Abstract
Since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, COVID-19) was first reported in China in December 2019, various variants have been identified in different areas of the world such as United Kingdom (alpha), South Africa (beta and omicron), Brazil (gamma), and India (delta). Some of SARS-CoV-2 variants, each of which is characterized by a unique mutation(s) in spike protein, are concerned due to their high infectivity and the capability to escape from neutralizing antibodies elicited by vaccinations. To identify peptide epitopes that are derived from SARS-CoV-2 viral proteins and possibly induce CD8+ T cell immunity, we investigated SARS-CoV-2-derived peptides that are likely to bind to major histocompatibility complex (MHC) class I molecules. We identified a total of 15 peptides that bind to human leukocyte antigen (HLA)-A*24:02, HLA-A*02:01, or HLA-A*02:06, and possibly induce cytotoxic T lymphocytes (CTLs); thirteen of them corresponded to ORF1ab polyprotein, one peptide to spike protein and the remaining one to membrane glycoprotein. CD8+ T cells that recognize these peptides were detected in peripheral blood samples in three individuals recovered from COVID-19 as well as non-infected individuals. Since most of these peptides are commonly conserved among other coronaviruses including SARS-CoV and/or MERS-CoV, these might be useful to maintain T cell responses to coronaviruses that are pandemic at present and will become the future threat. We could define pairs of TRA and TRB sequences of nine CTL clones that recognize SARS-CoV-2-derived peptides. We might use these SARS-CoV-2-derived peptide-reactive TCR sequences for investigating the history of SARS-CoV-2 infection.
Subject terms: Cellular immunity, Sequencing
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has become world-wide pandemic. Until December 21, 2021, ~273 million people have been confirmed the infection to COVID-19 and more than 5 million died of this disease.
In addition to intensive screening of infected individuals with PCR and/or antigen tests and subsequent isolation of corona-positive individuals, worldwide vaccination is fundamentally essential to overcome the present pandemic situation. Randomized trials demonstrated that Pfizer/BioNTech BNT162b2 and Moderna mRNA-1273 vaccines encoding the SARS-CoV-2 spike protein had ~95% protective ability against COVID-19 [1, 2]. Vaccine-elicited neutralizing antibodies inhibit the spike protein on SARS-CoV-2, which was originally reported in December 2019 in China, to attach to the host cells. While these vaccines against COVID-19 have been used widely, SARS-CoV-2 variants have continuously emerged in different areas of the world. Virus variants, alpha, beta, gamma, delta, and omicron, which were defined by a unique amino acid substitution(s) within the spike protein, were identified in the United Kingdom [3], South Africa [4, 5], Brazil [6], and India [7], respectively. Neutralizing activity against these SARS-CoV-2 variants tends to decrease as compared to that against the original reference strain in individuals who received vaccination [8–12]; therefore, a novel vaccine(s) that is not affected by mutations in the virus genome is required for the fight against SARS-CoV-2.
Although most of the studies have focused on the neutralizing antibodies against the spike protein, we here describe induction of CD8+ T cells that are expected to play crucial roles in virus clearance through killing of virus-infected cells. In fact, in SARS-CoV-2 infections, the presence of CD8+ T cells responding to SARS-CoV-2-derived peptides was shown to be associated with better clinical outcomes [13–15]. CD8+ T cells in COVID-19-recovered individuals could recognize not only spike protein-derived peptides but also other viral proteins including nucleocapsid phosphoprotein, membrane glycoprotein, and ORFs-derived peptides [16–18]. In previous studies, severe acute respiratory syndrome coronavirus (SARS-CoV)-specific memory T cells were detected in blood samples of SARS-recovered individuals after 6–11 years of the initial infection [19, 20]. T cell responses against the nucleocapsid phosphoprotein of SARS-CoV were maintained in individuals with the history of SARS-CoV infection 17 years after the outbreak of SARS in 2003 and these memory T cells showed cross-reactivity to SARS-CoV-2 [17]. These data indicate that memory T cells may also critically contribute to long-term protection from COVID-19.
In a present study, we demonstrate major histocompatibility complex (MHC) class I-binding epitope peptides derived from SARS-CoV-2 viral proteins to enhance antiviral CD8+ T cells immunity. To characterize cytotoxic T lymphocytes (CTLs) that recognize SARS-CoV-2-derived peptides and play a central role in T cell immunity, we further conduct T cell receptor (TCR) analysis for peptide-specific CTLs. The results should provide useful information for T cell responses against SARS-CoV-2 infection.
Materials and methods
Samples
The study protocol was approved by the Institutional Review Board of OncoTherapy Science and the written informed consent was obtained from each of peripheral blood mononuclear cells (PBMCs) donors.
PBMCs
For in vitro CTL induction, human leukocyte antigen (HLA)-A*24:02-positive PBMCs were isolated from blood of healthy volunteers by density gradient centrifugation using Ficoll-Paque PLUS (Cytiva) according to an industrial instruction manual. HLA-A*02:01- or HLA-A*02:06-positive PBMCs were purchased from Cellular Technology Limited (Cleveland, OH). For tetramer staining, HLA-A*24:02-positive PBMCs derived from COVID-19-recovered individuals with a prior positive SARS-CoV-2 PCR test were purchased from Precision For Medicine (Bethesda, MD). Healthy individuals-derived PBMCs that were collected before the SARS-CoV-2 pandemic (March 2018–May 2019) were purchased from Cellular Technology Limited.
Cell lines
TISI cells (HLA-A*24:02/-, lymphoblastoid cell) were purchased from the IHWG Cell and Gene Bank (Seattle, WA). T2 (HLA-A*02:01/-, lymphoblast), Jiyoye (HLA-A32, Burkitt’s lymphoma), and EB-3 (HLA-A3/Aw32, Burkitt’s lymphoma) cells were purchased from American Type Culture Collection (Manassas, VA). HEV0011 (HLA-A*02:06/-, B lymphocytes) cells were purchased from RIKEN Bioresource Research Center (Tsukuba, Japan). All cells were cultured in RPMI1640 media (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) and 1% antibiotic solution (Wako).
Peptides
SARS-CoV-2-derived 9-mer and 10-mer peptides were synthesized by Cosmo Bio Co., Ltd. (Otaru, Japan). The purity (>90%) and the sequences of peptides were confirmed by analytical HPLC and a mass spectrometry analysis. Peptides were dissolved in dimethyl sulfoxide at 20 mg/mL and stored at −80 °C until the use for in vitro CTL induction, an IFN-γ enzyme-linked immunospot (ELISPOT) assay and an IFN-γ enzyme-linked immunosorbent assay (ELISA).
in vitro CTL induction
Monocyte-derived dendritic cells (DCs) were generated from PBMCs. Monocytes were isolated from PBMCs by adherence to a plastic tissue culture dish (Becton Dickinson) and cultured in the presence of 1000 IU/mL of granulocyte-macrophage colony-stimulating factor (R&D System) and 1000 IU/mL of interleukin (IL)−4 (R&D System) in AIM-V medium (Invitrogen) supplemented with 2% human AB serum (SIGMA) for 7 days. On day 5, OK-432 (Chugai Pharmaceutical) was added in the culture medium to induce the maturation of DCs (final concentration: 0.1 KE/mL). Autologous CD8+ T cells (3.0 × 105 cells) purified from PBMCs with CD8-Positive Isolation Kit (Dynal) were cultured with DCs (1.5 × 104 cells) with 20 μg/mL of each peptide in AIM-V medium containing 2% human AB serum, 10 ng/mL of IL-7 (R&D System), and 30 ng/mL of IL-21 (Cell Genix). After 3 days of the culture, CD8+ T cells were restimulated with DCs and 20 μg/mL of each peptide. DCs were prepared by the same procedure described above. At day 7 of the culture, CD8+ T cells were further stimulated with 48 IU/mL of IL-2 (Novartis), 5 ng/mL of IL-7 (Novoprotein), and 5 ng/mL of IL-15 (Novoprotein) in AIM-V medium supplemented with 2% human AB serum. At day 11 of the culture, CD8+ T cell responses were examined by an IFN-γ ELISPOT assay.
IFN-γ ELISPOT assay
The human IFN-γ ELISPOT kit and AEC substrate set (BD Biosciences) were used to analyze CD8+ T cell responses to SARS-CoV-2-derived peptides. The ELISPOT assay was performed according to the industrial instruction manual. Briefly, TISI, T2, or HEV0011 cells were used as stimulator cells. These cells were cultured with or without 20 μg/mL of peptide overnight at 37 °C. To analyze CD8+ T cell responses, 500 μL of supernatant was removed from each well of culture plates for in vitro CTL induction. Subsequently 100 μL of cell suspensions and stimulator cells (2 × 104 cells) with or without each peptide were co-cultured in 96-well plates coated with anti-IFN-γ antibody. After 16–18 h incubation, IFN-γ spots were detected and counted using ImmunoSpot S6 analyzer (Cellular Technology Limited).
Limiting dilution
CD8+ T cells were diluted to 0.5 cell or 10 cells per well in 96-well round-bottom plates (Corning) and cultured with feeder cells in AIM-V medium containing 5% human AB serum, 30 ng/mL of anti-CD3 monoclonal antibody (clone UCHT1; BD Biosciences), and 150 IU/mL of IL-2. Jiyoye and EB-3 cells (1 × 104 cells each) were used as feeder cells after treating with 30 μg/mL of Mitomycin C (MEDAC) at 37 °C for 30 min. After 10 days of the culture, IL-2 was added to each well (final concentration: 150 IU/mL). After additional 4 days of the culture, IFN-γ ELISPOT was performed for screening CD8+ T cell responses against each peptide.
CTL expansion culture
CTLs that showed positive reactivity in the IFN-γ ELISPOT assay after limiting dilution were expanded. They were cultured with feeder cells in AIM-V medium containing 5% human AB serum and 40 ng/mL of anti-CD3 monoclonal antibody. Jiyoye and EB-3 cells (5 × 106 cells each) were used as feeder cells after treating with 30 μg/mL of Mitomycin C at 37 °C for 30 min. The half volume of culture medium was exchanged with fresh AIM-V containing 5% human AB serum and 72 IU/mL of IL-2 every 3 or 4 days. After 14 days of the culture, IFN-γ secretion from CTLs was measured by ELISA.
IFN-γ ELISA
ELISA were performed using IFN-γ ELISA kit (BD Biosciences) according to the industrial instruction manual. Briefly, CTLs (Responders) and TISI, T2, or HEV0011 cells (Stimulators) with or without each peptide were co-cultured in 200 μL of AIM-V medium supplemented with 5% human AB serum at several ratio of Responders to Stimulators (R/S ratio). Responders at 5 × 104, 2.5 × 104, 1.25 × 104, 0.625 × 104, or 0.3125 × 104 cells/well concentration were plated in 96-well round-bottom plates together with 1 × 104 Stimulators. After 16–18 h of the incubation, supernatants were collected for measurement of IFN-γ. IFN-γ production was calculated as pg/mL using a standard curve.
TCR sequencing
Total RNAs were extracted from peptide-specific CTL clones using RNeasy mini kit (QIAGEN). cDNAs were synthesized using SMARTScribe Reverse Transcriptase (Clontech). SARS-CoV-2-derived peptides-reactive TCR sequences were identified by Sanger sequencing. TCRα cDNA was amplified by PCR using a common forward primer for adaptor (5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTATCAACGCAGAGTGGCCAT-3') and a reverse primer specific for the constant region (5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGDBDHHCAGGGTCAGGGTTCTGGATA-3'). Sanger sequencing was performed using M13 forward primer (5'-TGTAAAACGACGGCCAGTG-3') and M13 reverse primer (5'-CAGGAAACAGCTATGACCAT-3') after TA cloning. TCRβ cDNA was amplified using a common forward primer for adaptor and a reverse primer specific for the constant region (5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGDVHDVTCTGATGGCTCAAACACAGC-3') [21].
in vitro stimulation
PBMCs (5 × 105 cells/mL) obtained from COVID-19-recovered individuals or healthy individuals without the history of SARS-CoV-2 infection were stimulated with 10 μg/mL of each peptide in mixture of 50% AIM-V and 50% RPMI1640 medium supplemented with 10% fetal bovine serum. After 4 days of the stimulation, PBMCs were restimulated with 10 μg/mL of each peptide. PBMCs were cultured for 12 days adding 120 IU/mL of IL-2 on days 5, 7, and 10.
Preparation of tetramers
SARS-CoV-2-derived peptides-loaded MHC class I (pMHCI) tetramers were generated by QuickSwitchTM Quant HLA-A*24:02 Tetramer Kit-PE (MBL International Corporation) according to an industrial instruction manual [22]. Briefly, 50 μL of tetramer folded with an irrelevant exchangeable peptide at 50 μg/mL was mixed with 1 μL of peptide at 1 mg/mL and 1 μL of Peptide Exchange Factor for 4 h at room temperature. Rate of peptide exchange was quantitated by flow cytometry. When irrelevant exchangeable peptides were exchanged for SARS-CoV-2-derived peptides at a rate of more than 75%, tetramers were used for staining PBMCs.
Tetramer staining
After in vitro stimulation, PBMCs were incubated with phycoerythrin (PE)-conjugated tetramer on ice for 30 min. Following wash in Dulbecco’s phosphate-buffered saline (DPBS, GIBCO) with 0.5% bovine serum albumin (BSA, Iwai Chemicals), the cells were stained with fluorescein isothiocyanate-conjugated anti-human CD8 antibody (clone RPA-T8, BD Biosciences), allophycocyanin-conjugated anti-human CD3 antibody (clone UCHT1, BD Biosciences), and phycoerythrin-Cy7 (PE-Cy7)-conjugated anti-human CD4 antibody (clone RPA-T4, BD Biosciences) on ice for 20 min. The cells were washed in DPBS with 0.5% BSA and then stained with DPBS containing 0.1 μg/mL 4',6-diamidino-2-phenylindole (DAPI, BD Biosciences). CD8+ T cells that bound to pMHCI tetramer were identified using SH800 (Sony Biotechnology). PBMCs were treated with 450 nM dasatinib (Cayman Chemical) for inhibiting TCR downregulation from surface of T cells before tetramer staining [23].
Results
Prediction and selection of MHC class I-binding peptides derived from SARS-CoV-2 viral proteins
The workflow for screening of SARS-CoV-2-derived MHC class I-binding peptides is shown in Fig. 1. Firstly, we predicted MHC class I-binding peptide epitopes derived from SARS-CoV-2 viral proteins including the structural proteins (spike, envelope, membrane glycoprotein, and nucleocapsid phosphoprotein) as well as the non-structural proteins (ORF1ab, ORF3a, ORF6, ORF7a, ORF8, and ORF10) as previously described [24]. Peptide-MHC affinity was calculated using NetMHC 4.0 server [25, 26]. We targeted HLA-A*24:02, HLA-A*02:01, and HLA-A*02:06 because they covered a large proportion (more than 80%) of Japanese populations [27].
Fig. 1.
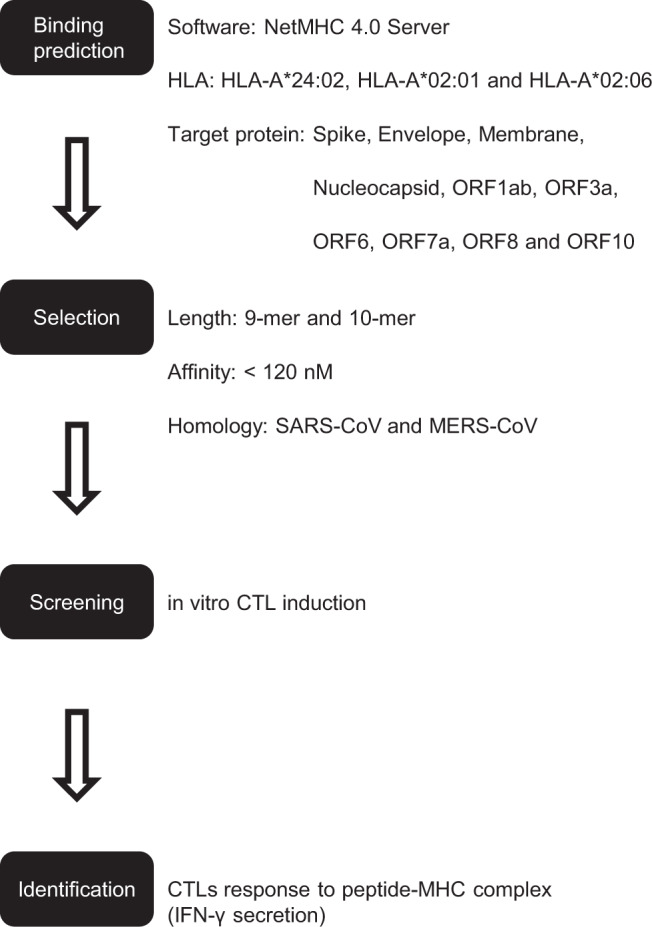
Flowchart for identifying MHC class I-binding peptides-candidates derived from SARS-CoV-2 viral proteins
We focused peptide epitopes that were conserved in either or both of SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) to develop common and universal vaccines that can be possibly effective for newly arising coronaviruses in the future.
We firstly selected 14 peptides (P01-P14) that were expected to bind to an HLA-A*24:02 molecule (predicted MHC affinity of <120 nM). Two of the 14 peptides (P06 and P11) were common among coronaviruses including SARS-CoV and MERS-CoV. The remaining 12 peptides were conserved in SARS-CoV. We also selected 15 peptides (P15-P28 and P08) that were expected to bind to both HLA-A*02:01 and HLA-A*02:06 molecules (predicted MHC affinity of <120 nM). They are commonly shared with SARS-CoV. All candidate peptides are shown in Table 1.
Table 1.
SARS-CoV-2-derived candidate peptides bind MHC class I (HLA-A*24:02, HLA-A*02:01, or HLA-A*02:06)
| ID | Protein | Sequence | Length (amino acids) | Starting position | Affinity (nM) | HLA | Conserved in |
|---|---|---|---|---|---|---|---|
| P01 | ORF1ab | TYASALWEI | 9 | 4090 | 18 | A*24:02 | SARS-CoV |
| P02 | S | PFAMQMAYRF | 10 | 897 | 39 | A*24:02 | SARS-CoV |
| P03 | ORF1ab | YDYLVSTQEF | 10 | 3811 | 41 | A*24:02 | SARS-CoV |
| P04 | ORF1ab | YYSQLMCQPI | 10 | 2560 | 44 | A*24:02 | SARS-CoV |
| P05 | ORF1ab | SYYSLLMPI | 9 | 4628 | 46 | A*24:02 | SARS-CoV |
| P06 | ORF1ab | AYANSVFNI | 9 | 5080 | 56 | A*24:02 |
SARS-CoV MERS-CoV |
| P07 | ORF1ab | RYKLEGYAF | 9 | 6676 | 62 | A*24:02 | SARS-CoV |
| P08 | ORF1ab | FTYASALWEI | 10 | 4089 |
69 21 5 |
A*24:02 A*02:01 A*02:06 |
SARS-CoV |
| P09 | ORF1ab | SYSLFDMSKF | 10 | 7039 | 85 | A*24:02 | SARS-CoV |
| P10 | ORF1ab | RYFKYWDQTY | 10 | 4677 | 90 | A*24:02 | SARS-CoV |
| P11 | ORF1ab | TAYANSVFNI | 10 | 5079 | 102 | A*24:02 |
SARS-CoV MERS-CoV |
| P12 | M | LWLLWPVTL | 9 | 54 | 108 | A*24:02 | SARS-CoV |
| P13 | ORF1ab | TYASALWEIQ | 10 | 4090 | 111 | A*24:02 | SARS-CoV |
| P14 | ORF1ab | YAYLRKHFSM | 10 | 5138 | 115 | A*24:02 | SARS-CoV |
| P15 | ORF1ab | SLFDMSKFPL | 10 | 7041 |
6 18 |
A*02:01 A*02:06 |
SARS-CoV |
| P16 | ORF1ab | LMYKGLPWNV | 10 | 6077 |
7 8 |
A*02:01 A*02:06 |
SARS-CoV |
| P17 | ORF1ab | FVLWAHGFEL | 10 | 6108 |
8 13 |
A*02:01 A*02:06 |
SARS-CoV |
| P18 | ORF1ab | YMPASWVMRI | 10 | 3654 |
12 103 |
A*02:01 A*02:06 |
SARS-CoV |
| P19 | ORF1ab | MMGFKMNYQV | 10 | 5982 |
18 77 |
A*02:01 A*02:06 |
SARS-CoV |
| P20 | ORF1ab | FELTSMKYFV | 10 | 6115 |
18 59 |
A*02:01 A*02:06 |
SARS-CoV |
| P21 | ORF1ab | MMILSDDAV | 9 | 5147 |
35 13 |
A*02:01 A*02:06 |
SARS-CoV |
| P22 | N | FGMSRIGMEV | 10 | 315 |
37 105 |
A*02:01 A*02:06 |
SARS-CoV |
| P23 | ORF1ab | TLMIERFVSL | 10 | 5245 |
42 48 |
A*02:01 A*02:06 |
SARS-CoV |
| P24 | ORF1ab | YVYNPFMIDV | 10 | 6160 |
51 15 |
A*02:01 A*02:06 |
SARS-CoV |
| P25 | ORF1ab | NVLAWLYAAV | 10 | 3466 |
72 11 |
A*02:01 A*02:06 |
SARS-CoV |
| P26 | ORF1ab | TLEPEYFNSV | 10 | 5740 |
79 22 |
A*02:01 A*02:06 |
SARS-CoV |
| P27 | ORF1ab | YSFLPGVYSV | 10 | 3098 |
81 23 |
A*02:01 A*02:06 |
SARS-CoV |
| P28 | ORF1ab | FMIDVQQWGF | 10 | 6165 |
84 29 |
A*02:01 A*02:06 |
SARS-CoV |
S spike protein, M membrane glycoprotein, N nucleocapsid phosphoprotein
Identification of MHC class I-binding peptides derived from SARS-CoV-2 (HLA-A*24:02)
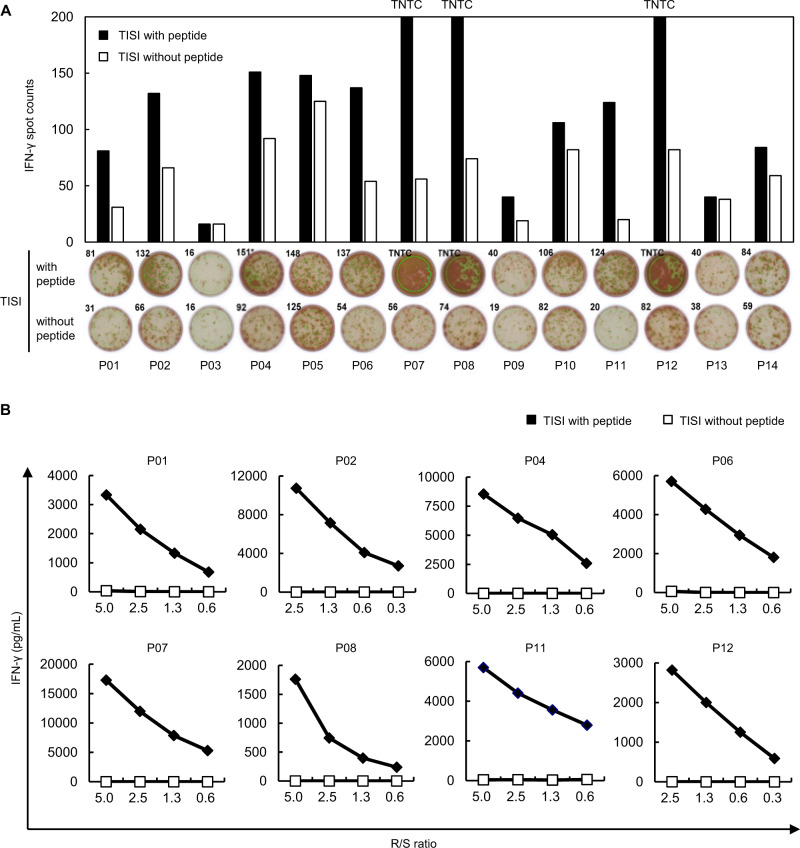
To examined CTL induction by peptides, we first used HLA-A*24:02-positive PBMCs isolated from two healthy donors. Briefly, autologous CD8+ T cells and monocyte-derived DCs were prepared from PBMCs. CD8+ T cells were cultured with DCs pulsed with each peptide. After 11 days of stimulation by each peptide, CD8+ T cells were cultured overnight with HLA-A*24:02-positive TISI cells that were pulsed with or without each peptide. CD8+ T cell responses to each peptide were analyzed by an IFN-γ ELISPOT assay. They were considered as positive when IFN-γ spot counts were 1.5 times or higher, compared with the peptide-negative control. Positive reactivities were detected in 9 of the 14 peptides examined (Fig. 2A).
Fig. 2.
Identification of HLA-A*24:02-binding peptides derived from SARS-CoV-2. A IFN-γ ELISPOT assay was performed after in vitro CTL induction. CD8+ T cells and HLA-A*24:02-positive TISI cells pulsed with each peptide were co-cultured overnight. TISI cells without the peptide were used as negative control. Bars represent the number of IFN-γ spots. CD8+ T cell responses were judged as positive when the number of IFN-γ spots were 1.5 times or higher than the negative control (P01, P02, P04, P06, P07, P08, P09, P11, and P12). Representative positive activities to P02, P06, P08, and P12 observed in PBMCs from two donors are shown. No positive response was observed for P03, P05, P10, P13, and P14 in two PBMCs. Spot counts higher than 200 are demonstrated as “too numerous to count (TNTC)”. B To confirm the response of CTLs to SARS-CoV-2-derived peptides, IFN-γ ELISA was performed. CTLs (Responders) were co-cultured overnight with TISI cells (Stimulators) pulsed with or without the peptide at the indicated ratio of Responders to Stimulators (R/S ratio). IFN-γ secretion was measured by ELISA. CTLs that revealed the peptide-specific IFN-γ secretion for eight peptides are shown. Similar results were obtained in independent experiments using three CTLs in the respective peptides
We subsequently conducted limiting dilution to establish peptide-specific CTL clones from the initial CD8+ T cell populations that showed positive reactivity in the IFN-γ ELISPOT assay. CD8+ T cells were diluted to 10 cells per well in 96-well round-bottom plates. After 14 days of the culture, peptide-specific CTLs were screened using an IFN-γ ELISPOT assay. CTL responses to each peptide were examined by an IFN-γ ELISA following the cell expansion. Peptide-specific IFN-γ secretion against eight peptides were observed when TISI cells were used as stimulator cells for presenting individual peptides (Fig. 2B). This indicates that these peptides were likely to be presented on the HLA-A*24:02 molecule and recognized by TCRs on CTLs.
Identification of MHC class I-binding peptides derived from SARS-CoV-2 (HLA-A*02:01 and HLA-A*02:06)
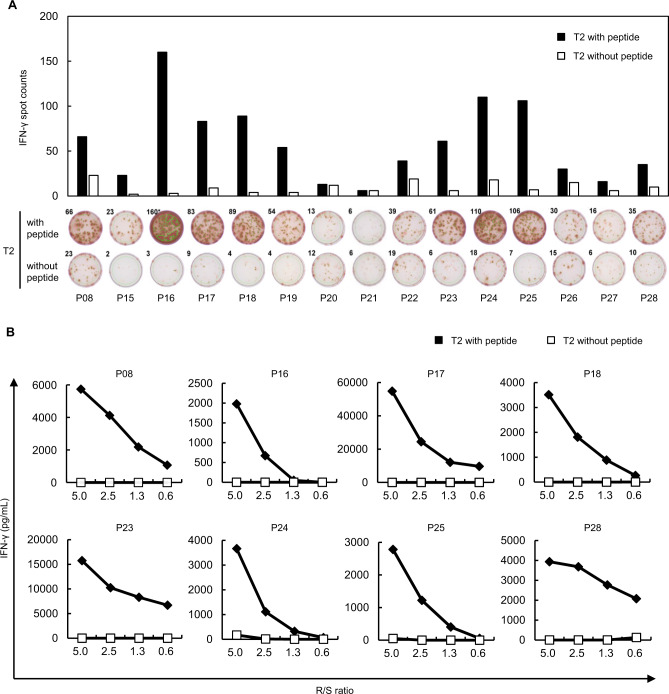
We similarly investigated CTL induction for 15 candidate epitopes (P15-P28 and P08 listed in Table 1) using HLA-A*02:01-positive donor-derived PBMCs. CD8+ T cells were cultured overnight with HLA-A*02:01-positive T2 cells pulsed with or without each peptide. Positive reactivity was detected by an IFN-γ ELISPOT assay for 13 of the 15 peptides examined (Fig. 3A). Then, we performed limiting dilution and further confirmed the peptide-specific IFN-γ secretion in expanded T cells stimulated with 8 of the 13 peptides (Fig. 3B). Since HLA-A*02:01 and HLA-A*02:06 have an only one amino acid substitution (F9Y) in a region of the peptide binding groove [28, 29], it has been indicated that some peptides bind to both HLA-A*02:01 and HLA-A*02:06 molecules [30, 31]. Hence, we investigated the cross-reactivity of these eight peptides to an HLA-A*02:06 molecule using HLA-A*02:06-positive donor-derived PBMCs. As we expected, we found the positive reactivity of seven peptides in an IFN-γ ELISPOT assay (Fig. S1A) as well as the peptide-specific IFN-γ secretions in an ELISA (Fig. S1B).
Fig. 3.
Identification of HLA-A*02:01-binding peptides derived from SARS-CoV-2. A IFN-γ ELISPOT assay was performed after in vitro CTL induction. CD8+ T cells and HLA-A*02:01-positive T2 cells pulsed with each peptide were co-cultured overnight. T2 cells without the peptide were used as negative control. Bars represent the number of IFN-γ spots. CD8+ T cell responses were judged as positive when the number of IFN-γ spots were 1.5 times or higher than the negative control. B To confirm the response of CTLs to SARS-CoV-2-derived peptides, IFN-γ ELISA was performed. CTLs (Responders) were co-cultured overnight with T2 cells (Stimulators) pulsed with or without the peptide at the indicated ratio of Responders to Stimulators (R/S ratio). IFN-γ secretion was measured by ELISA. CTLs that revealed the peptide-specific IFN-γ secretion for eight peptides are shown. Similar results were obtained in independent experiments using three CTLs in the respective peptides
TCR sequences recognizing SARS-CoV-2-derived peptides
SARS-CoV-2-derived peptide-specific CTL clones were established from CD8+ T cell population that showed positive reactivity to each of the peptides after in vitro CTL induction (Fig. S2A, B). Then DNA sequences of pairs of TCRα and TCRβ were examined using established CTL clones. We show pairs of TRA and TRB sequences that were likely to recognize SARS-CoV-2-derived peptides in Table 2.
Table 2.
SARS-CoV-2-derived peptide-reactive TCRs
| Peptide ID | Peptide-reactive TCRs | HLA | ||||
|---|---|---|---|---|---|---|
| Chain | V region | J region | CDR3 | |||
| P04 | Alpha | TRAV10 | TRAJ31 | CVVSALNARLMF | A*24:02 | |
| Beta | TRBV20-1 | TRBJ2-1 | CSARDYTSSYNEQFF | |||
| P06 | Alpha | TRAV1-2 | TRAJ32 | CAVRGSGGGATNKLIF | A*24:02 | |
| Beta | TRBV9 | TRBJ2–7 | CASSPSGPNYEQYF | |||
| P07 | Alpha | TRAV20 | TRAJ11 | CAVHFFPGYSTLTF | A*24:02 | |
| Beta | TRBV30 | TRBJ2–7 | CAGAGRGYEQYF | |||
| P08 | Alpha | TRAV26-1 | TRAJ5 | CIVSDMGRRALTF | A*24:02 | |
| Beta | TRBV4-1 | TRBJ2-1 | CASSFTGTSGSLGEQFF | |||
| P11 | Alpha | TRAV21 | TRAJ20 | CAVFNDYKLSF | A*24:02 | |
| Beta | TRBV7–9 | TRBJ2–7 | CASSSRTSGRSSYEQYF | |||
| P08 | Alpha | TRAV21 | TRAJ48 | CAVRRTYFGNEKLTF | A*02:01 | |
| Beta | TRBV20-1 | TRBJ2–7 | CSARGSDSYEQYF | |||
| P23 | Alpha | TRAV8-6 | TRAJ36 | CAITGANNLFF | A*02:01 | |
| Beta | TRBV20-1 | TRBJ1–5 | CSAEDRGSNSNQPQHF | |||
| P25 | 1 | Alpha | TRAV1-2 | TRAJ20 | CAVRDPRNDYKLSF | A*02:01 |
| Beta | TRBV27 | TRBJ1-1 | CATQVNTEAFF | |||
| 2 | Alpha | TRAV24 | TRAJ29 | CAFLSGNTPLVF | A*02:01 | |
| Beta | TRBV27 | TRBJ1–6 | CASSPAAYSPLHF | |||
Detection of SARS-CoV-2-derived peptides-reactive CD8+ T cells in PBMCs
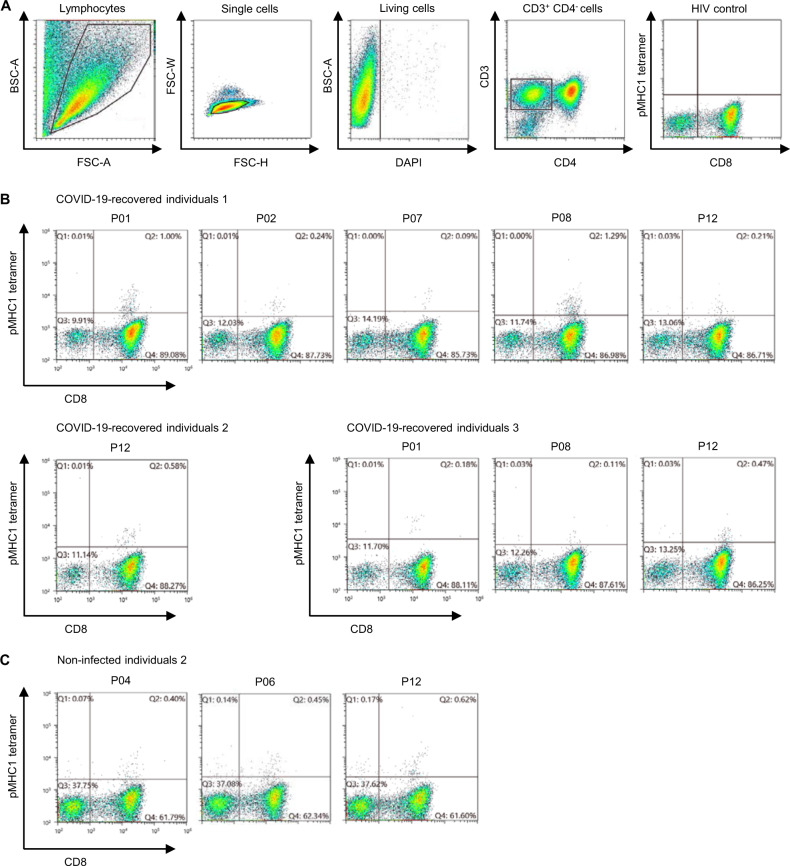
We analyzed HLA-A*24:02-positive PBMCs from individuals who recovered from COVID-19 (n = 3) and healthy individuals without any evident SARS-CoV-2 infection (n = 3) to examine CD8+ T cells that might recognize MHC class I-binding peptides we identified. After in vitro stimulation, PBMCs were stained with PE-conjugated pMHCI tetramers and analyzed by flow cytometry (Fig. 4A). pMHCI tetramer-binding CD8+ T cells were detected from three individuals recovered from COVID-19 (Fig. 4B). It is notable that we found CD8+ T cells recognizing SARS-CoV-2-derived peptides (P04, P06, and P12), which are conserved in other coronaviruses (Table S1), in one non-infected healthy individual as well (Fig. 4C).
Fig. 4.
Detection of CD8+ T cells that recognized SARS-CoV-2-derived MHC class I-binding peptides in PBMCs. A Gating strategy for detecting SARS-CoV-2-derived peptides-reactive CD8+ T cells. Firstly, lymphocytes population was separated by a forward scatter (FSC-A) and back scatter (BSC-A) gates. Doublets were excluded on forward scatter height (FSC-H) and forward scatter width (FSC-W). Living CD3+ CD4− cells were selected by staining with DAPI, and expression of CD3 and CD4. pMHC1 tetramer-binding CD8+ T cells were defined by gating based on HIV control tetramer staining. B HLA-A*24:02-positive PBMCs from three COVID-19-recovered individuals were stained using pMHCI tetramers after in vitro stimulation. CD8+ T cells that recognize SARS-CoV-2-derived peptides were detected from every individual. C HLA-A*24:02-positive PBMCs from three healthy individuals without any history of SARS-CoV-2 infection were stained using pMHCI tetramers after in vitro stimulation. Pre-existing CD8+ T cells that recognize SARS-CoV-2-derived peptides were detected from one of the three non-infected individuals
Impact of SARS-CoV-2 omicron variant on MHC class I-binding peptides
We investigated the influence of mutations found in SARS-CoV-2 omicron variant (BA.1), which is a new SARS-CoV-2 variant of concern and has been identified in 89 countries as of 16th of December, 2021, on 15 MHC class I-binding peptides identified in this study. We searched nonsynonymous substitutions and deletions in the GISAID database (https://www.gisaid.org) on 20th of December, 2021. We identified 132 mutations (76 mutations in ORF1ab polyprotein, 51 mutations in spike protein, and 5 mutations in membrane glycoprotein) in 9 or more (>0.1%) of 8993 sequences of omicron variant. Only one mutation at position 5086 (F5086Y) in ORF1ab may affect immunogenicity of P06 and P11, but other peptide sequences are conserved in the omicron variant (Table S2). This indicates that the peptides identified in this study may be used as a sort of universal peptides to induce CTLs against COVID-19 variants.
Discussion
We screened 29 SARS-CoV-2-derived epitope peptides (14 peptides for HLA-A*24:02 and 15 peptides for HLA-A*02:01) that were predicted to bind to MHC class I molecules for activation of CD8+ T cells and found that eight peptides each for HLA-A*24:02 and HLA-A*02:01 could activate CD8+ T cells isolated from PBMCs. We subsequently established CTL clones and confirmed their immune reactivities against these peptides presented on the expected HLA molecules. Among the eight peptides that activated CD8+ T cells derived from HLA-A*02:01 donors, seven peptides also activated CD8+ T cells derived from HLA-A*02:06-positive PBMCs. Interestingly, one peptide (P08) commonly induced CTLs derived from all of HLA-A*24:02-, HLA-A*02:01-, and HLA-A*02:06-positive PBMCs. Hence, a total of 15 peptides were likely to be presented on MHC class I molecules and activated CD8+ T cells. These 15 peptides corresponded to parts of spike protein, membrane glycoprotein, or ORF1ab polyprotein (Fig. S3).
Variant SARS-CoV-2 strains, alpha, beta, gamma, delta, and omicron, which are now classified as variants of concern by World Health Organization, rapidly expanded worldwide and are still expanding in some countries. SARS-CoV-2 variants of concern appear to escape at some extent from vaccinations using mRNA encoding the spike protein because they have mutations in domains recognized by neutralizing antibodies. Since multiple peptides identified from spike protein, membrane glycoprotein, and ORF1ab polyprotein in this study are well conserved among coronavirus species, it is likely that these regions are essential for virus survival or expansion. Hence, T cells induced by these peptides reported here might be less affected by mutations that may additionally occur in various virus proteins. In fact, mutations found in the omicron variant have little effect on immunogenicity of these peptides at present although we need to further monitor the variant sequences.
We confirmed the presence of CD8+ T cells that recognize SARS-CoV-2-derived peptides (P01, P02, P07, P08, and P12) in HLA-A*24:02-positive individuals recovered from COVID-19, indicating that CD8+ T cells were activated in vivo by coronavirus-derived peptides (possibly same as peptides identified in this study) presented on MHC class I molecules on antigen presenting cells including DCs in their body. It is notable that we found CD8+ T cells that recognized SARS-CoV-2-derived peptides (P04, P06, and P12) in one SARS-CoV-2-non-infected healthy individual. Since these peptides are conserved among other human coronaviruses, the presence of CD8+ T cells reacting to these peptides may reflect previous infections with these viruses, or this individual might have experienced non-symptomatic COVID-19. Understanding the effect of pre-existing SARS-CoV-2-derived peptide-reactive CD8+ T cells on non-symptomatic patients or prevention of COVID-19 should be important to consider the strategy for controlling pandemic or the future threat to newly arising coronaviruses.
Recently, one report indicated that a spike protein-derived polypeptide with adjuvant on nanoparticle, which was administrated by intramuscular route, successfully induced neutralizing antibodies and CD4+ T cell responses in rhesus macaques [32]. This report also supports the concept that SARS-CoV-2-derived epitope peptides might be a promising COVID-19 vaccine that would induce and maintain the T cell immunity against SARS-CoV-2 that will continuously cause new mutations. In addition, we found no significant homology with peptides possibly derived from human proteins by the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi), suggesting there is little possibility that our peptides might cause unintended immune reactions, which are unfavorable to our health condition.
Furthermore, T-Detect COVID Test (Adaptive Biotechnologies), a next-generation TCR sequencing-based test, was approved by FDA for identifying the recent or prior SARS-CoV-2 infection. Our peptide-reactive TCR sequences might be useful to investigate SARS-CoV-2 infection history or monitor the effectiveness in vaccinated individuals.
In this study, we have reported identification of 15 candidate vaccine peptides against COVID-19. Since T cell memories seem to be maintained in a longer period (few to several years) than antibodies and the peptides reported here are likely to avoid the influence of additional mutations that may occur in SARS-CoV-2 in the future, our peptides may enable us to provide the useful information to design the novel vaccine(s).
Supplementary information
Acknowledgements
We thank K. Kiyotani at Japanese Foundation for Cancer Research for helpful discussion about prediction and selection of MHC class I-binding peptides. We thank Y. Imai, M. Koyasu, and H. Yamashita for experimental support. This study was funded by the R&D budget of OncoTherapy Science, Inc.
Author contributions
Conceptualization: YN, TH. Investigation: M. Sakamoto, MH, M. Saito, YY, KT. Visualization: TH, M. Sakamoto, MH, M. Saito, YY, KT. Project administration: YN, TH. Supervision: YN. Writing—original draft: TH. Writing—review and editing: YN.
Competing interests
TH, M. Sakamoto, MH, M. Saito, YY, and KT are employees of OncoTherapy Science, Inc. YN is a stockholder and scientific advisor of OncoTherapy Science, Inc. TH and YN are listed as the inventors on pending patent application that is related to findings in this study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/22/2022
A Correction to this paper has been published: 10.1038/s10038-022-01024-1
Supplementary information
The online version contains supplementary material available at 10.1038/s10038-022-01013-4.
References
- 1.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl J Med. 2020;383:2603–15. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl J Med. 2021;384:403–16. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological. 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 4.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–43. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 5.Network for Genomic Surveillance in South Africa (NGS-SA). SARS-CoV-2 Sequencing Update 17 December 2021. Network for Genomic Surveillance in South Africa (NGS-SA). 2021. https://www.nicd.ac.za/wp-content/uploads/2021/12/Update-of-SA-sequencing-data-from-GISAID-17-Dec-21_Final.pdf.
- 6.Faria NR, Claro IM, Candido D, Moyses Franco LA, Andrade PS, Coletti TM, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2020. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586.
- 7.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–83.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl J Med. 2021;384:1466–8. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl J Med. 2021;385:187–9. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331–3. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–61.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–5. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130:2620–9. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–62. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 18.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–45. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh HL, Chia A, Chang CX, Leong HN, Ling KL, Grotenbreg GM, et al. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J Virol. 2011;85:10464–71. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–14. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyotani K, Park JH, Inoue H, Husain A, Olugbile S, Zewde M, et al. Integrated analysis of somatic mutations and immune microenvironment in malignant pleural mesothelioma. Oncoimmunology. 2017;6:e1278330. doi: 10.1080/2162402X.2016.1278330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poluektov Y, George M, Daftarian P, Delcommenne MC. Assessment of SARS-CoV-2 specific CD4 (+) and CD8 (+) T cell responses using MHC class I and II tetramers. Vaccine. 2021;39:2110–6. doi: 10.1016/j.vaccine.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lissina A, Ladell K, Skowera A, Clement M, Edwards E, Seggewiss R, et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods. 2009;340:11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–75.. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–7. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–17. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirata J, Hosomichi K, Sakaue S, Kanai M, Nakaoka H, Ishigaki K, et al. Genetic and phenotypic landscape of the major histocompatibility complex region in the Japanese population. Nat Genet. 2019;51:470–80.. doi: 10.1038/s41588-018-0336-0. [DOI] [PubMed] [Google Scholar]
- 28.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Buuren MM, Dijkgraaf FE, Linnemann C, Toebes M, Chang CX, Mok JY, et al. HLA micropolymorphisms strongly affect peptide-MHC multimer-based monitoring of antigen-specific CD8+ T cell responses. J Immunol. 2014;192:641–8. doi: 10.4049/jimmunol.1301770. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura S, Tsunoda T, Osawa R, Harada M, Watanabe T, Hikichi T, et al. Identification of an HLA-A2-restricted epitope peptide derived from hypoxia-inducible protein 2 (HIG2) PLoS One. 2014;9:e85267. doi: 10.1371/journal.pone.0085267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torikai H, Akatsuka Y, Miyauchi H, Terakura S, Onizuka M, Tsujimura K, et al. The HLA-A*0201-restricted minor histocompatibility antigen HA-1H peptide can also be presented by another HLA-A2 subtype, A*0206. Bone Marrow Transpl. 2007;40:165–74. doi: 10.1038/sj.bmt.1705689. [DOI] [PubMed] [Google Scholar]
- 32.Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–8. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.