Highlights
-
•
Knowledge of the genetic etiologies of cardiomyopathies has created novel opportunities for treatments.
-
•
Emerging treatments targets include gene therapy, myofilament function, protein quality control, and metabolism.
-
•
Consideration of the genetic cause, biophysical context, and cardiomyopathy disease stage will be critical to the success of these novel treatments.
Key Words: arrhythmogenic cardiomyopathy, dilated cardiomyopathy, genetics, hypertrophic cardiomyopathy, therapeutics
Abbreviations and Acronyms: AAV, adeno-associated virus; ACM, arrhythmogenic cardiomyopathy; ARVC, arrhythmogenic right ventricular cardiomyopathy; ATPase, adenosine triphosphatase; DCM, dilated cardiomyopathy; DMD, Duchenne muscular dystrophy; DNA, DNA; DSP, desmoplakin; FDA, U.S. Food and Drug Administration; GRT, gene replacement therapy; GST, gene silencing therapy; HCM, hypertrophic cardiomyopathy; HR, homologous recombination; LNP, lipid nanoparticle; LVOT, left ventricular outflow tract; RNA, RNA; TTR, transthyretin
Summary
The primary etiology of a diverse range of cardiomyopathies is now understood to be genetic, creating a new paradigm for targeting treatments on the basis of the underlying molecular cause. This review provides a genetic and etiologic context for the traditional clinical classifications of cardiomyopathy, including molecular subtypes that may exhibit differential responses to existing or emerging treatments. The authors describe several emerging cardiomyopathy treatments, including gene therapy, direct targeting of myofilament function, protein quality control, metabolism, and others. The authors discuss advantages and disadvantages of these approaches and indicate areas of high potential for short- and longer term efficacy.
Central Illustration
Genetic Architecture of Familial Cardiomyopathies
The 3 major subtypes of familial cardiomyopathies are hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and arrhythmogenic cardiomyopathy (ACM). Although these diagnoses have historically been made on the basis of established clinical criteria, new insights into genotype-phenotype correlations enable a clearer understanding of molecular etiology that can now be harnessed to target therapies more precisely.
The sarcomere is the primary genetic etiology for HCM, with 40% to 50% of patients carrying pathogenic variants in 1 of 8 sarcomere genes (1). Compared with patients without an identifiable genetic etiology (“genotype negative”), patients carrying pathogenic sarcomere variants present at younger ages, demonstrate greater hypertrophy, and have a higher risk for adverse events (1, 2, 3). There is a small number of nonsarcomere genes with established strong associations with HCM, including CSRP3, FHL1, PLN, FLNC, and ALPK3, that are rare causes of HCM (4, 5, 6), with some manifesting unique phenotypic features.
About 20% to 35% of patients with DCM have identifiable pathogenic variants (7). The genetic architecture for DCM is very diverse, with variants in genes encoding proteins across virtually every cardiac myocyte compartment (eg, nuclear membrane, sarcomere, Z-disc, desmosome, cytoskeleton) and with highly varied functionality (eg, contraction, structural integrity, ion channels, molecular chaperones) (8). Thanks to large, multicenter collaborative efforts, unique correlations among genotype, phenotype, and clinical outcomes are emerging, most notably for LMNA (9), DSP (10,11), RBM20 (12), and FLNC (13).
ACM can be left or right dominant, with the right-dominant form being classically recognized as arrhythmogenic right ventricular cardiomyopathy (ARVC). Classic ARVC is caused primarily by variants in the genes PKP2, JUP, DSG2, and DSC2, which encode the proteins of the cardiac desmosome. DSP variant carriers typically present with left-dominant DCM but may also present with ARVC. There are a small number of genes outside of the desmosome that represent rare causes of ARVC, including TMEM43, PLN, and DES (14). Most clinical cohorts have been dominated by PKP2 variant carriers.
For all of the aforementioned cardiomyopathy subtypes, the most common mode of inheritance is autosomal dominant. For each genetic variant that causes cardiomyopathy, classification as either a loss-of-function mechanism (typically from truncating variants) or gain-of-function mechanism (typically from missense variants) is critically important when considering genetics-based treatment approaches (see “Somatic Gene Therapy”).
Taken together, an increasingly precise understanding of cardiomyopathy etiology now underscores the importance of integrating genetic diagnosis with clinical assessment. Moreover, these observations highlight the importance of accounting for genetic diagnosis in the design of contemporary clinical trials. Additionally, it will be imperative to consider the variable extent and rate of cardiac remodeling, which is common to all forms of cardiomyopathy. This variability is related in part to background genetic modifiers, as suggested by reports of digenic contributors to cardiomyopathy and recent genomewide association studies (1,15, 16, 17, 18) and presents a major challenge to determining the optimal timing of novel therapies.
Current Therapies
There are several inhibitors of neurohormonal and adrenergic pathways with well-proven efficacy to reduce morbidity and mortality in patients with DCM associated with reduced systolic function (19). Recent trials of inhibitors of sodium-glucose cotransporter-2 have unexpectedly shown additive benefit in patients, independent of the presence of diabetes (20, 21, 22). None of these trials have included genetic information, and therefore it is unclear whether specific genetic subtypes of familial DCM derive differential benefit. For HCM and ACM, there is no currently proven disease-modifying benefit of any of these classes of drugs, and medical treatment of these conditions is limited to medications and procedures that provide symptomatic relief.
Emerging Therapies
Broadly speaking, new therapies under consideration to treat genetic cardiomyopathies fall into 2 categories: U.S. Food and Drug Administration (FDA)–approved drugs that are “repurposed” for new indications and completely novel classes of drugs or nonpharmacologic approaches. An example of the former is a trial of the angiotensin receptor blocker valsartan in early-stage HCM, the results of which are currently pending (23). This review will focus primarily on the latter category: novel pharmacologic and nonpharmacologic approaches to more precisely target specific genetic cardiomyopathies. We have organized the review by categorizing different approaches on the basis of their mechanisms of action and the stage of disease evolution each would be targeting (Central Illustration).
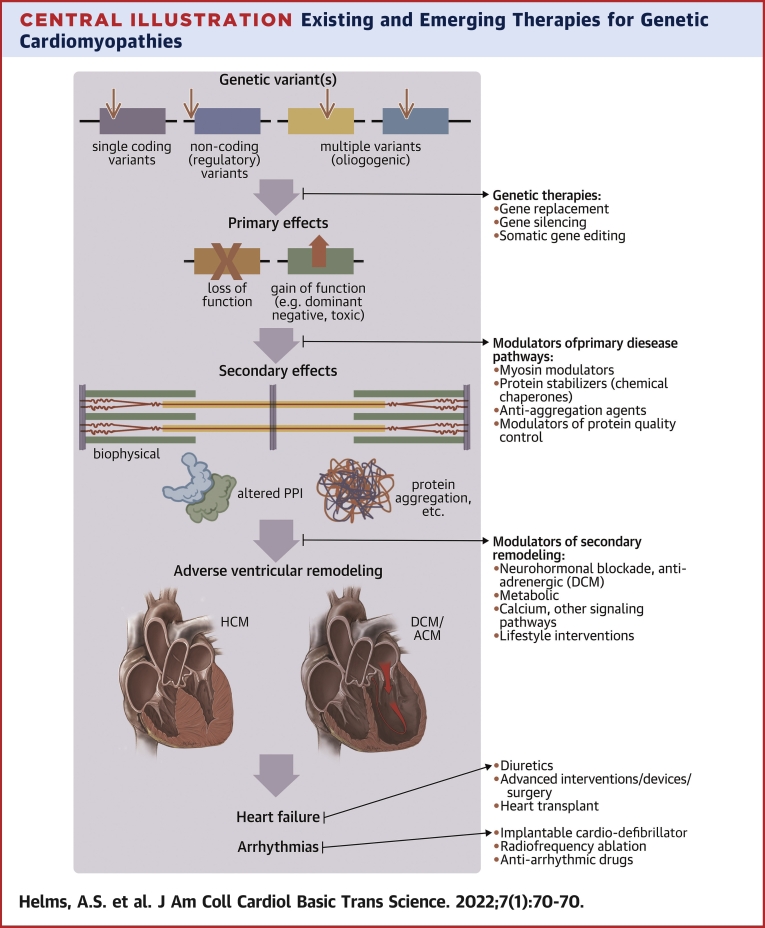
Central Illustration.
Existing and Emerging Therapies for Genetic Cardiomyopathies
Existing therapies are limited largely to treatment of symptomatic patients with established disease and adverse ventricular remodeling. Emerging therapies would target early effects of genetic and environmental triggers, ideally before disease is fully manifest. These include genetic therapies and modulators of primary and secondary disease pathways. Figure created with biorender.com. ACM = arrhythmogenic cardiomyopathy; DCM = dilated cardiomyopathy; HCM = hypertrophic cardiomyopathy.
Timing of Emerging Therapies
As the heart is a contracting organ, the persistent biomechanical work load, in the presence of sensitizing genetic variants, leads to progressive cardiac remodeling during aging. Importantly, the rate of progressive remodeling is highly variable across individuals. This phenotypic variability, driven by both genetic background and other clinical factors, is common to all forms of cardiomyopathy and presents a major challenge in identifying the optimal timing for potential treatment interventions (17,18). Supporting the value of careful disease staging, Ho et al (1) showed that patients with HCM with a young age of diagnosis had a much higher rate of adverse events than those presenting late in life, even when stratified by genetic subtype. Therefore, design and targeting of novel agents requires careful attention to timing to maximize the likelihood of success of clinical trials. Accurate disease staging is also important when considering the treatment target. Treatments such as gene therapy or myofilament modulators target the effects of pathogenic variants directly. These strategies may be most effective at earlier disease stages. Following extensive remodeling, these same treatments could potentially be harmful depending on the level of maladaptive remodeling that has occurred. For example, in HCM that has progressed to left ventricular systolic dysfunction, treatment with a medication to reduce myofilament activation may be detrimental by further lowering systolic function, though this has not yet been studied.
Somatic Gene Therapy
Gene therapies are appealing to treat a broad range of human disease because they target the direct molecular cause. Cardiac gene therapy could, in theory, be applied before a cardiomyopathy has even developed. All gene therapies require the delivery of nucleotide sequences directly to the target cells. Efficient and safe delivery has been the major hurdle. Gene therapy has been accomplished largely using viral vectors that have been coopted to deliver DNA (DNA) to target cells or by using techniques such as lipid nanoparticles (LNPs) to gain entry of RNA (RNA) into target cells. Early gene therapy attempts using viral vectors in the 1990s were largely unsuccessful because of limitations in efficacy and safety (24). Two classes of modern vectors have moved successfully past these limitations. Retroviral vectors (eg, lentivirus) have proved effective for immunodeficiency and hematopoietic disorders but permanently integrate into the genome and therefore pose a safety risk for cardiac gene therapy. In contrast, adeno-associated virus (AAV) is inherently replication deficient and nonintegrating. Modern AAV vectors have been developed to attain relatively high transduction levels with minimal toxicity (25). AAV has rapidly emerged as the dominant vector for somatic cell gene therapy with impressive responses in clinical trials for inherited retinal dystrophy and spinal muscular atrophy that have led to FDA-approved treatments (26,27). With some serotypes of AAV having natural tropism for the heart (eg, AAV9), AAV-based gene therapies are currently being evaluated for a range of cardiac diseases.
The most direct type of gene therapy is gene replacement therapy (GRT), in which a wild-type gene is expressed by a promoter within a viral vector to replace gene function in the setting of a loss-of-function variant (Figure 1). Loss-of-function variants are most often nonsense, splice, or frameshift that cause a shift in reading frame and premature termination codon (commonly referred to as “truncating” variants, even though actual truncated protein is rarely present, because of nonsense-mediated RNA decay of the mutant transcript). Loss of function is common in a number of cardiomyopathy genes, including MYBPC3, TTN, LMNA, DSP, PKP2, BAG3, and FLNC. The first AAV-based GRT clinical trial for cardiomyopathy is currently under way for Danon disease, caused by truncating variants in LAMP2.
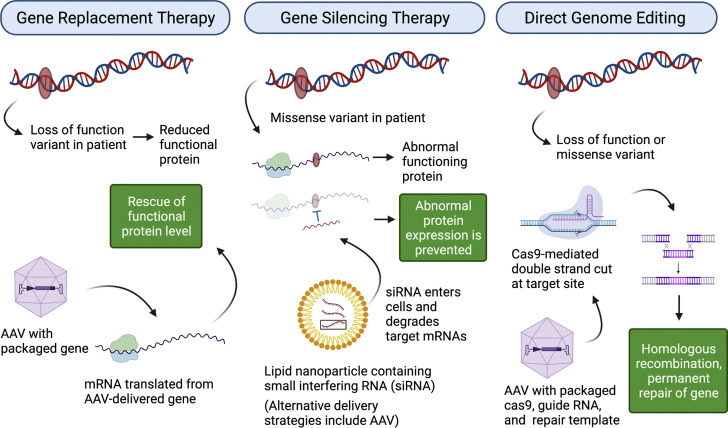
Figure 1.
Gene Therapy
With the gene replacement strategy, a wild-type gene is expressed by a promoter within a viral vector to replace gene function in the setting of a loss-of-function variant. Gene silencing is typically used to reduce expression of a mutant gene (referred to as allele-specific silencing) and applies primarily to missense variants (ie, single-nucleotide substitutions) that alter protein function. For direct genome editing, using CRISPR-Cas9, a targeted DNA cleavage can be directed to a precise location in the genome determined by the unique sequence of a guide RNA (RNA). Figure created with biorender.com. AAV = adeno-associated virus; mRNA = messenger RNA.
Expressing a wild-type gene may also be effective in the context of certain missense variants. For example, exogenous expression of a sarcomere protein can result in stoichiometric replacement of endogenous protein without overexpression (28). Therefore it is conceivable that GRT with a wild-type gene could outcompete a missense mutant sarcomere protein to drive its abundance below a certain threshold at which it would no longer significantly affect sarcomeric function. The same concept might be true of other cardiomyopathy-associated genes but has not been reported to date.
AAV-based GRT faces several hurdles: 1) the maximum gene size that can fit within the AAV vector is 4.7 kb, preventing application to larger genes; 2) because AAV is nonintegrating, the efficacy likely wanes after several years (the duration in humans is not yet known); 3) current AAV vectors require relatively high titers to attain sufficiently high cardiac transduction; 4) neutralizing antibodies are present in some individuals because of prior exposure to natural AAV and limit efficacy; and 5) future redosing of the AAV may be less effective because of induction of neutralizing antibodies elicited from the first dose. Technologies to circumvent the gene size requirement are being pursued, including trans-splicing approaches that use dual AAV vectors to express partial genes that splice together prior to translation in transduced cells (29). For some large genes, a shorter version of the gene is capable of partial rescue, reducing the required vector capacity. For example, in Duchenne muscular dystrophy (DMD), the expression of “mini” or “micro” DMD genes has shown efficacy in animal models (30,31) and is currently being tested in clinical trials (32). Recently, a trans-splicing approach was used for a 7-kb mini-DMD gene to effectively treat a canine model of DMD, demonstrating the potential for combined strategies to overcome packaging limits of AAV (33). Improvements in cardiac-specific targeting and transduction efficiency are expected through engineering of synthetic AAV capsids. Presumed waning efficacy will potentially be addressed by developing strategies to prevent neutralizing antibodies from developing that will enable more effective redosing and/or engineering of the AAV capsid (34, 35, 36).
Gene silencing therapy (GST) is used to reduce the expression of a mutant gene (referred to as allele-specific silencing) and applies primarily to missense variants (ie, single-nucleotide substitutions) that alter protein function. The primary approach for GST has been interfering RNA delivered by LNPs or AAV, and proof-of-concept studies have shown efficacy for cardiomyopathy-causing variants in MYH6 and MYL2 in mice (37,38). The major hurdle for GST is that separate interfering RNA–targeting constructs typically need to be developed for each individual variant. This hurdle is practically insurmountable with current technological throughput and also faces major obstacles from a clinical trial design perspective. Additionally, for many genes, the biologic tolerance for silencing of 1 allele (creating iatrogenic haploinsufficiency) over a human lifetime is not clear, and mouse models are inadequate to fully evaluate the potential long-term consequences of allele silencing because of their short life span. To avoid these potential long-term consequences, another approach is to combine silencing of the endogenous gene expression with GRT (as described previously); this strategy was recently shown to be feasible using lentiviral vectors to rescue KCNQ1-mediated long-QT syndrome in vitro, but it has not yet been demonstrated in vivo, for which AAV-mediated gene delivery would be required (39). So far, there has been 1 promising application for GST in cardiomyopathy that has reached clinical translation: transthyretin (TTR) amyloidosis. As the function of TTR appears to be largely dispensable, silencing constructs have been developed to degrade all TTR transcripts. Clinical trials for GST in TTR amyloidosis have recently shown efficacy, leading to the first approval for patisiran (silencing RNA therapy) for polyneuropathy using LNPs for delivery (40,41). A clinical trial of patisiran for TTR cardiomyopathy (APOLLO-B) is ongoing. LNP-based delivery of small interfering RNA into the heart has not yet been achieved, but this is an active area of study.
Direct genome editing is now conceivable because of the development of CRISPR-Cas9 technology (42). With CRISPR-Cas9, a targeted DNA cleavage can be directed to a precise location in the genome, determined by the unique sequence of a guide RNA. The technical feasibility of direct variant correction was recently shown using CRISPR-Cas9 genome editing in human embryos with a variant in MYBPC3 (43). However, as preimplantation genetic screening of embryos through an in vitro fertilization approach has been available for many years, there is no practical utility for direct embryonic genome editing for inherited cardiomyopathies. We will therefore focus in this section on the potential application of somatic genome editing.
Targeting with CRISPR-Cas9 alone results in error-prone repair called nonhomologous end joining, which invariably results in insertion or deletion variants at the targeted site, 2 thirds of which will shift the reading frame, resulting in premature truncation. To edit a specific nucleotide, homologous recombination (HR) can be achieved using a repair DNA template. Cas9 genes, guide RNA sequences, and HR templates can all, in theory, be delivered to the heart via AAV. The major obstacle to somatic CRISPR-Cas9 genome editing with current technology is that the rate of HR is very low, particularly in cardiomyocytes. Technologies to circumvent these limitations are an area of active investigation, including direct CRISPR-Cas9 base editing (44,45). If successful, this technology would have the potential for a complete cure, requiring only a single treatment. One important exception to the requirement for HR during CRISPR-Cas9 editing is the case of DMD. As large portions of the interior sequence of DMD are not required for muscle function, nonhomologous end joining can be targeted to create insertion/deletion variants that restore the reading frame without requiring HR. This strategy has been effective in rodent and dog models of DMD, and a clinical trial is in progress (46, 47, 48). Patients are candidates for this approach only if the their DMD variants are in specific exons for which the strategy has been developed. Additionally, this approach would not be translatable to most other genes, as the case of unnecessary interior sequence in the extremely large DMD gene is relatively unique (TTN may be an exception), and eligibility of patients with DMD depends on the specific locations of their variants.
Additional considerations for cardiac gene therapy
Gene therapy holds enormous potential for a complete or near cure for a host of different diseases. One recent and very promising example is for sickle cell disease and β-thalassemia, in which CRISPR-Cas9 gene editing of an erythroid enhancer region resulted in re-expression of fetal hemoglobin and a dramatic clinical response with elimination of vaso-occlusive episodes and need for transfusion (49). These diseases of the blood cell lineages are at the forefront, as gene editing of bone marrow–derived cells is more accessible and can be done ex vivo. In contrast, cardiac gene editing requires further technical development for effective delivery. Current applications rely on AAV9 vectors, which have inherent limitations related to packaging, need for high titers, immunogenicity, and possible off-target effects with unwanted delivery to other organs (eg, the liver). Nonviral vector delivery systems, including LNPs, are being actively pursued as vehicles for delivery of not only small interfering RNA, but also messenger RNA and DNA, to their target organs (50). LNP delivery of CRISPR-Cas9 reagents for base editing of PCSK9 in the liver was recently shown to be highly efficient, resulting in durable lowering of low-density lipoprotein cholesterol in nonhuman primates (45). Although specific targeting of LNPs to the heart has not yet been achieved, it is certainly a possible vehicle for future applications of cardiac gene therapy and could circumvent the obligate use of viral vectors for delivery of genetic material for gene editing or silencing. Regardless of the vehicle or method of delivery, the cost of genetic therapies will undoubtedly be a factor as well in their widespread clinical applicability.
Although still in the early days of cardiac gene therapy, and with lingering uncertainties about long-term safety and durability of response, patient selection for upcoming gene therapy trials will be critical. As it stands today, the optimal candidates for early-stage gene therapy trials are young patients with particularly severe and lethal cardiomyopathies for whom treatment options are limited or completely ineffective. Danon disease is a good example because of its rapid progression to heart failure and need for transplantation in pediatric and young adult male patients. Another example would be patients with biallelic variants in genes that cause complete loss of function, such as MYBPC3. The number of these patients worldwide is very small, but they have a severe neonatal DCM with very high morbidity and mortality, with no effective therapies other than cardiac transplantation (51). Patients who are carefully selected for high-risk criteria for progression to advanced heart failure, or those with already advanced disease who have no existing treatment options, are other potential target groups, although the latter group is less likely to demonstrate a treatment response.
Modulators of Primary Disease Pathways
In this section, we consider a primary disease pathway as a direct consequence of a genetic variant on the protein that it encodes. These effects would include biophysical changes, altered protein-protein interactions, or protein aggregation as a few specific examples (Central Illustration). We will discuss several well-established primary disease pathways for genetic cardiomyopathies for which pharmacologic approaches are emerging as novel therapies.
Direct contractile modulation
Hypercontractility in HCM and hypocontractility in DCM have been conceptualized as pathophysiologic hallmarks of disease, though this classification is likely oversimplified, and the specific mechanisms that lead to biophysical alterations may depend on the particular causative genetic variant (52,53). Inotropic therapy to improve contractility has long been explored as a potential therapy for systolic heart failure. Adrenergic receptor agonists and phosphodiesterase inhibitors are currently used in acute care settings and for bridging or palliation in end-stage disease (54). However, these agents cause arrhythmias and increase mortality, limiting their scope of clinical use (55,56). These adverse effects may be due in part to the fact that these agents target pathways upstream of the cardiac sarcomere, altering cyclin adenosine monophosphate and intracellular calcium handling. A high-throughput screen using a kinetic readout of myosin adenosine triphosphatase (ATPase) activity identified CK1827452/AMG-423 (omecamtiv mecarbil, Cytokinetics) as a compound that increases myosin ATPase activity (57) (Figure 2). Omecamtiv mecarbil was shown to selectivity target cardiac myosin without inducing changes in the calcium transient, and it resulted in improved cardiac contractility in preclinical heart failure models (57). Similarly, danicamtiv (MYK-491, MyoKardia) is a cardiac myosin modulator that enhances contractility and has demonstrated improved systolic function in preclinical models and in early clinical trials (NCT03447990, NCT03062956) (58). These compounds appear to work, at least in part, by activating the thin filament (59). A double-blind, placebo-controlled, randomized phase 3 trial of omecamtiv mecarbil demonstrated an 8% relative risk reduction in the primary composite outcome of heart failure events or death from cardiovascular causes (60). However, secondary endpoints were not met; specifically, no reduction in death from cardiovascular causes or improvements in total symptom score were observed. Given this somewhat modest result, it is unclear whether omecamtiv mecarbil will be integrated into the growing armamentarium of DCM treatments. A potential role could be in patients whose blood pressure is too low to enable significant up-titration of current goal-directed therapy.
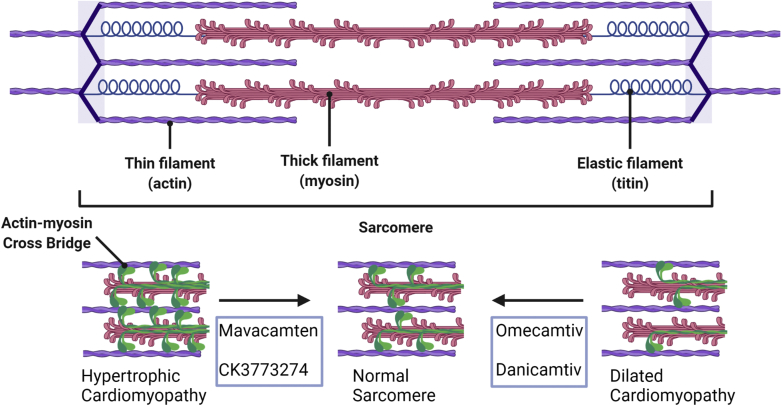
Figure 2.
Myosin Modulators
Omecamtiv was developed to improve contractile force for treatment of dilated cardiomyopathy. The mechanisms of this drug are complicated, but increased attachment time of myosin cross bridges to actin serves to activate the thin filament and increase contractility. Conversely, mavacamten and similar compounds were developed to decrease contractile force for treatment of hypertrophic cardiomyopathy. These compounds act by decreasing myosin ATPase activity, resulting in the formation of fewer myosin-actin cross bridges. Figure created with biorender.com.
Conversely, a strategy to decrease the ATPase rate of myosin has recently proved effective for the treatment of HCM. Originally identified using a high-throughput screen for molecules that decrease maximal actin-activated ATPase rate of myosin, MYK-461 (mavacamten, MyoKardia) is an allosteric modulator that stabilizes an autoinhibited state (super-relaxed state) of cardiac myosin (61, 62, 63). At the time of mavacamten’s development, it was unclear whether an increase in power output was a consistent molecular consequence of pathogenic sarcomeric variants that cause HCM. Thus, mavacamten was developed in part as an experimental tool to test if inhibiting excess sarcomere power output could ameliorate disease phenotypes (61). Mavacamten was subsequently shown to abrogate the hypertrophic phenotype in several mouse models of HCM resulting from different pathogenic MYH7 variants (61). A double-blind, randomized, placebo-controlled phase 3 trial tested the efficacy of mavacamten in 251 patients with HCM and symptomatic left ventricular outflow tract (LVOT) obstruction (EXPLORER-HCM [Clinical Study to Evaluate Mavacamten (MYK-461) in Adults With Symptomatic Obstructive Hypertrophic Cardiomyopathy]; NCT03470545). Compared with placebo, mavacamten improved exercise capacity, LVOT obstruction, New York Heart Association functional class, and health status (64). Furthermore, mavacamten treatment was associated with reductions in left ventricular hypertrophy and left atrial volumes, showing a favorable impact on cardiac remodeling (65). Mavacamten is the first disease-specific treatment for HCM and is currently awaiting FDA approval to treat patients with symptomatic, obstructive HCM. The utility of this therapy in nonobstructive HCM is less clear. A double-blind, placebo-controlled phase 2 dose-ranging study of mavacamten in patients with symptomatic nonobstructive HCM (MAVERICK-HCM [A Phase 2 Study of Mavacamten in Adults With Symptomatic Non-Obstructive Hypertrophic Cardiomyopathy]; NCT03442764) (66) revealed that biomarkers of cardiac wall stress (N-terminal pro–brain natriuretic peptide and cardiac troponin I) were significantly reduced by mavacamten, but the primary endpoint combining peak oxygen consumption and symptomatic alleviation was not different between the placebo and active treatment groups (66). Whether this therapy will offer symptomatic benefit in any subgroup of patients with nonobstructive HCM will require further investigation. Another allosteric modulator of myosin, CK3773274/CK-274 (Cytokinetics), which binds myosin at a site distinct from mavacamten, is also in development with a clinical trial currently enrolling (67). There is potential for these myosin modulators to favorably affect disease progression over the long term (Central Illustration), independent of symptoms or LVOT obstruction, as destabilization of the super-relaxed state may be an early driver of HCM pathogenicity (63,68,69). As this mechanism has been best described for variants in thick-filament genes, it remains to be seen whether patients with HCM carrying variants in thin-filament or nonsarcomere genes will derive the same benefit from myosin modulators, as these patients are under- or unrepresented in clinical trials at present.
Alternative mechanisms by which sarcomeric variants exert their effects include work stroke mechanics (stiffness), calcium-based activation, and altering the number of available myosin heads (70). These distinct molecular mechanisms may be targets for novel therapeutic strategies in the future. For example, pathogenic variants in TNNT2 and TPM1 affect calcium-based thin-filament regulation (71,72), which could be therapeutically targeted by increasing or decreasing troponin affinity for calcium (calcium sensitizers or desensitizers) for DCM or HCM, respectively (73, 74, 75).
Protein stabilizers (chemical chaperones)
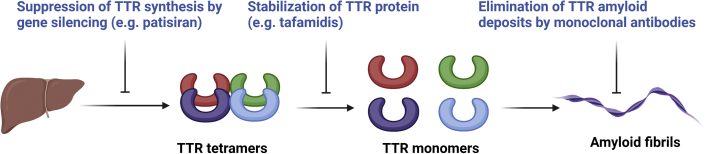
Cardiotoxic protein aggregation is implicated in a subset of cardiomyopathies. For example, in both wild-type and hereditary TTR cardiac amyloidosis, conformational instability in the tetrameric TTR results in the formation of amyloid fibrils (76). Among strategies to foster protein homeostasis are the use of chemical chaperones that bind to their respective protein target and restore normal function by preventing aggregation, degradation, and/or aberrant trafficking (77). Tafamidis, developed for the treatment of TTR amyloidosis, was discovered using a structurally based drug discovery approach to identify compounds that stabilize the TTR tetramer by binding a thyroxine-binding site and inhibiting amyloid formation (tafamidis meglumine, Fx-1006A) (78,79). Tafamidis was shown in a double-blind, placebo-controlled phase 3 clinical trial (ATTR-ACT [Safety and Efficacy of Tafamidis in Patients With Transthyretin Cardiomyopathy]; NCT01994889) to reduce all-cause mortality and cardiovascular-related hospitalizations in patients with TTR amyloid cardiomyopathy (80). The study population comprised patients with wild-type TTR (76%) and those with pathogenic variants in TTR (25%), and benefits appeared to be generalizable across these groups. Human monoclonal antibodies against the misfolded forms of TTR are also in early development, with 2 phase 1 clinical trials ongoing (81) (Figure 3).
Figure 3.
Cardiac TTR Amyloidosis Therapies
Gene silencing therapy, using a silencing RNA packaged in lipid nanoparticle and delivered to the liver, has been shown to be effective at treating the polyneuropathy associated with transthyretin (TTR) amyloidosis. Stabilization of the TTR tetramer with tafamidis is effective in treating both polyneuropathy and cardiomyopathy. Monoclonal antibodies to extract TTR amyloid deposits from tissues are in clinical trials. Figure created with biorender.com.
Protein aggregation is also thought to underlie, in part, the pathogenesis of other cardiomyopathies, including missense variants in FLNC (82,83). As specific proteins within these aggregates are identified, development of chemical chaperones or antibodies against misfolded fibrils may be become feasible and a more generalizable strategy to treat a broader range of cardiomyopathies (84).
Protein quality control modulators
Although many sarcomere genes are tolerant to a loss-of-function variation in 1 allele, for others, the remaining functional copy is inadequate to preserve normal function (85,86). Disruption of sarcomere stoichiometry through haploinsufficiency of one of its proteins is a common pathogenic mechanism in cardiomyopathies. For example, loss-of-function pathogenic variants in MYBPC3 lead to haploinsufficiency as the most common cause of familial HCM (87, 88, 89). The most common genetic causes of DCM are truncating variants in TTN, resulting in haploinsufficiency that sensitizes the ventricle to dilation and dysfunction (90). Truncating variants in desmosomal genes that cause ACM perturb stoichiometry at the intermediate filament to desmosome linkage (DSP) or the desmosome itself (PKP2, DSG2, DSC2) (10,91).
As an alternative to a gene replacement strategy, one potential therapeutic approach to address haploinsufficiency is to target protein quality control networks: the cell’s natural mechanism for maintaining protein hemostasis. The essential role protein quality control networks play in maintaining sarcomere homeostasis has been highlighted by the identification of pathogenic variants in molecular chaperones that lead to genetic cardiomyopathies including small heat shock protein (CRYAB) and heat shock protein 70 cochaperones BAG3 and DNAJC18 (92, 93, 94, 95, 96). Furthermore, recent genomewide association studies in HCM and DCM identified single-nucleotide variants near molecular cochaperones HSPB7 and BAG3 with opposing effects (protective vs detrimental) in HCM and DCM (17,18). Thus, there is strong genetic evidence implicating molecular chaperone regulation in the pathogenicity of cardiomyopathy and heart failure. A pharmacologic approach to target these chaperones may be an attractive therapeutic strategy. Specific Hsp70 chaperone-client interactions have been successfully targeted in a variety of preclinical models of cancer and neurodegenerative diseases to correct aberrations in protein homeostasis (97). Recently, MyBP-C has been identified as a client of HSP70 chaperones (98). Induced pluripotent stem cell–derived cardiomyocytes harboring MYBPC3 truncating variants demonstrate a compensatory response to maintain normal levels of MyBP-C, possibly by modulation of HSP70 chaperones to reduce the rate of protein degradation (99). Thus, targeting the HSP70–ubiquitin proteasome dependent pathway may be a viable therapeutic target for HCM associated with variants in MYBPC3. As other specific clients of HSP70 and its cochaperones are identified, chaperone modulation may be considered more broadly as a potential therapeutic option for cardiomyopathies (Central Illustration).
Modulators of secondary disease pathways
In this section, we consider secondary disease pathways as downstream consequences of the primary effects of a particular genetic variant. Some examples of these include alterations in cardiac metabolism and energy substrate utilization, calcium homeostasis, fibrosis, and the influence of environmental factors.
Metabolic modulation
The cardiomyopathic heart is energetically depleted, with lower rates of fatty acid oxidation and increased glycolysis being hallmarks of metabolic remodeling (100). These metabolic changes precede the development of overt structural remodeling, suggesting that they represent an early event in the pathogenesis of heart failure. Reliance on glucose likely results in a state of adenosine triphosphate depletion over time. This theory is supported by the failure of metabolic modulators that further inhibit fatty acid oxidation, perhexiline and trimetazidine, to improve functional status in clinical trials of patients with HCM (101). An intriguing hypothesis is that enhanced metabolism of ketone bodies in the heart (102) could be adaptive, by effectively bypassing the defect in fatty acid oxidation (103). One of the mechanisms by which sodium-glucose cotransporter-2 inhibitors work is by increasing circulating ketone levels, and preclinical studies have suggested that this may be a primary driver of cardioprotection (104). Further studies will be needed to test whether therapeutic ketosis could be a generalizable approach for treatment of different forms of cardiomyopathy.
Downstream signaling pathways
A number of other pathways are altered in the early phase of the response to the molecular and biophysical effects of pathogenic variants and may be targets for precision therapy (70). One example is calcium signaling, which can be perturbed directly by variants in calcium regulatory genes (eg, PLN) or indirectly across a range of DCM and HCM phenotypes. A reduction in SERCA2a abundance and uptake of calcium into the sarcoplasmic reticulum has been observed in both DCM and HCM (105,106), and activation of calcium/calmodulin-dependent protein kinase type II is observed in hearts from patients with sarcomeric HCM. Gene replacement for SERCA2a demonstrated efficacy for improving cardiac function in animal studies of heart failure, but a clinical trial of AAV-expressed SERCA2 failed to show significant clinical benefit (107, 108, 109). Nevertheless, targeting of calcium homeostasis through genetic or small-molecular therapeutics is an avenue worth pursuing, given its potential for broad applicability to different cardiomyopathy genotypes and phenotypes.
In desmosomal ACM, inhibition of Wnt/β-catenin signaling and activation of the Hippo pathway have been implicated in enhanced adipogenesis, but targeting of this core molecular pathway has not thus far translated to the clinic (110). Activation of inflammatory signaling has also been linked to fibrotic remodeling in ACM (111). Transforming growth factor–β signaling is noteworthy because it is among several pathways broadly implicated in fibrosis in HCM and DCM phenotypes and is blunted through angiotensin receptor inhibition.
Antifibrotic treatments could potentially be beneficial across a broad range of inherited cardiomyopathy subtypes. However, safely targeting fibrotic signaling has been challenging because fibrosis is vital for normal wound healing and repair responses (112). A promising strategy was recently coopted from cancer therapeutics: redirection of cytotoxic T cells to recognize a chimeric antigen receptor that specifically marks activated fibroblasts (via fibroblast activation protein) was able to specifically deplete activated fibroblasts and prevent remodeling in a mouse model of cardiac fibrosis induced by angiotensin II and phenylephrine infusion (113). To what extent fibrosis may represent a necessary salvage response to chronic injury sustained in the presence of cardiomyopathic variants versus a maladaptive response that can be interrupted remains an unanswered question with high priority for future work.
Environmental factors
Nongenetic factors can play an important modifying or additive role in the emergence and progression of genetic cardiomyopathies. For example, about 15% of women who develop peripartum cardiomyopathy have truncating variants in genes associated with DCM, most commonly TTN (114,115). As another example, diastolic hypertension confers an increased risk for developing HCM in both the presence and absence of a sarcomere gene variant (17). Obesity and exercise also play key modulatory roles in the level of physical functioning and clinical outcomes (116,117). Looking ahead to novel genetically targeted therapies, it will be critical to continue to incorporate lifestyle counseling and management of comorbid conditions to achieve maximal therapeutic benefit.
Conclusions
Since the first discovery of genetic etiologies more than 3 decades ago, we are now poised for a period of rapid translation to precision medicine–based treatments in genetic cardiomyopathies. Therapies that are very proximal to the underlying genetic cause, such as gene therapies and primary disease pathway modulators, have the advantage of being the most specific for a particular genetic etiology and have the greatest potential for impact on disease burden. Conversely, modulators of secondary disease pathways will be less specific and may target only one of many different consequences of the genetic variants that cause adverse cardiac remodeling. However, by targeting final common pathways, such therapies may be more broadly applicable and potentially closer to implementation with use of repurposed drugs. Optimal deployment of these treatments into clinical trials, and ultimately into real-world practice, will require careful consideration of patient selection, over a range from preclinical to advanced disease stages. Needless to say, balancing risks and benefits of different approaches will be of paramount importance as novel therapies continue to emerge. This is truly an exciting time of rapid discovery and translation to the clinical arena, facilitated by increasing partnerships between academic and industry entities and greatly enabled by patients eager to participate in early phase clinical trials, selflessly motivated by the opportunity to advance science and improve the lives of the next generation of patients living with genetic cardiomyopathies.
Funding Support and Author Disclosures
Dr Day has received support from the National Heart, Lung, and Blood Institute (NHLBI) and Bristol Myers Squibb; and is a consultant for Bristol Myers Squibb, Tenaya Therapeutics, BioMarin Therapeutics, Lexeo Therapeutics, and Pfizer. Dr Helms has received support from the National Heart, Lung, and Blood Institute, the National Science Foundation, Tenaya Therapeutics, Lexeo Therapeutics, and Bristol Myers Squibb; and is a consultant for Tenaya Therapeutics. Dr Thompson has received support from the National Heart, Lung, and Blood Institute and Merck Manuals.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Contributor Information
Adam S. Helms, Email: adamhelm@umich.edu.
Sharlene M. Day, Email: sharlene.day@pennmedicine.upenn.edu.
References
- 1.Ho C.Y.D.S., Ashley E.A., Michels M., et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy. Circulation. 2018;138(14):1387–1398. doi: 10.1161/CIRCULATIONAHA.117.033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos J.M., Will M.L., Gersh B.J., Kruisselbrink T.M., Ommen S.R., Ackerman M.J. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2014;89:727–737. doi: 10.1016/j.mayocp.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko C., Arscott P., Concannon M., et al. Genetic testing impacts the utility of prospective familial screening in hypertrophic cardiomyopathy through identification of a nonfamilial subgroup. Genet Med. 2018;20:69–75. doi: 10.1038/gim.2017.79. [DOI] [PubMed] [Google Scholar]
- 4.Walsh R., Buchan R., Wilk A., et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J. 2017;38:3461–3468. doi: 10.1093/eurheartj/ehw603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingles J., Goldstein J., Thaxton C., et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med. 2019;12 doi: 10.1161/CIRCGEN.119.002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes L.R., Garcia-Hernandez S., Lorenzini M., et al. Alpha-protein kinase 3 (ALPK3)-truncating variants are a cause of autosomal dominant hypertrophic cardiomyopathy. Eur Heart J. 2021;42:3063–3073. doi: 10.1093/eurheartj/ehab424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNally E.M., Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershberger R.E., Hedges D.J., Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S., Baldinger S.H., Gandjbakhch E., et al. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J Am Coll Cardiol. 2016;68:2299–2307. doi: 10.1016/j.jacc.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 10.Smith E.D., Lakdawala N.K., Papoutsidakis N., et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020;141:1872–1884. doi: 10.1161/CIRCULATIONAHA.119.044934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrke S., Lossnitzer D., Schöb M., et al. Use of cardiovascular magnetic resonance for risk stratification in chronic heart failure: prognostic value of late gadolinium enhancement in patients with non-ischaemic dilated cardiomyopathy. Heart. 2011;97:727–732. doi: 10.1136/hrt.2010.205542. [DOI] [PubMed] [Google Scholar]
- 12.Parikh V.N., Caleshu C., Reuter C., et al. Regional variation in RBM20 causes a highly penetrant arrhythmogenic cardiomyopathy. Circ Heart Fail. 2019;12 doi: 10.1161/CIRCHEARTFAILURE.118.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhtar M.M., Lorenzini M., Pavlou M., et al. Association of left ventricular systolic dysfunction among carriers of truncating variants in filamin C with frequent ventricular arrhythmia and end-stage heart failure. JAMA Cardiol. 2021;6(8):891–901. doi: 10.1001/jamacardio.2021.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James C.A., Jongbloed J.D.H., Hershberger R.E., et al. An international evidence based reappraisal of genes associated with arrhythmogenic right ventricular cardiomyopathy (ARVC) using the ClinGen framework. Circ Genom Precis Med. 2021;14(3) doi: 10.1161/CIRCGEN.120.003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marstrand P., Han L., Day S.M., et al. Hypertrophic cardiomyopathy with left ventricular systolic dysfunction: insights from the SHaRe registry. Circulation. 2020;141:1371–1383. doi: 10.1161/CIRCULATIONAHA.119.044366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang V.T., Arscott P., Helms A.S., Day S.M. Whole-exome sequencing reveals GATA4 and PTEN mutations as a potential digenic cause of left ventricular noncompaction. Circ Genom Precis Med. 2018;11 doi: 10.1161/CIRCGEN.117.001966. [DOI] [PubMed] [Google Scholar]
- 17.Harper A.R., Goel A., Grace C., et al. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat Genet. 2021;53(2):135–142. doi: 10.1038/s41588-020-00764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tadros R., Francis C., Xu X., et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat Genet. 2021;53(2):128–134. doi: 10.1038/s41588-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bueno H., Moura B., Lancellotti P., Bauersachs J. The year in cardiovascular medicine 2020: heart failure and cardiomyopathies. Eur Heart J. 2021;42(6):657–670. doi: 10.1093/eurheartj/ehaa1061. [DOI] [PubMed] [Google Scholar]
- 20.Zannad F., Ferreira J.P., Pocock S.J., et al. SGLT2 inhibitors in patients with heart failure with reduced ejection fraction: a meta-analysis of the EMPEROR-Reduced and DAPA-HF trials. Lancet. 2020;396:819–829. doi: 10.1016/S0140-6736(20)31824-9. [DOI] [PubMed] [Google Scholar]
- 21.McMurray J.J.V., Solomon S.D., Inzucchi S.E., et al. Dapagliflozin in patients with heart failure and reduced ejection fraction. N Engl J Med. 2019;381:1995–2008. doi: 10.1056/NEJMoa1911303. [DOI] [PubMed] [Google Scholar]
- 22.Packer M., Anker S.D., Butler J., et al. Cardiovascular and renal outcomes with empagliflozin in heart failure. N Engl J Med. 2020;383:1413–1424. doi: 10.1056/NEJMoa2022190. [DOI] [PubMed] [Google Scholar]
- 23.Ho C.Y., McMurray J.J.V., Cirino A.L., et al. The design of the Valsartan for Attenuating Disease Evolution in Early Sarcomeric Hypertrophic Cardiomyopathy (VANISH) trial. Am Heart J. 2017;187:145–155. doi: 10.1016/j.ahj.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. Gene therapy comes of age. Science. 2018;359(6372) doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 25.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol Ther. 2012;20(4):699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell S., Bennett J., Wellman J.A., et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendell J.R., Al-Zaidy S., Shell R., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 28.Westfall M.V., Rust E.M., Albayya F., Metzger J.M. Adenovirus-mediated myofilament gene transfer into adult cardiac myocytes. Methods Cell Biol. 1997;52:307–322. [PubMed] [Google Scholar]
- 29.Patel A., Zhao J., Duan D., Lai Y. Design of AAV vectors for delivery of large or multiple transgenes. Methods Mol Biol. 2019;1950:19–33. doi: 10.1007/978-1-4939-9139-6_2. [DOI] [PubMed] [Google Scholar]
- 30.Phelps S.F., Hauser M.A., Cole N.M., et al. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- 31.Scott J.M., Li S., Harper S.Q., et al. Viral vectors for gene transfer of micro-, mini-, or full-length dystrophin. Neuromuscul Disord. 2002;12(suppl 1):S23–S29. doi: 10.1016/s0960-8966(02)00078-0. [DOI] [PubMed] [Google Scholar]
- 32.Kornegay J.N., Li J., Bogan J.R., et al. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol Ther. 2010;18:1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan D. Systemic AAV micro-dystrophin gene therapy for duchenne muscular dystrophy. Mol Ther. 2018;26(10):2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122(1):23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velazquez V.M., Meadows A.S., Pineda R.J., Camboni M., McCarty D.M., Fu H. Effective depletion of pre-existing anti-AAV antibodies requires broad immune targeting. Mol Ther Methods Clin Dev. 2017;4:159–168. doi: 10.1016/j.omtm.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21(4):255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J., Wakimoto H., Seidman J.G., Seidman C.E. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaleta-Rivera K., Dainis A., Ribeiro A.J.S., et al. Allele-specific silencing ameliorates restrictive cardiomyopathy attributable to a human myosin regulatory light chain mutation. Circulation. 2019;140:765–778. doi: 10.1161/CIRCULATIONAHA.118.036965. [DOI] [PubMed] [Google Scholar]
- 39.Dotzler S.M., Kim C.S.J., Gendron W.A.C., et al. Suppression-replacement KCNQ1 gene therapy for type 1 long QT syndrome. Circulation. 2021;143:1411–1425. doi: 10.1161/CIRCULATIONAHA.120.051836. [DOI] [PubMed] [Google Scholar]
- 40.Coelho T., Adams D., Silva A., et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 41.Adams D., Gonzalez-Duarte A., O’Riordan W.D., et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 42.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma H., Marti-Gutierrez N., Park S.W., et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 44.Kantor A., McClements M.E., Maclaren R.E. CRISPR-Cas9 DNA base-editing and prime-editing. Int J Mol Sci. 2020;21(17):6240. doi: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musunuru K., Chadwick A.C., Mizoguchi T., et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 46.Amoasii L., Hildyard J.C.W., Li H., et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min Y.L., Li H., Rodriguez-Caycedo C., et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5(3) doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min Y.-L., Bassel-Duby R., Olson E.N. CRISPR correction of Duchenne muscular dystrophy. Ann Rev Med. 2019;70:239–255. doi: 10.1146/annurev-med-081117-010451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frangoul H., Altshuler D., Cappellini M.D., et al. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia. N Engl J Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 50.Wahane A., Waghmode A., Kapphahn A., Dhuri K., Gupta A., Bahal R. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules. 2020;25(12):2866. doi: 10.3390/molecules25122866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wessels M.W., Herkert J.C., Frohn-Mulder I.M., et al. Compound heterozygous or homozygous truncating MYBPC3 mutations cause lethal cardiomyopathy with features of noncompaction and septal defects. Eur J Hum Genet. 2015;23:922–928. doi: 10.1038/ejhg.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marston S.B. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J Cardiovasc Transl Res. 2011;4:245–255. doi: 10.1007/s12265-011-9266-2. [DOI] [PubMed] [Google Scholar]
- 53.Viswanathan S.K., Sanders H.K., McNamara J.W., et al. Hypertrophic cardiomyopathy clinical phenotype is independent of gene mutation and mutation dosage. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0187948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yancy C.W., Jessup M., Bozkurt B., et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62:e147–e239. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 55.O’Connor C.M., Gattis W.A., Uretsky B.F., et al. Continuous intravenous dobutamine is associated with an increased risk of death in patients with advanced heart failure: insights from the Flolan International Randomized Survival Trial (FIRST) Am Heart J. 1999;138:78–86. doi: 10.1016/s0002-8703(99)70250-4. [DOI] [PubMed] [Google Scholar]
- 56.Packer M., Carver J.R., Rodeheffer R.J., et al. for the PROMISE Study Research Group Effect of oral milrinone on mortality in severe chronic heart failure. N Engl J Med. 1991;325:1468–1475. doi: 10.1056/NEJM199111213252103. [DOI] [PubMed] [Google Scholar]
- 57.Malik F.I., Hartman J.J., Elias K.A., et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voors A.A., Tamby J.F., Cleland J.G., et al. Effects of danicamtiv, a novel cardiac myosin activator, in heart failure with reduced ejection fraction: experimental data and clinical results from a phase 2a trial. Eur J Heart Fail. 2020;22(9):1649–1658. doi: 10.1002/ejhf.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woody M.S., Greenberg M.J., Barua B., Winkelmann D.A., Goldman Y.E., Ostap E.M. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat Commun. 2018;9:3838. doi: 10.1038/s41467-018-06193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teerlink J.R., Diaz R., Felker G.M., et al. Cardiac myosin activation with omecamtiv mecarbil in systolic heart failure. N Engl J Med. 2020;384(2):105–116. doi: 10.1056/NEJMoa2025797. [DOI] [PubMed] [Google Scholar]
- 61.Green E.M., Wakimoto H., Anderson R.L., et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohde J.A., Roopnarine O., Thomas D.D., Muretta J.M. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc Natl Acad Sci U S A. 2018;115:E7486–E7494. doi: 10.1073/pnas.1720342115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson R.L., Trivedi D.V., Sarkar S.S., et al. Deciphering the super relaxed state of human beta-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc Natl Acad Sci U S A. 2018;115:E8143–E8152. doi: 10.1073/pnas.1809540115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olivotto I., Oreziak A., Barriales-Villa R., et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;396:759–769. doi: 10.1016/S0140-6736(20)31792-X. [DOI] [PubMed] [Google Scholar]
- 65.Saberi S., Cardim N., Yamani M., et al. Mavacamten favorably impacts cardiac structure in obstructive hypertrophic cardiomyopathy: EXPLORER-HCM cardiac magnetic resonance substudy analysis. Circulation. 2021;143:606–608. doi: 10.1161/CIRCULATIONAHA.120.052359. [DOI] [PubMed] [Google Scholar]
- 66.Ho C.Y., Mealiffe M.E., Bach R.G., et al. Evaluation of mavacamten in symptomatic patients with nonobstructive hypertrophic cardiomyopathy. J Am Coll Cardiol. 2020;75:2649–2660. doi: 10.1016/j.jacc.2020.03.064. [DOI] [PubMed] [Google Scholar]
- 67.Robertson L.A., Armas D.R., Robbie E., et al. A first in human study of the selective cardiac myosin inhibitor, CK-3773274. J Card Fail. 2019;25(8 suppl):S79–S80. [Google Scholar]
- 68.Toepfer C.N., Garfinkel A.C., Venturini G., et al. Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation. 2020;141:828–842. doi: 10.1161/CIRCULATIONAHA.119.042339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore J.R., Leinwand L., Warshaw D.M. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greenberg M.J., Tardiff J.C. Complexity in genetic cardiomyopathies and new approaches for mechanism-based precision medicine. J Gen Physiol. 2021;153(3) doi: 10.1085/jgp.202012662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clippinger S.R., Cloonan P.E., Greenberg L., Ernst M., Stump W.T., Greenberg M.J. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116:17831–17840. doi: 10.1073/pnas.1910962116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandra M., Rundell V.L., Tardiff J.C., Leinwand L.A., De Tombe P.P., Solaro R.J. Ca2+ activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am J Physiol Heart Circ Physiol. 2001;280:H705–H713. doi: 10.1152/ajpheart.2001.280.2.H705. [DOI] [PubMed] [Google Scholar]
- 73.Li M.X., Gelozia S., Danmaliki G.I., et al. The calcium sensitizer drug MCI-154 binds the structural C-terminal domain of cardiac troponin C. Biochem Biophys Rep. 2018;16:145–151. doi: 10.1016/j.bbrep.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwama Y., Takagi A. [Calcium sensitizer agents in heart failure therapy] Nihon Rinsho. 2007;65(suppl 5):61–65. [PubMed] [Google Scholar]
- 75.Zhang L., Nan C., Chen Y., et al. Calcium desensitizer catechin reverses diastolic dysfunction in mice with restrictive cardiomyopathy. Arch Biochem Biophys. 2015;573:69–76. doi: 10.1016/j.abb.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 76.Ruberg F.L., Grogan M., Hanna M., Kelly J.W., Maurer M.S. Transthyretin amyloid cardiomyopathy: JACC state-of-the-art review. J Am Coll Cardiol. 2019;73:2872–2891. doi: 10.1016/j.jacc.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Papp E., Csermely P. Chemical chaperones: mechanisms of action and potential use. Handb Exp Pharmacol. 2006;172:405–416. doi: 10.1007/3-540-29717-0_16. [DOI] [PubMed] [Google Scholar]
- 78.Miroy G.J., Lai Z., Lashuel H.A., Peterson S.A., Strang C., Kelly J.W. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc Natl Acad Sci U S A. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bulawa C.E., Connelly S., Devit M., et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci U S A. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maurer M.S., Schwartz J.H., Gundapaneni B., et al. Tafamidis treatment for patients with transthyretin amyloid cardiomyopathy. N Engl J Med. 2018;379:1007–1016. doi: 10.1056/NEJMoa1805689. [DOI] [PubMed] [Google Scholar]
- 81.Yadav J.D., Othee H., Chan K.A., Man D.C., Belliveau P.P., Towle J. Transthyretin amyloid cardiomyopathy-current and future therapies. Ann Pharmacother. 2021;55(12):1502–1514. doi: 10.1177/10600280211000351. [DOI] [PubMed] [Google Scholar]
- 82.Gianni D., Li A., Tesco G., et al. Protein aggregates and novel presenilin gene variants in idiopathic dilated cardiomyopathy. Circulation. 2010;121:1216–1226. doi: 10.1161/CIRCULATIONAHA.109.879510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian K., Gianni D., Balla C., et al. Cofilin-2 phosphorylation and sequestration in myocardial aggregates: novel pathogenetic mechanisms for idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2015;65:1199–1214. doi: 10.1016/j.jacc.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diteepeng T., Del Monte F., Luciani M. The long and winding road to target protein misfolding in cardiovascular diseases. Eur J Clin Invest. 2021;51(5) doi: 10.1111/eci.13504. [DOI] [PubMed] [Google Scholar]
- 85.Morrill S.A., Amon A. Why haploinsufficiency persists. Proc Natl Acad Sci U S A. 2019;116:11866–11871. doi: 10.1073/pnas.1900437116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marston S., Copeland O., Jacques A., et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 88.Glazier A.A., Thompson A., Day S.M. Allelic imbalance and haploinsufficiency in MYBPC3-linked hypertrophic cardiomyopathy. Pflugers Arch. 2019;471:781–793. doi: 10.1007/s00424-018-2226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Leary T.S., Snyder J., Sadayappan S., Day S.M., Previs M.J. MYBPC3 truncation mutations enhance actomyosin contractile mechanics in human hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2019;127:165–173. doi: 10.1016/j.yjmcc.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hinson J.T., Chopra A., Nafissi N., et al. Heart disease. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hall C.L., Sutanto H., Dalageorgou C., McKenna W.J., Syrris P., Futema M. Frequency of genetic variants associated with arrhythmogenic right ventricular cardiomyopathy in the genome aggregation database. Eur J Hum Genet. 2018;26:1312–1318. doi: 10.1038/s41431-018-0169-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang X., Bogomolovas J., Wu T., et al. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest. 2017;127:3189–3200. doi: 10.1172/JCI94310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Norton N., Li D., Rieder M.J., et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88:273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vasilescu C., Ojala T.H., Brilhante V., et al. Genetic basis of severe childhood-onset cardiomyopathies. J Am Coll Cardiol. 2018;72:2324–2338. doi: 10.1016/j.jacc.2018.08.2171. [DOI] [PubMed] [Google Scholar]
- 95.Vicart P., Caron A., Guicheney P., et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 96.Dominguez F., Cuenca S., Bilinska Z., et al. Dilated cardiomyopathy caused by BLC2-associated athanogene 3 (BAG3) mutations. J Am Coll Cardiol. 2018;72:2471–2481. doi: 10.1016/j.jacc.2018.08.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gestwicki J.E., Shao H. Inhibitors and chemical probes for molecular chaperone networks. J Biol Chem. 2019;294:2151–2161. doi: 10.1074/jbc.TM118.002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glazier A.A., Hafeez N., Mellacheruvu D., et al. HSC70 is a chaperone for wild-type and mutant cardiac myosin binding protein C. JCI Insight. 2018;3(11) doi: 10.1172/jci.insight.99319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Helms A.S., Tang V.T., O’Leary T.S., et al. Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy. JCI Insight. 2020;5 doi: 10.1172/jci.insight.133782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGarrah R.W., Crown S.B., Zhang G.F., Shah S.H., Newgard C.B. Cardiovascular Metabolomics. Circ Res. 2018;122:1238–1258. doi: 10.1161/CIRCRESAHA.117.311002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coats C.J., Pavlou M., Watkinson O.T., et al. Effect of trimetazidine dihydrochloride therapy on exercise capacity in patients with nonobstructive hypertrophic cardiomyopathy: a randomized clinical trial. JAMA Cardiol. 2019;4:230–235. doi: 10.1001/jamacardio.2018.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murashige D., Jang C., Neinast M., et al. Comprehensive quantification of fuel use by the failing and nonfailing human heart. Science. 2020;370:364–368. doi: 10.1126/science.abc8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Selvaraj S., Kelly D.P., Margulies K.B. Implications of altered ketone metabolism and therapeutic ketosis in heart failure. Circulation. 2020;141:1800–1812. doi: 10.1161/CIRCULATIONAHA.119.045033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nikolic M, Zivkovic V, Jovic JJ, et al. SGLT2 inhibitors: a focus on cardiac benefits and potential mechanisms. Heart Fail Rev. Published online February 3, 2021. 10.1007/s10741-021-10079-9 [DOI] [PubMed]
- 105.Helms A.S., Alvarado F.J., Yob J., et al. Genotype-dependent and -independent calcium signaling dysregulation in human hypertrophic cardiomyopathy. Circulation. 2016;134:1738–1748. doi: 10.1161/CIRCULATIONAHA.115.020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gianni D., Chan J., Gwathmey J.K., Del Monte F., Hajjar R.J. SERCA2a in heart failure: role and therapeutic prospects. J Bioenerg Biomembr. 2005;37:375–380. doi: 10.1007/s10863-005-9474-z. [DOI] [PubMed] [Google Scholar]
- 107.Hajjar R.J., Schmidt U., Matsui T., et al. Modulation of ventricular function through gene transfer in vivo. Proc Natl Acad Sci U S A. 1998;95:5251–5256. doi: 10.1073/pnas.95.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Byrne M.J., Power J.M., Preovolos A., Mariani J.A., Hajjar R.J., Kaye D.M. Recirculating cardiac delivery of AAV2/1SERCA2a improves myocardial function in an experimental model of heart failure in large animals. Gene Therapy. 2008;15:1550–1557. doi: 10.1038/gt.2008.120. [DOI] [PubMed] [Google Scholar]
- 109.Greenberg B., Butler J., Felker G.M., et al. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): a randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet. 2016;387:1178–1186. doi: 10.1016/S0140-6736(16)00082-9. [DOI] [PubMed] [Google Scholar]
- 110.Garcia-Gras E., Lombardi R., Giocondo M.J., et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gao S., Puthenvedu D., Lombardi R., Chen S.N. Established and emerging mechanisms in the pathogenesis of arrhythmogenic cardiomyopathy: a multifaceted disease. Int J Mol Sci. 2020;21(17):6320. doi: 10.3390/ijms21176320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davis J., Molkentin J.D. Myofibroblasts: trust your heart and let fate decide. J Mol Cell Cardiol. 2014;70:9–18. doi: 10.1016/j.yjmcc.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aghajanian H., Kimura T., Rurik J.G., et al. Targeting cardiac fibrosis with engineered T cells. Nature. 2019;573:430–433. doi: 10.1038/s41586-019-1546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ware J.S., Li J., Mazaika E., et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goli R., Li J., Brandimarto J., et al. Genetic and phenotypic landscape of peripartum cardiomyopathy. Circulation. 2021;143:1852–1862. doi: 10.1161/CIRCULATIONAHA.120.052395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fumagalli C., Maurizi N., Day S.M., et al. Association of obesity with adverse long-term outcomes in hypertrophic cardiomyopathy. JAMA Cardiol. 2020;5(1):65–72. doi: 10.1001/jamacardio.2019.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saberi S., Wheeler M., Bragg-Gresham J., et al. Effect of moderate-intensity exercise training on peak oxygen consumption in patients with hypertrophic cardiomyopathy: a randomized clinical trial. JAMA. 2017;317:1349–1357. doi: 10.1001/jama.2017.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]