Abstract
The corneal endothelium is critical for maintaining corneal clarity by mediating hydration through barrier and pump functions. Progressive loss of corneal endothelial cells during aging has been associated with the development of Fuchs endothelial corneal dystrophy (FECD), one of the main causes of cornea-related vision loss. The mechanisms underlying FECD development remain elusive. Single-cell RNA sequencing of isolated healthy human corneas discovered 4 subpopulations of corneal endothelial cells with distinctive signatures. Unsupervised clustering analysis uncovered nuclear enriched abundant transcript 1 (NEAT1), a long non-coding RNA (lncRNA), as the top expressed gene in the C0-endothelial subpopulation, but markedly downregulated in FECD. Consistent with human corneas, a UVA-induced mouse FECD model validated the loss of NEAT1 expression. Loss of NEAT1 function by an in vivo genetic approach reproduced the exacerbated phenotype of FECD by ablating corneal endothelial cells. Conversely, gain of function by a CRISPR-activated adenoviral delivery system protected corneas from UVA-induced FECD. Our findings provide novel mechanistic insights into the development of FECD, and targeting NEAT1 offers an attractive approach for treating FECD.
Keywords: single-cell transcriptome, corneal endothelium, lncRNA NEAT1, Fuchs endothelial corneal dystrophy, CRISPR-activated delivery system
Graphical abstract
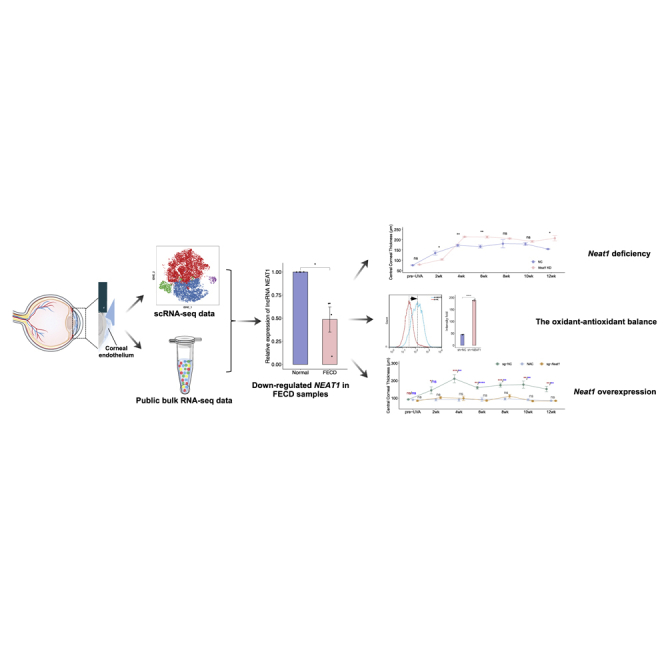
No curative therapy is available for effectively treating FECD, an age-related complex genetic disorder. Wang et al. identified NEAT1, a lncRNA strongly downregulated in FECD, as a key regulator in development of FECD, whose overexpression by the rAAV CRISPR system delayed the onset and progression of FECD.
Introduction
The cornea is a transparent avascular tissue that protects the eye against infections and contributes to focusing light on the retina. Its clarity depends on many factors, including the structural anatomy and physiology of corneal epithelium, stroma, and endothelium. The corneal endothelium exists as a monolayer of hexagonal cells situated in the posterior layer of the cornea, which plays a critical role in maintaining corneal clarity by mediating hydration through barrier and pump functions.1 Unlike mice, rabbits, and many other mammals, human corneal endothelial cells (CEnC) are post-mitotic cells that have limited proliferative capacity and typically do not proliferate in vivo; therefore, their density declines at an average of ∼0.6%/year.2,3 Damage to the corneal endothelium by several pathological conditions, such as Fuchs endothelial corneal dystrophy (FECD), and invasive cataract and glaucoma surgery can induce a decompensation of corneal endothelium and result in severe visual disturbance.4
The established view is that CEnCs are derived from cranial neural crest cells, which, following their migration into the eye, undergo a mesenchymal-epithelial transition to construct an integrated homologous monolayer. With advances in single-cell RNA sequencing (scRNA-seq),5 the single-cell transcriptional landscape has been revealed for multiple ocular tissues, including retina,6, 7, 8, 9, 10, 11, 12, 13 sclera,14 trabecular meshwork and ocular outflow,15,16 and limbal/corneal epithelium,17, 18, 19 while a comprehensive molecular atlas of corneal endothelium is still lacking. Moreover, several single-cell atlases on endothelial cells in various organs have been completed and have revealed the heterogeneous cell subpopulations that meet distinct physiological needs, such as kidneys,20 arteries, and veins.21 Although studies either directly or indirectly have shown that heterogeneity is present in cultured human CEnCs, to what extent the corneal endothelium differs in cellular state and functional commitments in vivo at the single-cell level remains largely unknown. Thus, to dissect the heterogeneity of CEnCs at the single-cell level will help reveal the unrecognized role of different cell subtypes in functional commitments, metabolic processes, and cellular associations with diseases.
The most common cause of CEnC loss and the leading indication for worldwide corneal transplantation is FECD. As the primary corneal endothelial dystrophy, FECD is an age-related complex genetic disorder with greater prevalence in females. Notably, FECD pathogenesis is hypothesized to involve an interaction between genetic and environmental factors as evidenced by the UVA (UVA) light-induced non-genetic mouse model.22 Although the characteristics of FECD are now well understood, corneal transplantation remains the only therapeutic choice. Nonetheless, several problematic issues have limited the feasibility of corneal transplantation, including shortage of donor corneas, recurrent allograft rejection, and graft failure in both acute and chronic phases. These challenges prompted researchers to seek potential novel therapeutics for the treatment of FECD.
In this study, we performed a scRNA-seq survey of 16,924 cells from 4 human corneal endothelium samples and compiled a transcriptomic atlas at an unprecedented resolution. Unbiased analyses identified 4 distinct clusters with unique transcriptional signals. Next, we used representative gene expression signatures as a guide to determine their functional commitment and metabolic heterogeneity. Furthermore, we investigated the association between FECD and pathogenic subtypes and identified a potentially pathogenetic signature with a preferential expression pattern of FECD-associated genes. Combined with bulk-seq data of FECD samples, we identified that the long non-coding RNA (lncRNA) NEAT1 was highly enriched in the FECD-associated subtype and was significantly decreased in FECD samples. We established a non-genetic FECD animal model induced by UVA and demonstrated that FECD progress was exacerbated by Neat1 knockdown (KD) and diminished by its overexpression, which implied a potential antioxidant role of Neat1 in the corneal endothelium. Overall, this study provided information on the cellular landscape of the human corneal endothelium and implicated lncRNA NEAT1 in FECD, providing critical sources and foundations for investigating the pathogenesis and potential therapeutic paradigm for corneal endothelial disease.
Results
Cellular heterogeneity of human corneal endothelium
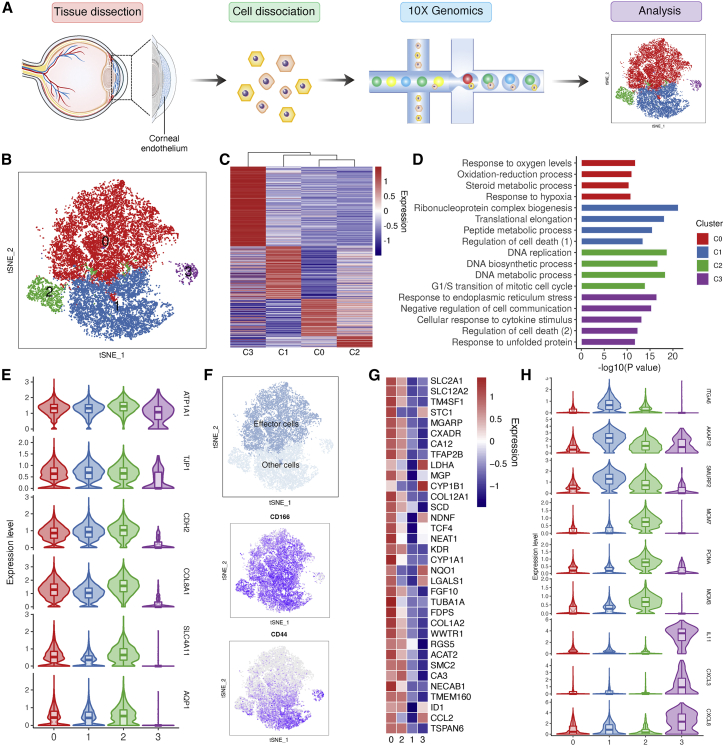
To explore the heterogeneity of human CEnCs, we isolated and sequenced cells from the corneal endothelium of healthy donors (Table S1) using the 10x Genomics platform (Figure 1A). Following preprocessing and quality control, a total of 16,942 cells passed filters with an average of 5,050 genes and 28,159 transcripts per cell (Figures S1A and S1B; Table S1). Canonical correlation analysis (CCA) was applied to remove potential batch or individual effects in the sequencing data (Figure S2). Upon unsupervised clustering, we divided the entire cell population into 4 clusters with distinct transcriptional profiles using t-distributed stochastic neighbor embedding (t-SNE) (Figures 1B and 1C; Table S2),23 and hierarchical similarities across clusters were obtained (Figure 1C). To characterize each endothelial subtype, we identified differentially expressed genes for each cluster. Gene Ontology (GO) analysis revealed that C0 was enriched in functions related to oxygen level response and oxidation-reduction process, C1 was related to translation elongation and peptide metabolism, C2 was associated with DNA replication and the cell cycle, and C3 was involved in the immune response and responses to unfolded protein (Figure 1D). For classical corneal endothelial markers, all of these clusters expressed ATP1A1 and TJP1, while for CDH2, COL8A1, SLC4A11, and AQP1, they were highly enriched in C0–C2, but diminished in C3 (Figure 1E). Although the heterogeneous cellular composition of the corneal endothelium in vivo has not been reported previously, the presence of heterogeneous cultured human CEnCs in vitro was unveiled by Kinoshita’s group, and a population called “effector cell” expressing CD166+/CD44− was identified for the treatment of corneal endothelial dysfunction.24,25 Interestingly, when we retrieved the distribution of these 2 markers in our single-cell data, we discovered that C0 approximately matched the surface phenotype of effector cells (Figure 1F). Next, we extracted top novel makers for this cluster, for instance, MGP (Matrix GLA protein), which plays a vital role in the development of the vascular endothelium;26 SLC12A2, which regulates the endothelial barrier function in the pulmonary microcirculation;27 SLC2A1, which encodes a major glucose transporter in the mammalian blood-brain barrier;28 and NQO 1, which has been reported to be involved in reactive estrogen metabolism in FECD pathogenesis.29 Notably, although the highest abundance of these markers was identified in C0, their specificity was poor due to the high similarity in transcriptional profiles between C0 and C2 (Figure 1G). In addition, we revealed representative markers for other clusters. Specifically, DNA replication-related genes, including MCM7, PCNA, and MCM3, were expressed in C2, while cytokine-related genes, including IL11, CXCL3, and CXCL8, were enriched in C3 (Figure 1H). We then performed histological staining to validate the existence of C2 and C3 in human corneal endothelium tissue and we observed a sporadic distribution of proliferating cell nuclear antigen-positive (PCNA+), C-X-C motif chemokine ligand 8-positive (CXCL8+), and IL-11+ cells in the corneal endothelium, implying a rare expression pattern of these genes in scRNA-seq data (Figures 2A and 2B). Overall, these results supported the existence of genetic diversity in the human corneal endothelium.
Figure 1.
Construction of human corneal endothelium atlas by scRNA-seq
(A) Flowchart of the experiments performed in this study. (B) t-SNE clustering of human corneal endothelial cells colored by 4 distinct clusters. (C) Heatmap for expression of differentially expressed genes (DEGs) in each subtype. (D) Representative GO terms of all DEGs in each cell type. (E) Expression of representative marker genes across clusters. (F) Definition and surface phenotype of “effector cells.” (G) Expression of top markers in “effector cells.” (H) Representative novel markers of C1–C3 cells.
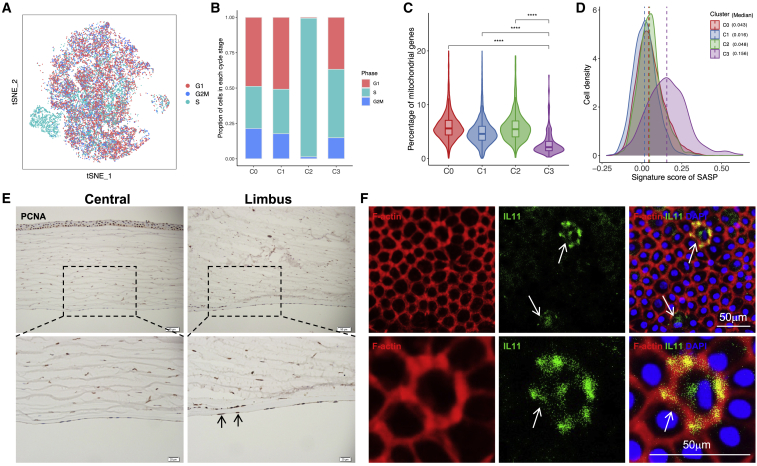
Figure 2.
Histological analysis of representative genes in C2 and C3 clusters
(A) t-SNE clustering of human corneal endothelial cells colored by cell-cycle phases. (B) Proportion of cell-cycle stages per cluster. (C) Percentage of mitochondrial genes in each cell cluster. ∗∗∗∗p < 0.0001 (2-sided Wilcoxon rank-sum tests). (D) Density plot showing the cell distribution with different SASP scores across cell clusters. (E) Immunohistochemical staining of PCNA validated the presence of C2 cluster in endothelium sections of human cornea. The PCNA+ cells in limbus are indicated by black arrows. (F) Immunofluorescence analysis of IL-11 showing the C3 cells in human cornea endothelium tissues.
Unique transcriptional signatures of 4 subtypes of CEnCs
The subtypes of CEnCs exhibited unique expression patterns that reflected their functional commitments. We attempted to characterize the subtype-specific functional roles of these cells. As previously reported, although human CEnCs retain proliferative capacity, the cell division of these cells is limited in vivo throughout life.30 Of note, we determined that genes related to DNA replication, such as PCNA, MCM3, and MCM7 were enriched in C2. For instance, PCNA, encoding a cell-cycle-related protein, is naturally expressed in cells about to proliferate.31 MCM3 and MCM7 belong to the mini-chromosome maintenance (MCM) family, known to have a role in the initiation and regulation of DNA replication during the cell cycle, and their expression is usually upregulated in proliferating cells and absent in terminally differentiated cells.32 Here, we surveyed the cell cycles in each subtype of CEnCs and found that the C2 presented an extraordinarily higher percentage of cells in the S phase, indicating a probability of higher proliferative capacity or activity for C2 cells (Figures 2A and 2B). However, it is noteworthy that MCM and PCNA proteins interact not only with S phase checkpoint regulators but also with critical components of DNA repair pathways;33,34 therefore, further in-depth investigations were needed to determine the bona fide functional identity of C2 cells. Considering that the corneal endothelium is a monolayer of mitochondria-rich cells, we then evaluated the content of mitochondrial genes and found a significantly lower percentage of mitochondrial genes in C3 cells (Figure 2C). When we assessed the senescent state of the corneal endothelium, we found a higher senescence-associated secretory phenotype (SASP) score in C3, which was known to contribute to aging by generating a low-grade inflammatory state (Figure 2D). These observations implied that C3 may exist as a senescence-like cluster with degenerating functions. Localization of PCNA and interleukin-11 (IL-11) expression to these cells was confirmed via immunofluorescence imaging (Figures 2E and 2F).
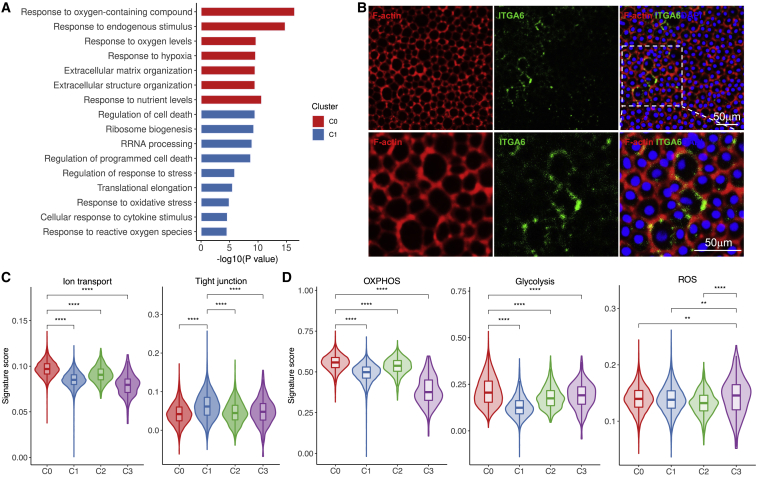
For C0 and C1 cells, we analyzed the biological function of each cell cluster using GO analysis of differentially expressed genes (DEGs), which revealed unique characteristics of these 2 subtypes. For instance, GO terms specific to C0 cells included “response to hypoxia” and “extracellular matrix organization,” while GO terms including “regulation of cell death” and “response to stress” were enriched for C1 cells (Figure 3A). We also surveyed the localization of C1 cells via immunofluorescence imaging of the top marker ITGA6 identified for this cluster (Figure 3B). In addition, CEnCs play vital roles in maintaining corneal clarity through barrier and pump functions. When we scored these clusters for ion transport and tight junction-related gene sets, we found that the highest score for ion transport was assigned to C0, while that for tight junction was assigned for C1 cells (Figures 3C and S4), which suggested the presence of different functional commitments across corneal endothelial clusters. Furthermore, we investigated the heterogeneity of multiple metabolic processes across corneal endothelial subtypes, and found the highest score of glycolysis and oxidative phosphorylation in C0, but a higher ROS score in C3 cells (Figures 3D and S4). Among these, the metabolic pathways of glycolysis were the primary sources of ATP that were necessary to maintain endothelial structure and pump fluid, suggesting the core role of C0 cells in corneal endothelial functions.
Figure 3.
Transcriptional characteristics of 4 endothelial cell subtypes
(A) Representative GO terms of all DEGs between C0 and C1 cells. (B) Immunofluorescence analysis of ITGA6 showing C1 cells in the human cornea endothelium. (C and D) Gene scoring analysis using curated molecular signatures for endothelial functions (C) and metabolic process (D). ∗∗p < 0.01; ∗∗∗∗p < 0.0001 (2-sided Wilcoxon rank-sum tests).
The diverse functional and metabolic involvement of cell subtypes prompted us to explore the cell-cell communications between them. Based on a ligand-receptor analysis, we determined that C0 possessed the highest frequency of putative interactions with other subtypes. We also noted that among the ligand-receptor links, fibroblast growth factor (FGF) accounted for approximately one-third of all communications (Figure S2). As reported, FGF was shown to efficiently reduce human corneal endothelial damage or to stimulate the proliferation and migration of human CEnCs in vitro.35,36 We took advantage of a pseudotime algorithm to reconstruct the trajectory of the corneal endothelium. We observed that C2 was positioned as an intermediate subcluster between C0 and C1, while C3 escaped the consistent trajectory of other subtypes (Figures S3A and S3B), suggesting a larger discrepancy in states with other cells.
Implication of lncRNA NEAT1 in FECD supported by single-cell and bulk-seq data
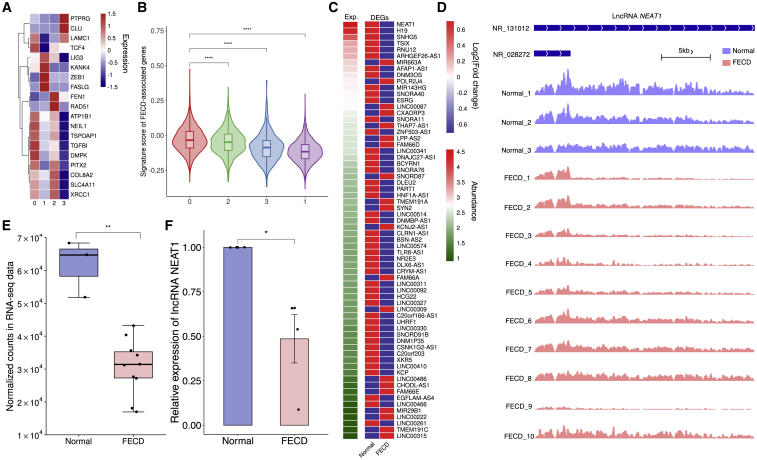
The advent of scRNA-seq enables the profiling of cellular heterogeneity in tissues and, more significantly, identifying pivotal disease-associated populations and major cell-type-specific causal genes. As the most common corneal endothelial dystrophy and the leading indication for corneal transplantation, previous studies have reproducibly linked genetic variants to FECD.37, 38, 39 Here, we first used single-cell transcriptomic atlas of human corneal endothelium to identify the subtype-specific expression patterns of FECD-associated genes (n = 21 genes; only well-documented genes with strong linkage to FECD were considered).37, 38, 39 We found that the highest percentage of FECD-associated genes were enriched in C0, such as TCF4, TGFB1, and COL8A2 (Figure 4A). Next, we calculated the signature scores of FECD-associated genes across subtypes (Figure 4B), and C0 scored the highest compared to other cells. As mentioned before, the C0 cells possessed a similar surface phenotype as “effector cells,” and played an essential role in endothelial function and metabolism. In addition, our results also indicated that C0 cells were likely more vulnerable to genetic perturbations and influenced FECD susceptibility.
Figure 4.
Downregulation of lncRNA NEAT1 in FECD
(A) Gene expression enrichment scores for FECD-associated genes. (B) Signature score of FECD-associated genes across 4 clusters. (C) Heatmap of differentially expressed lncRNA in FECD RNA-seq. (D) lncRNA NEAT1 IGV tracks showing significantly downregulated expression of 2 isoforms between normal and FECD libraries. (E) Boxplot of normalized counts of lncRNA NEAT1 in RNA-seq. (F) qPCR verified the downregulation of lncRNA NEAT1 (n = 4). ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001 (2-tailed t test).
To examine the signature profile of C0, we retrieved the top genes of C0 cells (Figure 1G), and the only lncRNA, NEAT1, a well-known lncRNA, with multiple functions in various biological processes and diseases,40, 41, 42, 43 aroused our interest. Notably, when we surveyed the abundance of these top genes, NEAT1 showed the highest expression across subtypes and a high abundance among the top genes (Figure S5). This prompted us to investigate whether NEAT1 played a vital regulatory role in endothelial homeostasis and FECD pathogenesis. We conducted an analysis of lncRNA profiles on public bulk RNA-seq data of corneal endothelium derived from normal (n = 3) and FECD (n = 10) individuals. The differentially expressed lncRNAs (with >1.5-fold change) were ranked according to their expression abundance in normal samples. We observed that NEAT1 possessed the highest abundance among all differential lncRNAs and was significantly downregulated in FECD samples (Figures 4C–4E), which was validated by quantitative real-time PCR (Figure 4F). These data demonstrated that NEAT1 was highly expressed in C0 cells of the healthy corneal endothelium, and an aberrant decline was detected in FECD samples. Next, we sought to further investigate the potential regulatory role of NEAT1 in FECD pathogenesis.
Silencing NEAT1-exacerbated FECD by impairing the oxidant-antioxidant balance in CEnCs
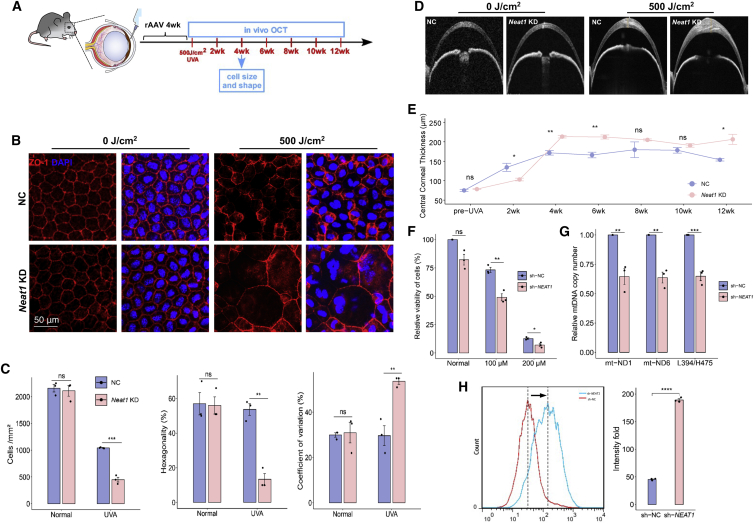
To investigate the regulatory capacity of NEAT1 in CEnCs, we attempted to explore whether NEAT1 expression influenced the progression of FECD by loss-of-function assays. Based on the susceptibility of the corneal endothelium to oxidative stress, we established a non-genetic FECD mouse model whose corneal exposure to UVA recapitulated the symptoms of FECD.22,44 Targeted irradiation of mouse corneas with UVA light induced the characteristic of late-onset FECD (Figure S6A), which was a readily achieved FECD model.22,44 Consistent with clinical samples, we detected significantly downregulated Neat1 in mouse corneal endothelium 4 weeks post-irradiation with varying doses (500, 750, and 1,000 J/cm2) (Figure S6B). To examine the impact of NEAT1 expression on FECD progression, we KD Neat1 expression in the mouse corneal endothelium by directly injecting Neat1-targeting short hairpin RNAs (shRNAs) into anterior chambers before UVA irradiation (Neat1 KD group) (Figures S7A and S7B), and then established a periodic experimental setup, as shown in Figure 5A. At the 4-week time point, zona occludens-1 (ZO-1) immunostaining was used to determine cell density and morphometric analysis was applied to assess progressive corneal endothelium damage. We observed that both groups of mice did not show any significant differences in cell density in the absence of UVA exposure, whereas the Neat1 KD group showed a sharp decline compared to negative controls (NCs) and presented exacerbated morphological changes, including increased cell size and loss of discernible cell borders (Figure 5B). Specifically, Neat1 KD exhibited a 57% lower cell density than NC mice; the hexagonality was 41% lower in Neat1 KD post-irradiation, and the coefficient of variation was greater by 24% in Neat1 KD as compared to NC, indicating that the corneal endothelium with Neat1 deficiency was more susceptible to UVA irradiation than normal counterparts, and promoted impairment of the corneal endothelium (Figures 5B and 5C).
Figure 5.
Knockdown (KD) of Neat1 expression in the mouse corneal endothelium and in human corneal endothelial cells
(A) Experimental timeline including intracameral injection of rAAV and FECD model construction. rAAV 4wk: UVA irradiation 4 weeks post-injection of virus. (B) Representative confocal images of whole mount of corneal endothelium detecting ZO-1 treated with control shRNA (NC) and Neat1 shRNA (Neat1 KD) in normal and FECD mouse. Scale bars, 50 μm. (C) ZO-1 immunostaining-based analysis for cell density, hexagonality, and coefficient of variation (n = 3). (D) Representative OCT images of mouse corneas at 3 months post-UVA. (E) OCT image-based CCT analysis from pre-UVA to 3 months post-UVA. n = 3 for each group. (F) Cell viability of HCEC upon NEAT1 KD with or without H2O2 treatment. (G) Mitochondrial abundance by comparative quantitative real-time-PCR upon KD of NEAT1. (H) Cellular ROS levels in NEAT1 KD HCEC upon H2O2 treatment. Quantitative data are shown as means ± SEs, n = 3. ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001 (2-tailed t test).
The central cornea thickness (CCT) is a hallmark of endothelial cell functional impairment. Post-irradiation, the effects of UVA on endothelium function were monitored by anterior segment–optical coherence tomography (OCT) at 2 weekly intervals (Figure 5A). Interestingly, despite the NC corneas showing endothelial functionally related edema with increased CCT compared to Neat1 deficiency corneas at the 2-week time point, the trend began to reverse after the 4th week. As described above, we detected striking Neat1-dependent variations in CCT values, and greater damage to endothelium function in Neat1 KD corneas remained at 12 weeks (Figures 5D and 5E). Furthermore, to exclude the potential effect of corneal epithelium on the functional evaluation of the corneal endothelium, we also examined the epithelial changes upon Neat1 silencing and UVA irradiation; no damage was detected (Figure S7C).
Accordingly, previous studies have linked an imbalance and dysregulation of the oxidative stress response with FECD pathogenesis.39 To identify the underlying biological process through which NEAT1 regulates cell function, we explored whether NEAT1 was involved in the oxidant-antioxidant homeostasis of CEnCs. We examined the effects of silencing NEAT1 in the human CEnC line (HCEC-B4G12) by NEAT1-targeting shRNAs, with KD efficiencies validated by quantitative real-time PCR (Figure S7D). We observed that KD of NEAT1 had little effect on the cell viability of CEnCs under normal conditions, whereas H2O2 treatment significantly decreased the viability of NEAT1-KD cells (Figure 5F). Oxidative stress plays a critical role in the subsequent accumulation of mitochondrial DNA (mtDNA) damage and contributes to FECD pathogenesis.45,46 Thus, we detected the mtDNA copy numbers and observed that NEAT1 KD resulted in a remarkable decline in mtDNA copy number, suggesting a downregulation of mtDNA-associated genes and impairment of mitochondrial function (Figure 5G).47 Furthermore, KD of NEAT1 also increased cellular reactive oxygen species (ROS) levels under oxidative conditions (Figure 5H), which have been shown to propagate oxidant-antioxidant imbalance in the pathogenesis of FECD.46 These data demonstrate that the deficiency of NEAT1 exacerbated the FECD progression, which may be regulated by impairing the oxidant-antioxidant balance in CEnCs.
FECD progress was diminished by Neat1 overexpression in vivo
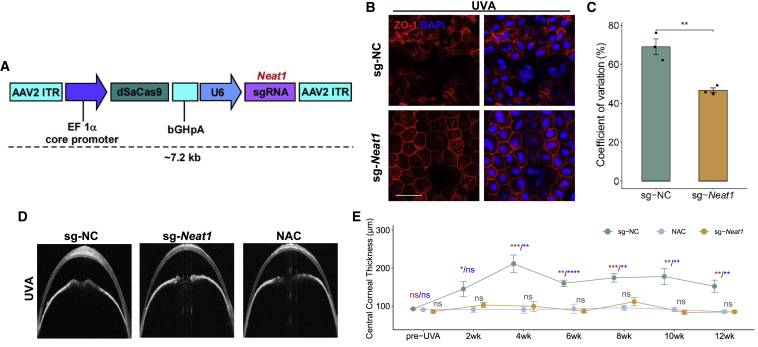
To determine the role of NEAT1 in protecting CEnCs from oxidative damage, we overexpressed Neat1 expression in mouse CEnCs and evaluated the counteracting effect of UVA irradiation on the progression of FECD. We adopted recombinant adeno-associated virus (rAAV) delivery of CRISPR activation (CRISPRa), a system that was shown to be suitable for activating genes with long sequences,48 to modulate the transcription of Neat1 in vivo (Figure 6A). Four weeks after rAAV injection into the anterior chamber, we found that the dSaCas9-Neat1 activator effectively upregulated Neat1 levels in the mouse cornea endothelium (Figure S8A). Next, we exposed the corneas of these mice to UVA irradiation following the established approach at 4 weeks post-irradiation. ZO-1 immunostaining was performed to determine cell density and morphometric analysis assessed the progressive CE damage. Contrary to Neat1 KD, the endogenous Neat1-activated group (sg-Neat1 group) retained the CEnC morphology of normal cell size, discernible cell borders, and decreased coefficient of variation (Figures 6B and 6C) compared with the NC group, although no significant differences were detected in cell density or percentage of hexagonality (Figure S8B). CCT measurements revealed an attenuated corneal thickness in the sg-Neat1 group, implying a greater functional preservation of CEnCs (Figure 6D).
Figure 6.
Neat1 overexpression attenuates oxidative damage in mouse corneal endothelium during FECD pathogenesis
(A) Neat1-targeting CRISPR-dCas9 activation system. (B) ZO-1 staining showed the corneal endothelium morphology after Neat1 overexpression during FECD. Scale bars, 50 μm. sg-NC, sgRNA− control; sg-Neat1, sgRNA-Neat1. (C) Coefficient of variation analysis from mouse corneal endothelium treated with sg-NC and sg-Neat1 rAAV (n = 3). (D) Comparison of OCT images from sg-NC, sg-Neat1, and NAC-treatment FECD mice. (E) Statistical analysis was used to analyze the effects of Neat1 and NAC treatment on CCT changes. n = 6 for each group. The red asterisk represents the difference between the sg-NC and NAC-treatment groups; the blue asterisk represents the difference between the sg-NC and sg-Neat1 groups; The black represents the difference between the sg-Neat1 and NAC groups. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant (2-tailed t test).
N-acetylcysteine (NAC), a derivative of l-cysteine and a precursor to glutathione, is known for its antioxidant and ROS-scavenging properties.49 Since NAC has been shown to be cytoprotective in the mouse model of early-onset FECD,22,50 we tested the therapeutic effects of the endogenously activated Neat1 (sg-Neat1 group) on FECD mice and took NAC-treated mice as the positive control. Twelve weeks of continuous CCT monitoring revealed a comparable cytoprotective effect of sg-Neat1 on UVA-exposed mouse CEnCs. Furthermore, to determine the potential side effects introduced by Neat1 overexpression, we examined the corneal epithelial integrity and intraocular pressure after Neat1 targeting by injected rAAV, and no significant changes were detected (Figures S8C and S8D). These data demonstrated that the lncRNA Neat1 played a critical role in protecting the mouse corneal endothelium from oxidative damage, which pointed toward the potential of our platform for the discovery of new intervention targets and therapeutic strategy for the alleviation of FECD.
Discussion
Traditionally, the corneal endothelium has long been considered to consist of a population of homogeneous cells. In this study, we performed a high-resolution single-cell survey of the human corneal endothelium for the first time, and revealed the heterogeneous cell composition with distinct transcriptional signals and functional commitments. Furthermore, based on FECD-association analysis across various subtypes and bulk-seq data of FECD samples, we identified a non-coding RNA NEAT1 that was highly expressed in FECD-associated subtypes in healthy corneal endothelium but was significantly decreased in FECD samples. In addition, we unveiled its potential antioxidant role in FECD models, which may provide a valuable foundation for novel intervention strategies for FECD.
This study revealed several novel insights. First, previous studies revealed that a subpopulation of cultured CEnCs was devoid of cell-state transition, a condition that may be applicable to cell-based therapy for corneal endothelial dysfunction.25 However, these studies largely focused on complex culture systems in vitro, and it is difficult to predict the in vivo applicability of the human corneal endothelium. Based on a combination of CD surface markers, our analysis revealed that the C0 cluster was highly similar to the “effector cells,” which also provides a more specific reference that may have implications for the identification of transplanting cells into the anterior chamber for corneal endothelial disorders.
Second, the lack of cell proliferation properties in the corneal endothelium resulted in an age-related decrease in cell density. CEnCs are not withdrawn from the cell cycle in vivo, but are arrested in G1 phase. Based on this cell-cycle standstill, throughout life, the division of endothelial cells either does not occur or occurs at a rate that is too slow to adequately replace dead cells. Thus, the regular mechanisms of repairing the corneal endothelium in response to cell loss mainly occurs via cell migration and enlargement. To perform cell-cycle analysis at the single-cell level, we calculated the proportion of cell-cycle stages for each cluster. Our study thus differed from previous approaches and revealed that most of the cells were in G1 phase, with the exception of the C2 cluster, which displayed an extraordinarily higher percentage of cells in S phase. In addition, PCNA, MCM3, and MCM7 were enriched in C2. E2F1, a transcription factor, was also significantly enriched in this cluster (data not shown). E2F1 belongs to the E2F family of transcription factors that regulate the expression of genes whose products are essential for progression through the mammalian cell cycle. In non-replicating cells, including CEnCs, retinoblastoma protein (pRb) tightly binds to E2F, preventing its activation.51 The regeneration of E2F1 activity is sufficient to induce S phase entry and overcomes a p16-mediated G1 block.52 Furthermore, our study clarified the location of C2 cells, and we observed that the peripheral regions of the cornea had more PCNA+ staining cells compared to central regions. Previous studies have speculated that a stem-like cell population tends to exist in the endothelial periphery.53 Meanwhile, MCM and PCNA were critical components of DNA repair pathways,33,34 and E2F1 as a DNA damage responsive protein also plays roles in DNA repair or apoptosis to respond to DNA damage.54 Overall, our data indicated that C2 cells represented the population with the potential in proliferation or DNA damage and repair, whether the C2 population possesses the characteristics of stem cells and what are their roles in proliferation or DNA repair remains to be further determined.
Third, we observed that NEAT1 was downregulated in the corneal endothelium of FECD patients and experimental animals. Previous studies have reported the roles of NEAT1 in regulating paraspeckle formation,55,56 viral infection,40 and tumorigenesis.41,42 In Parkinson’s disease, NEAT1 deprivation exacerbated leucine-rich repeat kinase 2 (LRRK2)-mediated oxidative stress-induced damage.57 In the vascular endothelium, NEAT1 can inhibit oxidative stress-induced cell injury via the miR-181d-5p/CDKN3 axis.58 However, its functional role in FECD pathogenesis remained unclear. Our results demonstrated that NEAT1 KD exacerbated FECD by impairing the oxidant-antioxidant balance in CEnCs, implying that NEAT1 is a potential target for FECD treatment to inhibit its progression. Given the relationship between NEAT1 and redox homeostasis, further studies were needed to define specific mechanisms by which NEAT1 influenced the transcriptional signatures and the ability of CEnCs to respond to external stimuli.
Fourth, in FECD mouse models, we elucidated that Neat1 overexpression via CRISPRa delivery is sufficient to achieve therapeutic efficacy against UVA irradiation in vivo and alleviated oxidative damage to the endothelium. In in vitro studies, human corneas or corneal endothelium have been demonstrated to have potential in efficient rAAV delivery.59, 60, 61 Successful transfection of different layers of the rabbit cornea by rAAV supported the potential of recombinant/exogenous protein delivery into corneal cells.62 Here, we adopted the dSaCas9 rAAV system, which can overcome the size limitations of rAAV48,63,64 and activate target genes in mouse corneal endothelium. This proof-of-concept study will contribute to the development of a non-invasive, specific, and potent rAAV CRISPR system for correcting transcriptional dysregulation in corneal endothelium. Importantly, during the 3-month observation period in the FECD mouse model, NAC achieved good therapeutic effects via daily feeding water treatment, and a comparable cytoprotective effect of sg-Neat1 was observed. In contrast, Neat1 overexpression using the CRISPRa system is a more feasible approach and may facilitate the development of CRISPR therapeutics targeting corneal disorders. For the FECD models used in our study, this non-genetic animal model based on corneal endothelium susceptibility to oxidative stress recapitulated the morphological and molecular changes in FECD, which overcame the lack of an in vivo model in studies on FECD pathophysiology and interventions. Nevertheless, it was necessary to point out that there were considerable differences between human and mouse corneal endothelium, especially in the capacity of regeneration and proliferation, that may pose some limitations to interpreting the results.
Overall, we provide here a broad survey of the single-cell transcriptional profile, which allowed us to systematically characterize the human corneal endothelium and to identify more specific markers of effector cells that may help to purify efficient cells for transplantation in patients with corneal endothelial dysfunction. Our findings may also facilitate the development of rAAV CRISPR system therapeutics against corneal endothelium-related diseases.
Materials and methods
Human tissue acquisition and processing
This study was approved by the ethics committee of Shandong Eye Institute, and participants provided informed consent. The study was carried out in strict accordance with the approved guidelines. For tissue dissociation and single-cell isolation, the corneal endothelium and posterior elastic layer complex were separated from the entire corneal tissue under stereoscope,65,66 and then placed in complete medium to stabilize overnight at 37°C. Subsequently, each sample was incubated with 5 mg/mL collagenase A (Roche, USA) for 1 h, and then was digested for another 30 min in Tryple solution (Sigma-Aldrich, USA). All dissociated cells were finally resuspended in Ca2+ free buffer (with 1% BSA) and passed through a 40-μm filter to remove the residual Descemet membrane and undissociated tissues. The isolated CEnCs were arrested and used and processed for cell viability testing, cell counting, and single-cell analysis.
For the collection of FECD specimens, the FECD corneal endothelium was immediately stored in corneal storage medium (Optisol-GS; Bausch & Lomb, USA) at 4°C after surgical removal. Normal donors were decade matched with FECD. Detailed sample information is provided in Table S1.
10x Genomics scRNA-seq
Single cells from each sample were independently processed into library generations on a 10x Genomics system. Briefly, the cells were partitioned into gel bead-in emulsions (GEM) generation, cDNA barcoding, and library preparation using the single-cell 3′ mRNA kit (V2; 10x Genomics) following the manufacturer’s directions. All of the libraries were subjected to quality tests (Fragment Analyzer 2100, Agilent Technologies, USA) and sequencing (Platform: Illumina NovaSeq 6000; read length: 150 bp, paired-end).
Data processing and downstream analysis
Transcripts were mapped to the human reference genome (GRCh38–3.0.0) following CellRanger pipeline (version 3.1.0), and read count matrices for each sample were generated by CellRanger count. Count data were imported into the Seurat R package (version 3.2.0),23 and quality control of each library was performed with the following steps: cells with <500 genes detected or a mitochondrial gene ratio of >20% were excluded, and genes expressed in <5 cells were removed; doublets were detected using the DoubletFinder package (version 2.0.2);67 considering the dissociation-induced artifacts for sensitive cells, cells expressing a previously published dissociation-induced gene signature were detected and removed if no other explainable marker genes were expressed.68
Next, a CCA algorithm was used to exclude batch effects during data integration.69 Normalization was performed using the LogNormalize function. For cell clustering, principal-component analysis (PCA) was performed on highly variable genes. The clustering at a resolution of 0.3 was performed for the top 16 PCs using the graph-based shared nearest-neighbor (SNN) method (FindClusters function). A total of 4 unsupervised cell clusters were obtained. Clustering results for individual or grouped samples were visualized using t-SNE. Cell types were classified based on differential expression analysis, with cluster-specific marker genes identified (FindMarkers function). GO enrichment analysis of cell-type-specific marker genes was performed with ToppGene online tools,70 and representative GO terms for each cell type were selected from the top 20 terms with p < 0.05.
Cell-cycle discrimination analysis
Assignment of cell-cycle phase of individual cells was performed in Seurat using cell-cycle-specific expression data.71 In brief, markers for G2/M and S phases were used for cell scoring. Cells with neither G2/M or S phase markers were regarded as being in G1 phase (CellCycleScoring function). The cells in each phase were quantified using the prop.table function.
Calculation of a signature score of a gene set
For gene scoring analysis, gene sets were acquired from the MSigDB database and are listed in Table S2. The AddModuleScore function in the Seurat R package was used to calculate the signature score of each gene set in each cell, and the 2-sided Wilcoxon rank-sum test was used for significance testing.
Cell-cell communication analysis
We used CellPhoneDB (version 1.1.0, https://github.com/Teichlab/cellphonedb)72 to systematically predict cell-cell communications based on ligand-receptor analysis with default parameters. Receptors or ligands expressed in at least 10% of cells of a given cell type and with a p < 0.05 were subsequently analyzed. For selected significant receptor-ligand pairs, we applied galluvial and the circlize R package to visualize interaction links.
RNA-seq data analysis
Public RNA-seq data of human corneal endothelia from FECD patients and normal people were acquired from the previous study (GEO: GSE112201).73 To avoid the batch effect and compare the expression of 2 isoforms of NEAT1 more precisely, we adopted samples only from the FECD patients with an expanded trinucleotide repeat of batch2. Sample RNA112 and RNA136 were excluded for abnormal correlation with other samples. Read pairs were aligned to the hg19 reference genome with STAR.74 Normalized read coverage of NEAT1 in each sample was visualized using the integrative genome browser (IGV).75 In addition, read counts for genes were normalized and calculated with DESeq276 to filter the DEGs between FECD and normal samples.
Immunofluorescence staining
Full-thickness corneal flat mounts were fixed in 4% paraformaldehyde and blocked with 5% normal serum for 30 min at room temperature. The samples were treated with primary antibodies overnight at 4°C and subsequently with secondary antibodies for 1 h at 37°C. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The flat mounts were examined and captured under confocal microscopy (LSM800, Zeiss, Germany). The antibodies used for immunofluorescence staining were anti-ZO-1 (1:100, Invitrogen, 40–2200), anti-F-actin (1:100, Abcam, ab176757), anti-IL-11 (1:100, Immumoway, YT2309), and anti-ITGA6 (1:100, Thermo Fisher, 710209), while Alexa Fluor 594-conjugated and 488-conjugated secondary antibodies were obtained from Life Technologies.
Immunohistochemical staining
The human cornea tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. After treatment with 3% H2O2, corneal sections were incubated with primary antibody overnight at 4°C and were stained with a horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at 37°C (MaiXin Biotechnology, China). All of the staining was examined under a Nikon fluorescence microscope (Nikon, Japan). Antibodies used for immunohistochemical staining included mouse anti-PCNA (1:40,000, Abcam, ab29).
Animals
Female C57BL/6 mice (age, 8 weeks; weight, 20–24 g) were purchased from Beijing Vital River Laboratory Animal Technology. The mice were housed in a pathogen-free environment at the Shandong Eye Institute animal facility. All of the animals were treated according to the guidelines established by the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. All of the procedures were approved by the Institutional Animal Care and Use Committee.
Construction and injection of Neat1-targeting rAAV
rAAV was constructed using inverted terminal repeat (ITR) from serotype 2 AAV and capsid protein from serotype 9. Mice were injected with rAAV, which mediated the efficient transduction of corneal endothelium at ∼3 × 1012 vector genomes per milliliter. rAAV containing the Neat1-shRNA, control shRNA target sequences, and Neat1-sgRNA and control sgRNA target sites were obtained from Genechem (China). All of the sequences are shown in Table S3. RNA was extracted from mouse corneal endothelium after at least 4 weeks to detect infection efficiency.
Intracameral injection was performed as previously described.77,78 Mice were systemically anesthetized with pentobarbital sodium (50 mg/kg, intraperitoneally) and topically anesthetized with 2% xylocaine. rAAV solution at 1 μL was injected using a 36G beveled needle (cat. no. NF36BV-2, World Precision Instruments, USA) mounted on a 10-μL microsyringe (cat. no. NanoFil 300,329, World Precision Instruments). An UltraMicroPump II (UMP2 and UMC4, World Precision Instruments) was used to control the volume of injection. The needle was entered at the limbus and we were careful not to traumatize the iris or the lens. Mice with injury to the iris or lens were excluded from the study. As the needle was withdrawn after injection, a cotton tip applicator was applied for ∼1 min to prevent virus/aqueous reflux.
FECD mouse model
The nongenetic FECD mouse model was obtained as previously described.22 Briefly, A UVA light-emitting diode (LED) light source (M365LP1, Thorlabs, USA) was focused to illuminate a 4-mm-diameter spot onto the mouse cornea. The energy was measured using a laser sensor (model L49 150A, Ophir, Israel), and the time of UVA exposure was adjusted to deliver the appropriate fluency (20 min 57 s for 500 J/cm2, 31 min 26 s for 750 J/cm2, 41 min 54 s for 1,000 J/cm2). The treated eye was irradiated, while the other untreated eye served as control.
In vivo imaging
After anesthetizing, mouse corneal images were taken using a slit-lamp biomicroscope with a camera (BQ900; HaagStreit, Switzerland). A solution of 0.25% fluorescein sodium was topically applied onto the lateral conjunctival sac of the mice to assess epithelial cell integrity by observing punctate staining under cobalt blue light. Anterior segment images were taken using anterior segment-OCT (OPTOVUE, USA). CCT was measured using inbuilt software. In vivo confocal microscopy (Heidelberg Engineering, Germany) was used to scan the cell morphology.
Analysis of cell density and morphology
CE images were captured from whole-mount ZO-1 immunostained slides and uploaded into Konan Analysis Suite software (Konan Medical, Japan). Automatic cell analysis was performed after the boundaries of the cells were determined. Cell density, coefficient of variation, and hexagonality were calculated, and the mean value for each was calculated from 3 images.
Cell line and lentivirus infection
A human CEnC line, HCEC-B4G12, was cultured in culture flasks coated with 10 mg/mL laminin (Sigma, USA) and 10 mg/mL chondroitin sulfate (Sigma). The medium consisted of serum-free human endothelial medium supplemented with 10 ng/mL basic FGF (bFGF). The cells were cultured at 37°C in a 5% CO2 incubator until 90% confluence, after which they were passaged using trypsin (Gibco, USA) and cultured for experiments in the same conditions described above. HCEC-B4G12 and Human-Endothelial-SFM (serum-free medium) were purchased from Creative Bioarray (USA), and FGF-2 was obtained from PeproTech (USA). The recombinant lentivirus used to infect HCEC-B4G12 for shRNA was purchased from Genechem and was used according to the manufacturer’s instructions. The target sequences are shown in Table S3. For CCK8 and ROS analysis, 48 h after infection, the cells were treated or untreated with H2O2 and then analyzed by microplate reader or flow cytometry 2 h later.
Quantitative real-time-PCR
Total RNA was extracted using TransZol Up Plus RNA Kit (Transgen, China). A total of 0.1–1 mg RNA was used as a template for reverse transcription with random hexamer primers using the cDNA Synthesis SuperMix Kit (Transgen). Quantitative real-time-PCR was conducted using SYBR qPCR Master Mix (Vazyme, China) on a Rotor-Gene Q system (Qiagen, Germany). Each experiment was repeated independently 3 times. The cycling conditions were 10 s at 95°C, followed by 40 two-step cycles (10 s at 95°C and 20 s at 60°C). Relative gene expression data were analyzed using the comparative CT method (ΔΔCT). All quantitative real-time-PCR primer pairs are listed in Table S4.
Statistical analysis
Data analysis was performed using R software (R Foundation for Statistical Computing, Austria). All data are expressed as means ± standard errors (SEs). The 2-tailed t test was used to analyze relationships between the 2 groups. Before analyzing the unpaired t test, we performed the D’Agostino-Pearson omnibus normality test to evaluate the normality of the variables using GraphPad Prism 7 software (GraphPad Software, USA). p < 0.05 was considered statistically significant.
Data and code availability
All of the data needed to evaluate the conclusions in the article are present in the text and the supplemental information. All of the raw and processed scRNA-seq files in this study have been deposited in the GSA database under accession code HRA000781. All other relevant data are within the article and supplemental information. This study did not generate any unique code.
Acknowledgments
The gene sequencing was conducted by OE Biotech (China). This work was partially supported by the Taishan Scholar Program (20150215 and 20161059), the Academic Promotion Program and Innovation Project of the Shandong First Medical University (2019ZL001 and 2019RC008), the Natural Science Foundation of Shandong Province (ZR2019ZD37 and ZR2019BH078), and the National Natural Science Foundation of China (81800876 and 82101092).
Author contributions
W.S. and Q.Z. contributed to the conception and design of the study. S.D. and B.Z. organized the database and performed the statistical analysis. Q.W., H.J., and X.Q. carried out the experiments. The manuscript was drafted by Q.W. and S.D. All of the authors contributed to manuscript revision and read and approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.01.005.
Contributor Information
Qingjun Zhou, Email: qjzhou2000@hotmail.com.
Weiyun Shi, Email: weiyunshi@163.com.
Supplemental information
▪▪▪
References
- 1.Bourne W.M., McLaren J.W. Clinical responses of the corneal endothelium. Exp. Eye Res. 2004;78:561–572. doi: 10.1016/j.exer.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Joyce N.C. Proliferative capacity of the corneal endothelium. Prog. Retin. Eye Res. 2003;22:359–389. doi: 10.1016/s1350-9462(02)00065-4. [DOI] [PubMed] [Google Scholar]
- 3.Joyce N.C. Proliferative capacity of corneal endothelial cells. Exp. Eye Res. 2012;95:16–23. doi: 10.1016/j.exer.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan D.T., Dart J.K., Holland E.J., Kinoshita S. Corneal transplantation. Lancet (London, England) 2012;379:1749–1761. doi: 10.1016/S0140-6736(12)60437-1. [DOI] [PubMed] [Google Scholar]
- 5.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 6.Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M., et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y., Wang X., Hu B., Mao Y., Chen Y., Yan L., Yong J., Dong J., Wei Y., Wang W., et al. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 2019;17:e3000365. doi: 10.1371/journal.pbio.3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Y.R., Shekhar K., Yan W., Herrmann D., Sappington A., Bryman G.S., van Zyl T., Do M.T.H., Regev A., Sanes J.R. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 2019;176:1222–1237.e22. doi: 10.1016/j.cell.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark B.S., Stein-O'Brien G.L., Shiau F., Cannon G.H., Davis-Marcisak E., Sherman T., Santiago C.P., Hoang T.V., Rajaii F., James-Esposito R.E., et al. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102:1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukowski S.W., Lo C.Y., Sharov A.A., Nguyen Q., Fang L., Hung S.S., Zhu L., Zhang T., Grünert U., Nguyen T., et al. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019;38:e100811. doi: 10.15252/embj.2018100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheaume B.A., Jereen A., Bolisetty M., Sajid M.S., Yang Y., Renna K., Sun L., Robson P., Trakhtenberg E.F. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 2018;9:2759. doi: 10.1038/s41467-018-05134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon M., Mohammadi S., Davila-Velderrain J., Goods B.A., Cadwell T.D., Xing Y., Stemmer-Rachamimov A., Shalek A.K., Love J.C., Kellis M., et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun. 2019;10:4902. doi: 10.1038/s41467-019-12780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Q., Dharmat R., Owen L., Shakoor A., Li Y., Kim S., Vitale A., Kim I., Morgan D., Liang S., et al. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun. 2019;10:5743. doi: 10.1038/s41467-019-12917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H., Chen W., Zhao F., Zhou Q., Reinach P.S., Deng L., Ma L., Luo S., Srinivasalu N., Pan M., et al. Scleral hypoxia is a target for myopia control. Proc. Natl. Acad. Sci. U S A. 2018;115 doi: 10.1073/pnas.1721443115. E7091–e7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Zyl T., Yan W., McAdams A., Peng Y.R., Shekhar K., Regev A., Juric D., Sanes J.R. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. U S A. 2020;117:10339–10349. doi: 10.1073/pnas.2001250117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel G., Fury W., Yang H., Gomez-Caraballo M., Bai Y., Yang T., Adler C., Wei Y., Ni M., Schmitt H., et al. Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. U S A. 2020;117:12856–12867. doi: 10.1073/pnas.2001896117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan N., Wang J., Wray B., Patel P., Yang W., Peng H., Lavker R.M. Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest. Ophthalmol. Vis. Sci. 2019;60:3570–3583. doi: 10.1167/iovs.19-27656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altshuler A., Amitai-Lange A., Tarazi N., Dey S., Strinkovsky L., Bhattacharya S., et al. Discrete limbal epithelial stem cell populations mediate corneal homeostasis and wound healing. Cell Stem Cell. 2021;28:1248–1261. doi: 10.1016/j.stem.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D.Q., Kim S., Li J.M., Gao Q., Choi J., Bian F., Hu J., Zhang Y., Li J., Lu R., et al. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul. Surf. 2021;20:20–32. doi: 10.1016/j.jtos.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumas S.J., Meta E., Borri M., Goveia J., Rohlenova K., Conchinha N.V., Falkenberg K., Teuwen L.A., de Rooij L., Kalucka J., et al. Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J. Am. Soc. Nephrol. 2020;31:118–138. doi: 10.1681/ASN.2019080832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalucka J., de Rooij L., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.A., Veys K., et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Liu C., Miyajima T., Melangath G., Miyai T., Vasanth S., Deshpande N., Kumar V., Ong Tone S., Gupta R., Zhu S., et al. Ultraviolet A light induces DNA damage and estrogen-DNA adducts in Fuchs endothelial corneal dystrophy causing females to be more affected. Proc. Natl. Acad. Sci. U S A. 2020;117:573–583. doi: 10.1073/pnas.1912546116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamuro J., Ueno M., Toda M., Sotozono C., Montoya M., Kinoshita S. Cultured human corneal endothelial cell aneuploidy dependence on the presence of heterogeneous subpopulations with distinct differentiation phenotypes. Invest. Ophthalmol. Vis. Sci. 2016;57:4385–4392. doi: 10.1167/iovs.16-19771. [DOI] [PubMed] [Google Scholar]
- 25.Hamuro J., Toda M., Asada K., Hiraga A., Schlotzer-Schrehardt U., Montoya M., Sotozono C., Ueno M., Kinoshita S. Cell homogeneity indispensable for regenerative medicine by cultured human corneal endothelial cells. Invest. Ophthalmol. Vis. Sci. 2016;57:4749–4761. doi: 10.1167/iovs.16-19770. [DOI] [PubMed] [Google Scholar]
- 26.Boström K., Zebboudj A.F., Yao Y., Lin T.S., Torres A. Matrix GLA protein stimulates VEGF expression through increased transforming growth factor-beta1 activity in endothelial cells. J. Biol. Chem. 2004;279:52904–52913. doi: 10.1074/jbc.M406868200. [DOI] [PubMed] [Google Scholar]
- 27.Simmons S., Erfinanda L., Bartz C., Kuebler W.M. Novel mechanisms regulating endothelial barrier function in the pulmonary microcirculation. J. Physiol. 2019;597:997–1021. doi: 10.1113/JP276245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng P.P., Romme E., van der Spek P.J., Dirven C.M., Willemsen R., Kros J.M. Glut1/SLC2A1 is crucial for the development of the blood-brain barrier in vivo. Ann. Neurol. 2010;68:835–844. doi: 10.1002/ana.22318. [DOI] [PubMed] [Google Scholar]
- 29.Miyajima T., Melangath G., Zhu S., Deshpande N., Vasanth S., Mondal B., Kumar V., Chen Y., Price M.O., Price F.W., Jr., et al. Loss of NQO1 generates genotoxic estrogen-DNA adducts in fuchs endothelial corneal dystrophy. Free Radic. Biol. Med. 2020;147:69–79. doi: 10.1016/j.freeradbiomed.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce N.C. Cell cycle status in human corneal endothelium. Exp. Eye Res. 2005;81:629–638. doi: 10.1016/j.exer.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Kawahira K. Immunohistochemical staining of proliferating cell nuclear antigen (PCNA) in malignant and nonmalignant skin diseases. Arch. Dermatol. Res. 1999;291:413–418. doi: 10.1007/s004030050431. [DOI] [PubMed] [Google Scholar]
- 32.Madine M.A., Khoo C.Y., Mills A.D., Laskey R.A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 33.Bailis J.M., Forsburg S.L. MCM proteins: DNA damage, mutagenesis and repair. Curr. Opin. Genet. Dev. 2004;14:17–21. doi: 10.1016/j.gde.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Essers J., Theil A.F., Baldeyron C., van Cappellen W.A., Houtsmuller A.B., Kanaar R., Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rieck P.W., Gigon M., Jaroszewski J., Pleyer U., Hartmann C. Increased endothelial survival of organ-cultured corneas stored in FGF-2-supplemented serum-free medium. Invest. Ophthalmol. Vis. Sci. 2003;44:3826–3832. doi: 10.1167/iovs.02-0601. [DOI] [PubMed] [Google Scholar]
- 36.Gospodarowicz D., Mescher A.L., Birdwell C.R. Stimulation of corneal endothelial cell proliferations in vitro by fibroblast and epidermal growth factors. Exp. Eye Res. 1977;25:75–89. doi: 10.1016/0014-4835(77)90248-2. [DOI] [PubMed] [Google Scholar]
- 37.Igo R.P., Jr., Kopplin L.J., Joseph P., Truitt B., Fondran J., Bardenstein D., Aldave A.J., Croasdale C.R., Price M.O., Rosenwasser M., et al. Differing roles for TCF4 and COL8A2 in central corneal thickness and fuchs endothelial corneal dystrophy. PLoS One. 2012;7:e46742. doi: 10.1371/journal.pone.0046742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitart V., Bencic G., Hayward C., Skunca Herman J., Huffman J., Campbell S., Bucan K., Navarro P., Gunjaca G., Marin J., et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum. Mol. Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 39.Ong Tone S., Kocaba V., Böhm M., Wylegala A., White T.L., Jurkunas U.V. Fuchs endothelial corneal dystrophy: the vicious cycle of Fuchs pathogenesis. Prog. Retin. Eye Res. 2021;80:100863. doi: 10.1016/j.preteyeres.2020.100863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S., Mosquera J.M., Pauwels J., Park K., Kossai M., et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhry H., Albukhari A., Morotti M., Haider S., Moralli D., Smythies J., Schödel J., Green C.M., Camps C., Buffa F., et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed A.S.I., Dong K., Liu J., Wen T., Yu L., Xu F., Kang X., Osman I., Hu G., Bunting K.M., et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc. Natl. Acad. Sci. U S A. 2018;115 doi: 10.1073/pnas.1803725115. E8660–e8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White T.L., Deshpande N., Kumar V., Gauthier A.G., Jurkunas U.V. Cell cycle re-entry and arrest in G2/M phase induces senescence and fibrosis in Fuchs Endothelial Corneal Dystrophy. Free Radic. Biol. Med. 2021;164:34–43. doi: 10.1016/j.freeradbiomed.2020.12.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azizi B., Ziaei A., Fuchsluger T., Schmedt T., Chen Y., Jurkunas U.V. p53-regulated increase in oxidative-stress--induced apoptosis in Fuchs endothelial corneal dystrophy: a native tissue model. Invest. Ophthalmol. Vis. Sci. 2011;52:9291–9297. doi: 10.1167/iovs.11-8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurkunas U.V. Fuchs endothelial corneal dystrophy through the prism of oxidative stress. Cornea. 2018;37(Suppl 1):S50–S54. doi: 10.1097/ICO.0000000000001775. [DOI] [PubMed] [Google Scholar]
- 47.Hartmann N., Reichwald K., Wittig I., Drose S., Schmeisser S., Luck C., Hahn C., Graf M., Gausmann U., Terzibasi E., et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell. 2011;10:824–831. doi: 10.1111/j.1474-9726.2011.00723.x. [DOI] [PubMed] [Google Scholar]
- 48.Lau C.H., Ho J.W., Lo P.K., Tin C. Targeted transgene activation in the brain tissue by systemic delivery of engineered AAV1 expressing CRISPRa. Molecular therapy. Nucleic Acids. 2019;16:637–649. doi: 10.1016/j.omtn.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun S.Y. N-acetylcysteine, reactive oxygen species and beyond. Cancer Biol. Ther. 2010;9:109–110. doi: 10.4161/cbt.9.2.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jun A.S., Meng H., Ramanan N., Matthaei M., Chakravarti S., Bonshek R., Black G.C., Grebe R., Kimos M. An alpha 2 collagen VIII transgenic knock-in mouse model of Fuchs endothelial corneal dystrophy shows early endothelial cell unfolded protein response and apoptosis. Hum. Mol. Genet. 2012;21:384–393. doi: 10.1093/hmg/ddr473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeGregori J., Leone G., Miron A., Jakoi L., Nevins J.R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. U S A. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukas J., Petersen B.O., Holm K., Bartek J., Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y., Sun H., Guo P., Hu M., Zhang Y., Tighe S., Chen S., Zhu Y. Characterization and prospective of human corneal endothelial progenitors. Int. J. Med. Sci. 2017;14:705–710. doi: 10.7150/ijms.19018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyagi S., Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 2009;28:3185–3195. doi: 10.1038/emboj.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naganuma T., Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simchovitz A., Hanan M., Niederhoffer N., Madrer N., Yayon N., Bennett E.R., Greenberg D.S., Kadener S., Soreq H. NEAT1 is overexpressed in Parkinson's disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. 2019;33:11223–11234. doi: 10.1096/fj.201900830R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M., Wang X., Yao J., Qiu Z. Long non-coding RNA NEAT1 inhibits oxidative stress-induced vascular endothelial cell injury by activating the miR-181d-5p/CDKN3 axis. Artif. Cells Nanomed. Biotechnol. 2019;47:3129–3137. doi: 10.1080/21691401.2019.1646264. [DOI] [PubMed] [Google Scholar]
- 59.Lai L., Lin K., Foulks G., Ma L., Xiao X., Chen K. Highly efficient ex vivo gene delivery into human corneal endothelial cells by recombinant adeno-associated virus. Curr. Eye Res. 2005;30:213–219. doi: 10.1080/02713680590927515. [DOI] [PubMed] [Google Scholar]
- 60.Sharma A., Ghosh A., Hansen E.T., Newman J.M., Mohan R.R. Transduction efficiency of AAV 2/6, 2/8 and 2/9 vectors for delivering genes in human corneal fibroblasts. Brain Res. Bull. 2010;81:273–278. doi: 10.1016/j.brainresbull.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Callaghan J., Crosbie D.E., Cassidy P.S., Sherwood J.M., Flügel-Koch C., Lütjen-Drecoll E., Humphries M.M., Reina-Torres E., Wallace D., Kiang A.S., et al. Therapeutic potential of AAV-mediated MMP-3 secretion from corneal endothelium in treating glaucoma. Hum. Mol. Genet. 2017;26:1230–1246. doi: 10.1093/hmg/ddx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J., Saghizadeh M., Tuli S.S., Kramerov A.A., Lewin A.S., Bloom D.C., Hauswirth W.W., Castro M.G., Schultz G.S., Ljubimov A.V. Different tropism of adenoviruses and adeno-associated viruses to corneal cells: implications for corneal gene therapy. Mol. Vis. 2008;14:2087–2096. [PMC free article] [PubMed] [Google Scholar]
- 63.Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parekh M., Peh G., Mehta J.S., Ramos T., Ponzin D., Ahmad S., Ferrari S. Passaging capability of human corneal endothelial cells derived from old donors with and without accelerating cell attachment. Exp. Eye Res. 2019;189:107814. doi: 10.1016/j.exer.2019.107814. [DOI] [PubMed] [Google Scholar]
- 66.Li W., Sabater A.L., Chen Y.T., Hayashida Y., Chen S.Y., He H., Tseng S.C. A novel method of isolation, preservation, and expansion of human corneal endothelial cells. Invest. Ophthalmol. Vis. Sci. 2007;48:614–620. doi: 10.1167/iovs.06-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van den Brink S.C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C.S., Robin C., van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 69.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wieben E.D., Aleff R.A., Tang X., Kalari K.R., Maguire L.J., Patel S.V., Baratz K.H., Fautsch M.P. Gene expression in the corneal endothelium of Fuchs endothelial corneal dystrophy patients with and without expansion of a trinucleotide repeat in TCF4. PLoS One. 2018;13:e0200005. doi: 10.1371/journal.pone.0200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goel M., Sienkiewicz A.E., Picciani R., Wang J., Lee R.K., Bhattacharya S.K. Cochlin, intraocular pressure regulation and mechanosensing. PLoS one. 2012;7:e34309. doi: 10.1371/journal.pone.0034309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bainbridge J.W., Stephens C., Parsley K., Demaison C., Halfyard A., Thrasher A.J., Ali R.R. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 2001;8:1665–1668. doi: 10.1038/sj.gt.3301574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
▪▪▪
Data Availability Statement
All of the data needed to evaluate the conclusions in the article are present in the text and the supplemental information. All of the raw and processed scRNA-seq files in this study have been deposited in the GSA database under accession code HRA000781. All other relevant data are within the article and supplemental information. This study did not generate any unique code.