Abstract
Brain network analysis is one efficient tool in exploring human brain diseases and can differentiate the alterations from comparative networks. The alterations account for time, mental states, tasks, individuals, and so forth. Furthermore, the changes determine the segregation and integration of functional networks that lead to network reorganization (or reconfiguration) to extend the neuroplasticity of the brain. Exploring related brain networks should be of interest that may provide roadmaps for brain research and clinical diagnosis. Recent electroencephalogram (EEG) studies have revealed the secrets of the brain networks and diseases (or disorders) within and between subjects and have provided instructive and promising suggestions and methods. This review summarized the corresponding algorithms that had been used to construct functional or effective networks on the scalp and cerebral cortex. We reviewed EEG network analysis that unveils more cognitive functions and neural disorders of the human and then explored the relationship between brain science and artificial intelligence which may fuel each other to accelerate their advances, and also discussed some innovations and future challenges in the end.
Keywords: Brain network analysis, Segregation and integration, Neuroplasticity, EEG pattern, Artificial intelligence
Introduction
The human brain is a more complex and dynamic network (Boccatetti et al. 2006). The corresponding structural and functional connectivity varies with lifespan (Betzel et al. 2016; Gilmore et al. 2018), cognitive activity (Liu et al. 2017b), memory, intelligence (Tang et al. 2010; Tanimizu et al. 2018), emotion (Chai et al. 2019; Li et al. 2019c), mental status, such as fatigue (Zhang et al. 2020c), and disorders. Brain network analysis has been demonstrated to be an effective tool that helpfully explores the connectivity patterns to uncover related features and phenomena concerning different brain functions and diseases.
Brain connectivity is usually divided into structural connectivity for the anatomical link, functional connectivity (FC) for statistical dependencies, and effective connectivity (EC) for casual interaction. FC and EC can reflect functional integration and anatomical segregation of the brain (Friston 2009) and have been suggested to be the accurate representations of the human brain. Moreover, EC is the more efficient connectivity basestone that is capable of predicting subjects’ neural states and future activities for the sake of its causality and directed information flow (Friston et al. 2003).
The alterations in brain network connectivity are usually expressed with related network features of affected regions, such as topological centrality (Zhuge and Zhang 2010), degeneracy (Friston and Price 2001), which can provide promising biosignatures for identifying, classifying, or predicting brain disorders (Du et al. 2018; Fornito et al. 2015) or neural responses (Si et al. 2020). Consequently, the brain needs to reconfigure its network organization dynamically, selectively, and adaptively when it confronts with changing cognitive demands to achieve an optimal balance between segregation and integration (Parr and Friston 2018) and unfolds its plasticity (Merzenich et al. 2014).
Frankly, it is one core task to construct a more reliable and time-varying FC or EC network. According to a mass of clinical-psychological and neurological studies (Anticevic et al. 2015; Brislin and Patrick 2019; Gratton et al. 2018; Wahbeh et al. 2016), signal patterns for network analysis mainly include functional magnetic resonance imaging (fMRI), positron emission tomography (PET), magnetoencephalogram (MEG), and electroencephalogram (EEG), etc. MEG readily suffers from environmental interferences (Brookes et al. 2011) and is highly expensive (Singh 2014), while PET is an invasive nuclear imaging technique and lacks high temporal resolution (Catana et al. 2012). Comparatively, high spatial-resolution fMRI (Buckner et al. 2009; Liang et al. 2012; Pasquale et al. 2016) and high temporal-resolution EEG (Brookes et al. 2011) are noninvasive and quite popular, but EEG is quite low cost and convenient.
Notwithstanding, fMRI is extremely helpful in characterizing the network connectivity in a specific cognitive task from different brain regions (Contreras et al. 2019), but just non-linear function of blood volume and deoxyhemoglobin (deoxygenated hemoglobin) content (Stephan and Friston 2010), that is hemodynamic model, which does not directly measure oscillatory behavior of the brain electrical activity time-sequentially (Vico Fallani et al. 2008), and is nonmovable. Although relatively poor spatial resolution, EEG can monitor the brain’s spontaneous electrical activity, as recorded from low-cost multiple electrodes precisely placed on the scalp, and possesses high temporal resolution with about one millisecond. Besides, EEG can also measure the mixture of several underlying base frequencies to reflect certain cognitive, affective, or attentional states. These frequencies vary slightly in individual factors, stimulus properties, and internal states, and have fruitful features, such as amplitude, latency, phase, frequency tag, and spectral peak, etc., which dynamically depict the variation of cognitive task, cortical regions, and thickness (van der Meij et al. 2016). Third, the brain-computer interface (BCI) auxiliary treatment and rehabilitating instruments, such as motor imagery-based (MI-BCI) (Zhang et al. 2020b) developed in recent decades (Kabbara et al. 2016; Zhang et al. 2016), highly rely on the EEG signals. Additionally, to identify reproducible large-scale networks across neural populations, EEG paves one hopeful way for high temporal dynamics of the network at source space (Li et al. 2019d; Sockeel et al. 2016). Analyzing such time-varying and nonstationary brain networks, EEG is one irreplaceable candidate in the view of the temporal- and spectral-phase domain and has been applied to demystify more and more psychological and mental functions (Li et al. 2020a; Zhang et al. 2020a).
Our current review collected papers of EEG-based network analysis and applications that focused on the EEG and time-variant EEG networks. Concretely, after introducing acquiring and preprocessing data concisely, we described some major methods of recent advances to construct stable network connectivity which can effectively capture the reliable relationships between networks and EEG recordings in sensor and source space and explored the reconfiguration mechanism of functional networks in a specific environment. Thereafter, we reviewed related studies that investigated the relationships among cognitive, emotion, diseases, and artificial intelligence (AI) which originates from brain network, and finally identified the role of EEG network analysis in all fields, and hoped this review may provide one promising roadmap of accelerating brain science and AI.
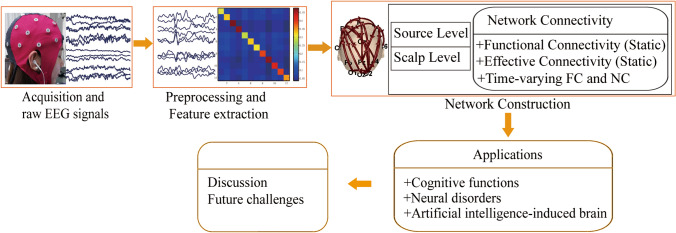
The rest of this review is structured as follows. Section II brings laconically out related work of collecting raw EEG signals, preprocessing, and extracting features. Section III reviews and generalizes some popular methods of network construction about different types of connectivity networks. Section IV revisits the applications of network analysis on cognition and diseases and also discusses the relationship between AI and the biological brain which is also an important part since AI is the fuel of brain science. Section V discusses our understanding of EEG network analysis, innovations, as well as its potential challenges in the future. Moreover, to help understand this review clearly, we gave its roadmap as Fig. 1.
Fig. 1.
The roadmap of our current review. Some subfigures are adapted from the material (Si et al. 2019)
Data and useful information
The acquisition of raw samples, preprocessing, and feature extraction cannot be been slighting those are the beginnings of the pipeline of network analysis on brain science compared with network construction, although it is not our concern in this review.
Acquisition of EEG data
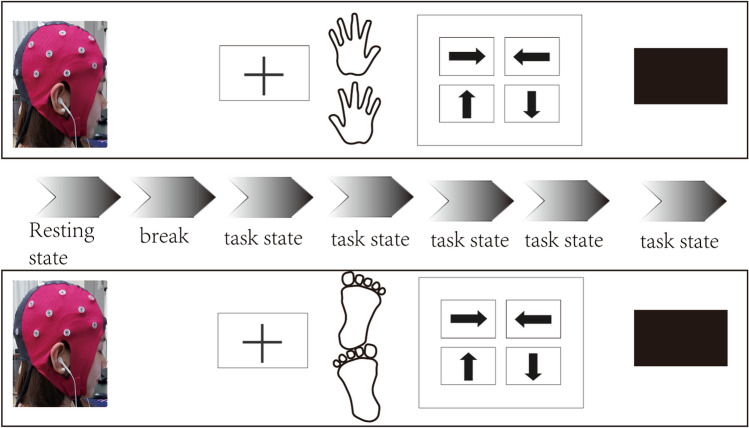
Before modeling of network connectivity, the acquisition of raw EEG samples must be accomplished. In terms of different specific tasks and requirements, researchers should design experimental tasks, and then recruit some related subjects and collect their data. For instance, Fig. 2 depicts the corresponding experimental procedure of MI to gather left and right hands and feet of subjects with cerebral stroke.
Fig. 2.
One experimental procedure about MI to gather left and right hands and feet of subjects with cerebral stroke
Preprocessing and feature extraction
The aim of preprocessing step is to get reliable and useful information of subjects, including baseline correction, bandpass filtering, the removal of some outliers and bad trials, data segmentation, and denoising with EEGLab (Delorme and Makeig 2004) and wavelet toolkit. Then, according to the target of the study, features, such as ERPs (P300, N100) (Li et al. 2018a), delta, gamma rhythms, or others, should be extracted from preprocessed data with primary component analysis (PCA) or other methods.
To understand the above three steps clearly, the technical contribution (Si et al. 2019) may provide more details.
Methods of network construction
The brain network topology changes adaptively and tempo-spatially (Jirsa et al. 2010) when responding to a certain environment or factor. This is called neuroplasticity of the brain, and this phenomenon is called reconfiguration or reorganization which needs to update the ongoing network connectivity with transient and heterogeneous (various) connections from resting-state or default mode network (DMN) connectivity (de Oliveira 2020; Wang et al. 2017).
A large number of academic advances have been published to back up the above statement according to long- and short-term alternations of network topology, where long-term changes are related to age, damage, intelligence, or diseases, and short-term changes characterize temporality and specificity (Wig 2017; Zhang et al. 2020c).
Review (Gilmore et al. 2018) tracks the brain development of childhood and remarks that structural and functional brain networks have matured and in latter childhood are much slower. Conversely, older adults show larger changes in network organization between resting-state and task and have increases in between-module connectivity, related to faster task performance and greater fractional anisotropy of the superior longitudinal fasciculus (Gallen et al. 2016). An opinion article (Barbey 2018) has expressed network topology and dynamics that originate from individual differences in general intelligence. Cell report (Griffis et al. 2019) surveys focal brain lesions reflect the network disconnections of white matter pathways rather than the destruction of gray matter regions. Damage to network hub regions, especially those connecting different subnetworks, have been found to cause the largest disturbances in network organization, lesions, and the significant alternations in global network topology regardless of lesion location (Aerts et al. 2016). Scientific experiments further verify that these diseases contribute to FC alternations (He et al. 2018; van den Heuvel and Sporns 2019) that visual, sensorimotor, auditory, and language resting-state connectivity networks are changed in longstanding type-1 diabetes with degree centrality (Joyce et al. 2010) and eigenvector centrality (Lohmann et al. 2010; Zhuge and Zhang 2010) mapping, but not disease progression (van Duinkerken et al. 2017).
Short-term change is the functional network-level integration altering dynamically and mostly spatially in terms of tasks and task difficulty, as well as the increased structural segregation (Cohen and D'Esposito 2016; Hearne et al. 2017; Simony et al. 2016; Wen et al. 2015). Short-term change in network connectivity results in short-term automatization of functional networks. Compared with long-term learning processes, short-term automatization (Mohr et al. 2016) is accompanied by decreasing activation of the frontoparietal network, indicating a release of high-level cognitive control, and segregation of the DMN from task-related networks. The short-term task automatization is activated by the brain’s ability to rapidly reconfigure its large-scale network organization involving complementary integration and segregation processes.
This finding (Guo et al. 2018) indicates that an external periodic visual stimulus can induce the modification of intrinsic oscillatory activities different from the resting-state activity at the network level. The further evidence uncovers how the brain reconfigures from rest idle to task state (Li et al. 2015a, 2020d; Song et al. 2019) and these factors on inter-subject’s reconfiguring variability (Li et al. 2020c), which guarantees the brain to efficiently process the information of the specific MI tasks (Caravaglios et al. 2015; Shine and Poldrack 2018; Zhang et al. 2019) (e.g., right or left MI) with ERD (Li et al. 2019d), or provides one stable and successful auditory control network for listening (Alavash et al. 2019). Moreover, the reconfiguring phenomenon also occurs in the DMN under task (Zuo et al. 2018).
The efficiency of brain reconfiguration differs across individuals. Higher intelligence leads to more efficiency in network reconfiguration (Cary et al. 2017; Hilger et al. 2020; Schultz and Cole 2016), and high-performing subjects exhibit more efficient brain connectivity which updates in the form of smaller changes in FC from idle-rest to task (Li et al. 2019a; Zhang et al. 2018). With higher reasoning ability, such a subject’s brain reorganization completes more immediately and efficiently (Hearne et al. 2017), and has more language fluency and more increasing language control network (Schultz and Cole 2016). Likewise, memory encoding performance impresses fatally on connectivity reorganization (Wu et al. 2019).
Concerning the above factors, network analysis is one reasonable and effective choice to explore how the brain reconfigures or reorganizes nowadays. Factually, network construction is one quite important step.
Network construction is to compute the network connectivity matrices from EEG time courses with statistics. Currently, the graphical theory is one major mathematic model of the complex network (Liu et al. 2017a). In this section, it is the concern of how to construct brain FC or EC networks. Generally, the FC network is undirected while the EC one is directed that can elucidate the information flow and transmission among brain regions, and accordingly, these methods of network construction will be divided into two classes.
FC networks
These construction methods of FC networks can be divided into bivariate and multivariate measures (Jalili 2016; Joudaki et al. 2012).
Correlation and coherence are bivariate measures and linear dependency. Correlation includes cross-correlation, Pearson’s correlation, and partial correlation. Cross-correlation measures the similarity between two series as a function of the lag/lead of one relative to the other, and is suitable to signal epochs of long-term EEG records (Chu et al. 2012).
Pearson’s correlation measures the temporal-domain linear dependency of one sensor on another and is impartial. Partial correlation measures the conditional dependency between two sensors that may reduce the prediction of indirect functional connectivity at the expensive cost compared with Pearson’s correlation (Jalili and Knyazeva 2011). While coherence measures the frequency-domain linear dependency between two sensors in a certain frequency whose derivatives are amplitude coherence, phase coherence (Liu and Zhang 2018), and imaginary coherence (Nolte et al. 2004; Sanchez Bornot et al. 2018).
The Pearson correlation coefficient between sensors l and k can be gained as
| 1 |
Partial correlation is obtained as
| 2 |
where is the covariance between sensors (or nodes) and , is the variance of , as the impartial correlation between and .
For coherence measure, the first step is computed the cross-spectrum between and ,
| 3 |
And then the coherence at each frequency can be normalized form of cross-spectrum Eq. (3).
| 4 |
where is noted as the epoch number in each brain status, as the frequency number, as complex-valued coefficients of the sensor pair , * is the transposition operation.
Phase order and synchronization likelihood are also bivariate but nonlinear dependencies between sensors. Phase order measures phase synchrony between two time series. Suppose the instantaneous phase of the time sequences of senor which has samples are written as
| 5 |
Then, the phase synchronization index is denoted as
| 6 |
where is the Hilbert transformation, is the imaginary unit.
Synchronization likelihood measures the conditional likelihood that the distance between two values differs in different moments for the same time course. First sequences are combined into state-space vector , the same as sequences .
| 7 |
where is the number of vectors, is the Theiler correction number for autocorrelation, and () is the step function.
S-estimator is a multivariate and related-entropy measure and gauges the inter-group synchronization between groups (sensors located in one network as a group) based on the eigenvalues of the correlation matrix formed by inter- and between-group correlation matrix (Joudaki et al. 2012; Yi et al. 2020). The S-estimator is defined as
| 8 |
where is the normalized eigenvalue of the correlation matrix of time series .
Its derivative is the S-Renyi estimator which is based on Renyi entropy and more robust (Sizemore and Bassett 2018). Compared with the front amplitude-synchrony ones, multivariate phase synchronization measures the mean phase coherence, which extends Eq. (6) with vectors and matrix. To compare with these methods, computational stimulation (Jalili 2016) has been done and demonstrated that coherence is more robust for increasing noise, but cannot capture the nonlinear interconnections as the same as correlation, compared with synchronization and phase order which are sensitive to volume conduction effect. More novel algorithms are also applied to construct reliable networks, for instance, visualization of the coherence matrix (Ji et al. 2018) for improving spatial performance, median coherence estimator robust against artifacts (Dukic et al. 2017).
EC network
EC network can articulate information flow (or causal influence of neuronal populations) with directions and time-stages for dynamical brain structural and functional organization under different tasks, and so it is widely used in clinical and neurological analysis. There are several popular methods to model EC networks.
Dynamical causal modeling
Compared with structural equation modeling for static dependencies of brain regions (Friston 2011), dynamical causal modeling (DCM) based on Bayesian framework was first proposed to analyze nonlinear brain network connectivity with a deterministic causal model of neuronal responses to external perturbation (Friston et al. 2003) for first fMRI data then EEG (David et al. 2006; Fastenrath et al. 2009), and reveals how neural activity is generated and neuronal variables fluctuate over separable timescales (Cooray et al. 2016). DCM can provide more promising and helpful psychological and neuropsychological signatures, such as on schizophrenia (Fogelson et al. 2014; Friston et al. 2016a; Zhou et al. 2018), Alzheimer’s disease (Penny et al. 2018), epileptic seizures (Bomela et al. 2020; Cooray et al. 2016), stroke (Bönstrup et al. 2016, 2018), drug-abstinent (Zhao et al. 2017), psychotic disorders (Díez et al. 2017) and so on.
DCM is computed according to EEG signals’ state equations and their responses,
| 9 |
where s denotes the neuronal states of cortical sources which must be transformed from scalp-level EEG signals, e denotes external stimuli, and θ denotes free parameters which can optimize the difference between the predicted and the observed EEG series.
And then the responses are convoluted to obtain its vectorized forms and associated likelihood,
| 10 |
Last, the Bayesian inference model is expressed as
| 11 |
where S is made of one block diagonal matrix with base components of the EEG responses, ε denotes the Gaussian white noises and its vector as which is convoluted () with the error’s temporal auto-correlation matrix, is the predicted cortical signal’s model. The optimized will be gained while the variational free energy of Eq. (11) is convergent and minimal.
Bayesian inversion in DCM serves to identify the structure of the brain connectivity network. Such convergence and speedup of the inversion issue should be got attention (Friston et al. 2003; Sengupta et al. 2014; Wang et al. 2013; Yao et al. 2018). Besides, Bayesian inference algorithms in group DCM analysis are mostly based on Laplace assumption which violates the robustness, but Bayesian model reduction (Friston et al. 2016b) is verified that makes an effect on the robustness, and group DCM with empirical Bayes (Friston et al. 2015) also differentiates the insignificant variability between subjects (Litvak et al. 2015) and estimates efficiently. To further uncover the relationships between intrinsic fluctuations in activity-dependent neuronal coupling and contextual factors, three-level hierarchical parametric empirical Bayes is proposed to assess such fluctuations in DCM connectivity parameters (van de Steen et al. 2019).
Granger causality and partial directed coherence
Granger causality (GC), based on linear vector autoregressive models of stochastic time-series data, precedes and predicts the effects among variables (Granger 1969) in both time and frequency domains. Partial directed coherence (PDC) is a GC measure in the frequency domain and can be combined with graph theory to analyze EC networks under different mental tasks (Huang et al. 2016) and resting-state brain data (Biazoli et al. 2013). To improve the robustness in estimating inaccuracy related to finite time-series samples, generalized PDC keeps the normalizations of PDC and achieves its variance stabilization (Baccala et al. 2008). PDC combined with multivariate empirical mode decomposition reveals that discriminates EC existence of bilateral hemisphere and contralateral lateralization during MI tasks (Liang et al. 2016). Compared with bivariate GC analysis, conditional multivariate Granger causality (cMVGC) is less sensitive to false indirect connections (Olejarczyk et al. 2017). Based on informatics theory for modeling EC networks, symbolic transfer entropy is verified to be more reliable and robust irrespective of sessions when comparing to vector autoregression or MVGC (Ye et al. 2020).
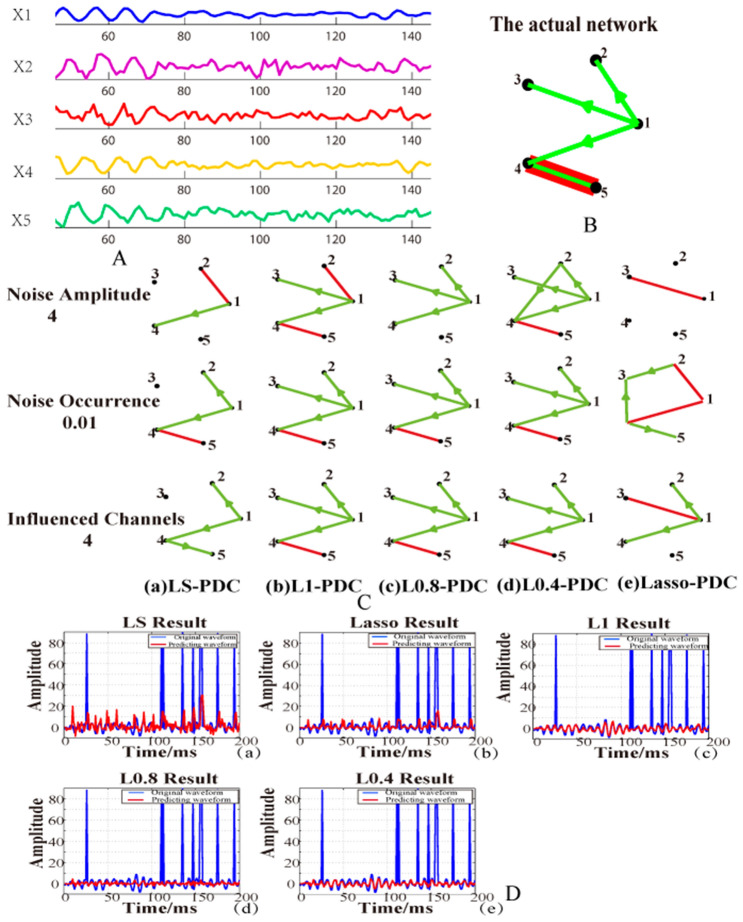
EEG recordings are usually fatally contaminated by artifacts or outliers (e.g., eye movement (Brunner et al. 2016)) which may lead to network distortion. These algorithms with EEG network analysis can suppress the influence of outliers and capture reliable causal relationship by Lp (0 < p < 1)-norm Granger Causality (Lp-GA) (Li et al. 2017), least absolute Lp (0 < p < 1) penalized sparse Granger (Bore et al. 2020), Lp-norm PDC (Li et al. 2018c) given the temporal and frequency domain respectively. There is an example to explain the network construction for EEG raw signal with outliers, shown in Fig. 3. The methods in these publications ( Bore et al. 2020; Li et al. 2018c) are derivatives from Lp (Li et al. 2017). Hence, we give some technical details here.
Fig. 3.
Lp norm PDC for EC network analysis. A Raw signals. B The actual network model, the green line for monodirectional connectivity, and the red one for bi-directional connectivity. C The connectivity networks are estimated under different artifacts or outliers by different PDC algorithms (least square PDC, Lasso-PDC, Lp-norm PDC, where p values are set as 1.0, 0.8, 0.4), and the results are shown as (C-a)–(C-e). D The valuation performances of these PDCs are displayed as (D-a)–(D-e).
Adapted from the reference (Li et al. 2018c)
Lp (0 < p < 1)-norm Granger causality for outlier removal in EEG recordings
Supposing brain signals as joint stationary random processes, whose observed values at time point are denoted as , . In the linear regressive model, each process can be predicted by its past information and past information of other variables, and described by
| 12 |
where is the maximum number of past observations in the model, and is the coefficient vector, which quantitatively describes the influence of the activity of on . is the covariance of the residuals between the expected and the predicted .
Let be the multivariate autoregressive (MVAR) coefficients, with being the number of time series and be the variables to be predicted for , with being the length of the signal.
Set , the design matrix, as
| 13 |
To clearly understand the causality linkage for the time series, taking a system with a three-time series as the example, the outlier covariance matrix can be written as
| 14 |
where all are the residuals estimated from the MVAR parameters. The noise covariance matrix from the restricted model of the system to measure the influence from on can be written by
| 15 |
where all are the noises estimated from the autoregressive model omitting . Then, the influence from the process on , conditioned on the process , is obtained by
| 16 |
and the statistical significance for both to cause and to cause can be determined by the F-statistic.
Then, we obtain the solutions of Eq. (12) from the optimization function defined in Lp () norm space to improve the GA’s robustness to the outlier effects as
| 17 |
where denotes the Lp () norm of a vector. The gradient for the function is
| 18 |
where the polarity function is set as
| 19 |
By the iteration of computing the gradient , we can find the optimal parameter and obtain the predicted process under different p.
Lp ()-GA is verified that it can also capture the intrinsic information of the time series rather than fitting the outliers.
Directed transfer function
Directed transfer function (DTF) is evolved from a multivariate connectivity estimator for GC analysis that can discriminate subtle variability between subjects with the neuronal disorder and healthy controls (Blinowska et al. 2017; Maharathi et al. 2016), but improper preprocessing may result in misleading results because DTF is insensitive to volume conduction (Kaminski and Blinowska 2014).
Time-varying network
The above methods of network construction are ignorant of the temporal-variability feature of neural signals. Markov chain is a mathematical model that can describe the transition from one state to another under certain probabilistic rules and just be suitable for such stochastic-like neural activities and useful for the construction of both FC and EC. The Markov-based framework can infer the time-varying networks from EEG data (Williams et al. 2018), and also unveil the fast sub-second network dynamics of EEG allied with fMRI data (Hunyadi et al. 2019), and microstates (Dimitriadis et al. 2015) and corresponding micro-FC networks (Duc and Lee 2020). However, the Markov-based model infers that the transitions between networks are not random (Vidaurre et al. 2017) with fMRI biomarker. If it is possible in the future, whether the EEG-based Markov model also attains the consistency of such transition nonrandom should be of interest.
The time-varying network is computed in term of normalized multivariate adaptive autoregressive (MVAAR) equations:
| 20 |
where is the coefficient matrix of the time-varying model, is the Gaussian noise. And then Eq. (20) is transformed into a frequency form, while is the form of in frequency domain, and is the order of the MVAAR model, and as the element of describe the flow direction between the ith and the jth element at moment as,
| 21 |
The directed causal flow between the ith and the jth element at moment and frequency is normalized as
| 22 |
For one single node, from which the total information flow can be obtained between the concerned frequency range as,
| 23 |
For the former any multivariate autoregressive model, if its parameters are estimated by Kalman filters, it belongs to time-varying modeling (Pagnotta and Plomp 2018). Different Kalman filters correlate with the dipole selection of EEG (Ghumare et al. 2018). To prevent fallacious signals, an adaptive Kalman filter (Rubega et al. 2019) can estimate accurately the parameters of the time-variant network model. To make the network topology robustness from noise, optimized Kalman filter proposed gets clearer and hidden information in real EEG recording (Pascucci et al. 2019). Time-varying network connectivity exactly reveals more latent information, such as, all states of behavioral microsleep (Toppi et al. 2016), epilepsy (Lehnertz et al. 2017), than static network connectivity.
Source-level or cortex connectivity networks
The above connectivity networks are modeled on the sensor or scalp space directly. But wide-area overlapping of scalp channels leads to volume conduction for EEG recordings (Brunner et al. 2016), which is also why it has a poor spatial resolution. Such that, sensor-level network connectivity cannot interpret the connectivity measure briefly and maybe is unreliable to analyze brain networks, and doesn’t allow to infer about interacting regions (Papadopoulou et al. 2019), or the scalp-level results are less reliable than source-level ones (Lai et al. 2018). Therefore, source-level or cortex connectivity and source localization are of interest and importance.
Publication (Athanasiou et al. 2017) demonstrates that cortex activation network connectivity makes the information flow on cortical areas more clear and finds the similarity of motor execution and MI. The coupling between cortical brain dynamics partially is due to white matter connectivity across multiple brain rhythms and may provide some evidence for segregation and integration at fast time scales for neural information processing (Chu et al. 2012). Recent studies also find that higher integration in the theta band and lower segregation in the alpha band during working memory (Dai et al. 2019), and the imbalance of brain segregation and integration for patients with disorders of consciousness (Rizkallah et al. 2019). By reconfiguring the EEG source-level functional network connectivity, there is a negative correlation between psychological resilience and functional network flexibility (Paban et al. 2019), and this research (Li et al. 2018d) also reveals some significant differences of functional network connectivity between- and within-subject groups. Additionally, some advances (Lai et al. 2018; Seeber et al. 2019) as well demonstrate that the effect of scalp-level topology analysis or diagnosis of neural disorders is similar to cortical- or subcortical-level ones by EEG source localization or reconstruction from high-density scalp recordings. Certainly, either source- or sensor-level network analysis depends on specific research and applications.
Before constructing source-level connectivity or source localization, preprocessing must be done, such as spatial filters or blind spatial source separation (Michel and Brunet 2019; Oosugi et al. 2017), which is the same as scalp-level connectivity. And then, the inverse problem is of concern. Notwithstanding, about three factors decide on the processing (Hassan et al. 2015): (1) the number of scalp electrodes; (2) EEG inverse problem and algorithm used to measure the undirected or directed connectivity; (3) frequency bands to estimate FC or EC among neocortical sources. As a result, low-, mid-, and high-density EEG must match appropriate modeling algorithms to improve the performance of source-space connectivity (Barzegaran and Knyazeva 2017; Hassan et al. 2015; La Foresta et al. 2019). EEG inverse problem is ill-posed for that the number of variables observed is remarkably smaller than causal factors (the number of points in the brain where this surface activity could come from). Hence, some additional constraints (e.g., statistical or physiological (Muñoz-Gutiérrez et al. 2018), and non-statistical ones (Asadzadeh et al. 2020)) should be posed to make sure the EEG inverse problem well-posed. For parametric inverse problems, the empirical Bayes framework with one data-driven estimator is the most popular to solve cost-function (Hu et al. 2018; Jatoi et al. 2016; Le Cam et al. 2017; López et al. 2014). Certainly, it is worth heeding that these problems include convergence, computational load, and global or local minima which affect the accuracy of source reconstruction or localization (Wipf and Nagarajan 2009).
Source can be reconstructed from the measured EEG data with channels and expressed as,
| 24 |
where is the lead field matrix and as the measurement noise. And the EEG inverse problem is to find the optimal estimated value with getting best by Bayesian inference.
Eventually, network connectivity well-constructed certainly furthers help scientists or doctors to explore more things in the brain, such as cognitive function, neural disorder.
Applications of network analysis
Cognition and network connectivity
Cognition subserves a set of mental processes referring to gaining knowledge and comprehension, which includes learning, thinking, remembering, judging, problems-solving, and attention, and relates to several brain regions, such as portions of the superior and lateral frontal cortex, medial parietal cortex, the cingulated and the insula (Petersen and Sporns 2015). Various cognitive activities are depicted by different cognitive networks, such as semantic network, synaptic network, informational network, and social network (Siew et al. 2019), which dynamically vary in anatomical segregation and functional integration. Therefore, one effective tool should be a need. Network connectivity has the great potential to reveal dynamic interdependencies between regions during cognitive activity with time-series EEG signals (Li et al. 2018a) (shown in Fig. 4).
Fig. 5.
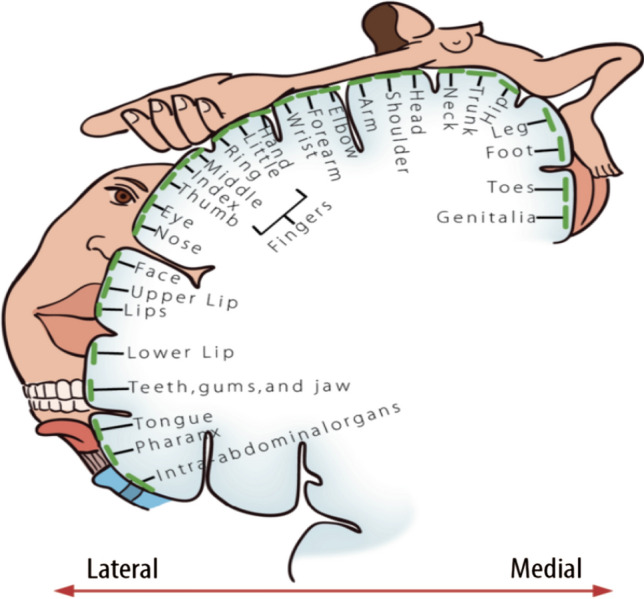
The mapping skeleton of the somatosensory cortex in the brain and the human body. The somatotopic map describes that the reverberatory cortical regions and the responding parts of the human body correspond to each other under tactile stimuli.
Adapted from the material (Privitera 2020)
Fig. 4.
One example of EEG network analysis on cognitive functions. a and b The event-related potentials (ERPs) of ST and TT sequences during the P300 task respectively, where P300 originates from the positive peak latency of ERP wave is so 300 ms. c P300 amplitude evoked by the ST and TT sequences. d Network topology of ST sequence, the red lines describe the stronger edges of T than that of S stimulus, while the line width does the quantitative variances of edge strengths. e Network attributes of two sequences, the blue bar denotes the network feature of the first stimulus, and the red one does that of the second one. f Network topology: the blue lines describe the weaker edges of T2 in TT than that of T in ST, and the line width does the quantitative variances of edge strengths under those conditions. g Network attributes, the red and blue bars describe the network attributes of T2 in two stimuli, respectively. h Task activation.
Adapted from the paper (Li et al. 2018a)
In these papers (Xu et al. 2014b; Zhang et al. 2013a, b), experiments demonstrate that periodic visual stimuli can activate parietal-occipital and frontal regions, and there exists obvious directed information flow between visual and frontal cortices (Li et al. 2015b, 2016), and so do further second harmonic responses of steady-state visual evoked potential (SSVEP) (Zhang et al. 2015). Musical experiment (Tian et al. 2013) uncovers that music is related to multi-oscillatory neural rhythms and tempo-transformation can indeed change the strength of theta and alpha power in bilateral occipital-parietal regions. Accordingly, visual and auditory stimuli can activate the occipital and parietal regions.
MI or movement involves multiple regions, such as the primary motor cortex (M1), supplementary motor area (SMA), premotor cortex (PMC), and dorsal-lateral prefrontal cortex (DLPFC), and correlates with the performances of related functional networks. Studies (Kim et al. 2018; Li et al. 2019c; Zhang et al. 2016) find that one more efficient frontal-parietal attention network will perform better on MI. Visual-motor coordination is an essential function of movement control which requires interactions of multiple brain regions to realize different visual-motor coordination states. Factually, changes between successive states and the smoothness of these changes further demonstrate that brain functional connectivity takes on such meta-stable dynamics (Li et al. 2020b; Liu et al. 2017b).
The emotional-recognition network realizes the combination of comprehensive activation and connection information for emotion recognition, which relates to neural rhythms, especially beta and gamma, or higher frequency bands in parietal, frontal, and occipital lobes (Li et al. 2019c). For the emotion response network, neural activity in emotional-response-related brain regions is found that it is significantly associated with prefrontal EEG asymmetry which can be measured with amplitude and entropy (Daly et al. 2019). Color stimuli also have significant impacts on the subject’s emotion and cognition, which results in forming a larger number of brain hubs and increasing frontal-parietal connectivity (Chai et al. 2019).
Word processing activates mainly semantic networks, also form similarity and synaptic networks. Hnazaee et al. (Fahimi Hnazaee et al. 2018) found that functional regions can orderly and differently engage in word processing depending on the type of information retrieved. And the higher-level abstract representation of info concepts activates bilateral anterior temporal lobes easily (Farahibozorg Feb/2018). In the word comprehension experiment, these results show that verifying features from the same modality (visual or auditory) network is faster than doing ones from across modalities, and integrating multimodal semantic network induces theta oscillation in the left anterior temporal lobe. For picture naming, there are six brain network states involved which are featured by high synchronization of gamma rhythm (30–45 Hz) and dynamically and transfer between each other (Giahi Saravani et al. 2019; Hassan et al. 2015).
Social concept representation and retrieval, domain-general semantic integration, and domain-specific integration of social semantic contents involve in Theory of Mind and discourse comprehension (Lin et al. 2018). Different discourse topics heavily can fire different brain regions. More detailed cognitive functions are described in Table 1.
Table 1.
Cognitive functions
| Cognitive function | Description | Involved neural regions |
|---|---|---|
| Attention and consciousness | Attention is the ability to choose and concentrate on relevant stimuli. Attention function correlates significantly with cognitive control or executive control (Mackie et al. 2013). Consciousness is a self-sustaining process with varying in vigilance and arousal, and a precondition to putting voluntary control on behavior. Attention is different from consciousness but strictly related to consciousness (Nani et al. 2019; Tallon-Baudry 2011) | The attention mainly activates sensory regions, and attentional control focuses on the posterior parietal cortex (lateral intraparietal area, superior parietal lobule) and prefrontal cortex (frontal eye field, and supplementary eye field, superior colliculus) (Yantis 2008), as well as dorsolateral prefrontal cortical regions (Sarter et al. 2001) |
| Working memory | Working memory is a cognitive system with a limited capacity that can maintain information provisionally. Working memory is important for reasoning and the guidance of decision-making and behavior (Eriksson et al. 2015). Multiple behavioral and health elements contribute to working memory (Moser et al. 2018) | Working memory can activate the fronto-parietal cortices and subcortices, such as the midbrain and cerebellum (Chai et al. 2018; Eriksson et al. 2015; Moser et al. 2018) |
| Learning and memory | Learning means to acquire new information or knowledge, it aims to memorize the info and knowledge. Learning & memory execute on three basic phases including encoding (acquisition and consolidation), storage, and retrieval (Gazzaniga et al. 2019). Short-term memory (STM) is different from long-term memory (LTM). STM (primary memory) is merely a temporary and short-lasting conscious maintenance while LTM (secondary memory) is maintained by stable and permanent changes in neural connections spread through the brain, including consolidation and storage (Brem et al. 2013) | The different aspects of learning and memory involve different brain systems, but the cerebellum, hippocampus, and amygdala play indispensable roles in the processes of learning and memory (Thompson and Kim 1996) |
| Sensation & Perception | Sense-perception is to select and identify information from the environment by sensory receptors—sense organs, such as eyes, nose, tongue, hands, and skin (Mesulam 1998; Morenko 2014). The sensorimotor contingencies (SCs) differentiate sensation from perception. If SC is determined by the feature of the visual appearance itself, it thus is taken for sensation. If SC is resolved by visual attributes, it is deemed as perception (O'Regan and Noë 2001) | The neural regions involved in sensory and perceptive processing are complex and transmodal. After haptic inputs are transferred into neural biological signals are sent through the thalamus to the primary somatosensory cortex for further processing (shown as Fig. 5) (Privitera 2020) |
| Speech & Language | Language is made up of social rules including semantics or meanings, make-new-word, grammar, and social context; while speech is the verbal means of communicating involving articulation, voice, fluency, and prosody. Broca’s area and Wernicke’s area are the primary neural regions, as well as angular gyrus and insula (Blank et al. 2002; Romeo et al. 2018) | Language is processed through two distinct pathways, i.e. the dorsal and the ventral stream, to realize phonological and semantic processing. Phonological processing is mainly made with the superior longitudinal fasciculus of the dorsal stream, while semantic processing is done dominantly with the ventral stream including inferior fronto-occipital fasciculus and the intra-temporal networks. Speech is probably related to the frontal aslant tract (Fujii et al. 2016) |
| Emotion | Emotion emerges when livings make sense of sensory inputs from the body and the world using empirical knowledge (Lindquist et al. 2012). Positive or negative emotion is activated in the left or right hemisphere that is in dispute. The asymmetry of emotional processing is still on the way (Alves et al. 2008) | Emotion is implemented by amygdale and affects cognition (e.g., perception, attention, memory, and decision-making), vice versa (Brosch et al. 2013; Gray et al. 2002; Salzman and Fusi 2010). Their relationship tie is the anterior cingulated cortex (Stevens et al. 2011) |
| Object identification | Object identification is the ability of discerning objects via a series of reflexive feedforward computations in the brain (DiCarlo et al. 2012). The invariant object recognition (relying on view-invariant diagnostic features) can be achieved by the human brain, not by advanced machine vision methods (Karimi-Rouzbahani et al. 2017; Roldan 2017) | Object recognition will fire the neural regions including mainly mid-temporal and temporopolar cortices, and right frontal regions, and the left occipitotemporal cortex which can speed up the prediction of objects (Rizkallah et al. 2018) |
| Executive control | Executive functions (EFs) are a set of cognitive processes that are necessary for the cognitive control of behavior to complete the chosen goals | EFs are located primarily in the frontal, parietal, and cerebellar. Temporal lobes are only related to EFs for the patient with dementia (Nowrangi et al. 2014) |
| Social cognition | Social cognition in humans characterizes psychological processes that permit us to infer or determine others’ imminent mental states (Adolphs 2009) | Social cognition depends on the medial prefrontal cortex, superior temporal sulcus, temporal-parietal junction, and precuneus, and attributes to affect social decision-makers (Lee and Harris 2013) |
Diseases and network connectivity
Neural disorders and diseases mean brain dysfunction which results in abnormal network connectivity. Different neural diseases take different abnormalities on their responding functional connectivity.
A seizure is a sudden and uncontrolled bioneural change in the brain, while epilepsy is a disorder. Clinical analysis reveals that children with epilepsy commonly have cognitive impairment (Kinney-Lang et al. 2019). To investigate how seizure is generated, Cooray (Cooray et al. 2016) used DCM and Bayesian belief updating to reveal that seizure dynamics change over time and space. The synchronization of network connectivity increases from interictal to preictal states during the transition of brain activity before epileptic seizures (Li et al. 2019e).
Psychogenic non-epileptic seizures (PNES) can appear outwardly like epileptic seizures, but not epilepsy and their cause is psychological (Alsaadi and Marquez 2005). For subjects with PNES or epilepsy, their network topology in the gamma band of the brain has decreased long linkage between the frontal region and posterior brain areas compared with healthy controls (Xu et al. 2014a; Xue et al. 2013), and but the spatial pattern of the network topology in beta band significantly differentiates from each other.
Alzheimer’s disease (AD) is a neurodegenerative disorder that has the characteristic of disturbance of higher cortical cognitive functions such as memory, comprehension, learning capacity, language, thinking, reasoning, and so forth (Tsolaki et al. 2014). EEG biomarkers of the network topology of AD’s subjects change greatly and obviously in different progress stages (preclinical, prodromal, and dementia for AD) (Dubois et al. 2016). In the preclinical stage of AD, the amyloid burden has one non-linear relationship with EEG metrics (e.g. frequency oscillations and spectral entropy) (Gaubert et al. 2019; Poil et al. 2013). In the prodromal stage, there is a higher α3/α2 (high alpha band/low alpha band) EEG power ratio in subjects with shrinkage and cutdown perfusion inside the temporoparietal projections which will result in AD dementia (Moretti 2016). From prodromal to dementia, the number of edges of AD subject’s connectivity networks gradually reduces and local–global efficiency loses (Franciotti et al. 2019). Measurements (Hata et al. 2016; La Foresta et al. 2019) also demonstrate that most cortical regions keep the phase desynchronization or disconnection for AD subjects, as the same as between the right dorsolateral prefrontal cortex and the right posterior-inferior parietal lobule. Their brain reproducibility and robustness have also been decreased greatly in alpha and beta bands of EEG signals with amplitude envelope correlation by leakage correction (AEC-c) functional connectivity which is a more effective measure in AD (Briels et al. 2020).
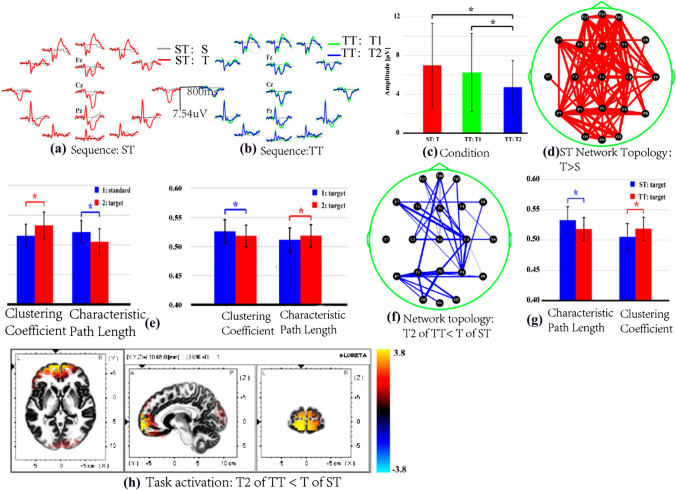
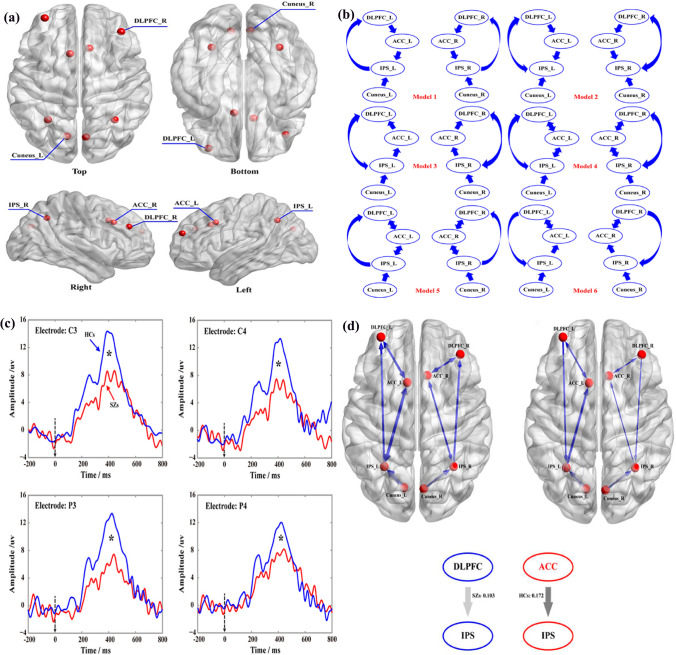
Schizophrenia (SZ) is a serious and chronic mental disorder that has psychotic and cognitive problems. People with schizophrenia have different brain structures, functions, and interactions among neurotransmitters compared with normal ones (Karlsgodt et al. 2010). DCM analysis (Li et al. 2018e) explained that SZ subjects showed the disconnectivity in their brain structure during the related cognitive process, which was found the dysfunctions among the anterior cingulated cortex, prefrontal cortex (PFC), DLPFC, and intraparietal sulcus, etc. Similarly, SZs’ functional connectivity alters (Liu et al. 2019; Naim-Feil et al. 2018; Yin et al. 2017). Therefore, the spatial patterns of these effective networks can differentiate SZs from healthy controls (Harmah et al. 2019; Li et al. 2019b) (shown as Fig. 6). Certainly, EEG network analysis is also applied to other popular brain disorders, shown in Table 2.
Fig. 6.
One example of EEG network analysis on neural disorders with DCM effective network. a The distribution of specific 8 DCM nodes from the top, bottom, left, and right. b Constructs 6 DCM connectivity from the 8 specific nodes via the existing P300 knowledge. c Averaged P300 waveforms for SZs and health controls (HCs), the red line for P300 of SZs, and the blue one for that of HCs. d The causal relationship in SZs and HCs with DCM. The top left model is for HCs and the top right one for SZs, the down graphs denote the direction of the information flow between two nodes and the strength of information flow.
Adapted from the reference (Li et al. 2018e)
Table 2.
Brain diseases and network connectivity
| Brain diseases | Responding network connectivity |
|---|---|
| Parkinson’s disease (PD) | PD results in cognitive and executive deficits related to changed functional brain connectivity among the old group (Gao and Wu 2016; Yi et al. 2017). The cognitive deficits reflect on the alpha rhythm, especially in frontotemporal regions (Hassan et al. 2017). The executive deficits contribute to frontoparietal connectivity decrease (Teramoto et al. 2016). While one scientific paper finds that those subjects with PD have higher bilateral gamma and left alpha2 rhythms, and alpha2-gamma coupling in the right posterior parietal compared with peer healthy controls (Bin Yoo et al. 2018) |
| Autism spectrum disorder (ASD) | ASD occurs mainly in children. The connectivity dramatically cuts down in alpha and theta band for children with ASD (Bosl et al. 2011; Zeng et al. 2017), and also low long-range connectivity (O'Reilly et al. 2017). Especially, the dynamic connectivity can be measured obviously in sensorimotor and advanced cognitive networks (Mash et al. 2019) |
| Attention-Deficit/Hyperactivity Disorder (ADHD) | Subjects with ADHD behave abnormally and pay inattention. Rhythmical experiments reveal that ADHDers have low clustering in hyperactivity while augmented segregation degree (Ghaderi et al. 2017; Michelini et al. 2019) |
| Amyotrophic Lateral Sclerosis (ALS) | ALS is one neurodegenerative illness that causes mainly motor cortex, and also cognitive networks (Dukic et al. 2019). Nodal assortativity of the alpha band in ALS patients is increased and the clustering coefficient also has greatly higher values in all neural frequencies (Fraschini et al. 2016; Iyer et al. 2015) |
| Auditory disorder | Tinnitus is one of the auditory disorders and still perceives the sound without external auditory stimuli. The auditory network with tinnitus has a comparatively different level of segregation and integration in most rhythm bands (Mohagheghian et al. 2019). The connectivity in the superior frontal cortex has various degrees of reduction for all frequency bands during the development stage of tinnitus (Lan et al. 2020). Sudden deafness also is one auditory disease that has inhibited the alpha2 band in the left frontal regions and strengthens the attention or emotional function networks (Cai et al. 2019) |
| Stroke | The connectivity of after-stroke patients has more new connections to unfold the neuroplasticity of the brain (Hordacre et al. 2018; Li et al. 2014). Moreover, the changing functional and structural topology can predict different deficits (Siegel et al. 2016) |
| Major depressive disorder (MDD) | MDD is one mental illness that is companied by a depressed mood. The resting-state connectivity indices (strength, clustering coefficient, path length, centrality, etc.) differ significantly from normal healthy controls (Saeedi et al. 2021; Shim et al. 2018). Simultaneously, MDD also affects MDD patients’ cognitive and motivational functions (Damborská et al. 2019) |
As such, the altering connectivity profiles in the brain can provide informative help to diagnose and treat patients (Contreras et al. 2015, 2017), such as ADHD (Cary et al. 2017), AD (Contreras et al. 2019), and so on.
Artificial intelligence and biological brain
AI is the simulation of human intelligence processes by machines that involves multiple brain cognitive skills: perceiving, learning, reasoning, and self-adjustment, etc., and therefore indeed is associated with human cognition according to the nature of brain cognition (van der Velde and Kamps 2010). AI has solved huge quantities of engineering questions and difficulties. Especially, in medical and neurological projects, AI is helping researchers and doctors to explore the complex brain.
Shreds of evidence verify that AI algorithms can estimate reliable causal relationships among multi-layer neural perceptrons in memory recognition tasks with considered time-lag and different initial conditions (Talebi et al. 2018), and detect strong synchronization and potential pre-seizure phenomena (Bomela et al. 2020). Massive amounts of papers on signal processing propose types of AI algorithms to solve EEG signal processing and network construction. The deep convolutional neural network, one of AI, can learn to extract useful rhythm features and decode specific task-related EEG signals (Schirrmeister et al. 2017; Zeng et al. 2018), thus effectively applies on feature-extraction, classification of EEG signals (Cecotti and Gräser 2011; Chandani 2017; Gao et al. 2020; Li et al. 2018b; Moon et al. 2020) and predict, evaluate EEG parameters (Ortolani et al. 2002), and monitor all states of patients with anesthesia (Acharya et al. 2018; Gu et al. 2019). AI can also dig hidden-deeper information to give one hand to doctors (Liu et al. 2017c) for diagnosis and treatment. For the sake of EEG signals with various and multiple rhythms, multi-scale neural networks were proposed to extract multiple frequency signatures (Raghu et al. 2020). Frankly, AI algorithms are boosting up network analysis in the brain science and neural medical field.
But AI has no capability of cooperating with self-understanding, self-control, self-consciousness, and self-motivation as the human brain does. The brain intelligence model is proposed to extend and advance contemporary AI in the light of human memory function (Lu et al. 2018). Additionally, the big gap between AI and neuroscience is the culture to communicate with each other which will be solved to extend them further (Chance et al. 2020). More reviews and reports remark and analyze that AI has been developing from brain science and also expedites it (Fan et al. 2020; Hassabis et al. 2017; Savage 2019; Shapshak 2018).
Discussion and future challenges
Network analysis is one vital tool in neuroscience and cognition, which opens the door to spy the brain and provide more helpful information for medical purposes. For a dynamic and time-variant brain, it is so important to select neural signal patterns. Certainly, EEG is nowadays an irreplaceable signal pattern for its high-temporal resolution to process time-series courses and explore more complex time-variant brain network dynamics. FC and EC of network connectivity can depict the segregation and integration of brain neuronal regions under specific tasks or neural disorders opposite to healthy controls. But EC with directed connectivity expresses more perfectly the causal relationship between different cortices, subcortices, and brainstem. To explore the secret of the brain neural system, it is the first place to construct useful and stable network connectivity. Although statistical or nonstatistical algorithms of network construction are plentiful, suitable ones are still on the way. On the other hand, EEG network analysis indeed is lifting the mysterious veils of the brain and its cognitive functions layer by layer, and having provided valuable biomarkers or signatures for brain neural diseases or auxiliary rehabilitation.
But raw EEG signals are filled with eye movement and electromyographic artifacts, it is one future trend to pursue interdisciplinary denoise advances, such as EEGDenoiseNet (Zhang et al. 2020a, b, c). Certainly, neural networks or AI algorithms for denoising EEG recordings need as huge data samples as possible. However, the subject resource is still a big matter. But the bad side gradually turns into a good one. Once the more secret of the brain is being unveiled, the farther and wider induced brain computation and applications go. After all, brain network and AI is the catalyzer for each other (Savage 2019).
The brain is also one subtle small-world and marvelous parallel system (Braga and Buckner 2017; Sigman and Dehaene 2008), future research would like to change focus on parallel subnetworks to deal with specific cognitive function or neural disorder, and find the relationship among these subnetworks or between a subnetwork and other secondary ones, which may uncover some unknown things. On the other side, graphic theory (Chen et al. 2018) is a mathematical tool but plays an indelible role in brain network analysis. Graphic challenges may activate the field of unknown brain functions or hidden information (Kao et al. 2017; Kepner et al. 2019): (1) Deep neural network or AI algorithm may work effects on graph representation and infer different types of the sparse subnetwork in a large whole network; (2) Subgraph block partition from one nonstationary and dynamic network may demystify the state-flow relationship among subnetwork and find the main or optimal subnetwork. Additionally, the brain neural system is a hierarchical and complex system, like an army, social group (Hilgetag and Goulas 2020), whether does control theory affect it? The answer is YES (Chen 2017; Gu et al. 2015; Lynn and Bassett 2019) and but is groping. In the future, the modeling and analysis of neural systems may be based on control theory and engineering which may unveil the mask of the neural-regulation mechanism of the brain and provide useful and accurate information for medical and auxiliary treatment.
Further to dig out more hidden information, recent network dynamics are developed for psychiatric illnesses which may play one important part in medical diagnosis and treatment (Durstewitz et al. 2020; Ouyang and Zhou 2020) based on the traditional network analysis. Zheng et al. (2020) also referred that the multiple-scale connectivity is the characteristics of brain network, and studied multiscale network analysis to recover the similarity among different brain cerebral regions under different scales. Additionally, the temporal feature runs through the whole brain neural system, Rabinovich et al. (2020) developed sequential network dynamics to study and analyze the cognitive function and neural diseases. Hence, the dynamics of the complex network will accelerate network analysis and network theory, and provide an interesting focus and direction to solve brain science in the future. In a nutshell, EEG network analysis has started and given its plentiful fruits by interdisciplinary technology in brain science.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (#61961160705, #U19A2082, #61901077), the Science and Technology Development Fund, Macau SAR (File no. 0045/2019/AFJ), the Project of Science and Technology Department of Sichuan Province (#2021YFSY0040, #2018JZ0073, #2020ZYD013), and the Key Research and Development Program of Guangdong Province, China (#2018B030339001).
Declaration
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cuihua Luo and Fali Li have contributed equally to this work.
References
- Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H, Subha DP. Automated EEG-based screening of depression using deep convolutional neural network. Comput Methods Programs Biomed. 2018;161:103–113. doi: 10.1016/j.cmpb.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Adolphs R. The social brain: neural basis of social knowledge. Annu Rev Psychol. 2009;60:693–716. doi: 10.1146/annurev.psych.60.110707.163514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts H, Fias W, Caeyenberghs K, Marinazzo D. Brain networks under attack: robustness properties and the impact of lesions. Brain. 2016;139:3063–3083. doi: 10.1093/brain/aww194. [DOI] [PubMed] [Google Scholar]
- Alavash M, Tune S, Obleser J. Modular reconfiguration of an auditory control brain network supports adaptive listening behavior. Proc Natl Acad Sci USA. 2019;116:660–669. doi: 10.1073/pnas.1815321116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaadi TM, Marquez AV. Psychogenic nonepileptic seizures. Am Fam Physician. 2005;72:849–856. [PubMed] [Google Scholar]
- Alves NT, Fukusima SS, Aznar-Casanova JA. Models of brain asymmetry in emotional processing. Psychol Neurosci. 2008;1:63–66. doi: 10.3922/j.psns.2008.1.010. [DOI] [Google Scholar]
- Anticevic A, Murray JD, Barch DM. Bridging levels of understanding in Schizophrenia through computational modeling. Clin Psychol Sci. 2015;3:433–459. doi: 10.1177/2167702614562041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadzadeh S, Yousefi Rezaii T, Beheshti S, Delpak A, Meshgini S. A systematic review of EEG source localization techniques and their applications on diagnosis of brain abnormalities. J Neurosci Methods. 2020;339:108740. doi: 10.1016/j.jneumeth.2020.108740. [DOI] [PubMed] [Google Scholar]
- Athanasiou A, Klados MA, Pandria N, Foroglou N, Kavazidi KR, Polyzoidis K, Bamidis PD. A systematic review of investigations into functional brain connectivity following spinal cord injury. Front Hum Neurosci. 2017;11:517. doi: 10.3389/fnhum.2017.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala LA, Sameshima K, Takahashi DY (2008) Generalized partial directed coherence. In: 2007 15th international conference on digital signal processing. Wiley, Cardiff, pp 163–166. 10.1109/ICDSP.2007.4288544
- Barbey AK. Network neuroscience theory of human intelligence. Trends Cogn Sci. 2018;22:8–20. doi: 10.1016/j.tics.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Barzegaran E, Knyazeva MG. Functional connectivity analysis in EEG source space: the choice of method. PLoS ONE. 2017;12:e0181105. doi: 10.1371/journal.pone.0181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Avena-Koenigsberger A, Goñi J, et al. Generative models of the human connectome. Neuroimage. 2016;124:1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazoli CE, Sturzbecher M, White TP, Dos Santos Onias HH, Andrade KC, de Araujo DB, Sato JR. Application of partial directed coherence to the analysis of resting-state EEG-fMRI data. Brain Connect. 2013;3:563–568. doi: 10.1089/brain.2012.0135. [DOI] [PubMed] [Google Scholar]
- Bin Yoo H, La Concha EOd, de Ridder D, Pickut BA, Vanneste S. The functional alterations in top-down attention streams of Parkinson's disease measured by EEG. Sci Rep. 2018;8:10609. doi: 10.1038/s41598-018-29036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank SC, Scott SK, Murphy K, Warburton E, Wise RJS. Speech production: Wernicke, Broca and beyond. Brain. 2002;125:1829–1838. doi: 10.1093/brain/awf191. [DOI] [PubMed] [Google Scholar]
- Blinowska KJ, Rakowski F, Kaminski M, de Vico Fallani F, Del Percio C, Lizio R, Babiloni C. Functional and effective brain connectivity for discrimination between Alzheimer's patients and healthy individuals: a study on resting state EEG rhythms. Clin Neurophysiol. 2017;128:667–680. doi: 10.1016/j.clinph.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Boccatetti S, Latora V, Moreno Y, Chavez M, Hwang D. Complex networks: structure and dynamics. Phy Rep. 2006;424:175–308. doi: 10.1016/j.physrep.2005.10.009. [DOI] [Google Scholar]
- Bomela W, Wang S, Chou C-A, Li J-S. Real-time inference and detection of disruptive EEG networks for epileptic seizures. Sci Rep. 2020;10:8653. doi: 10.1038/s41598-020-65401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönstrup M, Schulz R, Feldheim J, Hummel FC, Gerloff C. Dynamic causal modelling of EEG and fMRI to characterize network architectures in a simple motor task. Neuroimage. 2016;124:498–508. doi: 10.1016/j.neuroimage.2015.08.052. [DOI] [PubMed] [Google Scholar]
- Bönstrup M, Schulz R, Schön G, Cheng B, Feldheim J, Thomalla G, Gerloff C. Parietofrontal network upregulation after motor stroke. Neuroimage Clin. 2018;18:720–729. doi: 10.1016/j.nicl.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bore JC, Li P, Harmah DJ, Li F, Yao D, Xu P. Directed EEG neural network analysis by LAPPS (p≤1) penalized sparse Granger approach. Neural Netw. 2020;124:213–222. doi: 10.1016/j.neunet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- Bosl W, Tierney A, Tager-Flusberg H, Nelson C. EEG complexity as a biomarker for autism spectrum disorder risk. BMC Med. 2011;9:18. doi: 10.1186/1741-7015-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga RM, Buckner RL. Parallel interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron. 2017;95:457–471.e5. doi: 10.1016/j.neuron.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem A-K, Ran K, Pascual-Leone A. Learning and memory. Handb Clin Neurol. 2013;116:693–737. doi: 10.1016/B978-0-444-53497-2.00055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briels CT, Schoonhoven DN, Stam CJ, de Waal H, Scheltens P, Gouw AA. Reproducibility of EEG functional connectivity in Alzheimer's disease. Alzheimers Res Ther. 2020;12:68. doi: 10.1186/s13195-020-00632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brislin SJ, Patrick CJ. Callousness and affective face processing: clarifying the neural basis of behavioral-recognition deficits through use of brain ERPs. Clin Psychol Sci. 2019;7:1389–1402. doi: 10.1177/2167702619856342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes MJ, Hale JR, Zumer JM, Stevenson CM, Francis ST, Barnes GR, Owen JP, Morris PG, Nagarajan SS. Measuring functional connectivity using MEG: methodology and comparison with fcMRI. Neuroimage. 2011;56:1082–1104. doi: 10.1016/j.neuroimage.2011.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch T, Scherer KR, Grandjean D, Sander D. The impact of emotion on perception, attention, memory, and decision-making. Swiss Med Wkly. 2013;143:w13786. doi: 10.4414/smw.2013.13786. [DOI] [PubMed] [Google Scholar]
- Brunner C, Billinger M, Seeber M, Mullen TR, Makeig S. Volume conduction influences scalp-based connectivity estimates. Front Comput Neurosci. 2016;10:121. doi: 10.3389/fncom.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Li J, Chen Y, Chen W, Dang C, Zhao F, Li W, Chen G, Chen S, Liang M, Zheng Y. Inhibition of brain area and functional connectivity in idiopathic sudden sensorineural hearing loss with tinnitus, based on resting-state EEG. Front Neurosci. 2019;13:851. doi: 10.3389/fnins.2019.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaglios G, Muscoso EG, Di Maria G, Costanzo E. Patients with mild cognitive impairment have an abnormal upper-alpha event-related desynchronization/synchronization (ERD/ERS) during a task of temporal attention. J Neural Transm (Vienna) 2015;122:441–453. doi: 10.1007/s00702-014-1262-7. [DOI] [PubMed] [Google Scholar]
- Cary RP, Ray S, Grayson DS, Painter J, Carpenter S, Maron L, Sporns O, Stevens AA, Nigg JT, Fair DA. Network structure among brain systems in adult ADHD is uniquely modified by stimulant administration. Cereb Cortex. 2017;27:3970–3979. doi: 10.1093/cercor/bhw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catana C, Drzezga A, Heiss W-D, Rosen BR. PET/MRI for neurologic applications. J Nucl Med. 2012;53:1916–1925. doi: 10.2967/jnumed.112.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecotti H, Gräser A. Convolutional neural networks for P300 detection with application to brain-computer interfaces. IEEE Trans Pattern Anal Mach Intell. 2011;33:433–445. doi: 10.1109/TPAMI.2010.125. [DOI] [PubMed] [Google Scholar]
- Chai WJ, Abd Hamid AI, Abdullah JM. Working memory from the psychological and neurosciences perspectives: a review. Front Psychol. 2018;9:401. doi: 10.3389/fpsyg.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai MT, Amin HU, Izhar LI, Saad MNM, Abdul Rahman M, Malik AS, Tang TB. Exploring EEG effective connectivity network in estimating influence of color on emotion and memory. Front Neuroinform. 2019;13:66. doi: 10.3389/fninf.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance FS, Aimone JB, Musuvathy SS, Smith MR, Vineyard CM, Wang F. Crossing the cleft: communication challenges between neuroscience and artificial intelligence. Front Comput Neurosci. 2020;14:39. doi: 10.3389/fncom.2020.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandani M. Classification of EEG physiological signal for the detection of epileptic seizure by using DWT feature extraction and neural network. Int J Neurol Phys Ther. 2017;3:38–43. doi: 10.11648/j.ijnpt.20170305.11. [DOI] [Google Scholar]
- Chen G. Pinning control and controllability of complex dynamical networks. Int J Autom Comput. 2017;14:1–9. doi: 10.1007/s11633-016-1052-9. [DOI] [Google Scholar]
- Chen J, Wang H, Hua C, Wang Q, Liu C. Graph analysis of functional brain network topology using minimum spanning tree in driver drowsiness. Cogn Neurodyn. 2018;12:569–581. doi: 10.1007/s11571-018-9495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CJ, Kramer MA, Pathmanathan J, Bianchi MT, Westover MB, Wizon L, Cash SS. Emergence of stable functional networks in long-term human electroencephalography. J Neurosci. 2012;32:2703–2713. doi: 10.1523/JNEUROSCI.5669-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JR, D'Esposito M. The segregation and integration of distinct brain networks and their relationship to cognition. J Neurosci. 2016;36:12083–12094. doi: 10.1523/JNEUROSCI.2965-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JA, Goñi J, Risacher SL, Sporns O, Saykin AJ. The structural and functional connectome and prediction of risk for cognitive impairment in older adults. Curr Behav Neurosci Rep. 2015;2:234–245. doi: 10.1007/s40473-015-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JA, Goñi J, Risacher SL, Amico E, Yoder K, Dzemidzic M, West JD, McDonald BC, Farlow MR, Sporns O, Saykin AJ. Cognitive complaints in older adults at risk for Alzheimer's disease are associated with altered resting-state networks. Alzheimers Dement (Amst) 2017;6:40–49. doi: 10.1016/j.dadm.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras JA, Avena-Koenigsberger A, Risacher SL, et al. Resting state network modularity along the prodromal late onset Alzheimer's disease continuum. Neuroimage Clin. 2019;22:101687. doi: 10.1016/j.nicl.2019.101687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray GK, Sengupta B, Douglas PK, Friston K. Dynamic causal modelling of electrographic seizure activity using Bayesian belief updating. Neuroimage. 2016;125:1142–1154. doi: 10.1016/j.neuroimage.2015.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Li Y, Gan S, Du F. The reliability of estimating visual working memory capacity. Sci Rep. 2019;9:1155. doi: 10.1038/s41598-019-39044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly I, Williams D, Hwang F, Kirke A, Miranda ER, Nasuto SJ. Electroencephalography reflects the activity of sub-cortical brain regions during approach-withdrawal behaviour while listening to music. Sci Rep. 2019;9:9415. doi: 10.1038/s41598-019-45105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborská A, Tomescu MI, Honzírková E, Barteček R, Hořínková J, Fedorová S, Ondruš Š, Michel CM. EEG resting-state large-scale brain network dynamics are related to depressive symptoms. Front Psychiatry. 2019;10:548. doi: 10.3389/fpsyt.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O, Kiebel SJ, Harrison LM, Mattout J, Kilner JM, Friston KJ. Dynamic causal modeling of evoked responses in EEG and MEG. Neuroimage. 2006;30:1255–1272. doi: 10.1016/j.neuroimage.2005.10.045. [DOI] [PubMed] [Google Scholar]
- de Oliveira RMW. Neuroplasticity. J Chem Neuroanat. 2020;108:101822. doi: 10.1016/j.jchemneu.2020.101822. [DOI] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Sporns O, Romani GL, Corbetta M. A dynamic core network and global efficiency in the resting human brain. Cereb Cortex. 2016;26:4015–4033. doi: 10.1093/cercor/bhv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vico Fallani F, Astolfi L, Cincotti F, Mattia D, Tocci A, Salinari S, Marciani MG, Witte H, Colosimo A, Babiloni F. Brain network analysis from high-resolution EEG recordings by the application of theoretical graph indexes. IEEE Trans Neural Syst Rehabil Eng. 2008;16:442–452. doi: 10.1109/TNSRE.2008.2006196. [DOI] [PubMed] [Google Scholar]
- Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134:9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- DiCarlo JJ, Zoccolan D, Rust NC. How does the brain solve visual object recognition? Neuron. 2012;73:415–434. doi: 10.1016/j.neuron.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez Á, Ranlund S, Pinotsis D, Calafato S, Shaikh M, Hall M-H, Walshe M, Nevado Á, Friston KJ, Adams RA, Bramon E. Abnormal frontoparietal synaptic gain mediating the P300 in patients with psychotic disorder and their unaffected relatives. Hum Brain Mapp. 2017;38:3262–3276. doi: 10.1002/hbm.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis SI, Laskaris NA, Micheloyannis S. Transition dynamics of EEG-based network microstates during mental arithmetic and resting wakefulness reflects task-related modulations and developmental changes. Cogn Neurodyn. 2015;9:371–387. doi: 10.1007/s11571-015-9330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Fu Z, Calhoun VD. Classification and prediction of brain disorders using functional connectivity: promising but challenging. Front Neurosci. 2018;12:525. doi: 10.3389/fnins.2018.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Hampel H, Feldman HH, et al. Preclinical Alzheimer's disease: definition, natural history, and diagnostic criteria. Alzheimers Dement. 2016;12:292–323. doi: 10.1016/j.jalz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc NT, Lee B. Decoding brain dynamics in speech perception based on EEG microstates decomposed by multivariate Gaussian hidden Markov model. IEEE Access. 2020;8:146770–146784. doi: 10.1109/ACCESS.2020.3015292. [DOI] [Google Scholar]
- Dukic S, Iyer PM, Mohr K, Hardiman O, Lalor EC, Nasseroleslami B. Estimation of coherence using the median is robust against EEG artefacts. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:3949–3952. doi: 10.1109/EMBC.2017.8037720. [DOI] [PubMed] [Google Scholar]
- Dukic S, McMackin R, Buxo T, et al. Patterned functional network disruption in amyotrophic lateral sclerosis. Hum Brain Mapp. 2019;40:4827–4842. doi: 10.1002/hbm.24740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durstewitz D, Huys QJM, Koppe G. Psychiatric illnesses as disorders of network dynamics. Biol Psychiatry Cogn Neurosci Neuroimaging. 2020;S2451–9022(20):30019–30027. doi: 10.1016/j.bpsc.2020.01.001. [DOI] [PubMed] [Google Scholar]
- Eriksson J, Vogel EK, Lansner A, Bergström F, Nyberg L. Neurocognitive architecture of working memory. Neuron. 2015;88:33–46. doi: 10.1016/j.neuron.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahimi Hnazaee M, Khachatryan E, van Hulle MM. Semantic features reveal different networks during word processing: an EEG source Llocalization study. Front Hum Neurosci. 2018;12:503. doi: 10.3389/fnhum.2018.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Fang L, Wu J, Guo Y, Dai Q. From brain science to artificial intelligence. Engineering. 2020;6:248–252. doi: 10.1016/j.eng.2019.11.012. [DOI] [Google Scholar]
- Farahibozorg S (Feb/2018) Uncovering dynamic semantic network in the brain using novel approached for EEG/MEG connectome reconstruction. Dissertation, Selwyn College
- Fastenrath M, Friston KJ, Kiebel SJ. Dynamical causal modelling for M/EEG: spatial and temporal symmetry constraints. Neuroimage. 2009;44:154–163. doi: 10.1016/j.neuroimage.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Fogelson N, Litvak V, Peled A, Fernandez-del-Olmo M, Friston K. The functional anatomy of schizophrenia: a dynamic causal modeling study of predictive coding. Schizophr Res. 2014;158:204–212. doi: 10.1016/j.schres.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- Franciotti R, Falasca NW, Arnaldi D, Famà F, Babiloni C, Onofrj M, Nobili FM, Bonanni L. Cortical network topology in prodromal and mild dementia due to Alzheimer's disease: graph theory applied to resting state EEG. Brain Topogr. 2019;32:127–141. doi: 10.1007/s10548-018-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini M, Demuru M, Hillebrand A, Cuccu L, Porcu S, Di Stefano F, Puligheddu M, Floris G, Borghero G, Marrosu F. EEG functional network topology is associated with disability in patients with amyotrophic lateral sclerosis. Sci Rep. 2016;6:38653. doi: 10.1038/srep38653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- Friston KJ. Functional and effective connectivity: a review. Brain Connect. 2011;1:13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Price CJ. Dynamic representations and generative models of brain function. Brain Res Bull. 2001;54:275–285. doi: 10.1016/S0361-9230(00)00436-6. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/S1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Friston K, Zeidman P, Litvak V. Empirical Bayes for DCM: a group inversion scheme. Front Syst Neurosci. 2015;9:164. doi: 10.3389/fnsys.2015.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K, Brown HR, Siemerkus J, Stephan KE. The dysconnection hypothesis. Schizophr Res. 2016;176:83–94. doi: 10.1016/j.schres.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Litvak V, Oswal A, Razi A, Stephan KE, van Wijk BCM, Ziegler G, Zeidman P. Bayesian model reduction and empirical Bayes for group (DCM) studies. Neuroimage. 2016;128:413–431. doi: 10.1016/j.neuroimage.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Maesawa S, Ishiai S, Iwami K, Futamura M, Saito K. Neural basis of language: an overview of an evolving model. Neurol Med Chir (tokyo) 2016;56:379–386. doi: 10.2176/nmc.ra.2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallen CL, Turner GR, Adnan A, D'Esposito M. Reconfiguration of brain network architecture to support executive control in aging. Neurobiol Aging. 2016;44:42–52. doi: 10.1016/j.neurobiolaging.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L-L, Wu T. The study of brain functional connectivity in Parkinson's disease. Transl Neurodegener. 2016;5:18. doi: 10.1186/s40035-016-0066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Dang W, Wang X, Hong X, Hou L, Ma K, Perc M. Complex networks and deep learning for EEG signal analysis. Cogn Neurodyn. 2020;2020:1. doi: 10.1007/s11571-020-09626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubert S, Raimondo F, Houot M, Corsi M-C, Naccache L, Diego Sitt J, Hermann B, Oudiette D, Gagliardi G, Habert M-O, Dubois B, de Vico Fallani F, Bakardjian H, Epelbaum S. EEG evidence of compensatory mechanisms in preclinical Alzheimer's disease. Brain. 2019;142:2096–2112. doi: 10.1093/brain/awz150. [DOI] [PubMed] [Google Scholar]
- Gazzaniga MS, Ivry RB, Mangun GR. Cognitive neuroscience: the biology of the mind. New York: W.W. Norton & Company; 2019. [Google Scholar]
- Ghaderi AH, Nazari MA, Shahrokhi H, Darooneh AH. Functional brain connectivity differences between different ADHD presentations: impaired functional segregation in ADHD-combined presentation but not in ADHD-inattentive presentation. Basic Clin Neurosci. 2017;8:267–278. doi: 10.18869/nirp.bcn.8.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghumare EG, Schrooten M, Vandenberghe R, Dupont P. A time-varying connectivity analysis from distributed EEG sources: a simulation study. Brain Topogr. 2018;31:721–737. doi: 10.1007/s10548-018-0621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giahi Saravani A, Forseth KJ, Tandon N, Pitkow X. Dynamic brain interactions during picture naming. eNeuro. 2019 doi: 10.1523/ENEURO.0472-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Knickmeyer RC, Gao W. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 2018;19:123–137. doi: 10.1038/nrn.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424. doi: 10.2307/1912791. [DOI] [Google Scholar]
- Gratton C, Laumann TO, Nielsen AN, Greene DJ, Gordon EM, Gilmore AW, Nelson SM, Coalson RS, Snyder AZ, Schlaggar BL, Dosenbach NUF, Petersen SE. Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron. 2018;98:439–452.e5. doi: 10.1016/j.neuron.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JR, Braver TS, Raichle ME. Integration of emotion and cognition in the lateral prefrontal cortex. Proc Natl Acad Sci USA. 2002;99:4115–4120. doi: 10.1073/pnas.062381899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis JC, Metcalf NV, Corbetta M, Shulman GL. Structural disconnections explain brain network dysfunction after stroke. Cell Rep. 2019;28:2527–2540.e9. doi: 10.1016/j.celrep.2019.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Pasqualetti F, Cieslak M, Telesford QK, Yu AB, Kahn AE, Medaglia JD, Vettel JM, Miller MB, Grafton ST, Bassett DS. Controllability of structural brain networks. Nat Commun. 2015;6:8414. doi: 10.1038/ncomms9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Liang Z, Hagihira S. Use of multiple EEG features and artificial neural network to monitor the depth of Anesthesia. Sensors (Basel) 2019 doi: 10.3390/s19112499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Guo F, Zhang Y, Li F, Xia Y, Xu P, Yao D. Periodic visual stimulation induces resting-state brain network reconfiguration. Front Comput Neurosci. 2018;12:21. doi: 10.3389/fncom.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmah DJ, Li C, Li F, Liao Y, Wang J, Ayedh WMA, Bore JC, Yao D, Dong W, Xu P. Measuring the non-linear directed information flow in Schizophrenia by multivariate transfer entropy. Front Comput Neurosci. 2019;13:85. doi: 10.3389/fncom.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassabis D, Kumaran D, Summerfield C, Botvinick M. Neuroscience-inspired artificial intelligence. Neuron. 2017;95:245–258. doi: 10.1016/j.neuron.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Hassan M, Benquet P, Biraben A, Berrou C, Dufor O, Wendling F. Dynamic reorganization of functional brain networks during picture naming. Cortex. 2015;73:276–288. doi: 10.1016/j.cortex.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Hassan M, Chaton L, Benquet P, Delval A, Leroy C, Plomhause L, Moonen AJH, Duits AA, Leentjens AFG, van Kranen-Mastenbroek V, Defebvre L, Derambure P, Wendling F, Dujardin K. Functional connectivity disruptions correlate with cognitive phenotypes in Parkinson's disease. Neuroimage Clin. 2017;14:591–601. doi: 10.1016/j.nicl.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata M, Kazui H, Tanaka T, Ishii R, Canuet L, Pascual-Marqui RD, Aoki Y, Ikeda S, Kanemoto H, Yoshiyama K, Iwase M, Takeda M. Functional connectivity assessed by resting state EEG correlates with cognitive decline of Alzheimer's disease: an eLORETA study. Clin Neurophysiol. 2016;127:1269–1278. doi: 10.1016/j.clinph.2015.10.030. [DOI] [PubMed] [Google Scholar]
- He Y, Lim S, Fortunato S, Sporns O, Zhang L, Qiu J, Xie P, Zuo X-N. Reconfiguration of cortical networks in MDD uncovered by multiscale community detection with fMRI. Cereb Cortex. 2018;28:1383–1395. doi: 10.1093/cercor/bhx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne LJ, Cocchi L, Zalesky A, Mattingley JB. Reconfiguration of brain network architectures between resting-state and complexity-dependent cognitive reasoning. J Neurosci. 2017;37:8399–8411. doi: 10.1523/JNEUROSCI.0485-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger K, Fukushima M, Sporns O, Fiebach CJ. Temporal stability of functional brain modules associated with human intelligence. Hum Brain Mapp. 2020;41:362–372. doi: 10.1002/hbm.24807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag CC, Goulas A. 'Hierarchy' in the organization of brain networks. Philos Trans R Soc Lond B Biol Sci. 2020;375:20190319. doi: 10.1098/rstb.2019.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordacre B, Moezzi B, Ridding MC. Neuroplasticity and network connectivity of the motor cortex following stroke: a transcranial direct current stimulation study. Hum Brain Mapp. 2018;39:3326–3339. doi: 10.1002/hbm.24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yao D, Valdes-Sosa PA. Unified Bayesian estimator of EEG reference at infinity: rREST (regularized reference electrode standardization technique) Front Neurosci. 2018;12:297. doi: 10.3389/fnins.2018.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Ren A, Shang J, Lei Q, Zhang Y, Yin Z, Li J, von Deneen KM, Huang L. Combining partial directed coherence and graph theory to analyse effective brain networks of different mental tasks. Front Hum Neurosci. 2016;10:235. doi: 10.3389/fnhum.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyadi B, Woolrich MW, Quinn AJ, Vidaurre D, de Vos M. A dynamic system of brain networks revealed by fast transient EEG fluctuations and their fMRI correlates. Neuroimage. 2019;185:72–82. doi: 10.1016/j.neuroimage.2018.09.082. [DOI] [PubMed] [Google Scholar]
- Iyer PM, Egan C, Pinto-Grau M, Burke T, Elamin M, Nasseroleslami B, Pender N, Lalor EC, Hardiman O. Functional connectivity changes in resting-state EEG as potential biomarker for amyotrophic lateral sclerosis. PLoS ONE. 2015;10:e0128682. doi: 10.1371/journal.pone.0128682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili M. Functional brain entworks: does the choice of dependency estimator and binarization method matter? Sci Rep. 2016;6:29780. doi: 10.1038/srep29780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili M, Knyazeva MG. Constructing brain functional networks from EEG: partial and unpartial correlations. J Integr Neurosci. 2011;10:213–232. doi: 10.1142/S0219635211002725. [DOI] [PubMed] [Google Scholar]
- Jatoi MA, Kamel N, Lopez JD, Faye I, Malik AS (2016) MSP based source localization using EEG signals, pp 1–5. 10.1109/ICIAS.2016.7824074
- Ji C, Maurits NM, Roerdink JBTM. Data-driven visualization of multichannel EEG coherence networks based on community structure analysis. Appl Netw Sci. 2018;3:41. doi: 10.1007/s41109-018-0096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsa VK, Sporns O, Breakspear M, Deco G, McIntosh AR. Towards the virtual brain: network modeling of the intact and the damaged brain. Arch Ital Biol. 2010;148:189–205. doi: 10.4449/aib.v148i3.1223. [DOI] [PubMed] [Google Scholar]
- Joudaki A, Salehi N, Jalili M, Knyazeva MG. EEG-based functional brain networks: does the network size matter? PLoS ONE. 2012;7:e35673. doi: 10.1371/journal.pone.0035673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce KE, Laurienti PJ, Burdette JH, Hayasaka S. A new measure of centrality for brain networks. PLoS ONE. 2010;5:e12200. doi: 10.1371/journal.pone.0012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbara A, Khalil M, El-Falou W, Eid H, Hassan M. Functional brain connectivity as a new feature for P300 speller. PLoS ONE. 2016;11:e0146282. doi: 10.1371/journal.pone.0146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M, Blinowska KJ. Directed transfer function is not influenced by volume conduction-inexpedient pre-processing should be avoided. Front Comput Neurosci. 2014;8:61. doi: 10.3389/fncom.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao E, Gadepally V, Hurley M, Jones M, Kepner J, Mohindra S, Monticciolo P, Reuther A, Samsi S, Song W, Staheli D, Smith S (eds) (2017) Streaming graph challenge: stochastic block partition. In: 2017 IEEE high performance extreme computing conference (HPEC) Waltham USA 2017, pp 1–12. 10.1109/HPEC.2017.8091040
- Karimi-Rouzbahani H, Bagheri N, Ebrahimpour R. Invariant object recognition is a personalized selection of invariant features in humans, not simply explained by hierarchical feed-forward vision models. Sci Rep. 2017;7:14402. doi: 10.1038/s41598-017-13756-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Sun D, Cannon TD. Structural and functional brain abnormalities in Schizophrenia. Curr Dir Psychol Sci. 2010;19:226–231. doi: 10.1177/0963721410377601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepner J, Alford S, Gadepally V, Jones M, Milechin L, Robinett R, Samsi S (eds) (2019) Sparse deep neural network graph challenge. In: 2019 IEEE high performance extreme computing conference (HPEC) Waltham USA 2019, pp 1–7. 10.1109/HPEC.2019.8916336
- Kim YK, Park E, Lee A, Im C-H, Kim Y-H. Changes in network connectivity during motor imagery and execution. PLoS ONE. 2018;13:e0190715. doi: 10.1371/journal.pone.0190715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney-Lang E, Yoong M, Hunter M, Kamath Tallur K, Shetty J, McLellan A, Fm Chin R, Escudero J. Analysis of EEG networks and their correlation with cognitive impairment in preschool children with epilepsy. Epilepsy Behav. 2019;90:45–56. doi: 10.1016/j.yebeh.2018.11.011. [DOI] [PubMed] [Google Scholar]
- La Foresta F, Morabito FC, Marino S, Dattola S. High-density EEG signal processing based on active-source reconstruction for brain network analysis in Alzheimer’s disease. Electronics. 2019;8:1031. doi: 10.3390/electronics8091031. [DOI] [Google Scholar]
- Lai M, Demuru M, Hillebrand A, Fraschini M. A comparison between scalp- and source-reconstructed EEG networks. Sci Rep. 2018;8:12269. doi: 10.1038/s41598-018-30869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Li J, Chen Y, Chen W, Li W, Zhao F, Chen G, Liu J, Chen Y, Li Y, Wang C-D, Zheng Y, Cai Y. Alterations of brain activity and functional connectivity in transition from acute to chronic tinnitus. Hum Brain Mapp. 2020;42(2):485–494. doi: 10.1002/hbm.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam S, Ranta R, Caune V, Korats G, Koessler L, Maillard L, Louis-Dorr V. SEEG dipole source localization based on an empirical Bayesian approach taking into account forward model uncertainties. Neuroimage. 2017;153:1–15. doi: 10.1016/j.neuroimage.2017.03.030. [DOI] [PubMed] [Google Scholar]
- Lee VK, Harris LT. How social cognition can inform social decision making. Front Neurosci. 2013;7:259. doi: 10.3389/fnins.2013.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz K, Geier C, Rings T, Stahn K. Capturing time-varying brain dynamics. EPJ Nonlinear Biomed Phys. 2017;5:2. doi: 10.1051/epjnbp/2017001. [DOI] [Google Scholar]
- Li W, Li Y, Zhu W, Chen X. Changes in brain functional network connectivity after stroke. Neural Regen Res. 2014;9:51–60. doi: 10.4103/1673-5374.125330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Liu T, Wang F, Li H, Gong D, Zhang R, Jiang Y, Tian Y, Guo D, Yao D, Xu P. Relationships between the resting-state network and the P3: evidence from a scalp EEG study. Sci Rep. 2015;5:15129. doi: 10.1038/srep15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tian Y, Zhang Y, Qiu K, Tian C, Jing W, Liu T, Xia Y, Guo D, Yao D, Xu P. The enhanced information flow from visual cortex to frontal area facilitates SSVEP response: evidence from model-driven and data-driven causality analysis. Sci Rep. 2015;5:14765. doi: 10.1038/srep14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen B, Li H, Zhang T, Wang F, Jiang Y, Li P, Ma T, Zhang R, Tian Y, Liu T, Guo D, Yao D, Xu P. The time-varying networks in P300: a task-evoked EEG study. IEEE Trans Neural Syst Rehabil Eng. 2016;24:725–733. doi: 10.1109/TNSRE.2016.2523678. [DOI] [PubMed] [Google Scholar]
- Li P, Huang X, Li F, Wang X, Zhou W, Liu H, Ma T, Zhang T, Guo D, Yao D, Xu P. Robust Granger analysis in Lp norm space for directed EEG network analysis. IEEE Trans Neural Syst Rehabil Eng. 2017;25:1959–1969. doi: 10.1109/TNSRE.2017.2711264. [DOI] [PubMed] [Google Scholar]
- Li F, Yi C, Jiang Y, Liao Y, Si Y, Dai J, Yao D, Zhang Y, Xu P. Different contexts in the Oddball paradigm induce distinct brain networks in generating the P300. Front Hum Neurosci. 2018;12:520. doi: 10.3389/fnhum.2018.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Z, He H. Hierarchical convolutional neural networks for EEG-based emotion recognition. Cogn Comput. 2018;10:368–380. doi: 10.1007/s12559-017-9533-x. [DOI] [Google Scholar]
- Li P, Huang X, Zhu X, Liu H, Zhou W, Yao D, Xu P. Lp (p ≤ 1) norm partial directed coherence for directed network analysis of scalp EEGs. Brain Topogr. 2018;31:738–752. doi: 10.1007/s10548-018-0624-0. [DOI] [PubMed] [Google Scholar]
- Li F, Yi C, Jiang Y, Liao Y, Si Y, Yao D, Zhang Y, Xu P. The construction of large-scale cortical networks for P300 from scalp EEG. IEEE Access. 2018;6:68498–68506. doi: 10.1109/ACCESS.2018.2879487. [DOI] [Google Scholar]
- Li F, Wang J, Jiang Y, Si Y, Peng W, Song L, Jiang Y, Zhang Y, Dong W, Yao D, Xu P. Top-down disconnectivity in Schizophrenia during P300 tasks. Front Comput Neurosci. 2018;12:33. doi: 10.3389/fncom.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Yi C, Song L, Jiang Y, Peng W, Si Y, Zhang T, Zhang R, Yao D, Zhang Y, Xu P. Brain network reconfiguration during motor imagery revealed by a large-scale network analysis of Scalp EEG. Brain Topogr. 2019;32:304–314. doi: 10.1007/s10548-018-0688-x. [DOI] [PubMed] [Google Scholar]
- Li F, Wang J, Liao Y, Yi C, Jiang Y, Si Y, Peng W, Yao D, Zhang Y, Dong W, Xu P. Differentiation of schizophrenia by combining the spatial EEG brain network patterns of rest and task P300. IEEE Trans Neural Syst Rehabil Eng. 2019;27:594–602. doi: 10.1109/TNSRE.2019.2900725. [DOI] [PubMed] [Google Scholar]
- Li P, Liu H, Si Y, Li C, Li F, Zhu X, Huang X, Zeng Y, Yao D, Zhang Y, Xu P. EEG based emotion recognition by combining functional connectivity network and local activations. IEEE Trans Biomed Eng. 2019;66(10):2869–2881. doi: 10.1109/TBME.2019.2897651. [DOI] [PubMed] [Google Scholar]
- Li F, Peng W, Jiang Y, Song L, Liao Y, Yi C, Zhang L, Si Y, Zhang T, Wang F, Zhang R, Tian Y, Zhang Y, Yao D, Xu P. The dynamic brain networks of motor imagery: time-varying causality analysis of scalp EEG. Int J Neural Syst. 2019;29:1850016. doi: 10.1142/S0129065718500168. [DOI] [PubMed] [Google Scholar]
- Li F, Liang Y, Zhang L, Yi C, Liao Y, Jiang Y, Si Y, Zhang Y, Yao D, Yu L, Xu P. Transition of brain networks from an interictal to a preictal state preceding a seizure revealed by scalp EEG network analysis. Cogn Neurodyn. 2019;13:175–181. doi: 10.1007/s11571-018-09517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang L, Zhang F, Gu R, Peng W, Hu L. Demystifying signal processing techniques to extract resting-state EEG features for psychologists. Brain Sci Adv. 2020;6:189–209. doi: 10.26599/BSA.2020.9050019. [DOI] [Google Scholar]
- Li X, Mota B, Kondo T, Nasuto S, Hayashi Y. EEG dynamical network analysis method reveals the neural signature of visual-motor coordination. PLoS ONE. 2020;15:e0231767. doi: 10.1371/journal.pone.0231767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tao Q, Peng W, Zhang T, Si Y, Zhang Y, Yi C, Biswal B, Yao D, Xu P. Inter-subject P300 variability relates to the efficiency of brain networks reconfigured from resting- to task-state: evidence from a simultaneous event-related EEG-fMRI study. Neuroimage. 2020;205:116285. doi: 10.1016/j.neuroimage.2019.116285. [DOI] [PubMed] [Google Scholar]
- Li F, Cao Z, Xu P, Yi C, Liao Y, Jiang Y, Si Y, Song L, Zhang T, Yao D, Zhang Y. Reconfiguration of brain network between resting-state and P300 task. IEEE Trans Cogn Dev Syst. 2020 doi: 10.1109/TCDS.2020.2965135. [DOI] [Google Scholar]
- Liang X, Wang J, Yan C, Shu N, Xu K, Gong G, He Y. Effects of different correlation metrics and preprocessing factors on small-world brain functional networks: a resting-state functional MRI study. PLoS ONE. 2012;7:e32766. doi: 10.1371/journal.pone.0032766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Choi K-S, Qin J, Wang Q, Pang W-M, Heng P-A. Discrimination of motor imagery tasks via information flow pattern of brain connectivity. Technol Health Care. 2016;24(Suppl 2):S795–801. doi: 10.3233/THC-161212. [DOI] [PubMed] [Google Scholar]
- Lin N, Yang X, Li J, Wang S, Hua H, Ma Y, Li X. Neural correlates of three cognitive processes involved in theory of mind and discourse comprehension. Cogn Affect Behav Neurosci. 2018;18:273–283. doi: 10.3758/s13415-018-0568-6. [DOI] [PubMed] [Google Scholar]
- Lindquist KA, Wager TD, Kober H, Bliss-Moreau E, Barrett LF. The brain basis of emotion: a meta-analytic review. Behav Brain Sci. 2012;35:121–143. doi: 10.1017/S0140525X11000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Garrido M, Zeidman P, Friston K. Empirical Bayes for group (DCM) studies: a reproducibility study. Front Hum Neurosci. 2015;9:670. doi: 10.3389/fnhum.2015.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang P. Phase synchronization dynamics of neural network during seizures. Comput Math Methods Med. 2018;2018:1354915. doi: 10.1155/2018/1354915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li M, Pan Y, Lan W, Zheng R, Wu F-X, Wang J. Complex brain network analysis and its applications to brain disorders: a survey. Complexity. 2017;2017:1–27. doi: 10.1155/2017/8362741. [DOI] [Google Scholar]
- Liu T, Li F, Jiang Y, Zhang T, Wang F, Gong D, Li P, Ma T, Qiu K, Li H, Yao D, Xu P. Cortical dynamic causality network for auditory-motor tasks. IEEE Trans Neural Syst Rehabil Eng. 2017;25:1. doi: 10.1109/TNSRE.2016.2608359. [DOI] [PubMed] [Google Scholar]
- Liu Q, Farahibozorg S, Porcaro C, Wenderoth N, Mantini D. Detecting large-scale networks in the human brain using high-density electroencephalography. Hum Brain Mapp. 2017;38:4631–4643. doi: 10.1002/hbm.23688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang J, Dong X, Li Z, Shi X, Tong Y, Yang R, Wu J, Wang C, Yan T. Occipital alpha connectivity during resting-state electroencephalography in patients with ultra-high risk for psychosis and Schizophrenia. Front Psychiatry. 2019;10:553. doi: 10.3389/fpsyt.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann G, Margulies DS, Horstmann A, Pleger B, Lepsien J, Goldhahn D, Schloegl H, Stumvoll M, Villringer A, Turner R. Eigenvector centrality mapping for analyzing connectivity patterns in fMRI data of the human brain. PLoS ONE. 2010;5:e10232. doi: 10.1371/journal.pone.0010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López JD, Litvak V, Espinosa JJ, Friston K, Barnes GR. Algorithmic procedures for Bayesian MEG/EEG source reconstruction in SPM. Neuroimage. 2014;84:476–487. doi: 10.1016/j.neuroimage.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Li Y, Chen M, Kim H, Serikawa S. Brain intelligence: go beyond artificial intelligence. Mobile Netw Appl. 2018;23:368–375. doi: 10.1007/s11036-017-0932-8. [DOI] [Google Scholar]
- Lynn CW, Bassett DS. The physics of brain network structure, function, and control. Nat Rev Phys. 2019;1:318–332. doi: 10.1038/s42254-019-0040-8. [DOI] [Google Scholar]
- Mackie M-A, van Dam NT, Fan J. Cognitive control and attentional functions. Brain Cogn. 2013;82:301–312. doi: 10.1016/j.bandc.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharathi B, Loeb JA, Patton J. Estimation of resting state effective connectivity in epilepsy using direct-directed transfer function. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:716–719. doi: 10.1109/EMBC.2016.7590802. [DOI] [PubMed] [Google Scholar]
- Mash LE, Linke AC, Olson LA, Fishman I, Liu TT, Müller R-A. Transient states of network connectivity are atypical in autism: a dynamic functional connectivity study. Hum Brain Mapp. 2019;40:2377–2389. doi: 10.1002/hbm.24529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzenich MM, van Vleet TM, Nahum M. Brain plasticity-based therapeutics. Front Hum Neurosci. 2014;8:385. doi: 10.3389/fnhum.2014.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM. From sensation to cognition. Brain. 1998;121(Pt 6):1013–1052. doi: 10.1093/brain/121.6.1013. [DOI] [PubMed] [Google Scholar]
- Michel CM, Brunet D. EEG Source imaging: a practical review of the analysis steps. Front Neurol. 2019;10:325. doi: 10.3389/fneur.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelini G, Jurgiel J, Bakolis I, Cheung CHM, Asherson P, Loo SK, Kuntsi J, Mohammad-Rezazadeh I. Atypical functional connectivity in adolescents and adults with persistent and remitted ADHD during a cognitive control task. Transl Psychiatry. 2019;9:137. doi: 10.1038/s41398-019-0469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohagheghian F, Makkiabadi B, Jalilvand H, Khajehpoor H, Samadzadehaghdam N, Eqlimi E, Deevband MR. Computer-aided tinnitus detection based on brain network analysis of EEG functional connectivity. J Biomed Phys Eng. 2019;9:687–698. doi: 10.31661/jbpe.v0i0.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr H, Wolfensteller U, Betzel RF, Mišić B, Sporns O, Richiardi J, Ruge H. Integration and segregation of large-scale brain networks during short-term task automatization. Nat Commun. 2016;7:13217. doi: 10.1038/ncomms13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S-E, Chen C-J, Hsieh C-J, Wang J-L, Lee J-S. Emotional EEG classification using connectivity features and convolutional neural networks. Neural Netw. 2020;132:96–107. doi: 10.1016/j.neunet.2020.08.009. [DOI] [PubMed] [Google Scholar]
- Morenko A. Brain processes during the perception of sensory signals in men with high and low output α-frequencies. Ann Neurosci. 2014;21:144–149. doi: 10.5214/ans.0972.7531.210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti DV. Electroencephalography-driven approach to prodromal Alzheimer's disease diagnosis: from biomarker integration to network-level comprehension. Clin Interv Aging. 2016;11:897–912. doi: 10.2147/CIA.S103313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser DA, Doucet GE, Ing A, Dima D, Schumann G, Bilder RM, Frangou S. An integrated brain-behavior model for working memory. Mol Psychiatry. 2018;23:1974–1980. doi: 10.1038/mp.2017.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gutiérrez PA, Giraldo E, Bueno-López M, Molinas M. Localization of active brain sources from EEG signals using empirical mode decomposition: a comparative study. Front Integr Neurosci. 2018;12:55. doi: 10.3389/fnint.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim-Feil J, Rubinson M, Freche D, Grinshpoon A, Peled A, Moses E, Levit-Binnun N. Altered brain network dynamics in Schizophrenia: a cognitive electroencephalography study. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3:88–98. doi: 10.1016/j.bpsc.2017.03.017. [DOI] [PubMed] [Google Scholar]
- Nani A, Manuello J, Mancuso L, Liloia D, Costa T, Cauda F. The neural correlates of consciousness and attention: two sister processes of the brain. Front Neurosci. 2019;13:1169. doi: 10.3389/fnins.2019.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte G, Bai O, Wheaton L, Mari Z, Vorbach S, Hallett M. Identifying true brain interaction from EEG data using the imaginary part of coherency. Clin Neurophysiol. 2004;115:2292–2307. doi: 10.1016/j.clinph.2004.04.029. [DOI] [PubMed] [Google Scholar]
- Nowrangi MA, Lyketsos C, Rao V, Munro CA. Systematic review of neuroimaging correlates of executive functioning: converging evidence from different clinical populations. J Neuropsychiatry Clin Neurosci. 2014;26:114–125. doi: 10.1176/appi.neuropsych.12070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejarczyk E, Marzetti L, Pizzella V, Zappasodi F. Comparison of connectivity analyses for resting state EEG data. J Neural Eng. 2017;14:36017. doi: 10.1088/1741-2552/aa6401. [DOI] [PubMed] [Google Scholar]
- Oosugi N, Kitajo K, Hasegawa N, Nagasaka Y, Okanoya K, Fujii N. A new method for quantifying the performance of EEG blind source separation algorithms by referencing a simultaneously recorded ECoG signal. Neural Netw. 2017;93:1–6. doi: 10.1016/j.neunet.2017.01.005. [DOI] [PubMed] [Google Scholar]
- O'Regan JK, Noë A. A sensorimotor account of vision and visual consciousness. Behav Brain Sci. 2001;24:939–73. doi: 10.1017/S0140525X01000115. [DOI] [PubMed] [Google Scholar]
- O'Reilly C, Lewis JD, Elsabbagh M. Is functional brain connectivity atypical in autism? A systematic review of EEG and MEG studies. PLoS ONE. 2017;12:e0175870. doi: 10.1371/journal.pone.0175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolani O, Conti A, Di Filippo A, Adembri C, Moraldi E, Evangelisti A, Maggini M, Roberts SJ. EEG signal processing in anaesthesia. Use of a neural network technique for monitoring depth of anaesthesia. Br J Anaesth. 2002;88:644–648. doi: 10.1093/bja/88.5.644. [DOI] [PubMed] [Google Scholar]
- Ouyang G, Zhou C. Characterizing the brain's dynamical response from scalp-level neural electrical signals: a review of methodology development. Cogn Neurodyn. 2020;14:731–742. doi: 10.1007/s11571-020-09631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paban V, Modolo J, Mheich A, Hassan M. Psychological resilience correlates with EEG source-space brain network flexibility. Netw Neurosci. 2019;3:539–550. doi: 10.1162/netn_a_00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnotta MF, Plomp G. Time-varying MVAR algorithms for directed connectivity analysis: critical comparison in simulations and benchmark EEG data. PLoS ONE. 2018;13:e0198846. doi: 10.1371/journal.pone.0198846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou M, Friston K, Marinazzo D. Estimating directed connectivity from cortical recordings and reconstructed sources. Brain Topogr. 2019;32:741–752. doi: 10.1007/s10548-015-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, Friston KJ. The anatomy of inference: generative models and brain structure. Front Comput Neurosci. 2018;12:90. doi: 10.3389/fncom.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci D, Rubega M, Plomp G. Modeling time-varying brain networks with a self-tuning optimized Kalman filter. PLoS Comput Biol. 2019 doi: 10.1101/856179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny W, Iglesias-Fuster J, Quiroz YT, Lopera FJ, Bobes MA. Dynamic causal dodeling of preclinical autosomal-dominant Alzheimer's disease. J Alzheimers Dis. 2018;65:697–711. doi: 10.3233/JAD-170405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SE, Sporns O. Brain networks and cognitive architectures. Neuron. 2015;88:207–219. doi: 10.1016/j.neuron.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poil S-S, de Haan W, van der Flier WM, Mansvelder HD, Scheltens P, Linkenkaer-Hansen K. Integrative EEG biomarkers predict progression to Alzheimer's disease at the MCI stage. Front Aging Neurosci. 2013;5:58. doi: 10.3389/fnagi.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera AJ. Sensation and perception. In: Biswas-Diener R, Diener E, editors. Noba textbook series: PSYCHOLOGY. Champaign: DEF Publisher; 2020. [Google Scholar]
- Rabinovich MI, Zaks MA, Varona P. Sequential dynamics of complex networks in mind: consciousness and creativity. Phys Rep. 2020;883:1–32. doi: 10.1016/j.physrep.2020.08.003. [DOI] [Google Scholar]
- Raghu S, Sriraam N, Temel Y, Rao SV, Kubben PL. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw. 2020;124:202–212. doi: 10.1016/j.neunet.2020.01.017. [DOI] [PubMed] [Google Scholar]
- Rizkallah J, Benquet P, Kabbara A, Dufor O, Wendling F, Hassan M. Dynamic reshaping of functional brain networks during visual object recognition. J Neural Eng. 2018;15:56022. doi: 10.1088/1741-2552/aad7b1. [DOI] [PubMed] [Google Scholar]
- Rizkallah J, Annen J, Modolo J, Gosseries O, Benquet P, Mortaheb S, Amoud H, Cassol H, Mheich A, Thibaut A, Chatelle C, Hassan M, Panda R, Wendling F, Laureys S. Decreased integration of EEG source-space networks in disorders of consciousness. Neuroimage Clin. 2019;23:101841. doi: 10.1016/j.nicl.2019.101841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan SM. Object recognition in mental representations: directions for exploring diagnostic features through visual mental imagery. Front Psychol. 2017;8:833. doi: 10.3389/fpsyg.2017.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo RR, Segaran J, Leonard JA, Robinson ST, West MR, Mackey AP, Yendiki A, Rowe ML, Gabrieli JDE. Language exposure relates to structural neural connectivity in childhood. J Neurosci. 2018;38:7870–7877. doi: 10.1523/JNEUROSCI.0484-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubega M, Pascucci D, Queralt JR, van Mierlo P, Hagmann P, Plomp G, Michel CM. Time-varying effective EEG source connectivity: the optimization of model parameters. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:6438–6441. doi: 10.1109/EMBC.2019.8856890. [DOI] [PubMed] [Google Scholar]
- Saeedi A, Saeedi M, Maghsoudi A, Shalbaf A. Major depressive disorder diagnosis based on effective connectivity in EEG signals: a convolutional neural network and long short-term memory approach. Cogn Neurodyn. 2021 doi: 10.1007/s11571-020-09619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman CD, Fusi S. Emotion, cognition, and mental state representation in amygdala and prefrontal cortex. Annu Rev Neurosci. 2010;33:173–202. doi: 10.1146/annurev.neuro.051508.135256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Bornot JM, Wong-Lin K, Ahmad AL, Prasad G. Robust EEG/MEG based functional connectivity with the envelope of the imaginary coherence: sensor space analysis. Brain Topogr. 2018;31:895–916. doi: 10.1007/s10548-018-0640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarter M, Givens B, Bruno JP. The cognitive neuroscience of sustained attention: where top-down meets bottom-up. Brain Res Rev. 2001;35:146–160. doi: 10.1016/S0165-0173(01)00044-3. [DOI] [PubMed] [Google Scholar]
- Savage N. How AI and neuroscience drive each other forwards. Nature. 2019;571:S15–S17. doi: 10.1038/d41586-019-02212-4. [DOI] [PubMed] [Google Scholar]
- Schirrmeister RT, Springenberg JT, Fiederer LDJ, Glasstetter M, Eggensperger K, Tangermann M, Hutter F, Burgard W, Ball T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum Brain Mapp. 2017;38:5391–5420. doi: 10.1002/hbm.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DH, Cole MW. Higher intelligence is associated with less task-related brain network reconfiguration. J Neurosci. 2016;36:8551–8561. doi: 10.1523/JNEUROSCI.0358-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber M, Cantonas L-M, Hoevels M, Sesia T, Visser-Vandewalle V, Michel CM. Subcortical electrophysiological activity is detectable with high-density EEG source imaging. Nat Commun. 2019;10:753. doi: 10.1038/s41467-019-08725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta B, Friston KJ, Penny WD. Efficient gradient computation for dynamical models. Neuroimage. 2014;98:521–527. doi: 10.1016/j.neuroimage.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapshak P. Artificial intelligence and brain. Bioinformation. 2018;14:38–41. doi: 10.6026/97320630014038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim M, Im C-H, Kim Y-W, Lee S-H. Altered cortical functional network in major depressive disorder: a resting-state electroencephalogram study. Neuroimage Clin. 2018;19:1000–1007. doi: 10.1016/j.nicl.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine JM, Poldrack RA. Principles of dynamic network reconfiguration across diverse brain states. Neuroimage. 2018;180:396–405. doi: 10.1016/j.neuroimage.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Si Y, Wu X, Li F, Zhang L, Duan K, Li P, Song L, Jiang Y, Zhang T, Zhang Y, Chen J, Gao S, Biswal B, Yao D, Xu P. Different decision-making responses occupy different brain networks for information processing: a study based on EEG and TMS. Cereb Cortex. 2019;29:4119–4129. doi: 10.1093/cercor/bhy294. [DOI] [PubMed] [Google Scholar]
- Si Y, Li F, Duan K, Tao Q, Li C, Cao Z, Zhang Y, Biswal B, Li P, Yao D, Xu P. Predicting individual decision-making responses based on single-trial EEG. Neuroimage. 2020;206:116333. doi: 10.1016/j.neuroimage.2019.116333. [DOI] [PubMed] [Google Scholar]
- Siegel JS, Ramsey LE, Snyder AZ, Metcalf NV, Chacko RV, Weinberger K, Baldassarre A, Hacker CD, Shulman GL, Corbetta M. Disruptions of network connectivity predict impairment in multiple behavioral domains after stroke. Proc Natl Acad Sci USA. 2016;113:E4367–E4376. doi: 10.1073/pnas.1521083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siew CSQ, Wulff DU, Beckage NM, Kenett YN. Cognitive network science: a review of research on cognition through the lens of network representations, processes, and dynamics. Complexity. 2019;2019:1–24. doi: 10.1155/2019/2108423. [DOI] [Google Scholar]
- Sigman M, Dehaene S. Brain mechanisms of serial and parallel processing during dual-task performance. J Neurosci. 2008;28:7585–7598. doi: 10.1523/JNEUROSCI.0948-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simony E, Honey CJ, Chen J, Lositsky O, Yeshurun Y, Wiesel A, Hasson U. Dynamic reconfiguration of the default mode network during narrative comprehension. Nat Commun. 2016;7:12141. doi: 10.1038/ncomms12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP. Magnetoencephalography: basic principles. Ann Indian Acad Neurol. 2014;17:S107–S112. doi: 10.4103/0972-2327.128676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore AE, Bassett DS. Dynamic graph metrics: tutorial, toolbox, and tale. Neuroimage. 2018;180:417–427. doi: 10.1016/j.neuroimage.2017.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockeel S, Schwartz D, Pélégrini-Issac M, Benali H. Large-scale functional networks identified from resting-state EEG using spatial ICA. PLoS ONE. 2016;11:e0146845. doi: 10.1371/journal.pone.0146845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Lin H, Liu C, Jiang Y, Lin Y, Xue Q, Xu P, Wang Y. Transcranial magnetic stimulation to the middle frontal gyrus during attention modes induced dynamic module reconfiguration in brain networks. Front Neuroinform. 2019;13:22. doi: 10.3389/fninf.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Friston KJ. Analyzing effective connectivity with fMRI. Wiley Interdiscip Rev Cogn Sci. 2010;1:446–459. doi: 10.1002/wcs.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens FL, Hurley RA, Taber KH. Anterior cingulate cortex: unique role in cognition and emotion. J Neuropsychiatry Clin Neurosci. 2011;23:121–125. doi: 10.1176/jnp.23.2.jnp121. [DOI] [PubMed] [Google Scholar]
- Talebi N, Nasrabadi AM, Mohammad-Rezazadeh I. Estimation of effective connectivity using multi-layer perceptron artificial neural network. Cogn Neurodyn. 2018;12:21–42. doi: 10.1007/s11571-017-9453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallon-Baudry C. On the neural mechanisms subserving consciousness and attention. Front Psychol. 2011;2:397. doi: 10.3389/fpsyg.2011.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CY, Eaves EL, Ng JC, Carpenter DM, Mai X, Schroeder DH, Condon CA, Colom R, Haier RJ. Brain networks for working memory and factors of intelligence assessed in males and females with fMRI and DTI. Intelligence. 2010;38:293–303. doi: 10.1016/j.intell.2010.03.003. [DOI] [Google Scholar]
- Tanimizu T, Kono K, Kida S. Brain networks activated to form object recognition memory. Brain Res Bull. 2018;141:27–34. doi: 10.1016/j.brainresbull.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Morita A, Ninomiya S, Akimoto T, Shiota H, Kamei S. Relation between resting state front-parietal EEG coherence and executive function in Parkinson's disease. Biomed Res Int. 2016;2016:2845754. doi: 10.1155/2016/2845754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF, Kim JJ. Memory systems in the brain and localization of a memory. Proc Natl Acad Sci USA. 1996;93:13438–13444. doi: 10.1073/pnas.93.24.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Ma W, Tian C, Xu P, Yao D. Brain oscillations and electroencephalography scalp networks during tempo perception. Neurosci Bull. 2013;29:731–736. doi: 10.1007/s12264-013-1352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppi J, Astolfi L, Poudel GR, Innes CRH, Babiloni F, Jones RD. Time-varying effective connectivity of the cortical neuroelectric activity associated with behavioural microsleeps. Neuroimage. 2016;124:421–432. doi: 10.1016/j.neuroimage.2015.08.059. [DOI] [PubMed] [Google Scholar]
- Tsolaki A, Kazis D, Kompatsiaris I, Kosmidou V, Tsolaki M. Electroencephalogram and Alzheimer's disease: clinical and research approaches. Int J Alzheimers Dis. 2014;2014:349249. doi: 10.1155/2014/349249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Steen F, Almgren H, Razi A, Friston K, Marinazzo D. Dynamic causal modelling of fluctuating connectivity in resting-state EEG. Neuroimage. 2019;189:476–484. doi: 10.1016/j.neuroimage.2019.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. A cross-disorder connectome landscape of brain dysconnectivity. Nat Rev Neurosci. 2019;20:435–446. doi: 10.1038/s41583-019-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velde F, de Kamps M. Learning of control in a neural architecture of grounded language processing. Cogn Syst Res. 2010;11:93–107. doi: 10.1016/j.cogsys.2008.08.007. [DOI] [Google Scholar]
- van der Meij R, van Ede F, Maris E. Rhythmic components in extracranial brain signals reveal multifaceted task modulation of overlapping neuronal activity. PLoS ONE. 2016;11:e0154881. doi: 10.1371/journal.pone.0154881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duinkerken E, Schoonheim MM, IJzerman RG, Moll AC, Landeira-Fernandez J, Klein M, Diamant M, Snoek FJ, Barkhof F, Wink A-M. Altered eigenvector centrality is related to local resting-state network functional connectivity in patients with longstanding type 1 diabetes mellitus. Hum Brain Mapp. 2017;38:3623–3636. doi: 10.1002/hbm.23617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre D, Smith SM, Woolrich MW. Brain network dynamics are hierarchically organized in time. Proc Natl Acad Sci USA. 2017;114:12827–12832. doi: 10.1073/pnas.1705120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahbeh H, Goodrich E, Goy E, Oken BS. Mechanistic pathways of mindfulness meditation in Combat Veterans with posttraumatic stress disorder. J Clin Psychol. 2016;72:365–383. doi: 10.1002/jclp.22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-J, Hsieh I-F, Chen C-C. Accelerating computation of DCM for ERP in MATLAB by external function calls to the GPU. PLoS ONE. 2013;8:e66599. doi: 10.1371/journal.pone.0066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chung MK, Dentico D, Lutz A, Davidson R. Topological network analysis of electroencephalographic power maps. Connect Neuroimaging. 2017;10511:134–142. doi: 10.1007/978-3-319-67159-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Zhang D, Liang B, Zhang R, Wang Z, Wang J, Liu M, Huang R. Reconfiguration of the brain functional network associated with visual task demands. PLoS ONE. 2015;10:e0132518. doi: 10.1371/journal.pone.0132518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS. Segregated systems of human brain networks. Trends Cogn Sci. 2017;21:981–996. doi: 10.1016/j.tics.2017.09.006. [DOI] [PubMed] [Google Scholar]
- Williams NJ, Daly I, Nasuto SJ. Markov model-based method to analyse time-varying networks in EEG task-related data. Front Comput Neurosci. 2018;12:76. doi: 10.3389/fncom.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf D, Nagarajan S. A unified Bayesian framework for MEG/EEG source imaging. Neuroimage. 2009;44:947–966. doi: 10.1016/j.neuroimage.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang J, Chen M, Li S, Zhang Z, Kang C, Ding G, Guo T. Brain network reconfiguration for language and domain-general cognitive control in bilinguals. Neuroimage. 2019;199:454–465. doi: 10.1016/j.neuroimage.2019.06.022. [DOI] [PubMed] [Google Scholar]
- Xu P, Xiong X, Xue Q, Li P, Zhang R, Wang Z, Valdes-Sosa PA, Wang Y, Yao D. Differentiating between psychogenic nonepileptic seizures and epilepsy based on common spatial pattern of weighted EEG resting networks. IEEE Trans Biomed Eng. 2014;61:1747–1755. doi: 10.1109/TBME.2014.2305159. [DOI] [PubMed] [Google Scholar]
- Xu P, Xiong XC, Xue Q, Tian Y, Peng Y, Zhang R, Li PY, Wang YP, Yao DZ. Recognizing mild cognitive impairment based on network connectivity analysis of resting EEG with zero reference. Physiol Meas. 2014;35:1279–1298. doi: 10.1088/0967-3334/35/7/1279. [DOI] [PubMed] [Google Scholar]
- Xue Q, Wang Z-Y, Xiong X-C, Tian C-Y, Wang Y-P, Xu P. Altered brain connectivity in patients with psychogenic non-epileptic seizures: a scalp electroencephalography study. J Int Med Res. 2013;41:1682–1690. doi: 10.1177/0300060513496170. [DOI] [PubMed] [Google Scholar]
- Yantis S. The neural basis of selective attention: cortical sources and targets of attentional modulation. Curr Dir Psychol Sci. 2008;17:86–90. doi: 10.1111/j.1467-8721.2008.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Raman SS, Schiek M, Leff A, Frässle S, Stephan KE. Variational Bayesian inversion for hierarchical unsupervised generative embedding (HUGE) Neuroimage. 2018;179:604–619. doi: 10.1016/j.neuroimage.2018.06.073. [DOI] [PubMed] [Google Scholar]
- Ye S, Kitajo K, Kitano K. Information-theoretic approach to detect directional information flow in EEG signals induced by TMS. Neurosci Res. 2020;156:197–205. doi: 10.1016/j.neures.2019.09.003. [DOI] [PubMed] [Google Scholar]
- Yi G-S, Wang J, Deng B, Wei X-L. Complexity of resting-state EEG activity in the patients with early-stage Parkinson's disease. Cogn Neurodyn. 2017;11:147–160. doi: 10.1007/s11571-016-9415-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Chen C, Jiang L, Tao Q, Li F, Si Y, Zhang T, Yao D, Xu P. Constructing EEG large-scale cortical functional network connectivity based on brain atlas by S estimator. IEEE Trans Cogn Dev Syst. 2020 doi: 10.1109/TCDS.2020.2991414. [DOI] [Google Scholar]
- Yin Z, Li J, Zhang Y, Ren A, von Meneen KM, Huang L. Functional brain network analysis of schizophrenic patients with positive and negative syndrome based on mutual information of EEG time series. Biomed Sig Process and Contr. 2017;31:331–338. doi: 10.1016/j.bspc.2016.08.013. [DOI] [Google Scholar]
- Zeng K, Kang J, Ouyang G, Li J, Han J, Wang Y, Sokhadze EM, Casanova MF, Li X. Disrupted brain network in children with autism spectrum disorder. Sci Rep. 2017;7:16253. doi: 10.1038/s41598-017-16440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yang C, Dai G, Qin F, Zhang J, Kong W. EEG classification of driver mental states by deep learning. Cogn Neurodyn. 2018;12:597–606. doi: 10.1007/s11571-018-9496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu P, Guo D, Yao D. Prediction of SSVEP-based BCI performance by the resting-state EEG network. J Neural Eng. 2013;10:66017. doi: 10.1088/1741-2560/10/6/066017. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu P, Huang Y, Cheng K, Yao D. SSVEP response is related to functional brain network topology entrained by the flickering stimulus. PLoS ONE. 2013;8:e72654. doi: 10.1371/journal.pone.0072654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo D, Cheng K, Yao D, Xu P. The graph theoretical analysis of the SSVEP harmonic response networks. Cogn Neurodyn. 2015;9:305–315. doi: 10.1007/s11571-015-9327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Liu T, Li F, Li M, Liu D, Zhang R, He H, Li P, Gong J, Luo C, Yao D, Xu P. Structural and functional correlates of motor imagery BCI performance: insights from the patterns of fronto-parietal attention network. Neuroimage. 2016;134:475–485. doi: 10.1016/j.neuroimage.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Zhang T, Li M, Zhang L, Biswal B, Yao D, Xu P. The time-varying network patterns in motor imagery revealed by adaptive directed transfer function analysis for fMRI. IEEE Access. 2018;6:60339–60352. doi: 10.1109/ACCESS.2018.2875492. [DOI] [Google Scholar]
- Zhang T, Wang F, Li M, Li F, Tan Y, Zhang Y, Yang H, Biswal B, Yao D, Xu P. Reconfiguration patterns of large-scale brain networks in motor imagery. Brain Struct Funct. 2019;224:553–566. doi: 10.1007/s00429-018-1786-y. [DOI] [PubMed] [Google Scholar]
- Zhang S, Sun J, Gao X. The effect of fatigue on brain connectivity networks. Brain Sci Adv. 2020;6:120–131. doi: 10.26599/BSA.2020.9050008. [DOI] [Google Scholar]
- Zhang R, Li F, Zhang T, Yao D, Xu P. Subject inefficiency phenomenon of motor imagery brain-computer interface: influence factors and potential solutions. Brain Sci Adv. 2020;6:224–241. doi: 10.26599/BSA.2020.9050021. [DOI] [Google Scholar]
- Zhang L, Li Z, Zhang F, Gu R, Peng W, Hu L. Demystifying signal processing techniques to extract task-related EEG responses for psychologists. Brain Sci Adv. 2020;6:171–188. doi: 10.26599/BSA.2020.9050018. [DOI] [Google Scholar]
- Zhao Q, Li H, Hu B, Wu H, Liu Q. Abstinent heroin addicts tend to take risks: ERP and source localization. Front Neurosci. 2017;11:681. doi: 10.3389/fnins.2017.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Allard A, Hagmann P, Alemán-Gómez Y, Serrano MÁ. Geometric renormalization unravels self-similarity of the multiscale human connectome. Proc Natl Acad Sci USA. 2020;117:20244–20253. doi: 10.1073/pnas.1922248117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zeidman P, Wu S, Razi A, Chen C, Yang L, Zou J, Wang G, Wang H, Friston KJ. Altered intrinsic and extrinsic connectivity in schizophrenia. Neuroimage Clin. 2018;17:704–716. doi: 10.1016/j.nicl.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuge H, Zhang J. Topological centrality and its e-Science applications. J Am Soc Inf Sci. 2010;61:1824–2184. doi: 10.1002/asi.21353. [DOI] [Google Scholar]
- Zuo N, Yang Z, Liu Y, Li J, Jiang T. Core networks and their reconfiguration patterns across cognitive loads. Hum Brain Mapp. 2018;39(9):3546–3557. doi: 10.1002/hbm.24193. [DOI] [PMC free article] [PubMed] [Google Scholar]