glatt S., Zabel R., Vonkova I., Kumar A., Netz D. J., Pierik A. J., et al. (2015). Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2. Structure 23, 149–160. 10.1016/j.str.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Ganoza M. C. (1975). Identification of a Soluble Protein that Stimulates Peptide Bond Synthesis. Proc. Natl. Acad. Sci. 72, 4257–4260. 10.1073/pnas.72.11.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A. J., Farris M., Alefounder P., Williams P. H., Woodward M. J., O'Connor C. D. (2003). Co-ordination of Pathogenicity Island Expression by the BipA GTPase in Enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48, 507–521. 10.1046/j.1365-2958.2003.t01-1-03447.x [DOI] [PubMed] [Google Scholar]
- Gromadski K. B., Schümmer T., Strømgaard A., Knudsen C. R., Kinzy T. G., Rodnina M. V. (2007). Kinetics of the Interactions between Yeast Elongation Factors 1A and 1Bα, Guanine Nucleotides, and Aminoacyl-tRNA. J. Biol. Chem. 282, 35629–35637. 10.1074/jbc.m707245200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. R., Kinzy T. G. (2005). Translation Elongation Factor 1A Is Essential for Regulation of the Actin Cytoskeleton and Cell Morphology. Nat. Struct. Mol. Biol. 12, 772–778. 10.1038/nsmb979 [DOI] [PubMed] [Google Scholar]
- Guan X.-Y., Fung J. M.-W., Ma N.-F., Lau S.-H., Tai L.-S., Xie D., et al. (2004). Oncogenic Role ofeIF-5A2in the Development of Ovarian Cancer. Cancer Res. 64, 4197–4200. 10.1158/0008-5472.can-03-3747 [DOI] [PubMed] [Google Scholar]
- Guan X. Y., Sham J. S., Tang T. C., Fang Y., Huo K. K., Yang J. M. (2001). Isolation of a Novel Candidate Oncogene within a Frequently Amplified Region at 3q26 in Ovarian Cancer. Cancer Res. 61, 3806–3809. [PubMed] [Google Scholar]
- Gutierrez E., Shin B.-S., Woolstenhulme C. J., Kim J.-R., Saini P., Buskirk A. R., et al. (2013). eIF5A Promotes Translation of Polyproline Motifs. Mol. Cell 51, 35–45. 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamey J. J., Wienert B., Quinlan K. G. R., Wilkins M. R. (2017). METTL21B Is a Novel Human Lysine Methyltransferase of Translation Elongation Factor 1A: Discovery by CRISPR/Cas9 Knockout. Mol. Cell Proteomics 16, 2229–2242. 10.1074/mcp.m116.066308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamey J. J., Wilkins M. R. (2018). Methylation of Elongation Factor 1A: where, Who, and Why? Trends Biochem. Sci. 43, 211–223. 10.1016/j.tibs.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Hamey J. J., Winter D. L., Yagoub D., Overall C. M., Hart-Smith G., Wilkins M. R. (2016). Novel N-Terminal and Lysine Methyltransferases that Target Translation Elongation Factor 1A in Yeast and Human. Mol. Cell Proteomics 15, 164–176. 10.1074/mcp.m115.052449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauske-Abel H. M., Slowinska B., Zagulska S., Wilson R. C., Staiano-Coico L., Hanauske A.-R., et al. (1995). Detection of a Sub-set of Polysomal mRNAs Associated with Modulation of Hypusine Formation at the G1-S Boundary Proposal of a Role for eIF-5A in Onset of DNA Replication. FEBS Lett. 366, 92–98. 10.1016/0014-5793(95)00493-s [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Block R. (1973). Synthesis of Guanosine Tetra- and Pentaphosphate Requires the Presence of a Codon-specific, Uncharged Transfer Ribonucleic Acid in the Acceptor Site of Ribosomes. Proc. Natl. Acad. Sci. 70, 1564–1568. 10.1073/pnas.70.5.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K. (2015). Recent Functional Insights into the Role of (p)ppGpp in Bacterial Physiology. Nat. Rev. Microbiol. 13, 298–309. 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.-R., Zhao H.-Y., Li B.-K., Liu Y.-H., Liu M.-Z., Guan X.-Y., et al. (2011). Overexpression of eIF5A-2 Is an Adverse Prognostic Marker of Survival in Stage I Non-small Cell Lung Cancer Patients. Int. J. Cancer 129, 143–150. 10.1002/ijc.25669 [DOI] [PubMed] [Google Scholar]
- Henderson A., Hershey J. W. (2011). Eukaryotic Translation Initiation Factor (eIF) 5A Stimulates Protein Synthesis in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 108, 6415–6419. 10.1073/pnas.1008150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch S. J., Wang M., Zou S. B., Moon K. M., Foster L. J., Ibba M., et al. (2013). Divergent Protein Motifs Direct Elongation Factor P-Mediated Translational Regulation in Salmonella enterica and Escherichia coli . mBio 4, e00180–13. 10.1128/mBio.00180-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizli A. A., Chi Y., Swanger J., Carter J. H., Liao Y., Welcker M., et al. (2013). Phosphorylation of Eukaryotic Elongation Factor 2 (eEF2) by Cyclin A-cyclin-dependent Kinase 2 Regulates its Inhibition by eEF2 Kinase. Mol. Cell Biol 33, 596–604. 10.1128/mcb.01270-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W., Cunha C. E., Peske F., Konevega A. L., Wintermeyer W., Rodnina M. V. (2014a). GTP Hydrolysis by EF-G Synchronizes tRNA Movement on Small and Large Ribosomal Subunits. EMBO J. 33, 1073–1085. 10.1002/embj.201387465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W., Wintermeyer W., Rodnina M. V. (2014b). Synchronous tRNA Movements during Translocation on the Ribosome Are Orchestrated by Elongation Factor G and GTP Hydrolysis. BioEssays 36, 908–918. 10.1002/bies.201400076 [DOI] [PubMed] [Google Scholar]
- Holub M., Bezoušková S., Kalachová L., Weiser J. (2007). Protein Synthesis Elongation Factor Tu Present in Spores ofStreptomyces Coelicolor Can Be Phosphorylatedin Vitro by the Spore Protein Kinase. Folia Microbiol. 52, 471–478. 10.1007/bf02932106 [DOI] [PubMed] [Google Scholar]
- Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., et al. (2012). PhosphoSitePlus: a Comprehensive Resource for Investigating the Structure and Function of Experimentally Determined post-translational Modifications in Man and Mouse. Nucleic Acids Res. 40, D261–D270. 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. (1990). Both genes for EF-Tu in Salmonella typhimurium Are Individually Dispensable for Growth. J. Mol. Biol. 215, 41–51. 10.1016/s0022-2836(05)80093-2 [DOI] [PubMed] [Google Scholar]
- Hughes D. (2013). “Elongation Factors,” in Brenner's Encyclopedia of Genetics, 466–468. 10.1016/b978-0-12-374984-0.00468-x [DOI] [Google Scholar]
- Hummels K. R., Kearns D. B. (2020). Translation Elongation Factor P (EF-P). FEMS Microbiol. Rev. 44, 208–218. 10.1093/femsre/fuaa003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui T., Kobayashi S., Ashikari Y., Sato R., Endo W., Uematsu M., et al. (2016). Two Cases of Early-Onset Myoclonic Seizures with Continuous Parietal delta Activity Caused by EEF1A2 Mutations. Brain Development 38, 520–524. 10.1016/j.braindev.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Iwabuchi H., Sakamoto M., Sakunaga H., Ma Y. Y., Carcangiu M. L., Pinkel D., et al. (1995). Genetic Analysis of Benign, Low-Grade, and High-Grade Ovarian Tumors. Cancer Res. 55, 6172–6180. [PubMed] [Google Scholar]
- Jakobsson M. E., Davydova E., Małecki J., Moen A., Falnes P. Ø. (2015). Saccharomyces cerevisiae Eukaryotic Elongation Factor 1A (eEF1A) Is Methylated at Lys-390 by a METTL21-like Methyltransferase. PLoS One 10, e0131426. 10.1371/journal.pone.0131426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M. E., Małecki J., Falnes P. Ø. (2018). Regulation of Eukaryotic Elongation Factor 1 Alpha (eEF1A) by Dynamic Lysine Methylation. RNA Biol. 15, 314–319. 10.1080/15476286.2018.1440875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M. E., Małecki J., Nilges B. S., Moen A., Leidel S. A., Falnes P. Ø. (2017). Methylation of Human Eukaryotic Elongation Factor Alpha (eEF1A) by a Member of a Novel Protein Lysine Methyltransferase Family Modulates mRNA Translation. Nucleic Acids Res. 45, 8239–8254. 10.1093/nar/gkx432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskunas S. R., Lindahl L., Nomura M., Burgess R. R. (1975). Identification of Two Copies of the Gene for the Elongation Factor EF-Tu in E. coli . Nature 257, 458–462. 10.1038/257458a0 [DOI] [PubMed] [Google Scholar]
- Jeppesen M. G., Navratil T., Spremulli L. L., Nyborg J. (2005). Crystal Structure of the Bovine Mitochondrial Elongation Factor Tu·Ts Complex. J. Biol. Chem. 280, 5071–5081. 10.1074/jbc.m411782200 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O. P., Piper J., Tanner M., Stokke T., Chen L., et al. (1994). Detection and Mapping of Amplified DNA Sequences in Breast Cancer by Comparative Genomic Hybridization. Proc. Natl. Acad. Sci. 91, 2156–2160. 10.1073/pnas.91.6.2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp L. D., Lorsch J. R. (2004). The Molecular Mechanics of Eukaryotic Translation. Annu. Rev. Biochem. 73, 657–704. 10.1146/annurev.biochem.73.030403.080419 [DOI] [PubMed] [Google Scholar]
- Kemper W. M., Berry K. W., Merrick W. C. (1976). Purification and Properties of Rabbit Reticulocyte Protein Synthesis Initiation Factors M2Balpha and M2Bbeta. J. Biol. Chem. 251, 5551–5557. 10.1016/s0021-9258(17)33095-8 [DOI] [PubMed] [Google Scholar]
- Kenney J. W., Moore C. E., Wang X., Proud C. G. (2014). Eukaryotic Elongation Factor 2 Kinase, an Unusual Enzyme with Multiple Roles. Adv. Biol. Regul. 55, 15–27. 10.1016/j.jbior.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Khacho M., Mekhail K., Pilon-Larose K., Pause A., Côté J., Lee S. (2008). eEF1A Is a Novel Component of the Mammalian Nuclear Protein export Machinery. MBoC 19, 5296–5308. 10.1091/mbc.e08-06-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M. C., Ganoza M. C. (2001). Functional Interactions of anEscherichia Coliribosomal ATPase. Eur. J. Biochem. 268, 278–286. 10.1046/j.1432-1033.2001.01873.x [DOI] [PubMed] [Google Scholar]
- Kim K. K., Hung L.-W., Yokota H., Kim R., Kim S.-H. (1998). Crystal Structures of Eukaryotic Translation Initiation Factor 5A from Methanococcus Jannaschii at 1.8 A Resolution. Proc. Natl. Acad. Sci. 95, 10419–10424. 10.1073/pnas.95.18.10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J. R. P., Garland G., Pöyry T., Mead E., Vlahov N., Sfakianos A., et al. (2020). Control of Translation Elongation in Health and Disease. Dis. Model. Mech. 13, dmm043208. 10.1242/dmm.043208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Yuniati L., Magliozzi R., Low T. Y., Lim R., Bolder R., et al. (2012). Coupled Activation and Degradation of eEF2K Regulates Protein Synthesis in Response to Genotoxic Stress. Sci. Signal. 5, ra40. 10.1126/scisignal.2002718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Chen Y., Ero R., Ahmed T., Tan J., Li Z., et al. (2015). Structure of BipA in GTP Form Bound to the Ratcheted Ribosome. Proc. Natl. Acad. Sci. USA 112, 10944–10949. 10.1073/pnas.1513216112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa Y., Schwartzbach C. J., Liao H.-X., Mizumoto K., Kaziro Y., Miura K.-i., et al. (1991). Interactions of Bovine Mitochondrial Phenylalanyl-tRNA with Ribosomes and Elongation Factors from Mitochondria and Bacteria. Biochim. Biophys. Acta (Bba) - Gene Struct. Expr. 1090, 167–172. 10.1016/0167-4781(91)90097-6 [DOI] [PubMed] [Google Scholar]
- Kushner D. B., Lindenbach B. D., Grdzelishvili V. Z., Noueiry A. O., Paul S. M., Ahlquist P. (2003). Systematic, Genome-wide Identification of Host Genes Affecting Replication of a Positive-Strand RNA Virus. Proc. Natl. Acad. Sci. 100, 15764–15769. 10.1073/pnas.2536857100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha G. S., Oyeyemi B. F., Bhavesh N. S. (2019). Stringent Response Protein as a Potential Target to Intervene Persistent Bacterial Infection. Biochimie 165, 67–75. 10.1016/j.biochi.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Kyrpides N. C., Woese C. R. (1998). Universally Conserved Translation Initiation Factors. Proc. Natl. Acad. Sci. 95, 224–228. 10.1073/pnas.95.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam F. F., Jankova L., Dent O. F., Molloy M. P., Kwun S. Y., Clarke C., et al. (2010). Identification of Distinctive Protein Expression Patterns in Colorectal Adenoma. Prot. Clin. Appl. 4, 60–70. 10.1002/prca.200900084 [DOI] [PubMed] [Google Scholar]
- Lam W. W. K., Millichap J. J., Soares D. C., Chin R., McLellan A., FitzPatrick D. R., et al. (2016). Novel De Novo EEF1A2 Missense Mutations Causing Epilepsy and Intellectual Disability. Mol. Genet. Genomic Med. 4, 465–474. 10.1002/mgg3.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak J., Keilhauer E. C., Fürst M., Wuichet K., Gödeke J., Starosta A. L., et al. (2015). Arginine-rhamnosylation as New Strategy to Activate Translation Elongation Factor P. Nat. Chem. Biol. 11, 266–270. 10.1038/nchembio.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Surh Y.-J. (2009). eEF1A2 as a Putative Oncogene. Ann. New York Acad. Sci. 1171, 87–93. 10.1111/j.1749-6632.2009.04909.x [DOI] [PubMed] [Google Scholar]
- Lemke J. J., Sanchez-Vazquez P., Burgos H. L., Hedberg G., Ross W., Gourse R. L. (2011). Direct Regulation of Escherichia coli Ribosomal Protein Promoters by the Transcription Factors ppGpp and DksA. Proc. Natl. Acad. Sci. 108, 5712–5717. 10.1073/pnas.1019383108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A.-R. F., Kool M., et al. (2013). The eEF2 Kinase Confers Resistance to Nutrient Deprivation by Blocking Translation Elongation. Cell 153, 1064–1079. 10.1016/j.cell.2013.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévine A., Vannier F., Absalon C., Kuhn L., Jackson P., Scrivener E., et al. (2006). Analysis of the dynamicBacillus Subtilis Ser/Thr/Tyr Phosphoproteome Implicated in a Wide Variety of Cellular Processes. Proteomics 6, 2157–2173. 10.1002/pmic.200500352 [DOI] [PubMed] [Google Scholar]
- Li W., Atkinson G. C., Thakor N. S., Allas Ü., Lu C.-c., Chan K.-Y., et al. (2013). Mechanism of Tetracycline Resistance by Ribosomal protection Protein Tet(O). Nat. Commun. 4, 1477. 10.1038/ncomms2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Alafuzoff I., Soininen H., Winblad B., Pei J.-J. (2005). Levels of mTOR and its Downstream Targets 4E-BP1, eEF2, and eEF2 Kinase in Relationships with Tau in Alzheimer's Disease Brain. FEBS J. 272, 4211–4220. 10.1111/j.1742-4658.2005.04833.x [DOI] [PubMed] [Google Scholar]
- Li Z., Gonzalez P. A., Sasvari Z., Kinzy T. G., Nagy P. D. (2014). Methylation of Translation Elongation Factor 1A by the METTL10-like See1 Methyltransferase Facilitates Tombusvirus Replication in Yeast and Plants. Virology 448, 43–54. 10.1016/j.virol.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Li Z., Pogany J., Panavas T., Xu K., Esposito A. M., Kinzy T. G., et al. (2009). Translation Elongation Factor 1A Is a Component of the Tombusvirus Replicase Complex and Affects the Stability of the P33 Replication Co-factor. Virology 385, 245–260. 10.1016/j.virol.2008.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann C., Lindschau C., Vijgenboom E., Schröder W., Bosch L., Erdmann V. A. (1993). Prokaryotic Elongation Factor Tu Is Phosphorylated In Vivo . J. Biol. Chem. 268, 601–607. 10.1016/s0021-9258(18)54193-4 [DOI] [PubMed] [Google Scholar]
- Lipson R. S., Webb K. J., Clarke S. G. (2010). Two Novel Methyltransferases Acting upon Eukaryotic Elongation Factor 1A in Saccharomyces cerevisiae . Arch. Biochem. Biophys. 500, 137–143. 10.1016/j.abb.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Chen C., Zhang H., Kaur J., Goldman Y. E., Cooperman B. S. (2011). The Conserved Protein EF4 (LepA) Modulates the Elongation Cycle of Protein Synthesis. Proc. Natl. Acad. Sci. 108, 16223–16228. 10.1073/pnas.1103820108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Pan D., Pech M., Cooperman B. S. (2010). Interrupted Catalysis: the EF4 (LepA) Effect on Back-Translocation. J. Mol. Biol. 396, 1043–1052. 10.1016/j.jmb.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. C., Voisin V., Wang S., Wang D. Y., Jones R. A., Datti A., et al. (2014). Combined deletion of P ten and p53 in mammary epithelium accelerates triple‐negative breast cancer with dependency on e EF 2 K. EMBO Mol. Med. 6, 1542–1560. 10.15252/emmm.201404402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cash T. P., Jones R. G., Keith B., Thompson C. B., Simon M. C. (2006). Hypoxia-induced Energy Stress Regulates mRNA Translation and Cell Growth. Mol. Cell 21, 521–531. 10.1016/j.molcel.2006.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Bachran C., Gupta P., Miller-Randolph S., Wang H., Crown D., et al. (2012). Diphthamide Modification on Eukaryotic Elongation Factor 2 Is Needed to Assure Fidelity of mRNA Translation and Mouse Development. Proc. Natl. Acad. Sci. 109, 13817–13822. 10.1073/pnas.1206933109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hausmann S., Carlson S. M., Fuentes M. E., Francis J. W., Pillai R., et al. (2019). METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell 176 (3), 491–504. 10.1016/j.cell.2018.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2009). Eukaryotic Selenoproteins and Selenoproteomes. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1790, 1424–1428. 10.1016/j.bbagen.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Muñoz-Alonso M. J., García C., Sánchez-Murcia P. A., Martínez-Leal J. F., Domínguez J. M., et al. (2016). Translation Elongation Factor eEF1A2 Is a Novel Anticancer Target for the Marine Natural Product Plitidepsin. Sci. Rep. 6, 35100. 10.1038/srep35100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukash T. O., Turkivska H. V., Negrutskii B. S., El’skaya A. V. (2004). Chaperone-like Activity of Mammalian Elongation Factor eEF1A: Renaturation of Aminoacyl-tRNA Synthetases. Int. J. Biochem. Cell Biol. 36, 1341–1347. 10.1016/j.biocel.2003.11.009 [DOI] [PubMed] [Google Scholar]
- Malecki J., Aileni V. K., Ho A. Y. Y., Schwarz J., Moen A., Sørensen V., et al. (2017). The Novel Lysine Specific Methyltransferase METTL21B Affects mRNA Translation through Inducible and Dynamic Methylation of Lys-165 in Human Eukaryotic Elongation Factor 1 Alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389. 10.1093/nar/gkx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath H., Zhang H., Rehfeld F., Han J., Chang T.-C., Mendell J. T. (2019). Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection. Cell Rep. 29, 3134–3146. 10.1016/j.celrep.2019.10.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Inouye M. (1985). Characterization of the Lep Operon of Escherichia coli. Identification of the Promoter and the Gene Upstream of the Signal Peptidase I Gene. J. Biol. Chem. 260, 7206–7213. 10.1016/s0021-9258(17)39594-7 [DOI] [PubMed] [Google Scholar]
- Margus T., Remm M., Tenson T. (2007). Phylogenetic Distribution of Translational GTPases in Bacteria. BMC Genomics 8, 15. 10.1186/1471-2164-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M., Lobanov A. V., Manta B., Santesmasses D., Bofill A., Guigó R., et al. (2016). LokiarchaeotaMarks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems. Mol. Biol. Evol. 33, 2441–2453. 10.1093/molbev/msw122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M., Ridge P. G., Zhang Y., Lobanov A. V., Pringle T. H., Guigo R., et al. (2012). Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS One 7, e33066. 10.1371/journal.pone.0033066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M., Salinas G., Gabaldón T., Gladyshev V. N. (2019). Utilization of Selenocysteine in Early-Branching Fungal Phyla. Nat. Microbiol. 4, 759–765. 10.1038/s41564-018-0354-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M., Santesmasses D., Capella-Gutierrez S., Mateo A., Arnan C., Johnson R., et al. (2015). Evolution of Selenophosphate Synthetases: Emergence and Relocation of Function through Independent Duplications and Recurrent Subfunctionalization. Genome Res. 25, 1256–1267. 10.1101/gr.190538.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Granados C., Riechers S.-P., Stahl U., Lang C. (2008). Absence of See1p, a Widely conservedSaccharomyces Cerevisiaeprotein, Confers Both Deficient Heterologous Protein Production and Endocytosis. Yeast 25, 871–877. 10.1002/yea.1641 [DOI] [PubMed] [Google Scholar]
- Mateyak M. K., Kinzy T. G. (2013). ADP-ribosylation of Translation Elongation Factor 2 by Diphtheria Toxin in Yeast Inhibits Translation and Cell Separation. J. Biol. Chem. 288, 24647–24655. 10.1074/jbc.m113.488783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak M. K., Kinzy T. G. (2010). eEF1A: Thinking outside the Ribosome. J. Biol. Chem. 285, 21209–21213. 10.1074/jbc.r110.113795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B., Hershey J. W. B. (2015). The Translation Factor eIF5A and Human Cancer. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1849, 836–844. 10.1016/j.bbagrm.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister W. T., Barrett C. L. (1977). Roles of the Early Genes of Bacteriophage T7 in Shutoff of Host Macromolecular Synthesis. J. Virol. 23, 543–553. 10.1128/jvi.23.3.543-553.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulík K., Zhulanova E. (1995). Sequencing of the Tuf1 Gene and the Phosphorylation Pattern of EF-Tu1 during Development and Differentiation in Streptomyces Collinus Producing Kirromycin. Biochem. Biophysical Res. Commun. 213, 454–461. 10.1006/bbrc.1995.2153 [DOI] [PubMed] [Google Scholar]
- Misra S. K., Milohanic E., Aké F., Mijakovic I., Deutscher J., Monnet V., et al. (2011). Analysis of the Serine/threonine/tyrosine Phosphoproteome of the Pathogenic Bacterium Listeria Monocytogenes Reveals Phosphorylated Proteins Related to Virulence. Proteomics 11, 4155–4165. 10.1002/pmic.201100259 [DOI] [PubMed] [Google Scholar]
- Moore C. E. J., Mikolajek H., Regufe da Mota S., Wang X., Kenney J. W., Werner J. M., et al. (2015). Elongation Factor 2 Kinase Is Regulated by Proline Hydroxylation and Protects Cells during Hypoxia. Mol. Cel. Biol. 35, 1788–1804. 10.1128/mcb.01457-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J., Aoyagi S., Nanchi I., Nakatsuka S., Hirata E., Shibata S., et al. (2009). Overexpression of Eukaryotic Elongation Factor eEF2 in Gastrointestinal Cancers and its Involvement in G2/M Progression in the Cell Cycle. Int. J. Oncol. 34, 1181–1189. 10.3892/ijo_00000246 [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Cleveland J. L. (2016). Targeting the Polyamine-Hypusine Circuit for the Prevention and Treatment of Cancer. Amino acids 48, 2353–2362. 10.1007/s00726-016-2275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W. W., Zou S. B., Roy H., Xie J. L., Savchenko A., Singer A., et al. (2010). PoxA, yjeK, and Elongation Factor P Coordinately Modulate Virulence and Drug Resistance in Salmonella enterica . Mol. Cell 39, 209–221. 10.1016/j.molcel.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidig A., Yeung A. T., Rosay T., Tettmann B., Strempel N., Rueger M., et al. (2013). TypA Is Involved in Virulence, Antimicrobial Resistance and Biofilm Formation in Pseudomonas aeruginosa . BMC Microbiol. 13, 77. 10.1186/1471-2180-13-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novosylna O., Jurewicz E., Pydiura N., Goral A., Filipek A., Negrutskii B., et al. (2015). Translation Elongation Factor eEF1A1 Is a Novel Partner of a Multifunctional Protein Sgt1. Biochimie 119, 137–145. 10.1016/j.biochi.2015.10.026 [DOI] [PubMed] [Google Scholar]
- Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V. (2002). Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form. Cell 111, 721–732. 10.1016/s0092-8674(02)01086-3 [DOI] [PubMed] [Google Scholar]
- Ogle J. M., Ramakrishnan V. (2005). Structural Insights into Translational Fidelity. Annu. Rev. Biochem. 74, 129–177. 10.1146/annurev.biochem.74.061903.155440 [DOI] [PubMed] [Google Scholar]
- Oji Y., Tatsumi N., Fukuda M., Nakatsuka S.-I., Aoyagi S., Hirata E., et al. (2014). The Translation Elongation Factor eEF2 Is a Novel Tumor-Associated Antigen Overexpressed in Various Types of Cancers. Int. J. Oncol. 44, 1461–1469. 10.3892/ijo.2014.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ose T., Soler N., Rasubala L., Kuroki K., Kohda D., Fourmy D., et al. (2007). Structural Basis for Dynamic Interdomain Movement and RNA Recognition of the Selenocysteine-specific Elongation Factor SelB. Structure 15, 577–586. 10.1016/j.str.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Overhage J., Lewenza S., Marr A. K., Hancock R. E. W. (2007). Identification of Genes Involved in Swarming Motility Using a Pseudomonas aeruginosa PAO1 Mini-Tn 5-lux Mutant Library. J. Bacteriol. 189, 2164–2169. 10.1128/jb.01623-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., Lithgow G. J., et al. (2007). Inhibition of mRNA Translation Extends Lifespan in Caenorhabditis elegans . Aging cell 6, 111–119. 10.1111/j.1474-9726.2006.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-H., Johansson H. E., Aoki H., Huang B. X., Kim H.-Y., Ganoza M. C., et al. (2012). Post-translational Modification by β-Lysylation Is Required for Activity of Escherichia coli Elongation Factor P (EF-P). J. Biol. Chem. 287, 2579–2590. 10.1074/jbc.m111.309633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Nishimura K., Zanelli C. F., Valentini S. R. (2010). Functional Significance of eIF5A and its Hypusine Modification in Eukaryotes. Amino acids 38, 491–500. 10.1007/s00726-009-0408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Wolff E. C., Smit-McBride Z., Hershey J. W., Folk J. E. (1991). Comparison of the Activities of Variant Forms of eIF-4D. The Requirement for Hypusine or Deoxyhypusine. J. Biol. Chem. 266, 7988–7994. 10.1016/s0021-9258(18)92929-7 [DOI] [PubMed] [Google Scholar]
- Park S., Park J. M., Kim S., Kim J.-A., Shepherd J. D., Smith-Hicks C. L., et al. (2008). Elongation Factor 2 and Fragile X Mental Retardation Protein Control the Dynamic Translation of Arc/Arg3.1 Essential for mGluR-LTD. Neuron 59, 70–83. 10.1016/j.neuron.2008.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Karim Z., Yamamoto H., Kitakawa M., Qin Y., Nierhaus K. H. (2011). Elongation Factor 4 (EF4/LepA) Accelerates Protein Synthesis at Increased Mg2+ Concentrations. Proc. Natl. Acad. Sci. 108, 3199–3203. 10.1073/pnas.1012994108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L., Starosta A. L., Virumäe K., Atkinson G. C., Tenson T., Remme J., et al. (2012). Lys34 of Translation Elongation Factor eF-P Is Hydroxylated by YfcM. Nat. Chem. Biol. 8, 695–697. 10.1038/nchembio.1001 [DOI] [PubMed] [Google Scholar]
- Pereira S. F. F., Gonzalez R. L., Jr., Dworkin J. (2015). Protein Synthesis during Cellular Quiescence Is Inhibited by Phosphorylation of a Translational Elongation Factor. Proc. Natl. Acad. Sci. USA 112, E3274–E3281. 10.1073/pnas.1505297112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig P., Flower A. (2001). BipA Is Required for Growth of Escherichia coli K12 at Low Temperature. Mol. Gen. Genomics 266, 313–317. 10.1007/s004380100559 [DOI] [PubMed] [Google Scholar]
- Preukschas M., Hagel C., Schulte A., Weber K., Lamszus K., Sievert H., et al. (2012). Expression of Eukaryotic Initiation Factor 5A and Hypusine Forming Enzymes in Glioblastoma Patient Samples: Implications for New Targeted Therapies. PLoS One 7, e43468. 10.1371/journal.pone.0043468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G. (2019). Phosphorylation and Signal Transduction Pathways in Translational Control. Cold Spring Harb Perspect. Biol. 11, a033050. 10.1101/cshperspect.a033050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G. (2015). Regulation and Roles of Elongation Factor 2 Kinase. Biochem. Soc. Trans. 43, 328–332. 10.1042/bst20140323 [DOI] [PubMed] [Google Scholar]
- Qin S. L., Xie A. G., Bonato M. C., McLaughlin C. S. (1990). Sequence Analysis of the Translational Elongation Factor 3 from Saccharomyces cerevisiae . J. Biol. Chem. 265, 1903–1912. 10.1016/s0021-9258(19)39916-8 [DOI] [PubMed] [Google Scholar]
- Qin Y., Polacek N., Vesper O., Staub E., Einfeldt E., Wilson D. N., et al. (2006). The Highly Conserved LepA Is a Ribosomal Elongation Factor that Back-Translocates the Ribosome. Cell 127, 721–733. 10.1016/j.cell.2006.09.037 [DOI] [PubMed] [Google Scholar]
- Rajkovic A., Erickson S., Witzky A., Branson O. E., Seo J., Gafken P. R., et al. (2015). Cyclic Rhamnosylated Elongation Factor P Establishes Antibiotic Resistance in Pseudomonas aeruginosa . mBio 6, e00823. 10.1128/mBio.00823-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A., Hummels K. R., Witzky A., Erickson S., Gafken P. R., Whitelegge J. P., et al. (2016). Translation Control of Swarming Proficiency in Bacillus Subtilis by 5-Amino-Pentanolylated Elongation Factor P. J. Biol. Chem. 291, 10976–10985. 10.1074/jbc.m115.712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. (2002). Ribosome Structure and the Mechanism of Translation. Cell 108, 557–572. 10.1016/s0092-8674(02)00619-0 [DOI] [PubMed] [Google Scholar]
- Robertson E. S., Aggison L. A., Nicholson A. W. (1994). Phosphorylation of Elongation Factor G and Ribosomal Protein S6 in Bacteriophage T7-Infected Escherichia coli . Mol. Microbiol. 11, 1045–1057. 10.1111/j.1365-2958.1994.tb00382.x [DOI] [PubMed] [Google Scholar]
- Robichaud N., Sonenberg N., Ruggero D., Schneider R. J. (2019). Translational Control in Cancer. Cold Spring Harb Perspect. Biol. 11. 10.1101/cshperspect.a032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M. V. (2018). Translation in Prokaryotes. Cold Spring Harb Perspect. Biol. 10. 10.1101/cshperspect.a032664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M. V., Gromadski K. B., Kothe U., Wieden H.-J. (2005). Recognition and Selection of tRNA in Translation. FEBS Lett. 579, 938–942. 10.1016/j.febslet.2004.11.048 [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Serebryanik A. I., Ovcharenko G. V., El'Skaya A. V. (1994). ATPase Strongly Bound to Higher Eukaryotic Ribosomes. Eur. J. Biochem. 225, 305–310. 10.1111/j.1432-1033.1994.00305.x [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Wintermeyer W. (2009). Recent Mechanistic Insights into Eukaryotic Ribosomes. Curr. Opin. Cel. Biol. 21, 435–443. 10.1016/j.ceb.2009.01.023 [DOI] [PubMed] [Google Scholar]
- Ross W., Sanchez-Vazquez P., Chen A. Y., Lee J.-H., Burgos H. L., Gourse R. L. (2016). ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol. Cell 62, 811–823. 10.1016/j.molcel.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S., Hodson N., Griffiths G., Roberts I. S. (2000). Regulation of the Escherichia coli K5 Capsule Gene Cluster: Evidence for the Roles of H-NS, BipA, and Integration Host Factor in Regulation of Group 2 Capsule Gene Clusters in Pathogenic E. coli . J. Bacteriol. 182, 2741–2745. 10.1128/jb.182.10.2741-2745.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P., Eyler D. E., Green R., Dever T. E. (2009). Hypusine-containing Protein eIF5A Promotes Translation Elongation. Nature 459, 118–121. 10.1038/nature08034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid A., Arora G., Gupta M., Singhal A., Chakraborty K., Nandicoori V. K., et al. (2011). Interaction of Mycobacterium tuberculosis Elongation Factor Tu with GTP Is Regulated by Phosphorylation. J. Bacteriol. 193, 5347–5358. 10.1128/jb.05469-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vazquez P., Dewey C. N., Kitten N., Ross W., Gourse R. L. (2019). Genome-wide Effects onEscherichia Colitranscription from ppGpp Binding to its Two Sites on RNA Polymerase. Proc. Natl. Acad. Sci. USA 116, 8310–8319. 10.1073/pnas.1819682116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Tanaka T., Kushiro A., Hakoshima T., Tomita K.-i. (1991). Molecular Cloning, Nucleotide Sequence and Expression of thetufBgene Encoding Elongation Factor Tu fromThermus thermophilusHB8. FEBS Lett. 288, 98–100. 10.1016/0014-5793(91)81011-v [DOI] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr D., Kothe U., Wintermeyer W., Rodnina M. V. (2005). Control of Phosphate Release from Elongation Factor G by Ribosomal Protein L7/12. Embo j 24, 4316–4323. 10.1038/sj.emboj.7600884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Rodnina M. V., Wintermeyer W. (2009). Distinct Functions of Elongation Factor G in Ribosome Recycling and Translocation. Rna 15, 772–780. 10.1261/rna.1592509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R., Abdel‐Fattah W., Klassen R., Stark M. J. R. (2014). The Diphthamide Modification Pathway fromSaccharomyces Cerevisiae- Revisited. Mol. Microbiol. 94, 1213–1226. 10.1111/mmi.12845 [DOI] [PubMed] [Google Scholar]
- Schmeing T. M., Ramakrishnan V. (2009). What Recent Ribosome Structures Have Revealed about the Mechanism of Translation. Nature 461, 1234–1242. 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- Schmidl S. R., Gronau K., Pietack N., Hecker M., Becher D., Stülke J. (2010). The Phosphoproteome of the Minimal Bacterium Mycoplasma Pneumoniae. Mol. Cell Proteomics 9, 1228–1242. 10.1074/mcp.m900267-mcp200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller A. P., Green R. (2018). Roadblocks and Resolutions in Eukaryotic Translation. Nat. Rev. Mol. Cell Biol 19, 526–541. 10.1038/s41580-018-0011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller A. P., Wu C. C.-C., Dever T. E., Buskirk A. R., Green R. (2017). eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 66, 194–205. 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz I., Engel C., Niestroj A. J., Kehlen A., Rahfeld J.-U., Kleinschmidt M., et al. (2014). A Non-canonical Function of Eukaryotic Elongation Factor 1A1: Regulation of Interleukin-6 Expression. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1843, 965–975. 10.1016/j.bbamcr.2014.01.022 [DOI] [PubMed] [Google Scholar]
- Shah I. M., Dworkin J. (2010). Induction and Regulation of a Secreted Peptidoglycan Hydrolase by a Membrane Ser/Thr Kinase that Detects Muropeptides. Mol. Microbiol. 75, 1232–1243. 10.1111/j.1365-2958.2010.07046.x [DOI] [PubMed] [Google Scholar]
- Shan X., Yanping W., Yuwen M., Jue L., Can’e T., Aiping D., et al. (2020). Dimethylation of eEF1A at Lysine 55 Plays a Key Role in the Regulation of eEF1A2 on Malignant Cell Functions of Acute Myeloid Leukemia. Technology Cancer Res. Treat. 19, 1533033820914295. 10.1177/1533033820914295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T., Barjau J., Sohtome Y., Sodeoka M., Shinkai Y. (2014). Selenium-based S-Adenosylmethionine Analog Reveals the Mammalian Seven-Beta-Strand Methyltransferase METTL10 to Be an EF1A1 Lysine Methyltransferase. PLoS One 9, e105394. 10.1371/journal.pone.0105394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S., Janssen B. D., Hayes C. S., Fredrick K. (2010). Translation Factor LepA Contributes to Tellurite Resistance in Escherichia coli but Plays No Apparent Role in the Fidelity of Protein Synthesis. Biochimie 92, 157–163. 10.1016/j.biochi.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariah G., Todd P. K. (2021). Translational Control in Aging and Neurodegeneration. Wiley Interdiscip Rev RNA 12 (4), e1628. 10.1002/wrna.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogerson L., Engelhardt D. (1977). Dissimilarity in Protein Chain Elongation Factor Requirements between Yeast and Rat Liver Ribosomes. J. Biol. Chem. 252, 1471–1475. 10.1016/s0021-9258(17)40680-6 [DOI] [PubMed] [Google Scholar]
- Song G., Xu B., Shi H., Zhang Y., Zhang D., Cao X., et al. (2020). Conformational Activation of Ribosome Recycling by Intra- and Inter-molecular Dynamics of RRF. Int. J. Biol. macromolecules 160, 1212–1219. 10.1016/j.ijbiomac.2020.05.254 [DOI] [PubMed] [Google Scholar]
- Spahn C. M. T., Blaha G., Agrawal R. K., Penczek P., Grassucci R. A., Trieber C. A., et al. (2001). Localization of the Ribosomal protection Protein Tet(O) on the Ribosome and the Mechanism of Tetracycline Resistance. Mol. Cell 7, 1037–1045. 10.1016/s1097-2765(01)00238-6 [DOI] [PubMed] [Google Scholar]
- Stahl S., da Silva Mateus Seidl A. R., Ducret A., Kux van Geijtenbeek S., Michel S., Racek T., et al. (2015). Loss of Diphthamide Pre-activates NF-Κb and Death Receptor Pathways and Renders MCF7 Cells Hypersensitive to Tumor Necrosis Factor. Proc. Natl. Acad. Sci. USA 112, 10732–10737. 10.1073/pnas.1512863112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta A. L., Lassak J., Jung K., Wilson D. N. (2014). The Bacterial Translation Stress Response. FEMS Microbiol. Rev. 38, 1172–1201. 10.1111/1574-6976.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinchen W., Bange G. (2016). The Magic Dance of the Alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544. 10.1111/mmi.13412 [DOI] [PubMed] [Google Scholar]
- Su X., Lin Z., Lin H. (2013). The Biosynthesis and Biological Function of Diphthamide. Crit. Rev. Biochem. Mol. Biol. 48, 515–521. 10.3109/10409238.2013.831023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ge F., Xiao C.-L., Yin X.-F., Ge R., Zhang L.-H., et al. (2010). Phosphoproteomic Analysis Reveals the Multiple Roles of Phosphorylation in Pathogenic Bacterium Streptococcus Pneumoniae. J. Proteome Res. 9, 275–282. 10.1021/pr900612v [DOI] [PubMed] [Google Scholar]
- Sun Y., Du C., Wang B., Zhang Y., Liu X., Ren G. (2014). Up-regulation of eEF1A2 Promotes Proliferation and Inhibits Apoptosis in Prostate Cancer. Biochem. Biophysical Res. Commun. 450, 1–6. 10.1016/j.bbrc.2014.05.045 [DOI] [PubMed] [Google Scholar]
- Tomlinson V. A., Newbery H. J., Wray N. R., Jackson J., Larionov A., Miller W. R., et al. (2005). Translation Elongation Factor eEF1A2 Is a Potential Oncoprotein that Is Overexpressed in Two-Thirds of Breast Tumours. BMC cancer 5, 113. 10.1186/1471-2407-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler M. F., Summers S. M., Nguyen H.-T., Zacharia V. M., Hightower G. A., Smith J. T., et al. (2008). The Global, ppGpp-Mediated Stringent Response to Amino Acid Starvation in Escherichia coli . Mol. Microbiol. 68, 1128–1148. 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso F. J., Chakraburtty K., Nierhaus K. H. (1995). The Elongation Factor 3 Unique in Higher Fungi and Essential for Protein Biosynthesis Is an E Site Factor. J. Biol. Chem. 270, 20473–20478. 10.1074/jbc.270.35.20473 [DOI] [PubMed] [Google Scholar]
- Tsuboi M., Morita H., Nozaki Y., Akama K., Ueda T., Ito K., et al. (2009). EF-G2mt Is an Exclusive Recycling Factor in Mammalian Mitochondrial Protein Synthesis. Mol. Cell 35, 502–510. 10.1016/j.molcel.2009.06.028 [DOI] [PubMed] [Google Scholar]
- Ude S., Jürgen L., Starosta A. L., Kraxenberger T., Wilson D. N., Jung K. (2013). Translation Elongation Factor EF-P Alleviates Ribosome Stalling at Polyproline Stretches. Science 339 (6115), 82–85. 10.1126/science.1228985 [DOI] [PubMed] [Google Scholar]
- Van Dyke N., Pickering B. F., Van Dyke M. W. (2009). Stm1p Alters the Ribosome Association of Eukaryotic Elongation Factor 3 and Affects Translation Elongation. Nucleic Acids Res. 37, 6116–6125. 10.1093/nar/gkp645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M., Pani B., Griffiths L. A., Muchardt C., Abbott C. M., Singer R. H., et al. (2014). The Translation Elongation Factor eEF1A1 Couples Transcription to Translation during Heat Shock Response. Elife 3, e03164. 10.7554/eLife.03164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versées W. (2015). It Takes Two to Rule Translation Elongation. Structure 23, 7–9. 10.1016/j.str.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Voorhees R. M., Ramakrishnan V. (2013). Structural Basis of the Translational Elongation Cycle. Annu. Rev. Biochem. 82, 203–236. 10.1146/annurev-biochem-113009-092313 [DOI] [PubMed] [Google Scholar]
- Wang X., Xie J., Proud C. (2017). Eukaryotic Elongation Factor 2 Kinase (eEF2K) in Cancer. Cancers (Basel) 9. 10.3390/cancers9120162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang Y., Meyering-Voss M., Sprinzl M., Sigler P. B. (1997). Crystal Structure of the EF-Tu˙EF-Ts Complex from Thermus Thermophilus. Nat. Struct. Mol. Biol. 4, 650–656. 10.1038/nsb0897-650 [DOI] [PubMed] [Google Scholar]
- Wendrich T. M., Blaha G., Wilson D. N., Marahiel M. A., Nierhaus K. H. (2002). Dissection of the Mechanism for the Stringent Factor RelA. Mol. Cell 10, 779–788. 10.1016/s1097-2765(02)00656-1 [DOI] [PubMed] [Google Scholar]
- White J. T., Cato T., Deramchi N., Gabunilas J., Roy K. R., Wang C., et al. (2019). Protein Methylation and Translation: Role of Lysine Modification on the Function of Yeast Elongation Factor 1A. Biochemistry 58, 4997–5010. 10.1021/acs.biochem.9b00818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther K. S., Roghanian M., Gerdes K. (2018). Activation of the Stringent Response by Loading of RelA-tRNA Complexes at the Ribosomal A-Site. Mol. Cell 70, 95–105. 10.1016/j.molcel.2018.02.033 [DOI] [PubMed] [Google Scholar]
- Xie D., Ma N.-F., Pan Z.-Z., Wu H.-X., Liu Y.-D., Wu G.-Q., et al. (2008). Overexpression of EIF-5A2 Is Associated with Metastasis of Human Colorectal Carcinoma. Hum. Pathol. 39, 80–86. 10.1016/j.humpath.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Xie J., de Souza Alves V., von der Haar T., O'Keefe L., Lenchine R. V., Jensen K. B., et al. (2019). Regulation of the Elongation Phase of Protein Synthesis Enhances Translation Accuracy and Modulates Lifespan. Curr. Biol. 29 (5), 737–749. 10.1016/j.cub.2019.01.029 [DOI] [PubMed] [Google Scholar]
- Xie J., Mikolajek H., Pigott C. R., Hooper K. J., Mellows T., Moore C. E., et al. (2015). Molecular Mechanism for the Control of Eukaryotic Elongation Factor 2 Kinase by pH: Role in Cancer Cell Survival. Mol. Cel. Biol. 35, 1805–1824. 10.1128/mcb.00012-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Shen K., Lenchine R. V., Gethings L. A., Trim P. J., Snel M. F., et al. (2018). Eukaryotic Elongation Factor 2 Kinase Upregulates the Expression of Proteins Implicated in Cell Migration and Cancer Cell Metastasis. Int. J. Cancer 142, 1865–1877. 10.1002/ijc.31210 [DOI] [PubMed] [Google Scholar]
- Xie P. (2016). On the Pathway of Ribosomal Translocation. Int. J. Biol. macromolecules 92, 401–415. 10.1016/j.ijbiomac.2016.07.048 [DOI] [PubMed] [Google Scholar]
- Ximerakis M., Lipnick S. L., Innes B. T., Simmons S. K., Adiconis X., Dionne D., et al. (2019). Single-cell Transcriptomic Profiling of the Aging Mouse Brain. Nat. Neurosci. 22, 1696–1708. 10.1038/s41593-019-0491-3 [DOI] [PubMed] [Google Scholar]
- Xin H., Woriax V., Burkhart W., Spremulli L. L. (1995). Cloning and Expression of Mitochondrial Translational Elongation Factor Ts from Bovine and Human Liver. J. Biol. Chem. 270, 17243–17249. 10.1074/jbc.270.29.17243 [DOI] [PubMed] [Google Scholar]
- Xu C., Hu D.-m., Zhu Q. (2013). eEF1A2 Promotes Cell Migration, Invasion and Metastasis in Pancreatic Cancer by Upregulating MMP-9 Expression through Akt Activation. Clin. Exp. Metastasis 30, 933–944. 10.1007/s10585-013-9593-6 [DOI] [PubMed] [Google Scholar]
- Xu Y., Ruggero D. (2020). The Role of Translation Control in Tumorigenesis and its Therapeutic Implications. Annu. Rev. Cancer Biol. 4, 437–457. 10.1146/annurev-cancerbio-030419-033420 [DOI] [Google Scholar]
- Yamamoto H., Qin Y., Achenbach J., Li C., Kijek J., Spahn C. M. T., et al. (2014). EF-G and EF4: Translocation and Back-Translocation on the Bacterial Ribosome. Nat. Rev. Microbiol. 12, 89–100. 10.1038/nrmicro3176 [DOI] [PubMed] [Google Scholar]
- Yanagisawa T., Takahashi H., Suzuki T., Masuda A., Dohmae N., Yokoyama S. (2016). Neisseria Meningitidis Translation Elongation Factor P and its Active-Site Arginine Residue Are Essential for Cell Viability. PLoS One 11, e0147907. 10.1371/journal.pone.0147907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Li Z., Hao J., Qin Y. (2014). EF4 Knockout E. coli Cells Exhibit Lower Levels of Cellular Biosynthesis under Acidic Stress. Protein Cell 5, 563–567. 10.1007/s13238-014-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.-w., Yu M., Lee J. H., Chatnaparat T., Zhao Y. (2020). The Stringent Response Regulator (P) ppGpp Mediates Virulence Gene Expression and Survival in Erwinia Amylovora. BMC Genomics 21, 261. 10.1186/s12864-020-6699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S., Rasubala L., Ose T., Kohda D., Fourmy D., Maenaka K. (2005). Structural Basis for mRNA Recognition by Elongation Factor SelB. Nat. Struct. Mol. Biol. 12, 198–203. 10.1038/nsmb890 [DOI] [PubMed] [Google Scholar]
- Yu Y.-R., You L.-R., Yan Y.-T., Chen C.-M. (2014). Role of OVCA1/DPH1 in Craniofacial Abnormalities of Miller-Dieker Syndrome. Hum. Mol. Genet. 23, 5579–5596. 10.1093/hmg/ddu273 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Romero H., Salinas G., Gladyshev V. N. (2006). Dynamic Evolution of Selenocysteine Utilization in Bacteria: a Balance between Selenoprotein Loss and Evolution of Selenocysteine from Redox Active Cysteine Residues. Genome Biol. 7, R94. 10.1186/gb-2006-7-10-r94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Reynolds L., Crnogorac-Jurcevic T., Gillett C. E., Dublin E. A., Marshall J. F., et al. (2003). Combination of Microdissection and Microarray Analysis to Identify Gene Expression Changes between Differentially Located Tumour Cells in Breast Cancer. Oncogene 22, 3742–3748. 10.1038/sj.onc.1206428 [DOI] [PubMed] [Google Scholar]
- Zhu P., Liu Y., Zhang F., Bai X., Chen Z., Shangguan F., et al. (2018). Human Elongation Factor 4 Regulates Cancer Bioenergetics by Acting as a Mitochondrial Translation Switch. Cancer Res. 78, 2813–2824. 10.1158/0008-5472.can-17-2059 [DOI] [PubMed] [Google Scholar]
- Zou S. B., Hersch S. J., Roy H., Wiggers J. B., Leung A. S., Buranyi S., et al. (2012). Loss of Elongation Factor P Disrupts Bacterial Outer Membrane Integrity. J. Bacteriol. 194, 413–425. 10.1128/jb.05864-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S. B., Roy H., Ibba M., Navarre W. W. (2011). Elongation Factor P Mediates a Novel post-transcriptional Regulatory Pathway Critical for Bacterial Virulence. Virulence 2, 147–151. 10.4161/viru.2.2.15039 [DOI] [PMC free article] [PubMed] [Google Scholar]