Dear Editor,
Lipids are essential for all life on earth and play critical roles in energy storage and the formation of cellular membranes. Sphingosine-1-phosphate (S1P) is a naturally occurring bioactive lysophospholipid that regulates fundamental physiological processes by activating five G protein-coupled receptors (S1PR1–S1PR5). S1PRs are widely distributed on many cell types, including cells in the cardiovascular, immune, and central nervous systems.1 The multifunctional S1P-S1PR signaling is a driver of multiple diseases and S1PR1-targeted drugs have been approved as therapeutic strategies for the treatment of multiple sclerosis (MS) and autoimmune disorders. The first approved drug fingolimod (FTY720), after in-vivo phosphorylation to pFTY720, has cross reactivity with the S1PR family (S1PR1–3 and S1PR5), and subtype-selective modulators have been pursued as potential therapeutics. Siponimod is selective for S1PR1 and S1PR5 and has been approved for the treatment of secondary progressive MS. Molecules that lack S1PR3-targeting activity may be a useful strategy for the development of S1PR1 agonist drugs.2 Therefore, understanding the mechanism of S1P-S1PR signaling and identifying differences in the ligand selectivity of the S1PR family may assist in the development of drugs with improved safety profiles for the control of MS, cardiovascular and autoimmune disorders.
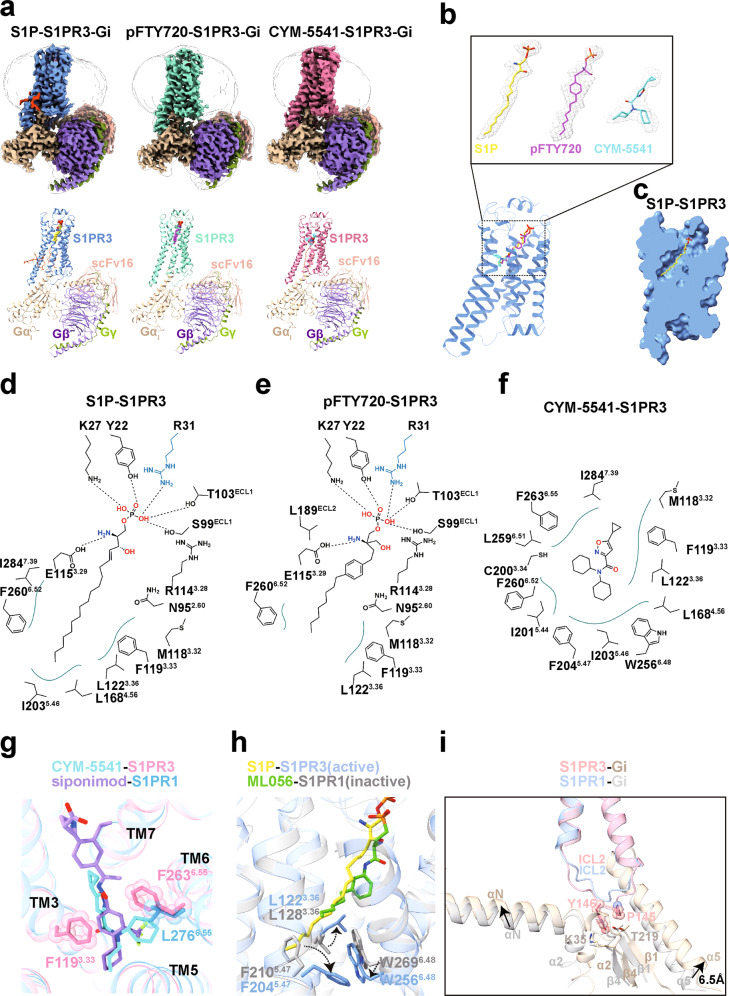
Here, we determined three cryo-electron microscopy (EM) structures of S1PR3–Gi complexes bound to the endogenous ligand S1P, pFTY720, and the selective agonist CMY-55413 at 3.1, 3.1, and 3.2 Å resolution, respectively (Fig. 1a, b; Supplementary information, Figs. S1–S3). The overall structures of these complexes had almost identical assembly modes to each other, which were similar to the structure of agonist-bound S1PR1–Gi complex in our companion manuscript.4 In S1PR3, the N-terminus adopts a capping helix that packs tightly against the extracellular loops, thus limiting the access of the ligands (Supplementary information, Fig. S4b). As observed in the structures of the lipid-activated GPCRs, an obvious gap between TM1 and TM7 provides the possibility for the ligands to enter the binding pocket.5 The charge distribution on the surface of S1PR3 indicated that the ligand access port is positively charged and this feature will help to position the phosphate head of ligands (Supplementary information, Fig. S4c).
Fig. 1. Structures of S1PR3–Gi complexes.
a The cryo-EM maps (upper panel) and structural models (lower panel) of endogenous lipid S1P-bound S1PR3–Gi–scFv16 complex, pFTY720-bound S1PR3–Gi–scFv16 complex and CMY-5541-bound S1PR3–Gi–scFv16 complex. The cryo-EM density maps colored according to different subunits are shown at 0.5, 0.209, and 0.473 contour levels, respectively. b Three agonists used in this study bound to orthosteric pocket of S1PR3. The cryo-EM density of the ligands presented as gray meshes allowed unambiguous identification of S1P (yellow), pFTY720 (magenta) and CYM-5541 (cyan). The density maps of the agonists are depicted at contour levels of 0.5, 0.209, and 0.473, respectively. c Cut away view of the binding pocket for lipid S1P-bound S1PR3. S1P binds deeply in a binding channel. d Diagram of S1P binding with S1PR3. The polar headgroup of S1P is observed to form polar interactions with residues from the N-terminus, ECL1 and TM3 of S1PR3. Polar interactions are highlighted as black dashed lines. e Diagram of pFTY720 binding with S1PR3. Polar interactions are highlighted as black dashed lines. f Diagram of CYM-5541 binding with S1PR3. CYM-5541 binds to the hydrophobic site of S1PR3. g Superimposition of CYM-5541-bound S1PR3 with siponimod-bound S1PR1. The ligand CYM-5541 forms direct interactions with F1193.33 and F2636.55, and as a result strengthens extracellular connection of TM bundles of S1PR3. F2636.55 in S1PR3 was replaced by L2766.55 in S1PR1. h Structural comparison of active S1PR3 and inactive S1PR1 reveals different conformations of L3.36, F5.47 and W6.48 side chains. i Structural superposition of S1PR3–Gi complex with S1PR1–Gi complex when S1PR3 and S1PR1 are aligned. The residues P145ICL2/34.52 and Y146ICL2/34.53 in S1PR3 are shown as sticks with sphere, whereas K35 and T219 in Gαi1 are shown as sticks with corresponding colors.
In the structure of the complex with the endogenous lipid S1P bound to S1PR3–Gi, the S1P molecule takes an extended conformation and makes multiple contacts with residues in the occluded orthosteric pocket of S1PR3 (Fig. 1c, d). S1P is an amphipathic molecule, and contains a polar head group and a hydrophobic acyl tail. The zwitterionic headgroup of S1P forms direct polar interactions with the surrounding residues Y22, K27 and R31 from N-terminus, S99 and T103 from ECL1, R1143.28 and E1153.29 from TM3, respectively. In addition, the acyl chain of S1P is accommodated into a deeply hydrophobic pocket, formed by TM3, TM5, TM6 and TM7 (Fig. 1d; Supplementary information, Fig. S5a). In the recently reported crystal structure of Fab-bound S1P–S1PR3 complex,6 the side-chain of residue R31 is exposed to extracellular region and forms direct interaction with Fab. In addition, the residues L3.36 and W6.48 in hydrophobic pocket are observed to display notable displacement in our structure (Supplementary information, Fig. S5b), due to Gi protein coupling. pFTY720, as a structural analog of S1P, was observed to bind in the orthosteric binding pocket in a nearly identical manner in the pFTY720–S1PR3 complex. In particular, the phosphorylated head helped to stabilize the pFTY720 recognition by S1PR3, while the phenyl ring contributed to the signal activation potency via potential hydrophobic contacts with TM3 and TM7. The rest of acyl chain of pFTY720 was extended into the deep channel, similar to the case with S1P (Fig.1e; Supplementary information, Fig. S5c). Furthermore, to validate these structural findings, we generated mutations of the residues involved in the recognition of the polar headgroup of S1P, and found that these substitutions had little effect on the receptor expression level but significantly decreased both S1P- and pFTY720-induced activation potency (Supplementary information, Fig. S5d–f). Further comparisons with the structure of siponimod-bound S1PR1, revealed that the critical residues involved in ligand headgroup recognition were highly conserved in both receptors. A notable difference is that residue R31 in S1PR3 had the potential to form an additional salt bridge with the phosphate group of S1P, whereas this arginine was absent in the equivalent position of S1PR1 (Supplementary information, Fig. S5g). Consistent with this observation, the activation effect of S1P toward S1PR3 was slightly stronger than that toward S1PR1 (Supplementary information, Fig. S5h).
Unlike the natural agonist S1P, the ligand CYM-5541, which does not have a polar headgroup, has been identified as an S1PR3 subtype-selective agonist. A previous study has demonstrated that CYM-5541 could activate S1PR3 as a full agonist.7 To examine the binding mode of CYM-5541 to S1PR3, we first determined the cryo-EM structure of the S1PR3–Gi complex bound to CYM-5541. The high-quality map allowed unambiguous modeling of CYM-5541 in the structure (Fig. 1b). CYM-5541 was observed to bind to the hydrophobic site of the receptor (Fig. 1f; Supplementary information, Fig. S6a) and the area it occupied overlapped with the acyl-chain of S1P, when compared with the S1P-bound S1PR3 structure. In addition, one of the cyclohexane-moieties of CYM-5541 was extended toward TM6, thus resulting in a considerable displacement of the side-chain of F2606.52 (Supplementary information, Fig. S6b).
The S1PR subtypes exhibit highly conserved sequence homology, particularly at the orthosteric binding pocket of S1PR1 and S1PR3. To further explore the ligand selectivity between S1PR1 and S1PR3, we performed structural superimposition of specific ligand-bound receptors. A notable difference was that L2766.55 in S1PR1 was replaced by F2636.55 in S1PR3, which resulted in a contraction of the orthosteric binding pocket in SP1R3 (Fig. 1g). We generated the F2636.55L mutant of SP1R3 as well as L2766.55F mutant of S1PR1 and performed cellular signaling assays. Consistent with previous functional studies,7,8 our results indicated that both the mutant receptors had reduced activation potency when bound to their respective receptor-selective agonists (Supplementary information, Fig. S7a, b). In addition, the F2636.55L mutant of S1PR3 displays increased potency to siponimod compared to wild-type S1PR3, and the L2766.55F mutant of S1PR1 shows increased potency to CYM-5541 compared to wild-type S1PR1, respectively (Supplementary information, Fig. S7c, d). Together, our results suggest that the key residue F/L6.55 should contribute to ligand selectivity in S1PR3 and S1PR1. The structures determined here provide a framework for understanding lipid signaling and could assist in the design and optimization of selective therapeutic modulators of S1PRs.
As indicated in our companion study,4 the polar headgroups of agonists targeting S1PR1 are believed to play an important role in receptor activation. Considering that the ligand CYM-5541 was reported to be the minimum moiety required by agonist that can activate S1PR3 so far, we investigated how CYM-5541 triggers signal transduction upon receptor activation. Our structural alignment and analysis indicated that the consensus motifs in S1PR3 adopt the nearly identical conformations as that in the activated S1PR1 (Supplementary information, Fig. S8). Interestingly, CYM-5541 appeared to tether TM3 and TM6 via its direct interactions with F1193.33 and F2636.55, respectively (Fig. 1g). This ligand-induced extracellular connecting of TM bundles thereby contributed to triggering the receptor activation upon agonist binding,9 in addition to subsequent conformational rearrangements of F5.47 and L3.36–W6.48 microswitches upon S1PR activation (Fig. 1h). Our functional assays showed that mutations of residues Y22, K27 and R31 at the N-terminal cap have little effect on the potency of CYM-5541 (Supplementary information, Fig. S6c), whereas the residues in the hydrophobic pocket play an important role in CYM-5541-induced S1PR3 activation (Supplementary information, Fig. S6d).
S1P-S1PR signaling modulates multi-organ pathophysiological processes by activating various intracellular effectors; in particular, S1PR1 couples exclusively with Gi, whereas S1PR3 signals through Gi, Gq, and G12/13 proteins. In general, the interface of S1PR3–Gi coupling resembles the overall architecture of similar GPCR–G proteins complexes.10 The main interface is mediated by the C-terminal α5 helix of the Gα with intracellular loops and the end of the TM6 of the receptor (Supplementary information, Fig. S9a). A comparison with the structures of S1PR1–Gi and S1PR5–Gi complexes indicated that the ICL2 region is folded into a distinct extended conformation in S1PR3. Particularly, the residues P145ICL2/34.52 and Y146ICL2/34.53 penetrate into an obvious groove that is formed by the αN–β1 hinge region and α2–β4 loop of the Gαi1 subunit, thus leading to a slight conformational change of the position of the αN in the S1PR3–Gi complex. A notable 6.5 Å-displacement of the extreme C-terminus of the α5-helix of Gαi1 occurred to move the C-terminus toward the TM5 of S1PR3, relative to the α5 in the structure of the S1PR1–Gi complex (Fig. 1i). In addition, the shorter ICL3 of S1PR3 stabilized the interface between the receptor and the Gi protein through van der Waals or hydrophobic contacts (Supplementary information, Fig. S9b). It appears that the ICL2 and ICL3 regions in GPCRs exhibit considerable structure and sequence diversity, and thus, the dynamic conformations of these regions in individual receptors contribute to the G protein coupling specificity.
In conclusion, the present study investigated the binding mode of the endogenous lipid S1P and revealed a detailed mechanism for the recognition of endogenous and synthetic agonists by S1PR3. Structural comparisons indicated that there were notable differences in the orthosteric sites between S1PR1 and S1PR3, which provides a basis for the ligand selectivity of these receptor subtypes. A previous study has indicated that CMY-5541 was a full agonist, as well as an allosteric modulator of S1PR3,7 herein, our structure deciphers that CMY-5541 bound to the same hydrophobic pocket as is occupied by the acyl chain of S1P in the S1PR3–Gi signaling complex. Interestingly, the structure of S1PR3 reveals more than one potential ligand-binding pockets in the orthosteric site (Supplementary information, Fig. S10). This provides a possibility to accommodate S1P in different conformations, as the alkyl chain of S1P is flexible, indicating the plasticity of the receptor. To understand the detailed allosteric mechanism of CYM-5541, additional structural information of S1PR3 bound simultaneously to CYM-5541 and S1P is required. In addition, the unusual conformation of the ICL2 region in the S1PR3–Gi complex provided evidence to understand the promiscuity and selective coupling mechanism of GPCRs with G proteins.
Supplementary information
Acknowledgements
Cryo-EM data were collected at West China Cryo-EM Center in Sichuan University and Cryo-EM Center in Southern University of Science and Technology (SUSTech), processed at Duyu High Performance Computing Center in Sichuan University. This work was supported by the Natural Science Foundation of China (3210110109 to W.Y., 3210110086 to L.C., 32070049 to Z.S.); Ministry of Science and Technology of China grant (2019YFA0508800 to Z.S.); Science and Technology department of Sichuan Province (2020YJ0208 to Z.S.).
Author contributions
W.W. and L.C. designed the expression constructs, purified the S1PR3–Gi–scFV16 complex; H.W. performed signaling assays with the assistance of Y.F.; W.W. and C.Z. prepared the final samples for data collection toward the structures with the assistance of Y.L., Y.F., X.W., H.F. and Y.C.; C.Z. prepared the cryo-EM grids, collected cryo-EM images, and performed map calculations under the supervision of Z.S.; C.Z. and L.C. built and refined the structure models under the supervision of P.F. and S.Y.; W.Y. and Z.S. supervised the overall project, and wrote the manuscript.
Data availability
All data produced or analyzed in this study are included in the main text or the supplementary materials. The cryo-EM density maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession numbers EMD-31346 and 7EW3 for the S1P–S1PR3 complex; EMD-31345 and 7EW2 for the pFTY720–S1PR1 complex; EMD-31347 and 7EW4 for the CYM-5541–S1PR1 complex.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Chang Zhao, Lin Cheng, Wei Wang, Heli Wang
Contributor Information
Wei Yan, Email: weiyan2018@scu.edu.cn.
Zhenhua Shao, Email: zhenhuashao@scu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00567-w.
References
- 1.Cartier A, Hla T. Science. 2019;366:eaar5551. doi: 10.1126/science.aar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan S, et al. ACS Med. Chem. Lett. 2013;4:333–337. doi: 10.1021/ml300396r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrero M, et al. Bioorg. Med. Chem. Lett. 2013;23:6346–6349. doi: 10.1016/j.bmcl.2013.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan, Y. et al. Cell Res. 10.1038/s41422-021-00566-x (2021).
- 5.Shao Z, et al. Nature. 2016;540:602–606. doi: 10.1038/nature20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda S, et al. Sci. Adv. 2021;7:eabf5325. doi: 10.1126/sciadv.abf5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo E, et al. ACS Chem. Biol. 2012;7:1975–1983. doi: 10.1021/cb300392z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Q, et al. Mol. Pharmacol. 2007;71:724–735. doi: 10.1124/mol.106.029223. [DOI] [PubMed] [Google Scholar]
- 9.Venkatakrishnan AJ, et al. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 10.Kang Y, et al. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data produced or analyzed in this study are included in the main text or the supplementary materials. The cryo-EM density maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession numbers EMD-31346 and 7EW3 for the S1P–S1PR3 complex; EMD-31345 and 7EW2 for the pFTY720–S1PR1 complex; EMD-31347 and 7EW4 for the CYM-5541–S1PR1 complex.