Abstract
Non-small-cell lung cancer (NSCLC) is a heterogeneous disease, and therapeutic management has advanced to identify various critical oncogenic mutations that promote lung cancer tumorigenesis. Subsequent studies have developed targeted therapies against these oncogenes in the hope of personalized treatment based on the tumor’s molecular genomics. This review presents a comprehensive review of the biology, new therapeutic interventions, and resistance patterns of two well-defined subgroups, tumors with KRAS and MET alterations. We also discuss the status of molecular testing practices for these two key oncogenic drivers, considering the progressive introduction of next-generation sequencing (NGS) and RNA sequencing in regular clinical practice.
Keywords: biomarker, directed therapy, genomic profiling, KRAS, MET, non-small-cell lung cancer, resistance, targeted therapy, tumor genomics
Keypoints
Lung cancer is no longer a single disease but a group of different diseases determined by a certain histologic type and a particular genetic makeup.
KRAS-mutated lung cancer is no longer considered an untargetable driver.
Multiple clinical trials are assessing the efficacy of KRAS-targeted therapy, with promising results.
Immune checkpoint inhibition is a good alternative to current therapies in the treatment of KRAS and MET-altered lung cancer.
Tyrosine kinase inhibitors are the most promising candidate therapies for treating MET-mutated tumors.
Introduction
With the estimation of 2.2 million incident cases and 1.8 million deaths, lung cancer is the second-most diagnosed cancer and the leading cause of cancer death in 2020, representing approximately 11% of cancers diagnosed and 18.0% deaths. 1 In the last decade, it has been recognized that lung cancer is made up of a group of molecularly and histologically heterogeneous subtypes. 2 Two major histologic subgroups are non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), which account for 76% and 13%, respectively, of all lung cancer cases in the United States. 3
Before 2002, limited treatment options, except for local removal and cytotoxic chemotherapy, were available. These factors contributed to the dismal outcomes of lung cancer. However, in the last two decades, quit-smoking campaigns and popularization of chest computational tomography (CT) scan screening in the United States and other countries changed the lung cancer prevalence pattern to where the number of non-smoking and early-stage lung cancer patients increased. 4 Besides, advances in targeted therapies and immune checkpoint inhibitors expanded lung cancer treatment options. A recent epidemiological analysis based on the Surveillance, Epidemiology, and End Results (SEER) Program national database showed that 2-year relative survival among patients with lung cancer improved substantially, from 26% among men with NSCLC diagnosed in 2001 to 35% among those with NSCLC diagnosed in 2014, a change largely attributable to the inclusion of more than 25 new therapeutic indications. 5 To confirm this finding, Liang et al. illustrate the lung cancer mortality of 12 representative countries (Canada, China, Japan, Singapore, Australia, the United Kingdom, Germany, Denmark, France, Italy, and Sweden) in different continents based on the global disease burden (GDB) database, finding a significant decrease in mortality, like that in the US cohort, since the approval of estimated glomerular filtration rate tyrosine kinase inhibitors (EGFR-TKIs). 6
Advances in the knowledge of pathways, technologies for detecting actionable genetic lesions, and newly developed drugs to block genomic drivers have allowed the oncology community to tailor the treatment options. 7 Several targetable major pathways have been identified in lung adenocarcinomas, such as EGFR, ALK, ROS1, Her2, MET, RET, BRAF, NTRK, and NRG1 fusions. Many drugs targeting these pathways have been developed and shown impressive clinical benefits. Some of them have now replaced chemotherapy as the first-line treatment, such as EGFR, ALK, ROS1, NTRK, MET, and RET inhibitors. 8 Nevertheless, while target therapy in NSCLC has provided disease control, the tumors inevitably develop drug resistance.
Nowadays, several molecular targets that were previously considered ‘unactionable’, such as KRAS, now have several therapies under consideration with promising early results. 9 Activating KRAS mutations are present in ~20–30% of patients with NSCLC. 10 The most prominent KRAS-mutant form in never-smokers is G12 V (56%), a form locked in a constitutively activated guanosine triphosphate (GTP)-bound state. In contrast, the dominant mutation among current/former smokers is G12 C (42%). 11 Until a few years ago, efforts to target KRAS directly have been unsuccessful. 12 However, several synthetic lethality screens have identified an indirect vulnerability in KRAS-mutant lung cancers. For example, in preclinical models, polo-like kinase 1, RhoA/Rho kinase, nuclear export XPO1 inhibitions have led to selective vulnerability of KRAS-mutant lung cancer.13,14 Previously, some preclinical models demonstrated that the pharmacological inhibition of upstream adapter protein SHP2 had rendered a response against the KRAS-G12 C variant. This RAS variant can cycle nucleotide to behave in a semi-constitutive manner and, hence, respond to ablation of upstream signaling. 15 This finding represented the first step for developing sotorasib (AMG-510) and adagrasib (MRTX-849). 16
The hepatocyte growth factor (HGF) ligand and MET receptor pathway has been known to promote cancer growth and invasion since about three decades now. 17 MET protein is a well-known driver oncogene. Three different types of genetic alterations have been seen: fusion, amplification, and mutation. MET exon 14 encodes for a juxtamembrane (JX) domain that is intracellular, containing a PKC phosphosite (S985), a caspase cleavage location (D1002), and an E3 ubiquitin ligase Casitas-B-lineage lymphoma (CBL) docking point (Y1003), which is involved in the downregulation of RTK activity. 18 Alterations in MET usually disrupt splicing sites along introns in the regions neighboring exon 14, including an intron 13 splice acceptor site and the donor location for intron 14. Also, mutations in exon 14 might provoke skipping of this one for the final mRNA molecule. The more frequent registered alterations include base substitutions, followed by indels (3–4% of NSCLC patients). 19 These MET genetic alterations in NSCLC were initially discussed by Ma et al. 20 and actively explored since then, enabling the elaboration of different potential targeted agents that could address this driver. In this comprehensive review, we summarize KRAS and MET characteristics in NSCLC and discuss an array of selective inhibitors, including small molecules and antibody-based approaches. We also discuss resistance biology for each scenario and the utility of immunotherapy.
Search strategy
The information was extracted from searches of the medical literature made in PUBMED, EMBASE, and the Cochrane Register of clinical trials from 2010. Besides, we searched the abstract registers of the ASCO, ESMO, WCLC, ELCC, and AACR meetings, emphasizing the last 5 years. Nine hundred forty-six references were found that were refined, selecting a total of 224 for the elaboration of this narrative review.
KRAS
KRAS biology in NSCLC
In the 1960s, an essential step toward the understanding of oncogenesis was made. The discovery of Murine Sarcoma Viruses (MSVs) during leukemia research led to the first general description of HRAS, KRAS, and NRAS genes. 21 The Kirsten Rat Sarcoma gene (KRAS) was initially described in 1969 by Kirsten et al. 22 He found that rats infected with a Murine Leukemia Virus would induce the production of an MSV with the ability to cause sarcoma in murine models. However, in these early days of research, the human KRAS homolog gene was not discovered yet.
The development of different laboratory techniques like DNA transfection and molecular cloning was important for studying human transforming DNA fragments.21,23 Different researchers discovered and repeatedly confirmed that murine and human cancer cells (induced and non-induced by oncogenic retroviruses) DNA fragments were capable of causing oncogenic transformation of NIH-3 T3 cell lines.24,25 While studying these tumor-derived DNA fragments, the orthologs for viral ras oncogenes (HRAS and KRAS) with certain point mutations were found. Shimizu et al. 26 reported a human homologue of v-Ki-ras in the human lung carcinoma cell line Calu-1 with a point mutation that caused an amino acid change at position 12 glycine for cysteine (G12 C). McCoy et al. also reported the presence of this gene in the human colon cancer cell line SW480. With the identification of the RAS gene family in humans, the era of molecular oncology in human cancer saw its beginning.
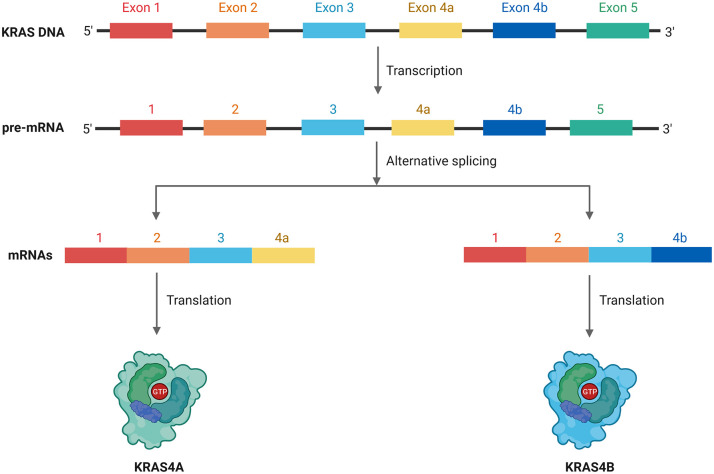
The KRAS gene is located in chromosome 12p12.1, it consists of six exons with four splice variants, from which two (KRAS4A and KRAS4B) are the most commonly expressed isoforms 27 (see Figure 1). KRAS4B comprises 188 amino acids, differing by only one residue to KRAS4A (189). Mutant variants of KRAS4B are widely present in human cancers with KRAS mutations, corresponding to the majority of KRAS-mutated proteins found in these tumors, followed by KRAS4A. 28 Even though these isoforms are similar in length, they only share 164 amino acids, starting from the N-terminal region. The C-terminal domain in these two isoforms is translated from two different exons, which creates a significant difference in protein post-translational changes, in which KRAS4A will receive a palmitoylation and farnesylation (creating dual targeting motifs for membrane binding). At the same time, KRAS4B will only be given the farnesyl radical.27,28 These are critical differences with great importance regarding targeted therapy.
Figure 1.
KRAS is characterized by having multiple splicing isoforms, arising from a pool of 6 exons. The two most common isoforms are KRAS4A (in which the exon 4a is conserved) and KRAS4B (in which the exon 4b is conserved). Most of genetic alterations related to KRAS occur in the KRAS4B isoform.
According to the Human Protein Atlas, KRAS is expressed in almost all cell types in the human body, with a moderate expression in bronchi and almost no overt protein expression in lung parenchyma (RNA expression is present). KRAS is a guanosine triphosphate hydrolase (GTPase) with a deficient intrinsic activity, a subfamily member of small GTPases. 29 Upon activation, KRAS (and all the Ras family members) will act as a molecular switch, triggering different signaling cascades related to cell proliferation, differentiation, cytoskeleton dynamics, and vesicle trafficking, and secretion as well as protein translocation to membranes. 30
Under hemostatic conditions, the KRAS protein will experience two different activity states. When bound to GTP, KRAS will be in its active form, and when bound to GDP, it will shut off. As all GTPases, KRAS will be dependent on the GTP-GDP cycle, which will be catalyzed by the presence of nearby guanine nucleotide exchange factors (GEFs) GTPase-activating proteins (GAPs), both also known as regulator factors.31,32 The activation of KRAS usually follows the stimulation of certain nearby receptor tyrosine kinases (RTKs) in the cell membrane; a common example would be the epidermal growth factor receptor (EGFR). When EGFR is stimulated by its ligands (namely EGF), certain conformational changes will induce the autophosphorylation of the intracellular domains of this receptor, and recruitment of adaptor proteins will occur. These adaptor proteins are usually multi-domain complexes, one of those domains is GEF. Some known adaptor proteins with RAS GEFs are CNRASGEF, RASGEF1A, RASGRF2, RASGRP1, RASGRP4, and SOS1. 33
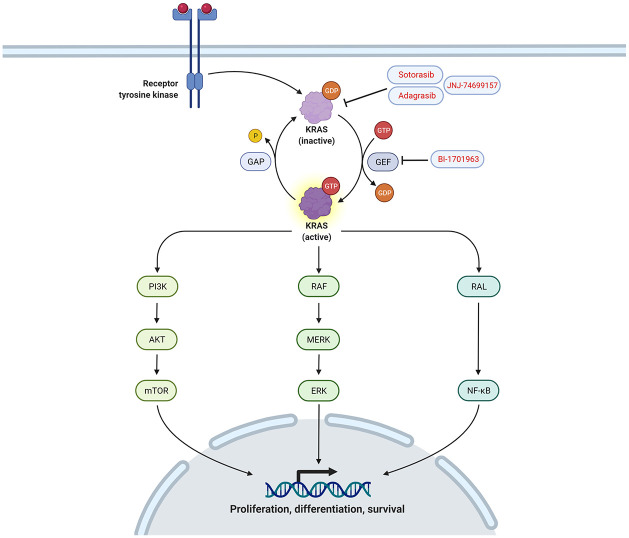
When KRAS is in its GDP-bound state, a GEF domain will interact with it, inducing the release of GDP and the quick attachment of a new cytosolic GTP molecule, thus, switching on KRAS. Once activated, Ras proteins will trigger a cascade of intracellular signaling mechanisms via three main effector pathways mediated by RALGDS, PI3K/Akt/mTOR, and the most important one, the MAPK pathway in which Raf (another subset of retroviral-derived oncogenes) MEK and ERK proteins are involved. An overview of KRAS signaling pathways and the current targeted therapies is shown in Figure 2.
Figure 2.
Under normal conditions, KRAS drives the controlled activation of different proliferation and growth intracellular pathways. When activated by a receptor tyrosine kinase, KRAS releases GDP and binds GTP. When bound to GTP, KRAS will trigger the PI3 K/Akt/mTOR, MAPK, and NF-kB pathways. When mutated, KRAS will become permanently active, inducing uncontrolled cell proliferation. Drugs like sotorasib, adagrasib, and JNJ-74699157 permanently bind to the inactivated form of KRAS-G12 C, keeping KRAS from being activated by GTP. BI-1701963 inhibits the GEF protein SOS1, one of the main guanosine exchanging factors related to Ras proteins.
Most of the mutant variants of KRAS involve conformational changes in the protein that makes it impossible for GAPs to induce the release of GTP from KRAS; therefore, KRAS remains indefinitely in its active state, behaving as a potent driver of cancer.34 –37 Mutations in KRAS are the most common genetic abnormalities seen in human cancer, with a prevalence of approximately 30%. 38 In the case of pancreatic adenocarcinoma, KRAS mutations might be present in more than 90% of cases. Other cancers with a high prevalence of these mutations are colorectal cancer (CRC), stomach cancer, endometrial cancer, and lung cancer, especially in lung adenocarcinoma and with less frequency in lung squamous cell carcinoma. 39 KRAS mutations in lung cancer are the most frequent oncogenic driver in western countries, accounting for about 20–25% of lung adenocarcinomas and 10–15% in Asian countries.40,41 As occurs with other targetable driver mutations, The Cancer Genome Atlas and the Clinical Lung Cancer Genome Project have shown that KRAS alterations are present almost exclusively in adenocarcinomas rather than squamous cell cancers.42,43 An interesting particularity of KRAS mutations in lung cancer is that they are closely related with a patient’s positive smoking history, while other driver mutations like EGFR, BRAF, ROS1, and ALK are usually seen in nonsmokers 44 ; thus, only 5–10% of all KRAS-mutated lung carcinomas occur in patients with no history of tobacco consumption. 45
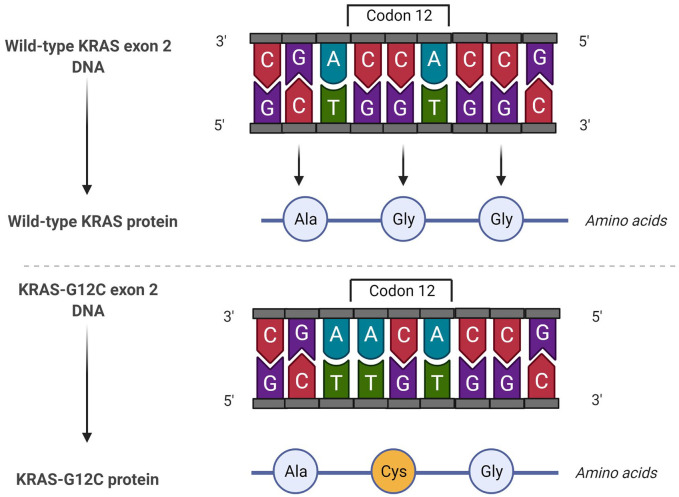
Approximately 95% of KRAS mutations are in codon 12. Variant KRAS-G12 C is the most common, corresponding to 39% of cases (Figure 3), followed by G12 V (18–21%) and KRAS-G12D (17–18%). Redig et al. analyzed a cohort of KRAS-mutant lung cancer patients and compared the prevalence of different mutant variants regarding smoking status. They found that transition mutation (guanine > adenine) were more frequent in never-smokers than in current or past smokers, in which transversion mutations (G > T) were more prevalent. 46
Figure 3.
This illustration depicts a single nucleotide polymorphism of guanine to thymine in codon 12 of exon 2 of KRAS, induces a change in the translated amino acid sequence of glycine to cysteine, generating a constitutively active form of KRAS.
Heterogeneity of KRAS-mutant lung cancer
Unlike other common genetic abnormalities in lung cancer, KRAS-mutant lung cancer presents with high heterogeneity, both from biological and clinical perspectives. According to Ferrer et al., 47 three different features might be involved in this heterogeneity: (1) the presence of concomitant genetic alterations, (2) the different KRAS point mutations, and (3) the mutant KRAS allelic content. Approximately nine different mutant variants of KRAS have been identified in codons 12, 13, and 61. Different studies have demonstrated that specific point mutations generate a protein with different affinities for certain effector proteins, making that particular effector a key oncogenic pathway in the bearing tumor. A variety of in vitro studies have shown that KRAS-G12 C or KRAS-G12 V had inferior levels of activated (phosphorylated) Akt. Simultaneously, an increase in RAL activation was seen compared to wild-type KRAS cell lines. 48
On the contrary, G12D mutant cell lines demonstrated a higher affinity toward the PI3 K-Akt-mTOR pathway. Another experimental analysis showed that KRAS-G12 C was also associated with higher levels of ERK1/2 phosphorylation (member of the MAPK pathway) than KRAS-G12D. These data were further confirmed by assessing the effectiveness of a MEK inhibitor in murine models of KRAS-G12 C tumors versus KRAS-G12D, showing that G12 C tumors were significantly more sensitive to MEK inhibition, with higher levels of response rate and with a higher progression-free survival (PFS). These biological differences might seem trivial; however, the dependence on certain effector pathways results critical for treatment choices, as there are no currently available targeted therapies for KRAS, but some molecules are approved for different members of its effector cascades.
The activation of different transcriptional programs related to specific mutant variants has been an independent predictor of response to certain targeted therapies and a predictor of poor overall survival (OS). 47 Ihle et al. 49 analyzed 215 tissue samples from lung cancer patients enrolled in a targeted therapy clinical trial and found that patients harboring KRAS-G12 C or KRAS G12 V variants have a worse PFS compared to patients with other variants or with a wild-type KRAS (p = 0.046). Nadal et al. studied 179 patients with resected lung adenocarcinoma. KRAS mutation harboring was related with increased disease-free survival (DFS) compared to KRAS wild-type patients (p = 0.009). They also found that patients with the KRAS-G12 C variant had a significantly lower DFS than other mutants or wild-type patients (p < 0.001). A worse OS was also seen in KRAS-G12 C patients (p = 0.003). The prognosis of KRAS-mutant variants has not been validated yet in other standard clinical settings like advanced-stage disease. 11
Another important determinant of biological behavior is the allelic content. An anabolic metabolism is a common feature of KRAS-mutant cells. This metabolic state is usually dependent on the presence of Ras mutations. 50 Some evidence suggests that this metabolic regulation and cell adaption to oxidative stress vary significantly between patients with KRAS-mutant NSCLC. 47 This difference is in part explained by the presence of co-occurring mutations, but it has been demonstrated that allelic content is also a determinant of metabolic regulation and redox management. Kerr et al. analyzed KRAS-mutant cell lines and KRAS-mutant murine lung cancer models. They found an increased glycolytic activity, glutathione biosynthesis, increased antioxidant capacity, and a higher metastatic potential (especially in G12D homozygous specimens) than those cell lines or tumors that were heterozygous for a KRAS-mutant variant. 51 These investigators also found that mice bearing homozygous KRAS mutations had a lower OS than its heterozygous peers (p = 0.0045, exact survival days per group were not mentioned by authors). 51
KRAS and co-occurring mutations
The presence of co-occurring mutations is also a determinant of heterogeneity between KRAS-mutant tumors. A wide array of research suggests that co-occurring genomic abnormalities have an impact in biological behaviors, clinical prognosis, and sensitivity to certain targeted therapies. 52 Skoulidis et al. performed an integrative analysis of the genomics, transcriptomics, and proteomics of samples of KRAS-mutant lung adenocarcinoma from patients with early-stage and chemorefractory disease. They described three clusters of patients defined by co-occurring genetic alterations in STK11/LKB1, which the authors called KL subgroup; TP53 (KP subgroup) and CDKN2A/B (KC subgroup). 53 A metabolic reprogramming and adaption characterized the KL subgroup to oxidative stress mediated by HIF1-α. This subgroup of tumors also presented with deficient expression of programmed death ligand 1 (PD-L1) and a decreased concentration of tumor-infiltrating lymphocytes (TILs), indicating that this type of tumor does not depend on immune evasion to thrive.
On the contrary, the KP subgroup presented with a high infiltration of inflammatory cells, immunoediting, and higher levels of PD-L1 expression (a mean of 56.3% positivity) even compared with wild-type KRAS LUAC (32.3%). The KC subgroup showed enrichment of gene expression signatures typical of upper and lower gastrointestinal neoplastic conditions. 53 Furthermore, Skoulidis et al. also performed an interesting analysis in which they assessed the objective response rate (ORR) of patients with lung adenocarcinoma receiving anti-PD1 therapy in clinical trials. They analyzed the results according to their proposed genetic subgroups. They found that ORR for the KL, KP, and K-only (KRAS mutations with no co-occurring alterations) groups was 7.4%, 35.7%, and 28.6%, respectively (p < 0.001) for the Stand Up to Cancer study cohort. 54 The authors also analyzed patients from the Checkmate-057 nivolumab clinical trial finding the respective ORRs of 0, 57.1, and 18.2% (p = 0.047). 54
Next-generation sequencing and RNA-seq for KRAS
Different sequencing and gene amplification techniques have been used in the diagnosis of tumors harboring certain driver mutations. Some molecular techniques commonly used to detect these mutations include direct sequencing, amplification refractory mutation system (ARMS), droplet digital PCR (ddPCR) and lately next-generation sequencing (NGS; mainly in research settings). Of all the techniques, NGS presents significant advantages such as capturing a broader spectrum of mutations than Sanger sequencing (the most common method por EGFR mutation analysis), avoiding analysis bias as capillary sequencing depends on pre-knowledge of the gene or locus under study, while NGS analysis is unselective and even new mutations and alterations can be discovered. 55 In 2018, Jing et al. analyzed 112 lung cancer samples using NGS. According to their results, 10 patients (8.93%) had KRAS mutations and some few patients with two or three concomitant mutations with other genes like EGFR, NRAS, and PIK3CA. From the KRAS-mutant samples, the most common variant was G12D followed by G12C and G12V. 56 Furthermore, there are some kits that are currently used in certain clinical settings for the analysis of multiple genes of importance of patients with lung cancer, either from tissue samples or from liquid biopsy. Nacchio et al. analyzed 194 liquid biopsies of patients with advanced NSCLC that were treatment-naïve, using an NGS panel called SiRe. About 36 patients presented with KRAS mutations either in exon 2 or 3 and no concomitant alterations. In this cohort KRAS-G12 C alteration was the most common (91.7%). 57
RNA-Seq is an interesting molecular technique that uses NGS methods to deliver a signature transcriptomic analysis of a certain sample and analyze gene expression under certain circumstances in the scenario of a certain driver mutation. With RNA-Seq, a differential gene expression analysis can show how different in terms of gene expression, are tumors of a certain tissue with different driver alterations, 58 revealing new prognostic markers based on expression on also characterizing better the population comprising the tumor (in the case of single cell analysis). Maroni et al. used single cell RNA-Seq (sc-RNA-Seq) to identify a particular cluster of epithelial cells within KRAS-mutant NSCLC tumors that was significantly associated with low OS. They also showed that this particular cluster identified in human samples had a counterpart in mice and that both epithelial cell clusters expressed BMI-1. These researchers also tested the efficacy of a drug called PTC596 (a BMI-1 inhibitor) in treating tumors in an NSG mice xenograft model of KRAS-mutant lung cancer using A549 cells. Their results showed that tumor size decreased considerably in the mice that received PTC596 versus those that received only the therapy vehicle (p = 9.70 × 10–5). 59 RNA-Seq has also been used in lung cancer as a prognostic tool. Yang et al. analyzed 516 lung cancer samples downloaded from The Cancer Genome Atlas website. They analyzed the differential expression of KRAS isoforms (KRAS4A and KRAS4B) in KRAS-mutant and KRAS wild-type patients and correlated the findings with OS. The authors found that when KRAS4A was increased in expression or proportion, OS was considerably lower (p = 0.0149 for expression and 3.18 × 10–3 for proportion). 60
Novel therapies for KRAS-mutant lung cancer
Even though KRAS was one of the first oncogenes discovered, there is no approved targeted therapy yet. In the last decades, different scientific publications and studies have concluded KRAS as an undruggable target. The first attempts to create small molecules that would inhibit KRAS binding to GTP were considered an impossible mission because of the high affinity of KRAS for GTP and the concomitant high concentration of GTP in cells.61,62 Other different approaches have been investigated with good in vitro results but with no in vivo effectiveness. 63 We will discuss a set of novel therapies that show promise in the treatment of KRAS-mutant LUAC.
BI-1701963
As mentioned above, the key regulators of the KRAS GTPase activity are GEFs and GAPs; therefore, targeting these proteins might induce an indirect blockade of KRAS its effector pathways. BI-1701963 is a small oral molecule developed by Boehringer Ingelheim as a ‘pan-KRAS inhibitor’. This molecule selectively inhibits SOS1, a quite common adaptor protein with an Ras GEF domain. By inhibiting SOS1, KRAS would not exit its GDP-bound inactivated status, shutting down the whole pathway. It is called a pan-KRAS inhibitor because as it is not targeting the protein itself, it does not matter what mutant variant certain tumor might have, all variants will be equally blocked. In 2020, at the American Association for Cancer Research Annual Meeting, data regarding the effectiveness of BI-1701963 was presented. The investigators shared that BI-1701963 combined with an MEK inhibitor candidate (BI-3406) could reduce GTP-loaded KRAS formation and inhibit the MAPK pathway signaling. These observations were stable across different mutant variants, including the most common G12C, G12V, and G12D, and G13D oncoproteins. The authors previously showed that BI-1701963 and the MEK inhibitor trametinib could elicit tumor regressions in vivo xenografts of murine models with KRAS-driven tumors. Currently, BI-1701963 is the first GEF-KRAS inhibitor reaching clinical trials and can be found at clinicaltrials.gov by its identifier: NCT04111458.
Sotorasib
As it was reviewed, the most common KRAS-mutant variant is G12C. Sotorasib is a small molecule developed by Amgen that acts as a specific inhibitor of the KRAS-G12 C variant. Sotorasib selectively binds to the P2 pocket of the switch II domain of KRAS-G12 C, generating an irreversible inhibition (mediated by covalent bonding) of the inactive GDP-bound KRAS. Preclinical studies showed that this drug inhibited almost totally detectable ERK phosphorylation, the last agent in the MAPK pathway. 64 In September 2020, phase-I clinical trial of sotorasib in patients with KRAS-G12 C-mutant cancer was published in the New England Journal of Medicine. A total of 129 patients were studied, from which 59 had NSCLC, 42 had CRC, and 28 had other tumors. From the subgroup of NSCLC, the authors found that 32.2% of patients had an objective response (complete or partial), and 88.1% achieved disease control (objective response or stable disease). Median PFS was 6.3 months. It is essential to say that these patients had a median of three prior therapy lines (with a range of 0–11). Responses were also seen in colorectal, pancreatic, endometrial cancers, and melanoma. In May 2021, sotorasib earned accelerated approval by the US Food and Drug Administration (FDA).
JNJ-74699157
Like sotorasib, JNJ-74699157 is a KRAS-G12 C inhibitor that also achieved clinical testing in 2019, manufactured by Johnson & Johnson. Its phase-I clinical trial is registered at clinicaltrials.gov with the identifier NCT04006301. To date, recruitment has been completed with 10 patients enrolled. JNJ-74699157 is a small molecule for oral administration that is directed against the P2 pocket in the switch-II region of KRAS-G12C. No preclinical data are available regarding this molecule. 65
Adagrasib
Initially coined as MRTX849, adagrasib is also an oral administrated small molecule directed against KRAS-G12 C developed by Mirati Therapeutics. It is a tetrahydro-pyridopyrimidine that irreversibly covalent inhibits KRAS. In vitro analyses confirmed that lung and pancreatic cell lines (H358 and PaCa-2, respectively) when exposed to adagrasib, present with almost a complete inhibition of the MAPK pathway shown by deficient levels of ERK phosphorylation. Preclinical studies evidenced that upon adagrasib administration, tumor regression was achieved in 65% of cell lines and patient-derived xenograft models from multiple tumor types. 66 During the 2019 AACR-NCI-EORTC joint conference, results regarding a phase-I/II clinical trial of patients with KRAS-G12 C tumors (mainly NSCLC and CRC) showed that 4 out of 12 patients assessed achieved a partial response (PR), and eight presented with stable disease. Adagrasib is also being tested in combination therapy with inhibitors of other effector pathway agents like TKIs, mTOR inhibitors, or cell cycle inhibitors. Currently, there are four clinical trials registered at clinicaltrials.gov, in which adagrasib is being evaluated in combination with different drugs like pembrolizumab, docetaxel, cetuximab, afatinib, and TNO155 (and SHP2 inhibitor).66,67 Recently, Tanaka et al. defined for the first time the mechanistic spectrum of acquired resistance to adagrasib in a patient who developed polyclonal acquired resistance with the emergence of 10 heterogeneous resistance alterations in serial cell-free DNA spanning four genes (KRAS, NRAS, BRAF, and MAP2K1), all of which converge to reactivate RAS-MAPK signaling. Notably, a novel KRASY96D mutation affecting the switch-II pocket, to which adagrasib and other inactive-state inhibitors bind, was identified that interferes with crucial protein–drug interactions and confers resistance to these inhibitors in engineered and patient-derived KRAS-G12C cancer models. Interestingly, a novel, functionally distinct tri-complex KRAS-G12C active-state inhibitor RM-018 retained the ability to bind and inhibit KRAS-G12C/Y96D and could overcome resistance. 68
Targeting KRAS neoantigens
The development of mutations in different human cancer driver genes is usually followed by the appearance of neoantigens, which are essentially a short oligopeptide that is not naturally expressed in healthy cells. Therefore, the presentation of these neoantigens via class-I HLA molecules could induce immune responses against that particular epitope. 69 Certain epitopes of cancer can be synthesized in the laboratory and can be administered to patients with tumors bearing these same epitopes, eliciting an immune response against the tumor. This kind of therapeutic approach is what we know as a cancer vaccine. 70 Arbelaez et al. developed long synthetic peptides (SLPs) against neoepitopes of G12C, G12V, and G12D KRAS-mutant variants. They also conjugated cationic lipoplexes to every SLP to facilitate the delivery of these peptides to secondary lymph organs, eliciting activation of CD4+ T-cells but of CD8+ T-cells as well. The authors found that when alone, SLPs can induce activation of CD4+ T-cells alone, with little response on the tumoral burden; however, when lipoplexes were added, tumor regression was seen in different murine models in a CD8+ T-cell-dependent manner. The authors also tested the use of SLP + lipoplexes + immune checkpoint inhibitors, achieving profound responses in the studied mice. 71 Wan et al. tested two different peptide vaccines for KRAS-G12D in CT26 mice models. They achieved tumor regression in 50% of the mice treated.
Furthermore, they tested the vaccine with a preventive approach and found that 87.5% of mice were tumor-free after receiving a tumor xenograft. 72 To date, different clinical trials for KRAS-mutant cancers using peptide vaccines are running. Some of these trials are accompanied by immune checkpoint blockade, and others are cell-based, as autologous dendritic cells can also be loaded in vitro with the studied peptides and then reinfused in the patient. This approach has the potential of bearing better results, as dendritic cells will rapidly activate the cognate T cell receptors (TCRs) that identify the peptide attached to a particular HLA molecule.
KRAS and tumor immunity
As it is mentioned before, certain KRAS variants are characterized by a strong modulation of the immune system. It has been demonstrated that mutant KRAS can induce a constellation of inflammatory changes in its microenvironment, modifying the tumor niche by eliciting the silencing of immune responses. KRAS-mutant cells can induce the secretion of particular cytokine and chemokine profiles via the activation of the PI3K pathway, the MAPK pathway, and the subsequent activation of NF-κB. IL-6 and TGF-β seem to be critical in KRAS-mediated immunoediting. The increased levels of TGF-β in the microenvironment can promote T-cell regulators (Treg) recruitment and induce the polarization of macrophages to the M2 type. 73 Another important regulator of immune responses in KRAS-mutant tumors is the expression of PD-L1 by malignant cells. As discussed above, the KRAS-mutant subgroup of patients with co-occurring mutations in TP53 (KP subgroup) present with high levels of expression of PD-L1. Different clinical studies have assessed the efficacy of immune checkpoint inhibition using anti-PD-1, anti-PD-L1, and anti-CTLA4 antibodies in KRAS-mutant NSCLC. Even though not statistically significant differences were found in ORRs between KRAS-mutants and KRAS-wild-type, a correlation for better outcomes was perceived in the subgroup of KRAS-mutant with a high level of PD-L1 expression (>50%).
Currently, different approaches of cellular immunotherapy are under investigation for the treatment of KRAS-mutant LUAC. Researchers are using cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and autologous TILs to treat these malignancies. A very novel approach in the field of cellular therapies is the use of genetically modified T-cells. Using genetic engineering techniques like CRIPS/Cas and TALEN, 74 the endogenous TCR can be silenced, and then, transduction of a new laboratory-designed TCR construct can be done for another expression on T-cells and induction of a specific response against cells presenting certain HLA-epitope complexes derived from KRAS-mutant variants.75 –77 The administration of synthetic peptides can also enhance this approach.
KRAS has played a crucial role in understanding cancer and oncogenesis since its initial discovery in retroviruses. Even though heavy research has focused on developing effective therapies, none of the different approaches studied were effective. However, the last 5 years have yielded motivating and inspiring data regarding the treatment of these complex malignancies. It is a cause of joy to see how scientists are putting all their efforts without rest, for the sake of KRAS-mutant bearing cancer patients. Cellular therapies and targeted therapies arise as the promised land of tumoral treatment after almost four decades of intense dedication.
MET
MET biology
MET (mesenchymal–epithelial transition; also known as HGF receptor, AUTS9, RCCP2, and DFNB97), is a proto-oncogene located on chromosome 7q31. It codifies for a protein of 170 kD which is an RTK, with a highly glycosylated extracellular α-subunit and a transmembrane β-subunit, which are linked by a disulfide bond. MET is a molecule that is essential for the survival and function of normal cells; in particular, this latter protein has crucial roles in embryogenesis, 78 organ development, 79 and regeneration. 80
The extracellular subunit contains a semaphorin domain, a cysteine-rich MET-related domain, and four immunoglobulin-plexin transcription domains. The intracellular part contains a JX domain, an intracellular tyrosine kinase domain that mediates MET-associated signaling, and a tail on the C-terminal. 81 The JX domain contains a serine residue (Ser985), which when phosphorylated performs inhibition of the receptor kinase activity. 82 There have been identified two ligands for MET, the mammalian HGF and the scatter factor, along with their splicing isoforms and a bacterial leucine-rich surface protein named Internalin B. 83
HGF binding to MET induces homodimerization and phosphorylation of the Y1234 and Y1235 tyrosine residues, located within the tyrosine kinase domain’s catalytic loop. 84 Subsequently, tyrosine residues 1349 and 1356, located in the carboxy-terminal tail, undergo phosphorylation. When phosphorylated, these two residues form a unique tandem SH2 recognition motif (Y1349VHVX3Y1356VNV). 85 This SH2 recognition motif induces the recruitment of signaling effectors that include the adaptor proteins GRB2, 86 SHC, 87 CRK, and CRKL, 88 the effector molecules PI3K, PLCγ, SRC, 85 SHIP-2, 85 and STAT-3. 89 Furthermore, MET is also associated with GRB2-associated binding protein 1 (GAB1), 90 a multi-adapter protein, bound to and phosphorylated by MET, creating binding sites for other adaptors and effectors downstream. GAB1 can be directly bound to MET, or indirectly using GRB2. Additional tyrosine residues are involved in MET signaling. Phosphorylated Y1313 binds and activates PI3 K, promoting cellular viability and motility. Likewise, phosphorylated Y1365 is involved in the regulation of cell morphogenesis. 91
The downstream response to MET activation relies on stereotypical signaling modulators common to many RTKs. For activation of the Mitogen-activated protein kinase (MAPK) cascades, MET activation stimulates the activity of the rat sarcoma viral oncogene homolog (RAS) guanine nucleotide exchanger Son of Sevenless (SOS) via binding with SHC and GRB2, 92 leading to the activation of RAS. This downstream information leads to the indirect activation of v-raf murine sarcoma viral oncogene homolog B1 (RAF) kinases, which can subsequently activate the MAPK effector kinase MEK and finally MAPK, which can then translocate to the nucleus to activate transcription factors responsible for regulating a large number of genes. In the context of MET signaling, this results in phenotypes, such as cell proliferation, cell motility, and cell cycle progression. 93 SRC homology 2 domain–containing phosphatase-2 (SHP2) can also link MET signaling to the MAPK cascade, as sequestration of SHP2 to GAB1 is responsible for extending the duration of MAPK phosphorylation. 94
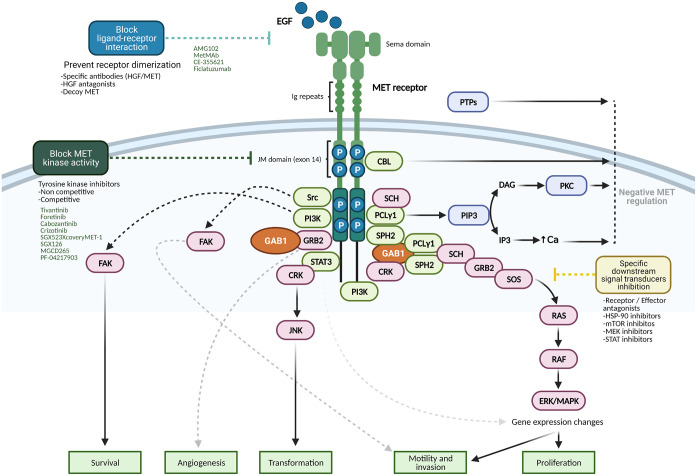
The other major arm of MET signaling is the PI3 K/Akt signaling axis. The p85 subunit of PI3K can bind either directly to MET or indirectly through GAB1, which signals through AKT/protein kinase B. This axis is primarily responsible for the cell survival response to MET signaling. 95 Transformation downstream of the MET receptor is mediated by the phosphorylation of Janus kinase 1 (JNK), which occurs via binding to CRK. STAT3 has also been implicated in transformation, although its proposed mechanism is controversial. Cellular migration is also mediated downstream of MET by focal adhesion kinase (FAK), which is localized to cellular adhesion complexes. FAK is activated through phosphorylation by SRC family kinases, which have been shown to associate directly with MET. 85 The MET–SRC–FAK interaction leads to cell migration and the promotion of anchorage-independent growth. 96 Figure 4 depicts the intracellular signaling of MET.
Figure 4.
MET signaling adaptors and mediators, as well as signaling pathways are depicted. Therapeutic intervention strategies to block and inhibit MET receptor oncogenic signaling cascade include blocking ligand-receptor interaction, preventing receptor dimerization, blocking MET kinase intrinsic activity, and inhibiting specific downstream signal transducers. DAG, diacylglycerol; HGF, hepatocyte growth factor; IP3, inositol triphosphate; PIP3, phosphatidylinositol (3,4,5)-triphosphate.
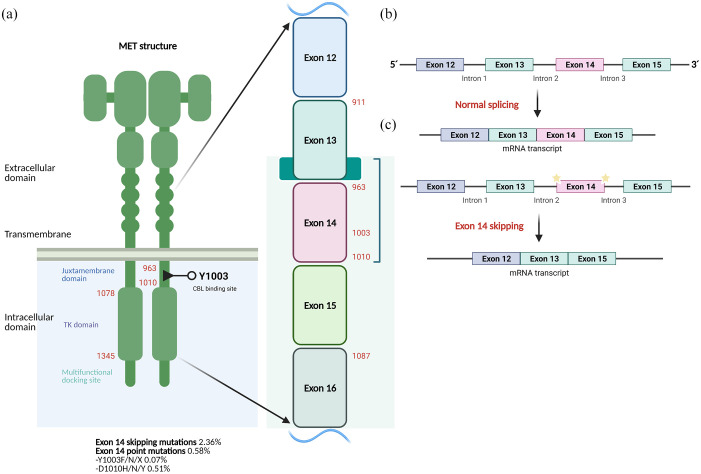
Negative regulation of the MET receptor is crucial for its tightly controlled activity and can occur through several mechanisms. The Y1003 site, located in the JX domain, is a negative regulatory site for MET signaling that acts by recruiting c-CBL (casitas B-lineage lymphoma). 97 Regulation of MET signaling is also accomplished via its binding to various protein tyrosine phosphatases (PTPs), including the receptor-type PTPs density enhanced phosphatase 1 (dEP1) (or PTPrI) and leukocyte common antigen-related molecule (LAR) (or PTPrF), and the non-receptor PTPs PTP1B and T-cell protein tyrosine phosphatase (TCPTP). These PTPs modulate MET signaling by dephosphorylation of either the tyrosines in the MET kinase domain (in the case of PTP1b and TCPTP) or the docking tyrosines (in the case of dEP1). Finally, binding of PLCγ to MET results in the activation of protein kinase C (PKC), which can then negatively regulate MET receptor phosphorylation and activity. 98 Independently of PKC activation, an increase in intracellular calcium levels can also lead to negative MET regulation. 98 Figure 5 depicts the exon structure of MET.
Figure 5.
(a) The structure of MET and frequency of MET exon 14 alterations in lung adenocarcinoma. (b) Normal MET splicing leads to the biosynthesis of the normal receptor that can be targeted by E3-ubiquitin ligase cCBL and directed for lisosomal degradation. (c) Mutations in the splice junctions of MET exon 14 can lead to exon 14 skipping resulting in the mature MET receptor that lacks juxtamembrane regulatory domain. Consequently, the receptor cannot be targeted by cCBL, impairing its lysosomal degradation thereby leading to de accumulation of the protein and increase receptor activity.
Crosstalk between MET and other RTKs has also been studied in great depth because of its potential importance in developing resistance. MET has also been shown by multiple studies to interact directly with the EGFR, allowing activation of MET after stimulation of cells with the EGFR ligands EGF or transforming growth factor (TGF)-α. 99 Stimulation of cells expressing both MET and EGFR with EGF resulted in phosphorylation of MET, and stimulation with ligands for both receptors resulted in synergistic activation of downstream modulators, indicating mutual activation of these two pathways. 100 Evidence also exists for MET interaction with the other EGFR family members ERBB2 and ERBB3 (for erythroblastic leukemia viral oncogene homologs B2 and B3), causing transactivation of both receptors. 101 Interaction of MET with the closely related RON (recepteur d’origine nantais) receptor has also been shown to cause transphosphorylation of the MET receptor in the absence of HGF. 102 Interestingly, it was recently shown that transactivation of RON by MET may be a feature of cancer cells that are ‘addicted’ to MET signaling. 103 Recently, transactivation between MET and both platelet-derived growth factor receptor (PDGFR) and Axl was found to play a role in bladder cancer. 104
MET gene mutations in lung cancer
Activating mutations in RTKs play a critical role in oncogenesis. 105 A set of different gene mutations in the MET sequence have been found to promote lung cancer evolution. Somatic intronic mutations in the JX domain, with further loss of CBL-E3 ligase binding, was characterized to demonstrate elevated MET expression and prolonged ligand-dependent MET activation. Normally, introns flanking MET exon 14 are spliced out, resulting in an mRNA transcript with an intact exon 14 that contains the CBL-E3 ligase binding site. Mutations may disrupt splicing sites, resulting in aberrant splicing and consequently, skipping of exon 14. As a result of absent binding to CBL-E3 ligase, there is decreased polyubiquitination which translates in delayed downregulation, and sustained MET activation.105,106 More than half of these are indel mutations, many of which have been described recently. This heterogeneity and variability in splicing generate a diagnostic challenge, which requires the correct implementation of analytical methods with high sensitivity and specificity. Approximately 4% of lung adenocarcinomas carry alterations in MET exon 14. The mutation rates were 2.6% in adenocarcinoma, 4.8% in adenosquamous carcinoma, and 31.8% in sarcomatoid carcinoma. 107 Conversely, MET exon 14 mutation was not detected in squamous cell carcinoma, large-cell carcinoma, and lymphoepithelioma-like carcinoma. Besides, MET exon 14 mutation occurred mutually exclusively with known driver mutations but tended to coexist with MET amplification or copy number gain. Similarly, low-level MET amplification and polysomy might occur in the background of EGFR or KRAS mutation whereas high-level amplification (MET/CEP7 ratio ⩾ 5) was mutually exclusive to the major driver genes except MET exon 14 mutation. 108 In vitro, both small molecule TKI and MET-directed monoclonal antibodies have been found to be active in cell lines harboring MET exon 14 alterations. 109 In most reports, the most frequent exon 14 alteration was splice donor mutation and PRs to either selective or multi-targeted kinase inhibitors. In most reports, the most frequent exon 14 alterations were splice donor mutation and PRs to either selective or multi-targeted kinase inhibitors.108 –113
MET gene amplification in lung cancer
In about 4% of TKI-naïve cases of NSCLC, MET amplification has been identified as a primary oncogenic event, and as a secondary in ≈20% in EGFR-mutated NSCLC with acquired resistance to TKIs (especially after the use of osimertinib).114,115 In addition, the reported frequency of MET CNG (copy number gain) in NSCLC ranges from 0.7 to 21%, depending on the technique used and the cut-point for positivity. Increased copy numbers of the MET gene can be detected by fluorescence in situ hybridisation (FISH), reverse transcriptase-PCR, or NGS. MET gene amplification is expressed as the level of gene copy number gain and MET: CEP7 ratio. A MET: CEP7 ratio > 2 and the presence of five signals or more per cell for copy number gain, are considered positive for MET amplification. 116 MET amplification is associated with poor prognosis in patients with NSCLC. 114 Recently, Kim et al. conducted a systematic review to determine MET and GCN amplification’s prognostic value. From the integration of 7,647 patients (21 studies), a worse prognosis was found for patients harboring high MET CNG (hazard ratio (HR) = 1.45, 95% confidence interval (CI) = 1.16–1.80; p = 0.001). Subgroup analyzes showed that high MET CNG significantly correlated with a poor prognosis in patients with adenocarcinoma (HR = 1.41, 95% CI = 1.11–1.79; p = 0.005) and in Asian populations (HR = 1.58, 95% CI = 1.32–1.88; p < 0.00001). 117
Recently, in a small series, the ORR to crizotinib (a MET/ALK/ROS1 inhibitor) dramatically differed between cases with different MET/CEP7 ratios (ratio ⩾ 1.8 to ⩽2.2 ORR 0%; ratio > 2.2 to <5; ORR 17%; ratio ⩾ 5; ORR = 67%). 118 In EGFR-mutant NSCLC with acquired resistance to EGFR-TKIs, the ORR to the combination of an EGFR inhibitor and capmatinib (INC280) was 0% among those with a mean MET/cell of <5% and 40% among those with mean MET/cell ⩾ 5. 119 MET exon 14 alterations harbor concurrent high-level MET copy number gain in 20% of cases. 120 In patients with EGFR mutations, secondary MET amplification leads to acquired EGFR-TKI resistance due to transactivation of ErbB3 signaling. 121 Based on this, different clinical trials are exploring a combination of MET and EGFR-TKI in patients with mutant EGFR (TATTON and INSIGHT trials).
HGF and HGF receptor over-expression
Previous studies have noted that over-expression of MET was positively associated with vascular and lymphatic invasion, which led to a higher risk of cancer relapse and more advanced stage among NSCLC patients.122,123 From therapy’s experience, MET positivity was closely related to radioresistance and chemo-resistance (by activating the focal adhesion kinase—FAK and downregulating the expression of different apoptosis factors), hence correlated with unfavorable outcomes. 124 In terms of pathogenesis, HGF could facilitate tumor metastasis through MET/HGF pathways by inducing the epithelial–mesenchymal transition (EMT) process.125,126 Also, MET over-expression was related to the prognosis of patients that harbored various EGFR status as MET and EGFR shared signal molecules in downstream pathways. 127 Thus, MET over-expression could affect patients’ efficacy that received EGFR-TKIs as a result.128,129 Interestingly, p-MET expression, which represents the activation level of MET, did not impact the survival of NSCLC.
MET gene fusion and rearrangement
MET was first identified when the oncogenic chromosomal rearrangement Tpr-Met was induced in a sarcoma cell line. 130 Although MET fusion gene products are not frequently found, they have recently been documented in lung adenocarcinoma. Stransky et al. demonstrated translocation events involving MET across different tumor types. Specifically, in lung adenocarcinoma, fusion of the dimerization motif to intact kinase domain led to generation of a chimeric fusion protein, KIF5B–MET. 131
NGS and RNA-seq in lung cancer with MET alterations
As for KRAS, NGS testing is being slowly introduced in the diagnosis of MET alterations, especially in METex14 mutations. Samples might be tested from solid tissue or can also be assessed from liquid biopsies when not sufficient solid tissue is available or when it is estimated that is collection might be delayed for 2 weeks or more. 132 In the case of NSCLC, whole-genome and even whole-exome sequencing are not recommended as most of the data will be clinically irrelevant. Therefore, there is a targeted approach for testing that looks for actionable druggable targets this is what we know as target enrichment. The process of target enrichment is also important because sequencing depth needs to be high to successfully detect low-frequency allele variants. There are namely two types of NGS techniques used in different tumor genetic detection assays. The first one is amplicon-based methods, in which a set of primers are used to flank the genetic region of interest and allow amplification; however, this method has shown to be ineffective because of a large proportion of allele dropout that results in false-negative results. 133 These allele dropouts are the consequence of single nucleotide polymorphisms and short indels in the primer binding sequences, or it can even be a deletion of a whole genomic region that might harbor the primer binding sites. Some clinical kits made for NGS of MET exon 14 mutations testing have not been able to be optimized. 134 Poirot et al. 135 analyzed 191 NSCLC tumor samples using two amplicon-based detection kits, they also did an in silico analysis of eight commercial NGS kits for mutation detection. They found that the amplicon-based methods yielded a considerably high proportion of false-negative results and that the commercially available kits could not be able to detect more than 63% of the literature-reported MET exon 14 mutations based on primer design. The hybrid capture library method bypasses the weaknesses of the amplicon method using fragmented regions of genomic DNA to which nucleotide probes will bind; however, these probes, which would be the analogs to primers, will bind to regions surrounding the area of interest, avoiding allele dropout due to primers not binding. Furthermore, the hybridization probes used in this method are considerably longer than primers, making them more tolerant to the presence of mismatches in the binding sites. 136 The MSK-IMPACT and the FoundationOne CDx kits, which are reliable diagnostic tools, utilize the hybrid capture tool and are also equipped with a complimentary bioinformatic software that reliably detect a wide group of MET exon 14 alterations without the use of RNA-based testing. 132
The use of RNA sequencing has not yet been introduced as a standard practice, this is because of RNA-seq only detect the direct result of the MET exon 14 skipping, which is the fusion of exons 13 and 15. Nevertheless, RNA-seq might be useful in the detection of METex14 in patient with non-canonical mutations intronic mutations. A lot of different technical challenges are faced when using RNA-seq as a complimentary diagnostic tool. First, it is complicated to extract enough high-quality RNA from patients’ samples, especially when samples are scarce and when they are derived from formalin-fixed, paraffin-embedded specimens. When the RNA used does not have an adequate quality control, accurate interpretation of negative results cannot be done. Also, false-positive results might arise from low basal levels of alternative splicing, showing that there is fusion of exons 13 and 15 when there is no MET exon 14 alterations. 137
MET inhibitors in NSCLC
Tyrosine kinase inhibitors
MET TKIs are broadly classified according to the binding site and mechanism. 138 Type-I inhibitors are adenosine triphosphate (ATP)-competitive and bind to tyrosine 1230 in the activation loop. Type-Ia inhibitors (i.e., crizotinib) interact with the glycine residue G1163, resulting in more significant off-target actions, while type-Ib inhibitors such as capmatinib, tepotinib, and savolitinib present with stronger interactions with Y1230 and no interaction with G1163. 17 Type-II inhibitors, form which cabozantinib is a great example, bind directly to the adenine binding site of ATP, with an extension to the hydrophobic back pocket, with potency depending on the activation state of MET protein.17,139 Table 1 summarizes the most significant results of the studies conducted with MET inhibitors.
Table 1.
Results for current clinical trials running for MET-mutated lung cancer. .
| Study | Phase | Drug | N | Line | MET alterarion | ORR | PFS | DOR |
|---|---|---|---|---|---|---|---|---|
| MET exon 14 mutation and de novo MET amplification (NSCLC) | ||||||||
| PROFILE 1001 NCT00685195 | I | Crizotibib 250 mg BID | 69 | First 26 > second: 43 | MET exon 14 skipping | 32% | 71 months (95% CI: 5.4–9.1) | 9.1 months (95% CI: 6.4–12.7) |
| GEOMETRY mono -1 NCT02414139 | II | Camatinib 400 mg BID | 28 | First line | MET exon 14 skipping | BIRC: 67.9% Inv: 60.7% | 9.69 months (95% CI: 5.52–13.86) | 11.14 months (95% CI: 5.55–NR) |
| 69 | Second/third line | BIRC: 40.6% Inv: 42% | 5.42 months (95% CI: 4.17–6.97) | 9.72 months (95% CI: 5.55–12.98) | ||||
| VISION NCT02864992 | II | Tepotinib 500 mg/day | 87 | First: 33 second: 31 > second: 23 | MET exon 14 skipping | BIRC: liquid biopsy: 50% tissue biopsy: 45.1% | BIRC: liquid biopsy: 9.5 months (95% CI: 6.7–NR) tissue biopsy: 10.8 months (95% CI: 6.3–NR) | Not reported |
| First: 17 second: 13 > second: 4 | Inv: liquid biopsy: 55.3% tissue biopsy: 54.9% | Inv: liquid biopsy: 9.5 months (95% CI: 5.3–21.1) tissue biopsy: 12.2 months (95% CI 6.3–NR) | ||||||
| NCT02897479 | II | Savolitinib 600 mg/day | 34 | Second: 21 > second: 5 | MET exon 14 skipping | 12/31 (38.7%) | Not reported | 34 weeks (range: 16–96) |
| METROS NCT02499614 | II | Crizotibib 250 mg BID | 26 | — | MET exon 14 skipping MET/CEP7 ratio > 2.2 |
7 (27%) | 4.4 months (95% CI: 3.0–5.8) | Not reported |
| NCT00585195 | I | Crizotibib 250 mg BID | 3 | — | Low: ⩾1.8 ⩽ 2.2 copies | 1 (33.3%) | 18 months (95% CI: 0.8–14.0) | 12.1 months (95% CI: 12.1–12.1) |
| 14 | — | Medium: >2.2 to <4 copies | 2 (14.3%) | 1.9 months (95% CI: 1.3–5.5) | 3.7 months (95% CI: 3.7–3.7) | |||
| 20 | 8 (40%) | 6.7 months (95% CI: 3.4–7.4) | 5.5 months (95% CI: 3.3–25.8) | |||||
| MET amplification in EGFR-TKI resistance (NSCLC) | ||||||||
| NCT01610336 | II | Gefitinib 250 mg daily Capmatinib 400 mg BID |
41 | Post first/second | Copy number < 4 | 5 (12%) | Copy number ⩾ 65.49 months (95% CI: 4.21–7.29) | Not reported |
| 18 | Generation | Copy number ⩾ 4 < 6 | 4 (22%) | |||||
| 36 | EGFR-TKI | Copy number ⩾6 | 17 (47%) | |||||
| 4 | Second: 86 | IHC: 0 | 1 (25%) | |||||
| 2 | IHC: +1 | 0 (0%) | IHC: +35.45 months (95% CI: 3.71–7.10) | |||||
| 16 | >second: 75 | IHC: +2 | 3 (19%) | |||||
| 78 | IHC: +3 | 25 (32%) | ||||||
| TATTON NCT02143466 | Ib | Savolitinib 300 mg or 600 mg Osimertinib 80 mg/daily |
51 | Post first/second generation EGFR-TKI T790M- | EGFR mutant and MET amplification (FISH copies ⩾ 5 or MET/CEP7 ratio ⩾ 2, NGS 20% tumor cells and ⩾ copies), or MET over-expression (3+) | 33 (65%) | 9.0 months (95% CI: 5.5–11.9) | 9.0 months (95% CI: 6.1–22.7) |
| 18 | Post first/second generation EGFR-TKI T790M+ | 12 (67%) | 11.0 months (95% CI: 4.0–NR) | 12.4 /95% (95% CI: 2.8–NR) | ||||
| 69 | Post third-generation EGFR-TKI | 21 (30%) | 5.4 months (95% CI: 4.1–8.0) | 7.9 months (95% CI 4.0–10.5) 8.0 months (95% CI: 4.5–NR) | ||||
| 36 | Post first/second generation EGFR-TKI T790M- | 23 (64%) | 9.1 months (95% CI: 5.4–12.9) | 8.0 months (95% CI: 4.5–NR) | ||||
| MET kinase domain mutant/amplified (pRCC) | ||||||||
| NCT02127710 | II | Savolitinib 600 mg/daily | 44 | First: 26 > First: 18 | MET mutation, polysomy, amplification | 4 (18%) | 6.2 months (65% CI: 4.1–7.0) | Range: 2.4–16.4 months |
| 46 | First: 23 > First:23 | MET-independent | 0 (0%) | 1.4 months (95% CI: 1.4–2.7) | ||||
| 19 | First: 11 > First: 8 | Unknown MET status | 0 (0%) | |||||
| NCT00726323 | II | Foretinib | 74 | First: 60 ⩾ First: 14 | MET mutation MET amplification Chromosome 7 polysomy | 10 (13.5%) | 9.3 months (95% CI: 6.9–12.9) | 18.5 months |
BID, Twice a day; BIRC, Blinded Independent Review Committee; CEP7, Chromosome 7; CI, confidence interval; DOR, duration of response; EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridisation; IHC, immunohistochemistry; MET, mesenchymal–epithelial transition; NGS, next-generation sequencing; NR, Not reported; NSCLC, non-small-cell lung cancer; ORR, objective response rate; PFS, progression-free survival; pRCC, Papillary renal cell carcinoma; TKI, Tyrosine Kinase Inhibitor.
Type-II MET inhibitors
Capmatinib (TABRECTA). Capmatinib (INC280) is an oral, type-Ib MET inhibitor with high potency and high selectivity. Preclinical studies have demonstrated that capmatinib can block MET phosphorylation and activate key downstream signaling mediators in MET-dependent cell lines. 140 Moreover, different pleiotropic effects were seen on other signaling pathways including EGFR and HER3. During the dose-escalation part of the phase-1 trial (NCT01324479), 38 patients received capmatinib, starting at a dose of 100 mg BID in a capsule vehicle. 141 NSCLC patients were not included in this cohort. Dose-limiting toxicities (DLTs) for grade-3 fatigue and grade-3 bilirubin increase occurred at doses of 200 mg BID and 250 mg BID, respectively. Maximum tolerated dose (MTD) was not reached.
No DLTs were identified at 600 mg BID capsule formulation. In addition, 400 mg BID tablets were comparable in terms of tolerance and exposure. This dose was expected to achieve and sustain MET inhibition; therefore, this dose became the recommended Phase-2 dose (RP2D). Overall, capmatinib showed a good tolerance profile, with very few adverse events (AEs) that included nausea (32%), hyporexia (29%), vomiting (29%), and fatigue (26%). The most common grade 3 or 4 AE were fatigue (8%), ALT increase (5%), and hypophagia (5%). 141
There were two dose-expansion cohorts included in the primary trial that included advanced NSCLC patients. 142 The first cohort was comprised of MET dysregulated NSCLC, in which patients with MET over-expression and amplification were included. The second cohort enrolled EGFR wild-type NSCLC with MET over-expression identified by immunohistochemistry (IHC). A post hoc analysis further assessed MET status using gene copy number (GCN) and amplification by fluorescence in situ hybridization (FISH) and MET mutations by NGS. Overall, 55 subjects were included (26 for the first cohort and 29 for the second). Overall response rate (ORR) was 20% (95% CI: 10.4–33.0), with a particularly high response rate of 47% for 15 patients harboring MET GCN ⩾ 6 (n = 15). Remarkably, all patients with METex14 alterations achieved an objective response, including one patient with complete response (CR). The most frequently seen all grade capmatinib-related AEs included nausea (42%), peripheral edema (33%), and vomiting (31%), and no grade-3 or -4 capmatinib-related AEs occurred in >10% of patients. 142
GEOMETRY mono-1 (NCT02414139) is a phase-II single-arm, multi-center, multi-cohort trial evaluating the efficacy of capamatinib at a dose of 400 mg BID in the treatment of advanced NSCLC with wild-type EGFR and ALK. This trial is comprised of seven cohorts that are individually and centrally prescreened for MET status and past therapies. 143 Cohorts 1b, 2, and 3 included patients that had received prior therapy and have an MET amplification.
Enrollment in these cohorts was finalized early due to futility. Currently, preliminary results for METex14-altered NSCLC patients, regardless of GCN have been reported. These patients were divided in to two groups, cohort 4 for patients with 1–2 prior treatment lines (n = 69), and cohort 5b, that included treatment-naïve patients (n = 28). Primary endpoint was ORR, and secondary endpoint was duration of response (DOR). For cohort 4, ORR was 41% (95% CI: 27.6–51.6), with a median DOR (mDOR) of 9.7 months (95% CI: 5.5–13.0). For cohort 5b, ORR was found to be 68% (95% CI: 47.6–84.1), and the mDOR was 12.6 months (95% CI: 5.5–25.3). A small number of subjects affected by brain metastases were included in both cohorts, with an overall intracranial response of 54%, complete intracranial response was achieved in several cases. 143 The study included 334 patients in total. The observed safety profile was similar with earlier trials of capamatinib.
Most common all-grade capmatinib-related AEs were peripheral edema (42%), nausea (33%), creatinine increase (20%), vomiting (19%), fatigue (14%), hyporexia (13%), and diarrhea (11%), with the majority of them being grade 1–2. In 4.5% of patients, pneumonitis was seen, with grade-3 pneumonitis in 1.8% of patients and one death (0.3%). Treatment discontinuation was performed in 8 patients (2.4%) due to pneumonitis. Hepatotoxicity was diagnosed in 13% of patients, with a grade 3 or 4 severity seen in 6% and treatment discontinuation of 0.9%. Based on these results, the US FDA conceded accelerated approval for capmatinib (TabrectaTM) for NSCLC patients with an METex14 alteration detected by a companion diagnostic tool (FoundationOne CDx assay).
Even though capmatinib showed promising results, drug resistance was seen in patients with MET exon 14 mutations. Currently, the mechanisms for this drug resistance are not completely known and need to be further characterized. Secondary resistance to type-I MET inhibitors have been seen for MET mutations in D1228 and Y1230, through in vitro screening and mutagenesis assays.144,145 Different case reports have described these resistance mutations during crizotinib therapy for METex14 altered tumors.146 –149 In vitro experiments with MET-amplified cell lines exposed with capmatinib suggest that activation of EGFR signaling occurs, with further recruitment of downstream mediators like PIK3CA.144,150 –152 Similarly, preclinical data suggest that KRAS signaling may be upregulated in MET exon 14–mutated neoplasms, and this expression of mutant KRAS can induce resistance to MET-directed therapy. 153 Different genetic alterations are positively selected under MET-directed therapy. A case series of 20 patients under MET-directed TKI therapy found acquired MET resistance mutations, MET mutant allele amplification, new KRAS mutations, and amplifications of different drivers (KRAS, EGFR, HER3, and BRAF) after post-therapy NGS. 154 Acquisition of MET D1228 N mutations with additional HER3 and EGFR amplification was confirmed with NGS for one patient, and EGFR amplification with HER3 gain in a second patient. 154
An extensive series of 298 patients with MET exon 14 altered NSCLC showed a prevalence of concurrent MDM2 amplification of 35%, CDK4 amplification in 21%, EGFR amplification in 6%, and KRAS mutations in 3%. 155 Furthermore, simultaneous MET amplification was found in 15%, which was associated with a higher tumor mutational burden (TMB). Co-ocurring RAS-MAPK pathway genetic alterations in genes like KRAS and NF1 were also associated with a decreased response to MET TKI therapy in a case series of 289 patients. 156 All the potential implications of the mentioned genomic alterations over capmatinib-associated response and resistance are yet to be validated.
The patterns of clinical progression after capmatinib are described. In particular, brain metastases occur with greater frequency in oncogenic-driven NSCLC; however, it has not been possible to establish the MET exon 14 mutation population’s risk. 157 In a series of 34 patients with MET exon 14 altered NSCLC, brain metastases were diagnosed in 21% of patients, being the second-most common metastatic location after bone. In other descriptive study that evaluated 71 patients, the incidence of brain metastatic lesions was 37%. 158 As described previously, only a small subset of patients with brain metastases were included on GEOMETRY mono-1. However, intracranial responses were seen in over half, 75 suggesting moderate intracranial activity for capmatinib.
Lately, Dagogo-Jack et al. 154 reported the results of a phase-II trial that evaluated the use of capmatinib in patients with MET-Altered lung cancer previously treated with an MET inhibitor. A total of 20 patients were enrolled between May 2016 and November 2019, including 15 patients with MET skipping alterations and five patients with MET amplification. All patients had received crizotinib, and three had also received other MET-directed therapies. Two patients (10%) achieved an objective response to capmatinib, and 14 had SD, yielding a disease control rate (DCR) of 80%. Among five patients who discontinued crizotinib for intolerance, the DCR was higher (83%), including two cases with tumor shrinkage close to 30%. Intracranial DCR among four patients with measurable brain metastases was 100%, with no intracranial objective responses. Overall, the median PFS and OS were 5.5 (95% CI: 1.3–11.0) and 11.3 (95% CI: 5.5–NR) months, respectively. MET D1228 and Y1230 mutations and MAPK alterations were recurrently detected in post crizotinib, pre capmatinib plasma, NGS evaluation. Besides, new and persistent MET mutations and MAPK pathway alterations were detected at capmatinib progression. 159
Tepotinib (TEPMETKO). Tepotinib (EMD1214063) is also a type-Ib MET inhibitor with a high selectivity and potency profile. In preclinical studies, tepotinib was able to inhibit HGF-induced MET phosphorylation in cancer cell lines with a mean of IC50 of 3 nM. Tepotinib also induced clearance of human tumors in xenograft animal models. These results were seen regarding of MET activation (HGF-dependent or -independent).160,161
The first-in-human phase-I trial (registered with NCT01014936) of tepotinib in 149 patients with different advanced solid malignant neoplasms (17 cases of primary lung cancer) was conducted. There was not characterization of a maximum-tolerated dose with a daily dose of 1400 mg. The recommended dose for the following phase-II study (RP2D) was determined as 500 mg QD, as modeling data supported that this dose would be enough to achieve ⩾90% MET inhibition in ⩾90% of patients. 162
The VISION study (registered as NCT02864992), was a multi-center, open-label, and multi-cohort phase-II trial, clinically meaningful efficacy was seen with tepotinib in patients with advanced NSCLC with MET exon 14 mutations. Three cohorts were included as follows: cohort A were patients with skipping mutations; cohort B included subjects with MET amplification; and cohort C, which is still enrolling subjects with skipping alterations for confirmatory analysis of cohort A results. Until January 2020, 152 patients with MET exon 14 skipping tumors, that were diagnosed on tissue or liquid biopsy had received tepotinib (500 mg PO), from which 99 subjects (89 with adenocarcinoma) were eligible for analysis of outcomes. ORR was 46% (95% CI: 36.4–56.8; all were PRs) with a DCR of 65.7%. The median PFS was 8.5 months, and the median duration of OS was 17.1 months (95% CI: 12.0–26.8); however, data were immature at the timepoint of analysis. 163
AEs presented in 89% of the safety population. The most frequently seen treatment-related AE of grade 3 or higher were peripheral edema (7% prevalence), leading to a reduction in drug dose in 16% of subjects and a medication interruption in about 18%.
In March 2020, tepotinib received approval in Japan by the Ministry of Health, Labor and Welfare and fast track approval by the US FDA. Furthermore, ArcherMET® CDx was also approved to detect MET exon 14 mutations in advanced NSCLC patients from blood and tissue samples for consideration of treatment with tepotinib. 164
Since MET amplification is a mechanism of acquired resistance to EGFR after first-line osimertinib therapy, the hypothesis of adding tepotinib could overcome MET-related osimertinib resistance. Information on the combination of tepotinib plus gefitinib demonstrated improved patients’ outcomes with EGFR mutations who developed MET amplification as a resistance mechanism compared to chemotherapy (INSIGHT NCT01982955). The PFS was 16.6 versus 4.2 months (HR: 0.13; 90% CI: 0.04–0.43), and the OS was 37.3 versus 13.1 months (HR: 0.08; 90% CI: 0.01–0.51) for the TKIs combo and chemotherapy, respectively. 165 INSIGHT 2 (NCT03940703) is an ongoing global (approximately 100 sites in 15 countries), open-label, phase-II trial of tepotinib plus osimertinib in patients with advanced EGFR-mutant NSCLC designed to establish ORR by investigator assessment, DOR, PFS, OS, and pharmacokinetics. 166
Recently, Pudelko et al. assessed the short- and long-term effect of tepotinib on MET downstream signaling and the phosphorylation status of other than MET RTKs and non-RTKs on the parental tepotinib-resistant cells (EBC1 and EBC1-TR1). They observed activation of several RTKs, including ERBB (EGFR, ErbB2, and ErbB3), FGFR3, AXL, RET, DDR1, and M-CSFR in both cell lines. EBC1-TR1 cells displayed elevated levels of phosphorylated AXL and EGFR compared to EBC1 parental cells and increased phosphorylation of ERK1/2, AKT, c-Jun, and YES. 167 Also, the inhibition of Src homology 2-domain-containing phosphatase 2 (SHP2) delayed the emergence of tepotinib resistance and synergized with tepotinib in treatment-naive and tepotinib-resistant cells as well as in xenograft models. 168
Savolitinib. Savolitinib (AZD6094, volitinib, HMPL-504; AstraZeneca) is a potent and selective (>650 folds selectivity compared to other 265 kinases) type-Ib selective MET inhibitor that has shown anti-tumor activity in the preclinical setting and entered phase-I and -II studies. In a phase-I trial developed in patients with NSCLC, preliminary anti-tumor activity was observed in those with increased MET gene copy number, gene amplification, or high MET protein expression. 169 Preliminary results from a phase-II clinical trial (registered as NCT02897479) conducted in China showed high efficacy and safety of savolitinib in subjects diagnosed with pulmonary sarcomatoid carcinoma and other types of MET exon 14 NSCLC. The latest data disclose from this trial 170 showed that MET treatment-naïve patients (n = 70), from which 57.1% had NSCLC, had an ORR of 47.5% (95% CI: 34.6–60.7), and a DCR of 93.4% (95% CI: 84.1–98.2). Approximately 58% of subjects received treatment for 6 months or more. Median PFS was 6.8 months (95% CI: 4.2–13.8). Adverse events that led to treatment withdrawal were seen in 14.3% subjects. Hypersensitivity and hepatic injury and were the most common manifestations with an incidence of 2.9% each. The former study also demonstrated that savolitinib was able to enter the blood–brain barrier (BBB) and be effective in patients bearing central nervous system metastases. A drug application for savolitinib for the treatment of MET exon 14 NSCLC is currently under review by the China National Medical Products Administration. 18
Bozitinib. Bozitinib (APL-101, PLB1001, and CBT101) is a highly selective and specific MET inhibitor (8 nM) bearing a great activity in animal cancer models, including tumors like lung, hepatocellular, pancreatic, and gastric carcinoma. 171 Bozitinib was shown to have a higher apparent permeability as well as a lower efflux rate when compared with other MET inhibitors like foretinib, crizotinib, and cabozantinib. In an in vitro cellular experiment, bozitinib showed a higher specificity for MET inhibition and could permeate the BBB in murine specimens. Hu et al. analyzed the genetic variants of 188 IDH1/IDH2 + glioblastoma subjects and found MET exon 14 mutations in about 14% (95% CI: 8.0–23.5) of patients, with a positive association for a worse prognosis. In a phase-I clinical trial (registered as NCT02978261), bozitinib was evaluated in GBM harboring PTPRZ1-MET fusions and/or MET exon 14 (n = 6), two achieved PR, and two SD. 172 In addition, two had PD, and the TKI was tolerated with few side effects, recommending bozitinib as monotherapy with a dosage of 300 mg BID.
Different studies with bozitinib are already running. The NCT03175224 is a phase-I/II open-label, international, multi-center, trial assessing the pharmacokinetics, preliminary efficacy, and safety of bozitinib in NSCLC patients with MET exon 14 and MET dysregulated advanced solid malignant neoplasms. Another phase-II study (NCT04258033) recently started in China included 185 participants with advanced MET-dysregulated NSCLC with the objective of assessing efficiency and safety of bozitinib. 18
TPX-0022. A type-I kinase inhibitor with a macrocyclic structure that designed to inhibit MET, CSF1R, and SRC with enzymatic kinase inhibition IC50 values of 0.14, 0.71, and 0.12 nM, respectively. 173 In MET-amplified gastric cancer cell lines, TPX-0022 inhibited cell division, with IC50 < 0.2 nM, which is comparable to capmatinib. Compared to crizotinib, TPX-0022 had a >10-fold potency. TPX-0022 also demonstrated tumor growth inhibition by modification of the tumor microenvironment via induction of differentiation of tumor-associated macrophages into an M1 phenotype, as well as increasing cytotoxicity of T-cells. 173
The first-in-human dose-escalation phase I clinical trial, SHIELD-1 (NCT03993873), involving patients with advanced solid tumors harboring MET alterations (exon 14 deletions, MET amplification, fusion, or oncogenic kinase domain mutations) was recently finished. At the data cutoff, 22 heavily pretreated patients, with a median of 3 prior therapy lines (range: 1–6), were enrolled. The median patient age was 63 years (range: 44–84), and 15 patients (68.2%) had an ECOG performance status of 1. There were 13 patients (59.1%) with NSCLC, 4 (18.2%) with gastric/gastroesophageal junction (GEJ) cancer, 4 (18.2%) with CRC, and 1 (4.5%) with glioblastoma. Most AEs were grade 1 or 2 in severity. The most common all-grade treatment-emergent AEs were dizziness (55%), attributed to off-target TRK inhibition, increased lipase (32%), and fatigue (32%). Five patients (23%) experienced AEs that led to dose reduction, and 2 (9%) experienced AEs that led to dose discontinuation. The most common all-grade treatment-related AEs were dizziness (46%), increased lipase (23%), and increased amylase (18%). Of the 10 patients who were TKI naive, five achieved PR. Three responding patients had gastric/GEJ cancer, 1 had CRC, and 1 had NSCLC. 174
Type II MET small molecule inhibitors
Cabozantinib (CABOMETYX). Cabozantinib (also known as Cometriq, XL184, BMS-907351, Exelixis) is a type-II MET inhibitor (IC50 value of 1.3 nM) with activity over a wide array of molecular targets, including VEGFR2, FLT3, c-KIT, AXL and RET. Cabozantinib is currently approved by the FDA for the treatment of advanced medullary thyroid carcinoma, advanced clear-cell renal-cell carcinoma and for hepatocellular carcinoma after relapse with sorafenib. Starting in 2005, cabozantinib was the first oral MET inhibitor entering clinical trials. With IC50 values of 4, 5, and 14.6 nM, cabozantinib is able to inhibit MET-activating kinase domain mutations in the residues Y1248 C/H, D1246 N, and K1262R, respectively. In murine experiments, cabozantinib drastically changed tumor histology, resulting in tumoral regression, reduced endothelial proliferation and increased apoptotic rate. Tumor growth inhibition was dose-dependent in breast, lung, and glioma models. 175 A previous study carried out in an unselected population showed an ORR at week 12 of 10%, including six patients with confirmed PR. The overall DCR at week 12 was 38%, and objective tumor regression was observed in 64%. 176
Although clinical trials of cabozantinib in MET exon 14 alterations have not been published yet, multiple case studies suggest a good safety profile and effectiveness of this multikinase inhibitor.111,177,178 An Italian phase-II trial (CABinMET, NCT03911193) currently evaluates cabozantinib in patients with MET-mutated NSCLC.
Merestinib (LY2801653). Merestinib is a multi-targeted TKI that can inhibit MET, RON, AXL, MER receptor tyrosine kinase (MERTK), TIE-2, TIE-1, ROS1, and discoidin domain RTK 1 (DDR1). The in vitro IC50 of merestinib against MET is 4.7 nM and the cell-based IC50 is 35–52 nM, depending on the cell lines utilized. Inhibition of tumor growth and metastasis in NSCLC by merestinib, an inhibitor of several kinases, including MET. 179
Treatment with merestinib inhibited MET signaling’s constitutive activation and resulted in inhibition of cell proliferation, anchorage-independent growth, migration, and invasion. In addition, in the H1993 NSCLC cell line, which harbors MET amplification and also over-expression of RON, merestinib was superior to crizotinib in terms of cellular growth inhibitory activity (9.28 nM versus 45.4 nM, respectively). 139 Recently, Recondo et al. reported a case of a patient with MET exon 14 skipping who developed PD on crizotinib, with a resistance MET mutation of Y1230 C detected in tumor and liquid biopsies at progression. This patient achieved PR when switched to merestinib. 154 These results might suggest a possible introduction of merestinib as a therapeutic alternative for patients with MET exon 14. In the first-in-human phase I trial, merestinib was evaluated for tolerability and safety in 186 patients with one of three different non-NSCLC tumors. Approximately 32% of subjects enrolled had a SD as the best response with a recommended dose of 120 mg QD with acceptable exposure and safety. 180
Glesatinib (MGCD265). Glesatinib (MGCD265) is an orally bioavailable, small molecule, multi-targeted tyrosine kinase inhibitor with potential antineoplastic activity. Glesatinib binds to and inhibits the phosphorylation of several RTKs, including the MET receptor, Tek/Tie-2 receptor, VEGFR types 1, 2, and 3, and RO. 181 Inhibition of these RTKs and their downstream signaling pathways may inhibit tumor angiogenesis and tumor cell proliferation. Preclinical studies showed a dose-dependent inhibition in tumoral growth with glesatinib, with an IC50 of 80 nM, in the H1299 NSCLC cell line. 182
Studies in a gastric cancer xenograft model revealed that, in addition to the typically reported cellular activities, glesatinib combined with erlotinib disrupted the glycolysis pathway, suggesting a novel mechanism of action for this drug. Glesatinib has been studied in various advanced solid tumors, including NSCLC, as a monotherapy and in combination with either docetaxel or erlotinib. In an ongoing phase-1 study in patients with MET-positive or AXL-rearranged advanced solid tumors, glesatinib demonstrated preliminary single-agent activity with all three patients with MET dysregulated NSCLC. For example, a patient with MET exon 14 NSCLC achieved response with glesatinib following a relapse under crizotinib, including a considerable size decrease of a liver metastatic lesion positive for MET Y1230 H as well as the inability to detect this alteration in plasma DNA. 18
Antibody-based therapies
Unlike small molecule inhibitors that are ATP-competitive by inhibiting MET kinase domain, immunotherapy targeting the HGF/MET pathway alter signaling by inhibition of the interaction between receptor and ligand. Different than small molecule inhibitors that usually target multiple RTKs, monoclonal antibodies specifically target the MET protein, an event proven in preclinical and clinical studies in progress. Because antibody-based therapies’ mechanism of action is disrupting HGF/MET interaction, most trials enroll patients with MET over-expression, not restricted to MET exon 14.
Targeting MET with monoclonal antibodies has been challenging. In the randomized phase-III of the monoclonal antibody onartuzumab (MetMAb) in unselected patients with NSCLC, adding onartuzumab to erlotinib in patients previously treated with chemotherapy had a detrimental effect on OS; this therapy also showed no benefit in an exploratory analysis of patients with MET-amplified tumors by FISH (>5 copies). 183 Similarly, the addition of onartuzumab to chemotherapy treatment for patients with advanced NSCLC, gastrointestinal tumors, and glioblastoma was not effective. 184 A similar compound emibetuzumab (LY2875358) was initially evaluated, but its development was later stopped. 185 Rilotumumab, a monoclonal antibody directed against soluble HGF, also resulted in deleterious outcomes combined with chemotherapy in patients with advanced gastric and esophagogastric tumors. 186 These early trials lacked adequate biomarker enrichment and patient selection and MET expression by IHC has not proven to be a reliable indicator of MET oncogenic dependency. The leading cause of the ineffectiveness of combining monoclonal METs with chemotherapy in unselected patients is unclear. 187 However, one of the hypotheses is that MET inhibitors may dysregulate immune-mediated cytotoxicity by altering the tumor microenvironment. For instance, MET inhibition can impair interferon-gamma induction of programmed cell death ligand 1 (PD-L1) expression in vitro and decrease neutrophil anti-tumor activity.188,189
Telisotuzumab vedotin. Telisotuzumab vedotin (ABBV-399, ABT-700; ABBVie) is an antibody-drug conjugate comprised of telisotuzumab, which is a monoclonal antibody that targets MET RTK, conjugated to a cytotoxic agent called monomethyl auristatin E (MMAE) using a valine-citrulline (vc) peptide linker (vc-MMAE; vedotin). After the conjugate binds to MET, internalization and intracellular enzymatic cleavage occurs, releasing MMAE in the cytosol, where it binds to tubulin monomers, inhibiting tubulin polymerization, inducing a G2/M phase arrest with further cancer cell apoptosis. 190 Telisotuzumab vedotin can antagonize MET signaling in both HGF-dependent and -independent manners and inhibit tumor growth driven by MET over-expression, amplification, or autocrine HGF stimulation.191,192
In the first in-human trial of ABBV-399 (NCT02099058), 46 patients were enrolled. Approximately 60% of the subjects presented with NSCLC positive for MET alterations.
Sixteen patients with MET-positive NSCLC were treated with a dose of 2.4–3.0 mg/kg, from which 3 (18.8%; 95% CI: 4.1–45.7) achieved a PR (DOR, 4.8 months and PFS 5.7 months; 95% CI: 1.2–15.4). Only one patient with NSCLC of squamous histology was confirmed to bear MET exon 14 and achieved PD as the best response. 193
Lung-MAP S1400 K was designed to evaluate the response to telisotuzumab vedotin in patients with MET–positive squamous cell NSCLC. 194 In this trial, patients with previously treated SCC with–positive tumors (H score ⩾ 150, Ventana SP44 assay) were enrolled into two cohorts (cohort 1, immune checkpoint inhibitor-naive, and cohort 2, immune checkpoint inhibitor refractory). Telisotuzumab vedotin 2.7 mg/kg was administered intravenously every 3 weeks until disease progression or unacceptable toxicity, response assessments were performed every 6 weeks, and the primary endpoint was the response. Forty-nine patients (14% of screened patients) were assigned to S1400 K, 28 patients enrolled (15 in Cohort 1 and 13 in Cohort 2), and 23 were eligible. S1400 K closed on December 21, 2018, owing to a lack of efficacy. Overall, two responses (ORR of 9%; 95% CI: 0–20%) were reported in cohort 1 (1 complete and one unconfirmed PR), whereas 10 patients had SD, with a DCR of 52%. The median OS and PFS were 5.6 and 2.4 months, respectively. There were three grade-5 events (2 pneumonitis in cohort 2, and 1 bronchopulmonary hemorrhage in cohort 1). 194
Recently, the findings from the phase-2 trial (NCT03539536) were presented in a poster at the American Association for Cancer Research Annual Meeting 2021. 195 The study explored the safety and efficacy of telisotuzumab vedotin with a tubulin inhibitor MMAE, in previously treated patients with MET–positive advanced NSCLC. The ongoing phase-2 study expects to enroll about 233 patients across two stages and has an estimated completion date of January 2025. During the open-label, single-arm study, patients received telisotuzumab vedotin at 1.9 mg/kg intravenously every 14 days. Patients with non-squamous histology were separated into cohorts by EGFR mutations and then into subgroups based on level of MET expression; intermediate MET was defined as staining on ⩾25% to <50% of tumor cells at 3+ intensity, and high MET was considered ⩾50% staining at 3+ intensity. For patients with squamous histology, MET positivity was defined as staining on ⩾75% of tumor cells at 1+ intensity. The results included 93 evaluable patients from stage 1 of the study. MET amplification was reported in 8.6% and MET exon 14 skipping mutations were observed in 4.3% of patients, all in the non-squamous EGFR wild-type (WT) cohort. Patients had received a median of 2 prior therapies (range: 1–4), which included platinum-based therapies in most patients, immunotherapy for most patients with EGFR WT, and EGFR tyrosine kinase inhibitors for all the patients with EGFR mutations. 195 For the non-squamous EGFR WT cohort (n = 37), the ORR was 35.1% (95% CI: 20.2–52.5%). For the MET-high subgroup (n = 13), the ORR was 53.8% (95% CI: 25.1–80.8%) and 25.0% (95% CI: 9.8–46.7%) for the MET-intermediate subgroup which in turn had a DOR of 6.9 months. For the EGFR-mutant group (n = 30), the ORR was 13.3% (95% CI: 3.8–30.7%), with responses only in those with high MET (n = 22). Moreover, for the squamous cohort (n = 21), the ORR was 14.3% (95% CI: 3.0–36.3%; all responses were PR) and the DOR was 4.4 months. 195 An ongoing phase 1/1b trial (NCT02099058) is evaluating telisotuzumab vedotin in patients with NSCLC with MET over-expression and in a subgroup (cohort E), the use of the biconjugate plus osimertinib. 196
Amivantamab (JNJ-61186372, JNJ-6372). MET amplification is a well-known mechanism of resistance to EGFR-TKI therapy. Different antibodies targeting the interaction HGF/MET have been evaluated in TKI-resistant EGFR-mutant NSCLC, including drugs like onartuzumab,183,197 ficlatuzumab, 198 rilotumumab, 199 and emibetuzumab. 200 Bispecific antibodies for EGFR and MET have also been studied in NSCLC harboring both EGFR and MET mutations. Some examples are amivantamab and LY3164530. Amivantamab (JNJ-61186372, JNJ-6372; Janssen) is an EGFR-MET bispecific IgG 1 monoclonal antibody with an active Fc backbone that can target both activating and resistant EGFR mutations and MET mutations and amplification. 201
In preclinical studies, the combination of amivantamab with Lazertinib (YH25448), an oral, highly potent, irreversible, third-generation, mutant-selective, and wild-type-sparing EGFR-TKI, demonstrates synergistic inhibition of tumor growth. The phase-1 CHRYSALIS study (NCT02609776) recently present the safety and early efficacy results of the combination in EGFR-mutant patients progressing on Osimertinib. Seventy-one patients received the combination with an ORR of 43.5% (95% CI: 23.2–65.5), including 10 PRs, and nine patients with SD. The median treatment duration was 8.2 months (0.5–10.7), and most treatment-related AEs were grade 1–2, with grade ⩾3 reported in 7%.
A first-in-human, open-label, multi-center, phase-I trial on amivantamab (NCT02609776) demonstrated efficacy (ORR 36%) with an acceptable safety profile in patients with EGFR exon 20 ins NSCLC treated with multiple previous lines. 202 Based on this study, FDA-granted breakthrough therapy designation for amivantamab to treat patients with metastatic NSCLC with EGFR exon 20 insertion mutations, whose disease has progressed on or after platinum-based chemotherapy. Recently, the CHRYSALIS Study showed robust clinical activity and durable responses in patients with metastatic or unresectable NSCLC and EGFR Exon 20 insertion mutations. 202 The clinical development program for amivantamab in untreated advanced EGFR-mutated NSCLC includes the Phase-3 MARIPOSA and PAPILLON combination trials.203,204 Emibetuzumab (LY3164530) is another bispecific antibody for MET and EGFR receptors, consisting of an IgG4 to MET and a single-chain variable fragment (scFv) to EGFR fused to the N-terminus of each heavy chain. An ORR of 10.3% was seen in the first-in-human study (NCT02221882), with the presence of toxicities commonly associated with EGFR inhibition. 205
Sym-015. Sym-015 is a combination of two humanized IgG1 monoclonal antibodies, (Hu9006 and Hu9338), that recognize non-overlapping epitopes in the Sema domain of MET, which prevents HGF from binding.206,207 An open-label, phase Ia/IIa trial (registered as NCT02648724) for sym-015, included 12 patients with MET exon 14 NSCLC, treated with a P2 dose of 18 mg/kg on cycle 1 day 1 followed by 12 mg/kg Q2 W. Three subjects developed PR and five SD. Sym-015 showed a good level of tolerance with the P2 dose with a good response in NSCLC harboring MET exon 14 skipping mutations. 208
Immunotherapy in patients with METex14
Unlike EGFR/ALK-positive NSCLC, which are not characterized for expression of PD-L1, MET exon 14 tumors express remarkable levels of PD-L1. Two previous studies, developed in China and in the United States, showed that 41% and 69% of MET exon 14 NSCLC had a had a PD-L1 expression of ⩾50%, respectively.209,210 Both studies results showed a considerably higher expression when compared with a larger cohort of 1398 patients with NSCLC (no specific subgroup) (20.9%). 211 Awad et al. reported a large cohort of 1387 MET exon 14 NSCLC demonstrating that they express considerably higher levels of PD-L1 compared to wild-type NSCLC (48% vs 29%). Even though PD-L1 expression is higher in MET exon 14 NSCLC, TMB distribution in these group of tumors was lower than general NSCLC (3.6 vs 7.0 Mut/Mb). 212
A study with 298 MET exon 14 NSCLC reported an average TMB of 6.9 Mut/Mb, compared with 10.7 Mut/Mb for unselected NSCLC. 155
To date, the information on the use of immunotherapy in patients with MET alterations, and especially with the MET exon 14 mutation, is limited. Case reports and case series suggest that immunotherapy might not be effective for METex14 NSCLC despite a high PD-L1 expression. One possible hypothesis underlying the inferior response to checkpoint inhibition is the low TMB. Baba et al. 208 reported a patient with 95% PD-L1 METex14 NSCLC that had no response to pembrolizumab. Reis et al. 209 reported two similar cases.
In a small study of patients with MET exon 14 NSCLC (N = 25), of whom 13 received an immune checkpoint inhibitor in the second-line setting, six patients had prolonged PFS (>18 months). Of these six patients, five showed responses within the first four months of treatment, four patients had a PR, and two had a CR. PD-L1 levels were ⩾20% for four of six patients; however, these data must be interpreted carefully because the outcomes for the other seven patients are not described. 213 In a retrospective study published by Sabari et al., 24 patients with MET exon 14 cancers received either treatment with one agent (n = 22) or concomitant immunotherapy, including 11 patients on first line, with an ORR of 17% (95% CI: 6–36%) and a median PFS of 1.9 (95% CI: 1.7–2.7) months. Responses to immunotherapy were not predictable by PD-L1 expression (including PD-L1 levels ⩾50%) nor TMB. 209 This ORR was similar to the response rate of 14% observed in the OAK trial with atezolizumab, which had an unselected, previously treated patient population (N = 425). 214 In contrast, a preclinical study revealed a role for the HGF / MET pathway in neutrophil recruitment and function and suggested that MET co-treatment may improve responses to cancer immunotherapy in patients with MET-dependent tumors. 215 In an in vitro study of a gastric cancer cell line (Hs746 T) harboring both MET exon 14 and MET amplification, it was found that MET pathway and PD-L1 expression can suppress immune cell function.216,217 The COSMIC-021218 trial is a multi-center phase Ib clinical trial to evaluate the safety and efficacy of cabozantinib in combination with atezolizumab in patients with multiple tumor types, including NSCLC. The dose-escalation phase of this study determined the optimal dose of cabozantinib to be 40 mg daily combined with atezolizumab.219 In the ASCO Annual Meeting 2020, Neal et al. reported the results from cohort 7 of NSCLC with unknown MET status patients after prior immunotherapy. In the 30-patient cohort, ORR was calculated as 27%, mean time to response was 1.4 months, median DOR was 5.7 months, DCR was 83%, and median PFS was 4.2 (95% CI: 2.7–7) months.
Other studies explore the efficacy and safety of MET TKI combined with immunotherapy. These studies including capmatinib + pembrolizumab (NCT04139317) and capmatinib + nivolumab (NCT02323126). In 2020, enrollment for the NCT04323436 trial started. This is a double-blind, placebo-controlled, randomized study evaluating the efficacy and safety of capmatinib + spartalizumab (an anti-PD-1 antibody) versus capmatinib + placebo as first-line treatment for advanced MET exon 14 NSCLC patients. The primary endpoints of NCT04323436 trial are ORR and PFS. 18
Conclusion
As genomic understanding of cancer improves, the knowledge regarding lung cancer have created so many different categories, that lung cancer is no longer a single disease, but a group of heterogenous neoplastic disorders that differ slightly or even drastically in their genetic makeup. During the last decade, the revolution of targeted therapy and immunotherapy have increased considerably the number of clinical trials and therapies approved for specific types of driver-mutated lung cancer. The availability of a targeted approach to KRAS is just a few years away from current therapy. Some TKIs have improved the survival of patients harboring MET alterations, however, a wide array of clinical trials are running, with promising results in the near future.
Footnotes
Author contributions: Juan Esteban Garcia-Robledo: Methodology; Resources; Software; Writing – original draft; Writing – review & editing.
R Rosell: Writing – review & editing.
Alejandro Ruiz-Patiño: Visualization; Writing – review & editing.
Carolina Sotelo: Writing – review & editing.
Oscar Arrieta: Writing – review & editing.
Zyanya Lucia, Zatarain-Barrón: Writing – review & editing.
Camila Ordóñez-Reyes: Writing – review & editing.
Jaller: Writing – review & editing.
Leonardo Rojas: Writing – review & editing.
Alessandro Ruso: Writing – review & editing.
Diego de Miguel Pérez: Writing – review & editing.
Christian Rolfo: Writing – review & editing.
Andrés F., Cardona: Conceptualization; Formal analysis; Methodology; Project administration; Resources; Software; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing.
Conflict of interest statement: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: AFC discloses financial research support from Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb, Foundation Medicine, Roche Diagnostics, Termo Fisher, Broad Institute, BioNTech, Amgen, Flatiron Health, Teva Pharma, Rochem Biocare, Bayer, INQBox and The Foundation for Clinical and Applied Cancer Research – FICMAC. In addition, he was linked and received honoraria as an advisor, participate in speakers’ bureau. He gave expert testimony to EISAI, Merck Serono, Jannsen Pharmaceutical, Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb, Pfizer, Novartis, Celldex Therapeutics, Foundation Medicine, Eli Lilly, Guardant Health, Illumina, and Foundation for Clinical and Applied Cancer Research – FICMAC. OA reports personal fees from Pfizer, grants and individual fees from AstraZeneca, grants and individual fees from Boehringer-Ingelheim, Lilly, Merck, Bristol-Myers Squibb, Roche, outside the submitted work. CR reports relation with Mylan, Archer Biosciences, Oncopass, Inivata, Merck Serono Novartis, MSD, Boehringer-Ingelheim, Guardant Health, etc AstraZeneca as part of Speakers’ Bureau. Also, he received research funding from Pfizer and had uncompensated Relationships with OncoDNA, Biomark, and Guardant Health. LR received honoraria as an advisor and participate in speakers’ bureau from Merck Sharp & Dohme, Boehringer-Ingelheim, Roche, Bristol-Myers Squibb, Astra Zeneca and Eli Lilly. In addition, he was linked and received honoraria as researcher.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Juan Esteban Garcia-Robledo  https://orcid.org/0000-0003-2912-152X
https://orcid.org/0000-0003-2912-152X
Alejandro Ruíz-Patiño  https://orcid.org/0000-0003-1274-9273
https://orcid.org/0000-0003-1274-9273
Andrés F. Cardona  https://orcid.org/0000-0003-3525-4126
https://orcid.org/0000-0003-3525-4126
Contributor Information
Juan Esteban Garcia-Robledo, Division of Hematology/Oncology, Mayo Clinic, Scottsdale, AZ, USA.
Rafael Rosell, Cancer Biology and Precision Medicine Program, Germans Trias i Pujol Research Institute (IGTP)/Dr. Rosell Oncology Institute (IOR), Quirón-Dexeus University Institute, Barcelona, Spain.
Alejandro Ruíz-Patiño, Direction of Research and Education, Luis Carlos Sarmiento Angulo Cancer Treatment and Research Center (CTIC), Bogotá, Colombia; Foundation for Clinical and Applied Cancer Research (FICMAC), Bogotá, Colombia Molecular Oncology and Biology Systems Research Group (Fox-G), Universidad El Bosque, Bogotá, Colombia; Foundation for Clinical and Applied Cancer Research (FICMAC), Bogotá, Colombia Molecular Oncology and Biology Systems Research Group (Fox-G), Universidad El Bosque, Bogotá, Colombia.
Carolina Sotelo, Foundation for Clinical and Applied Cancer Research (FICMAC), Bogotá, Colombia Molecular Oncology and Biology Systems Research Group (Fox-G), Universidad El Bosque, Bogotá, Colombia.
Oscar Arrieta, Thoracic Oncology Unit and Personalized Oncology Laboratory, National Cancer Institute (INCan), México City, México.
Lucia Zatarain-Barrón, Thoracic Oncology Unit and Personalized Oncology Laboratory, National Cancer Institute (INCan), México City, México.
Camila Ordoñez, Foundation for Clinical and Applied Cancer Research (FICMAC), Bogotá, Colombia Molecular Oncology and Biology Systems Research Group (Fox-G), Universidad El Bosque, Bogotá, Colombia.
Elvira Jaller, Department of Internal Medicine, Universidad El Bosque, Bogotá, Colombia.
Leonardo Rojas, Foundation for Clinical and Applied Cancer Research (FICMAC), Bogotá, Colombia Department of Clinical Oncology, Clínica Colsanitas, Bogotá, Colombia Clinical and Translational Oncology Group, Clínica del Country, Bogotá, Colombia.
Alessandro Russo, Medical Oncology Unit, A.O. Papardo, Messina, Italy Marlene and Stewart Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore, MD, USA.
Diego de Miguel-Pérez, Marlene and Stewart Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore, MD, USA.
Christian Rolfo, Marlene and Stewart Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, Baltimore, MD, USA.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. DOI: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2. Travis WD, Brambilla E, Burke AP, et al. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol 2015; 10: 1240–1242. [DOI] [PubMed] [Google Scholar]
- 3. Tolwin Y, Gillis R, Peled N. Gender and lung cancer – SEER-based analysis. Ann Epidemiol 2020; 46: 14–19. [DOI] [PubMed] [Google Scholar]
- 4. Jemal A, Ma J, Rosenberg PS, et al. Increasing lung cancer death rates among young women in southern and midwestern states. J Clin Oncol 2012; 30: 2739–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howlader N, Forjaz G, Mooradian MJ, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med 2020; 383: 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang W, Liu J, He J. Driving the improvement of lung cancer prognosis. Cancer Cell 2020; 38: 449–451. [DOI] [PubMed] [Google Scholar]
- 7. Yuan M, Huang LL, Chen JH, et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther 2019; 4: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajurkar S, Mambetsariev I, Pharaon R, et al. Non-small cell lung cancer from genomics to therapeutics: a framework for community practice integration to arrive at personalized therapy strategies. J Clin Med 2020; 9: 1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fakih M, O’Neil B, Price TJ, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRAS G12C inhibitor, in advanced solid tumors. J Clin Oncol 2019; 37: 3003. [Google Scholar]
- 10. Collisson EA, Taylor B, Campbell J, et al. Comprehensive molecular profiling of lung adenocarcinoma: the cancer genome atlas research network. Nature 2014; 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 2015; 10: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCormick F. KRAS as a therapeutic target. Clin Cancer Res 2015; 21: 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J, McMillan E, Kim HS, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 2016; 538: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J, Hu K, Guo J, et al. Suppression of KRas-mutant cancer through the combined inhibition of KRAS with PLK1 and ROCK. Nat Commun 2016; 7: 11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nichols RJ, Haderk F, Stahlhut C, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 2018; 20: 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghimessy A, Radeczky P, Laszlo V, et al. Current therapy of KRAS-mutant lung cancer. Cancer Metastasis Rev 2020; 39: 1159–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012; 12: 89–103. [DOI] [PubMed] [Google Scholar]
- 18. Hong L, Zhang J, Heymach JV, et al. Current and future treatment options for MET exon 14 skipping alterations in non-small cell lung cancer. Ther Adv Med Oncol. Epub ahead of print 15 February 2021. DOI: 10.1177/1758835921992976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015; 5: 850–859. [DOI] [PubMed] [Google Scholar]
- 20. Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005; 65: 1479–1488. [DOI] [PubMed] [Google Scholar]
- 21. Weiss RA. A perspective on the early days of RAS research. Cancer Metastasis Rev 2020; 39: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirsten WH, Schauf V, McCoy J. Properties of a murine sarcoma virus. Bibl Haematol 1970; 36: 246–249. [DOI] [PubMed] [Google Scholar]
- 23. Schäfer R, Griegel S, Schwarte I, et al. Transforming activity of DNA fragments from normal human lymphocytes results from spontaneous activation of a c-Ha-ras1 gene. Mol Cell Biol 1985; 5: 3617–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krontiris TG, Cooper GM. Transforming activity of human tumor DNAs. Proc Natl Acad Sci U S A 1981; 78: 1181–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pulciani S, Santos E, Lauver AV, et al. Transforming genes in human tumors. J Cell Biochem 1982; 20: 51–61. [DOI] [PubMed] [Google Scholar]
- 26. Shimizu K, Birnbaum D, Ruley MA, et al. Structure of the Ki-ras gene of the human lung carcinoma cell line Calu-1. Nature 1983; 304: 497–500. [DOI] [PubMed] [Google Scholar]
- 27. Rásó E. Splice variants of RAS – translational significance. Cancer Metastasis Rev 2020; 39: 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsai FD, Lopes MS, Zhou M, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A 2015; 112: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hunter JC, Manandhar A, Carrasco MA, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res 2015; 13: 1325–1335. [DOI] [PubMed] [Google Scholar]
- 30. Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017; 170: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer 2003; 3: 459–465. [DOI] [PubMed] [Google Scholar]
- 32. Li H, Yao XQ, Grant BJ. Comparative structural dynamic analysis of GTPases. PLoS Comput Biol 2018; 14: e1006364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vigil D, Cherfils J, Rossman KL, et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy. Nat Rev Cancer 2010; 10: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 35. Gibbs JB, Sigal IS, Poe M, et al. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A 1984; 81: 5704–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGrath JP, Capon DJ, Goeddel DV, et al. Comparative biochemical properties of normal and activated human ras p21 protein. Nature 1984; 310: 644–649. [DOI] [PubMed] [Google Scholar]
- 37. Sweet RW, Yokoyama S, Kamata T, et al. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature 1984; 311: 273–275. [DOI] [PubMed] [Google Scholar]
- 38. Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol 2016; 58: 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salgia R, Pharaon R, Mambetsariev I, et al. The improbable targeted therapy: KRAS as an emerging target in non-small cell lung cancer (NSCLC). Cell Rep Med 2021; 2: 100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. El Osta BE, Behera M, Kim S, et al. Characteristics and outcomes of patients (pts) with metastatic KRAS mutant lung adenocarcinomas: lung cancer mutation consortium (LCMC) database. J Clin Oncol 2017; 35: 9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013; 24: 2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seidel D, Zander T, Heukamp LC, et al. A genomics-based classification of human lung tumors. Sci Transl Med 2013; 5: 209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu QG, Zhang SM, Ding XX, et al. Driver genes in non-small cell lung cancer: characteristics, detection methods, and targeted therapies. Oncotarget 2017; 8: 57680–57692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012; 18: 6169–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Redig AJ, Chambers ES, Lydon CA, et al. Genomic complexity in KRAS mutant non-small cell lung cancer (NSCLC) from never/light-smokers v smokers. J Clin Oncol 2016; 34: 9087. [Google Scholar]
- 47. Ferrer I, Zugazagoitia J, Herbertz S, et al. KRAS-mutant non-small cell lung cancer: from biology to therapy. Lung Cancer 2018; 124: 53–64. [DOI] [PubMed] [Google Scholar]
- 48. Muñoz-Maldonado C, Zimmer Y, Medová M. A comparative analysis of individual ras mutations in cancer biology. Front Oncol 2019; 9: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012; 104: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kimmelman AC. Metabolic dependencies in RAS-driven cancers. Clin Cancer Res 2015; 21: 1828–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016; 531: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang H, Liang SQ, Schmid RA, et al. New horizons in KRAS-mutant lung cancer: dawn after darkness. Front Oncol 2019; 9: 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015; 5: 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 2018; 8: 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed 2013; 98: 236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jing C, Mao X, Wang Z, et al. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her-2 and TP53 mutations in patients with non-small cell lung cancer. Mol Med Rep 2018; 18: 2191–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nacchio M, Sgariglia R, Gristina V, et al. KRAS mutations testing in non-small cell lung cancer: the role of liquid biopsy in the basal setting. J Thorac Dis 2020; 12: 3836–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet 2019; 20: 631–656. [DOI] [PubMed] [Google Scholar]
- 59. Maroni G, Bassal MA, Krishnan I, et al. Identification of a targetable KRAS-mutant epithelial population in non-small cell lung cancer. Commun Biol 2021; 4: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang IS, Kim S. Isoform specific gene expression analysis of KRAS in the prognosis of lung adenocarcinoma patients. BMC Bioinformatics 2018; 19: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B 2019; 9: 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Janes MR, Zhang J, Li LS, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 2018; 172: 578–589.e17. [DOI] [PubMed] [Google Scholar]
- 63. Patricelli MP, Janes MR, Li LS, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 2016; 6: 316–329. [DOI] [PubMed] [Google Scholar]
- 64. Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019; 575: 217–223. [DOI] [PubMed] [Google Scholar]
- 65. Goebel L, Müller MP, Goody RS, et al. KRasG12C inhibitors in clinical trials: a short historical perspective. RSC Med Chem 2020; 11: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hallin J, Engstrom LD, Hargis L, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020; 10: 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Romero D. Two new agents target KRAS G12C. Nat Rev Clin Oncol 2020; 17: 6. [DOI] [PubMed] [Google Scholar]
- 68. Tanaka N, Lin JJ, Li C, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov Candisc 2021; 11: 1913–1922. DOI: 10.1158/2159-8290.cd-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jiang T, Shi T, Zhang H, et al. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol 2019; 12: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Apostolopoulos V. Cancer vaccines: research and applications. Cancers 2019; 11: 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arbelaez CA, Estrada J, Gessner MA, et al. A nanoparticle vaccine that targets neoantigen peptides to lymphoid tissues elicits robust antitumor T cell responses. Npj Vaccines 2020; 5: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wan Y, Zhang Y, Wang G, et al. Recombinant KRAS G12D protein vaccines elicit significant anti-tumor effects in mouse CT26 tumor models. Front Oncol 2020; 10: 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cullis J, Das S, Bar-Sagi D. Kras and tumor immunity: friend or foe? Cold Spring Harb Perspect Med 2018; 8: a031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Garcia-Robledo JE, Barrera MC, Tobón GJ. CRISPR/Cas: from adaptive immune system in prokaryotes to therapeutic weapon against immune-related diseases: CRISPR/Cas9 offers a simple and inexpensive method for disease modeling, genetic screening, and potentially for disease therapy. Int Rev Immunol 2020; 39: 11–20. [DOI] [PubMed] [Google Scholar]
- 75. Berdien B, Mock U, Atanackovic D, et al. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther 2014; 21: 539–548. [DOI] [PubMed] [Google Scholar]
- 76. Tran E, Robbins PF, Lu YC, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016; 375: 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rive CM, Yung E, Hughes C, et al. Recombinant T cell receptors specific for HLA-A*02:01-restricted neoepitopes containing KRAS codon 12 hotspot mutations. bioRxiv 2020. DOI: 10.1101/2020.06.15.149021. [DOI] [Google Scholar]
- 78. Uehara Y, Minowa O, Mori C, et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 1995; 373: 702–705. [DOI] [PubMed] [Google Scholar]
- 79. Bladt F, Riethmacher D, Isenmann S, et al. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 1995; 376: 768–771. [DOI] [PubMed] [Google Scholar]
- 80. Huh CG, Factor VM, Sánchez A, et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A 2004; 101: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu X, Newton RC, Scherle PA. Developing c-MET pathway inhibitors for cancer therapy: progress and challenges. Trends Mol Med 2010; 16: 37–45. [DOI] [PubMed] [Google Scholar]
- 82. Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol 2011; 3(Suppl. 1): S7–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shen Y, Naujokas M, Park M, et al. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 2000; 103: 501–510. [DOI] [PubMed] [Google Scholar]
- 84. Rodrigues GA, Park M. Autophosphorylation modulates the kinase activity and oncogenic potential of the Met receptor tyrosine kinase. Oncogene 1994; 9: 2019–2027. [PubMed] [Google Scholar]
- 85. Ponzetto C, Bardelli A, Zhen Z, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 1994; 77: 261–271. [DOI] [PubMed] [Google Scholar]
- 86. Fixman ED, Fournier TM, Kamikura DM, et al. Pathways downstream of Shc and Grb2 are required for cell transformation by the Tpr-Met oncoprotein. J Biol Chem 1996; 271: 13116–13122. [DOI] [PubMed] [Google Scholar]
- 87. Pelicci G, Giordano S, Zhen Z, et al. The motogenic and mitogenic responses to HGF are amplified by the Shc adaptor protein. Oncogene 1995; 10: 1631–1638. [PubMed] [Google Scholar]
- 88. Garcia-Guzman M, Dolfi F, Zeh K, et al. Met–induced JNK activation is mediated by the adapter protein Crk and correlates with the Gab1 – Crk signaling complex formation. Oncogene 1999; 18: 7775–7786. [DOI] [PubMed] [Google Scholar]
- 89. Zhang YW, Wang LM, Jove R, et al. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene 2002; 21: 217–226. [DOI] [PubMed] [Google Scholar]
- 90. Weidner KM, Di Cesare S, Sachs M, et al. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 1996; 384: 173–176. [DOI] [PubMed] [Google Scholar]
- 91. Maulik G, Madhiwala P, Brooks S, et al. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. J Cell Mol Med 2002; 6: 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Graziani A, Gramaglia D, dalla Zonca P, et al. Hepatocyte growth factor/scatter factor stimulates the Ras-guanine nucleotide exchanger. J Biol Chem 1993; 268: 9165–9168. [PubMed] [Google Scholar]
- 93. Paumelle R, Tulasne D, Kherrouche Z, et al. Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway. Oncogene 2002; 21: 2309–2319. [DOI] [PubMed] [Google Scholar]
- 94. Maroun CR, Naujokas MA, Park M. Membrane targeting of Grb2-associated binder-1 (Gab1) scaffolding protein through Src myristoylation sequence substitutes for Gab1 pleckstrin homology domain and switches an epidermal growth factor response to an invasive morphogenic program. Mol Biol Cell 2003; 14: 1691–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xiao GH, Jeffers M, Bellacosa A, et al. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 2001; 98: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hui AY, Meens JA, Schick C, et al. Src and FAK mediate cell-matrix adhesion-dependent activation of met during transformation of breast epithelial cells. J Cell Biochem 2009; 107: 1168–1181. [DOI] [PubMed] [Google Scholar]
- 97. Petrelli A, Gilestro GF, Lanzardo S, et al. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature 2002; 416: 187–190. [DOI] [PubMed] [Google Scholar]
- 98. Gandino L, Longati P, Medico E, et al. Phosphorylation of serine 985 negatively regulates the hepatocyte growth factor receptor kinase. J Biol Chem 1994; 269: 1815–1820. [PubMed] [Google Scholar]
- 99. Jo M, Stolz DB, Esplen JE, et al. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem 2000; 275: 8806–8811. [DOI] [PubMed] [Google Scholar]
- 100. Puri N, Salgia R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. J Carcinog 2008; 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bachleitner-Hofmann T, Sun MY, Chen CT, et al. HER kinase activation confers resistance to MET tyrosine kinase inhibition in MET oncogene-addicted gastric cancer cells. Mol Cancer Ther 2008; 7: 3499–3508. [DOI] [PubMed] [Google Scholar]
- 102. Follenzi A, Bakovic S, Gual P, et al. Cross-talk between the proto-oncogenes Met and Ron. Oncogene 2000; 19: 3041–3049. [DOI] [PubMed] [Google Scholar]
- 103. Benvenuti S, Lazzari L, Arnesano A, et al. Ron kinase transphosphorylation sustains MET oncogene addiction. Cancer Res 2011; 71: 1945–1955. [DOI] [PubMed] [Google Scholar]
- 104. Yeh CY, Shin SM, Yeh HH, et al. Transcriptional activation of the Axl and PDGFR-α by c-Met through a ras- and Src-independent mechanism in human bladder cancer. BMC Cancer 2011; 11: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of Met in lung cancer. Cancer Res 2006; 66: 283–289. [DOI] [PubMed] [Google Scholar]
- 106. Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009; 4: 5–11. [DOI] [PubMed] [Google Scholar]
- 107. Liu X, Jia Y, Stoopler MB, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol 2016; 34: 794–802. [DOI] [PubMed] [Google Scholar]
- 108. Tong JH, Yeung SF, Chan AW, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin Cancer Res 2016; 22: 3048–3056. [DOI] [PubMed] [Google Scholar]
- 109. Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in Non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 2016; 34: 721–730. [DOI] [PubMed] [Google Scholar]
- 110. Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008; 14: 2895–2899. [DOI] [PubMed] [Google Scholar]
- 111. Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring met mutations causing exon 14 skipping. Cancer Discov 2015; 5: 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jenkins RW, Oxnard GR, Elkin S, et al. Response to crizotinib in a patient with lung adenocarcinoma harboring a MET splice site mutation. Clin Lung Cancer 2015; 16: e101–e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jorge SE, Schulman S, Freed JA, et al. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer 2015; 90: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009; 20: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fuchs V, Roisman L, Kian W, et al. The impact of osimertinib’ line on clonal evolution in EGFRm NSCLC through NGS-based liquid biopsy and overcoming strategies for resistance. Lung Cancer 2021; 153: 126–133. [DOI] [PubMed] [Google Scholar]
- 116. Noonan SA, Berry L, Lu X, et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J Thorac Oncol 2016; 11: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kim JH, Kim HS, Kim BJ. Prognostic value of MET copy number gain in non-small-cell lung cancer: an updated meta-analysis. J Cancer 2018; 9: 1836–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Camidge DR, Ou SHI, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014; 32: 8001. [Google Scholar]
- 119. Qin S, Chan SL, Sukeepaisarnjaroen W, et al. A phase II study of the efficacy and safety of the MET inhibitor capmatinib (INC280) in patients with advanced hepatocellular carcinoma. Ther Adv Med Oncol. Epub ahead of print 11 December 2019. DOI: 10.1177/1758835919889001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Santarpia M, Massafra M, Gebbia V, et al. A narrative review of MET inhibitors in non-small cell lung cancer with MET exon 14 skipping mutations. Transl Lung Cancer Res 2021; 10: 1536–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–1043. [DOI] [PubMed] [Google Scholar]
- 122. Wang X, Song N, Zhang Y, et al. Coexpression of c-Met and Notch-1 correlates with poor prognosis in resected non-small-cell lung cancer. Tumour Biol 2015; 36: 7053–7059. [DOI] [PubMed] [Google Scholar]
- 123. Olivero M, Rizzo M, Madeddu R, et al. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br J Cancer 1996; 74: 1862–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Breindel JL, Haskins JW, Cowell EP, et al. EGF receptor activates MET through MAPK to enhance non-small cell lung carcinoma invasion and brain metastasis. Cancer Res 2013; 73: 5053–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu F, Song S, Yi Z, et al. HGF induces EMT in non-small-cell lung cancer through the hBVR pathway. Eur J Pharmacol 2017; 811: 180–190. [DOI] [PubMed] [Google Scholar]
- 126. Begemann D, Anastos H, Kyprianou N. Cell death under epithelial–mesenchymal transition control in prostate cancer therapeutic response. Int J Urol 2018; 25: 318–326. [DOI] [PubMed] [Google Scholar]
- 127. Kim IH, Lee IH, Lee JE, et al. Clinical significance of C-MET overexpression and epidermal growth factor receptor mutation in platinum-based adjuvant chemotherapy outcome in surgically resected lung adenocarcinoma. Ann Surg Oncol 2017; 24: 770–777. [DOI] [PubMed] [Google Scholar]
- 128. Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016; 380: 494–504. [DOI] [PubMed] [Google Scholar]
- 129. Ma G, Deng Y, Chen W, et al. The prognostic role of MET protein expression among surgically resected non-small cell lung cancer patients: a meta-analysis. Front Oncol 2019; 9: 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984; 311: 29–33. [DOI] [PubMed] [Google Scholar]
- 131. Stransky N, et al. The landscape of kinase fusions in cancer. Nat Commun. 2014; 5: 4846. DOI: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Socinski MA, Pennell NA, Davies KD. MET exon 14 skipping mutations in non–small-cell lung cancer: an overview of biology, clinical outcomes, and testing considerations. JCO Precis Oncol 2021; 5: 653–663. DOI: 10.1200/po.20.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Backes AC, Zech B, Felber B, et al. Small-molecule inhibitors binding to protein kinase. Part II: the novel pharmacophore approach of type II and type III inhibition. Expert Opin Drug Discov 2008; 3: 1427–1449. [DOI] [PubMed] [Google Scholar]
- 134. Reungwetwattana T, Liang Y, Zhu V, et al. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: the why, the how, the who, the unknown, and the inevitable. Lung Cancer 2017; 103: 27–37. [DOI] [PubMed] [Google Scholar]
- 135. Poirot B, Doucet L, Benhenda S, et al. MET exon 14 alterations and new resistance mutations to tyrosine kinase inhibitors: Risk of inadequate detection with current amplicon-based NGS panels. J Thorac Oncol 2017; 12: 1582–1587. [DOI] [PubMed] [Google Scholar]
- 136. Bang YJ, Su WC, Schuler M, et al. Phase 1 study of capmatinib in MET-positive solid tumor patients: dose escalation and expansion of selected cohorts. Cancer Sci 2020; 111: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Schuler M, Berardi R, Lim WT, et al. Molecular correlates of response to capmatinib in advanced non-small-cell lung cancer: clinical and biomarker results from a phase I trial. Ann Oncol 2020; 31: 789–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wolf J, Seto T, Han JY, et al. Capmatinib in MET exon 14–mutated or MET -amplified non–small-cell lung cancer. N Engl J Med 2020; 383: 944–957. [DOI] [PubMed] [Google Scholar]
- 139. Qi J, McTigue MA, Rogers A, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res 2011; 71: 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tiedt R, Degenkolbe E, Furet P, et al. A drug resistance screen using a selective MET inhibitor reveals a spectrum of mutations that partially overlap with activating mutations found in cancer patients. Cancer Res 2011; 71: 5255–5264. [DOI] [PubMed] [Google Scholar]
- 141. Heist RS, Sequist LV, Borger D, et al. Acquired resistance to crizotinib in NSCLC with MET exon 14 skipping. J Thorac Oncol 2016; 11: 1242–1245. [DOI] [PubMed] [Google Scholar]
- 142. Jin W, Shan B, Liu H, et al. Acquired mechanism of crizotinib resistance in NSCLC with MET exon 14 skipping. J Thorac Oncol 2019; 14: e137–e139. [DOI] [PubMed] [Google Scholar]
- 143. Zhang Y, Yin J, Peng F. Acquired resistance to crizotinib in advanced lung adenocarcinoma with MET exon 14 skipping. Lung Cancer 2017; 113: 69–71. [DOI] [PubMed] [Google Scholar]
- 144. Ou SHI, Young L, Schrock AB, et al. Emergence of preexisting MET Y1230C mutation as a resistance mechanism to crizotinib in NSCLC with MET exon 14 skipping. J Thorac Oncol 2017; 12: 137–140. [DOI] [PubMed] [Google Scholar]
- 145. Kim S, Kim TM, Kim DW, et al. Acquired resistance of MET-amplified non-small cell lung cancer cells to the MET inhibitor capmatinib. Cancer Res Treat 2019; 51: 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. McDermott U, Pusapati RV, Christensen JG, et al. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res 2010; 70: 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Jamme P, Fernandes M, Copin MC, et al. Alterations in the PI3K pathway drive resistance to MET inhibitors in NSCLC harboring MET exon 14 skipping mutations. J Thorac Oncol 2020; 15: 741–751. [DOI] [PubMed] [Google Scholar]
- 148. Suzawa K, Offin M, Lu D, et al. Activation of KRAS mediates resistance to targeted therapy in MET exon 14–mutant non–small cell lung cancer. Clin Cancer Res 2019; 25: 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Recondo G, Bahcall M, Spurr LF, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14–mutant NSCLC. Clin Cancer Res 2020; 26: 2615–2625. [DOI] [PubMed] [Google Scholar]
- 150. Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol 2016; 11: 1493–1502. [DOI] [PubMed] [Google Scholar]
- 151. Rotow JK, Gui P, Wu W, et al. Co-occurring alterations in the RAS–MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer. Clin Cancer Res 2020; 26: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Remon J, Besse B. Brain metastases in oncogene-addicted non-small cell lung cancer patients: incidence and treatment. Front Oncol 2018; 8: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Awad MM, Leonardi GC, Kravets S, et al. Impact of MET inhibitors on survival among patients with non-small cell lung cancer harboring MET exon 14 mutations: a retrospective analysis. Lung Cancer 2019; 133: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Dagogo-Jack I, Moonsamy P, Gainor JF, et al. A phase 2 study of capmatinib in patients with MET-altered lung cancer previously treated with a MET inhibitor. J Thorac Oncol 2021; 16: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bladt F, Faden B, Friese-Hamim M, et al. EMD 1214063 and EMD 1204831 constitute a new class of potent and highly selective c-Met inhibitors. Clin Cancer Res 2013; 19: 2941–2951. [DOI] [PubMed] [Google Scholar]
- 156. Medová M, Pochon B, Streit B, et al. The novel ATP-competitive inhibitor of the MET hepatocyte growth factor receptor EMD1214063 displays inhibitory activity against selected MET-mutated variants. Mol Cancer Ther 2013; 12: 2415–2424. [DOI] [PubMed] [Google Scholar]
- 157. Falchook GS, Kurzrock R, Amin HM, et al. First-in-man phase I trial of the selective MET inhibitor tepotinib in patients with advanced solid tumors. Clin Cancer Res 2020; 26: 1237–1246. [DOI] [PubMed] [Google Scholar]
- 158. Paik PK, Felip E, Veillon R, et al. Tepotinib in non–small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med 2020; 383: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Markham A. Tepotinib: first approval. Drugs 2020; 80: 829–833. [DOI] [PubMed] [Google Scholar]
- 160. Wu YL, Cheng Y, Zhou J, et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med 2020; 8: 1132–1143. [DOI] [PubMed] [Google Scholar]
- 161. Smit EF, Felip E, Karachaliou N, et al. 1415TiP INSIGHT 2: tepotinib + osimertinib in patients (pts) with EGFR-mutant NSCLC having acquired resistance to first-line osimertinib due to MET amplification (METamp). Ann Oncol 2020; 31: S894. [Google Scholar]
- 162. Pudelko L, Jaehrling F, Stroh C, et al. Abstract 3774: Unraveling mechanisms of resistance to tepotinib and future treatment options. Cancer Res. 2020; 80(16 Supplement): 3774–3774. doi:10.1158/1538-7445.AM2020-3774 [Google Scholar]
- 163. Pudelko L, Jaehrling F, Reusch C, et al. SHP2 inhibition influences therapeutic response to tepotinib in tumors with MET alterations. iScience 2020; 23: 101832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Jia H, et al. Discovery of (S)-1-(1-(Imidazo[1,2- a ]pyridin-6-yl)ethyl)-6-(1-methyl-1 H -pyrazol-4-yl)-1 H -[1,2,3]triazolo[4,5- b ]pyrazine (volitinib) as a highly potent and selective mesenchymal-epithelial transition factor (c-Met) inhibitor in clinical development. J Med Chem 2014; 57: 7577–7589. [DOI] [PubMed] [Google Scholar]
- 165. Lu S, Fang J, Li X, et al. Phase II study of savolitinib in patients (pts) with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations (METex14+). J Clin Oncol 2020; 38: 9519. [Google Scholar]
- 166. Shih J, Zhong B, Shi H, et al. Abstract 2096: bozitinib, a highly selective inhibitor of cMet, demonstrates robust activity in gastric, lung, hepatic and pancreatic in vivo models. Cancer Res 2017; 77: 2096. [Google Scholar]
- 167. Hu H, Mu Q, Bao Z, et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell 2018; 175: 1665–1678.e18. [DOI] [PubMed] [Google Scholar]
- 168. Deng W, Zhai D, Rogers E, et al. Abstract 1325: TPX-0022, a polypharmacology inhibitor of MET/CSF1R/SRC inhibits tumor growth by promoting anti-tumor immune responses. Cancer Res 2019; 79: 1325. [Google Scholar]
- 169. Hong D, Bazhenova L, Cho B, et al. First-in-human safety, pharmacokinetics, and preliminary efficacy of TPX-0022, a novel inhibitor of MET/SRC/CSF1R in patients with advanced solid tumors harboring genetic alterations in MET. Eur J Cancer 2020; 138: S1. [Google Scholar]
- 170. Yakes FM, Chen J, Tan J, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther 2011; 10: 2298–2308. [DOI] [PubMed] [Google Scholar]
- 171. Hellerstedt BA, Vogelzang NJ, Kluger HM, et al. Results of a phase ii placebo-controlled randomized discontinuation trial of cabozantinib in patients with non–small-cell lung carcinoma. Clin Lung Cancer 2019; 20: 74–81. [DOI] [PubMed] [Google Scholar]
- 172. Wang SXY, Zhang BM, Wakelee HA, et al. Case series of MET exon 14 skipping mutation-positive non-small-cell lung cancers with response to crizotinib and cabozantinib. Anticancer Drugs 2019; 30: 537–541. [DOI] [PubMed] [Google Scholar]
- 173. Klempner SJ, Borghei A, Hakimian B, et al. Intracranial activity of cabozantinib in MET exon 14–positive NSCLC with brain metastases. J Thorac Oncol 2017; 12: 152–156. [DOI] [PubMed] [Google Scholar]
- 174. Wu W, Bi C, Credille KM, et al. Inhibition of tumor growth and metastasis in non-small cell lung cancer by LY2801653, an inhibitor of several oncokinases, including MET. Clin Cancer Res 2013; 19: 5699–5710. [DOI] [PubMed] [Google Scholar]
- 175. He AR, Cohen RB, Denlinger CS, et al. First-in-human phase I study of merestinib, an oral multikinase inhibitor, in patients with advanced cancer. Oncologist 2019; 24: e930–e942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Morgillo F, Amendola G, Della Corte CM, et al. Dual MET and SMO negative modulators overcome resistance to EGFR inhibitors in human nonsmall cell lung cancer. J Med Chem 2017; 60: 7447–7458. [DOI] [PubMed] [Google Scholar]
- 177. Engstrom LD, Aranda R, Lee M, et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation-mediated resistance to type I MET inhibitors in nonclinical models. Clin Cancer Res 2017; 23: 6661–6672. [DOI] [PubMed] [Google Scholar]
- 178. Spigel DR, Edelman MJ, O’Byrne K, et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non–small-cell lung cancer: METLung. J Clin Oncol 2017; 35: 412–420. [DOI] [PubMed] [Google Scholar]
- 179. Bendell JC, Hochster H, Hart LL, et al. A phase ii randomized trial (GO27827) of first-line FOLFOX plus bevacizumab with or without the MET inhibitor onartuzumab in patients with metastatic colorectal cancer. Oncologist 2017; 22: 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Wakelee H, Zvirbule Z, De Braud F, et al. Efficacy and safety of onartuzumab in combination with first-line bevacizumab- or pemetrexed-based chemotherapy regimens in advanced non-squamous non–small-cell lung cancer. Clin Lung Cancer 2017; 18: 50–59. [DOI] [PubMed] [Google Scholar]
- 181. Catenacci DVT, Tebbutt NC, Davidenko I, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2017; 18: 1467–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Finisguerra V, Prenen H, Mazzone M. Preclinical and clinical evaluation of MET functions in cancer cells and in the tumor stroma. Oncogene 2016; 35: 5457–5467. [DOI] [PubMed] [Google Scholar]
- 183. Martin V, Chiriaco C, Modica C, et al. Met inhibition revokes IFNγ-induction of PD-1 ligands in MET-amplified tumours. Br J Cancer 2019; 120: 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Finisguerra V, Di Conza G, Di Matteo M, et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 2015; 522: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Doronina SO, Toki BE, Torgov MY, et al. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat Biotechnol 2003; 21: 778–784. [DOI] [PubMed] [Google Scholar]
- 186. Wang J, Anderson MG, Oleksijew A, et al. ABBV-399, a c-Met antibody-drug conjugate that targets both MET-amplified and c-Met-overexpressing tumors, irrespective of MET pathway dependence. Clin Cancer Res 2017; 23: 992–1000. [DOI] [PubMed] [Google Scholar]
- 187. Yao HP, Tong XM, Hudson R, et al. MET and RON receptor tyrosine kinases in colorectal adenocarcinoma: molecular features as drug targets and antibody-drug conjugates for therapy. J Exp Clin Cancer Res 2020; 39: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Strickler JH, et al. First-in-human phase I, dose-escalation and -expansion study of telisotuzumab vedotin, an antibody–drug conjugate targeting c-Met, in patients with advanced solid tumors. J Clin Oncol 2018; 36: 3298–3306. [DOI] [PubMed] [Google Scholar]
- 189. Waqar SN, Redman MW, Arnold SM, et al. A phase ii study of telisotuzumab vedotin in patients with c-MET-positive stage IV or recurrent squamous cell lung cancer (LUNG-MAP sub-study S1400K, NCT03574753). Clin Lung Cancer 2020; 22: 170–177. DOI: 10.1016/j.cllc.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Camidge DR, Moiseenko F, Cicin I, et al. Abstract CT179: telisotuzumab vedotin (teliso-v) monotherapy in patients with previously treated c-Met + advanced non-small cell lung cancer. In: American association for cancer research annual meeting 2021, 10–15 April 2021. DOI: 10.1158/1538-7445.am2021-ct179. [DOI] [Google Scholar]
- 191. Camidge DR, Goldman J, Cole G, et al. 1414TiP evaluating telisotuzumab vedotin in combination with osimertinib in patients with advanced non-small cell lung cancer: a phase I/Ib study cohort. Ann Oncol 2020; 31: S894. [Google Scholar]
- 192. Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013; 31: 4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tan EH, Lim WT, Ahn MJ, et al. Phase 1b trial of ficlatuzumab, a humanized hepatocyte growth factor inhibitory monoclonal antibody, in combination with gefitinib in Asian patients with NSCLC. Clin Pharmacol Drug Dev 2018; 7: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Tarhini AA, Rafique I, Floros T, et al. Phase 1/2 study of rilotumumab (AMG 102), a hepatocyte growth factor inhibitor, and erlotinib in patients with advanced non–small cell lung cancer. Cancer 2017; 123: 2936–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Rosen LS, Goldman JW, Algazi AP, et al. A first-in-human phase I study of a bivalent MET antibody, emibetuzumab (LY2875358), as monotherapy and in combination with erlotinib in advanced cancer. Clin Cancer Res 2017; 23: 1910–1919. [DOI] [PubMed] [Google Scholar]
- 196. Moores SL, Chiu ML, Bushey BS, et al. A novel bispecific antibody targeting EGFR and cMet is effective against EGFR inhibitor-resistant lung tumors. Cancer Res 2016; 76: 3942–3953. [DOI] [PubMed] [Google Scholar]
- 197. Park K, John T, Kim SW, et al. Amivantamab (JNJ-61186372), an anti-EGFR-MET bispecific antibody, in patients with EGFR exon 20 insertion (exon20ins)-mutated non-small cell lung cancer (NSCLC). J Clin Oncol 2020; 38: 9512. [Google Scholar]
- 198. A study of amivantamab and lazertinib combination therapy versus osimertinib in locally advanced or metastatic non-small cell lung cancer (MARIPOSA), 2020, https://clinicaltrials.gov/ct2/show/NCT04487080
- 199. A study of combination amivantamab and carboplatin-pemetrexed therapy, compared with carboplatin-pemetrexed, in participants with advanced or metastatic non-small cell lung cancer characterized by epidermal growth factor receptor (EGFR) exon 20 insertions, 2020, https://www.clinicaltrials.gov/ct2/show/NCT04538664
- 200. Patnaik A, Gordon M, Tsai F, et al. A phase I study of LY3164530, a bispecific antibody targeting MET and EGFR, in patients with advanced or metastatic cancer. Cancer Chemother Pharmacol 2018; 82: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Grandal MM, Havrylov S, Poulsen TT, et al. Simultaneous targeting of two distinct epitopes on MET effectively inhibits MET- and HGF-driven tumor growth by multiple mechanisms. Mol Cancer Ther 2017; 16: 2780–2791. [DOI] [PubMed] [Google Scholar]
- 202. Poulsen TT, Grandal MM, Skartved NJØ, et al. Sym015: a highly efficacious antibody mixture against met-amplified tumors. Clin Cancer Res 2017; 23: 5923–5935. [DOI] [PubMed] [Google Scholar]
- 203. Camidge DR, Janku F, Martinez-Bueno A, et al. Safety and preliminary clinical activity of the MET antibody mixture, Sym015 in advanced non-small cell lung cancer (NSCLC) patients with MET amplification/exon 14 deletion (MET Amp/Ex14∆). J Clin Oncol 2020; 38: 9510. [Google Scholar]
- 204. Sabari JK, Leonardi GC, Shu CA, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann Oncol 2018; 29: 2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Xu Z, Li H, Dong Y, et al. Incidence and PD-L1 expression of MET 14 skipping in Chinese population: a non-selective NSCLC cohort study using RNA-based sequencing. Onco Targets Ther 2020; 13: 6245–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Hong L, Negrao MV, Dibaj SS, et al. Programmed death-ligand 1 heterogeneity and its impact on benefit from immune checkpoint inhibitors in NSCLC. J Thorac Oncol 2020; 15: 1449–1459. [DOI] [PubMed] [Google Scholar]
- 207. Awad MM, Lee J, Madison R, et al. Characterization of 1,387 NSCLCs with MET exon 14 (METex14) skipping alterations (SA) and potential acquired resistance (AR) mechanisms. J Clin Oncol 2020; 38: 9511. [Google Scholar]
- 208. Baba K, Tanaka H, Sakamoto H, et al. Efficacy of pembrolizumab for patients with both high PD-L1 expression and an MET exon 14 skipping mutation: a case report. Thorac Cancer 2019; 10: 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Reis H, Metzenmacher M, Goetz M, et al. MET expression in advanced non–small-cell lung cancer: effect on clinical outcomes of chemotherapy, targeted therapy, and immunotherapy. Clin Lung Cancer 2018; 19: e441–e463. [DOI] [PubMed] [Google Scholar]
- 210. Mayenga M, Assié JB, Monnet I, et al. Durable responses to immunotherapy of non-small cell lung cancers harboring MET exon-14–skipping mutation: a series of 6 cases. Lung Cancer 2020; 150: 21–25. [DOI] [PubMed] [Google Scholar]
- 211. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet (London, England) 2017; 389: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Glodde N, Bald T, van den Boorn-Konijnenberg D, et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity 2017; 47: 789–802.e9. [DOI] [PubMed] [Google Scholar]
- 213. Ahn HK, Kim S, Kwon D, et al. MET receptor tyrosine kinase regulates the expression of co-stimulatory and co-inhibitory molecules in tumor cells and contributes to PD-L1-mediated suppression of immune cell function. Int J Mol Sci 2019; 20: 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Agarwal N, Vaishampayan U, Green M, et al. Phase Ib study (COSMIC-021) of cabozantinib in combination with atezolizumab: results of the dose escalation stage in patients (pts) with treatment-naïve advanced renal cell carcinoma (RCC). Ann Oncol 2018; 29: viii308. DOI: 10.1093/annonc/mdy283.081 [Google Scholar]
- 215. Feng Y, Thiagarajan PS, Ma PC. MET signaling: novel targeted inhibition and its clinical development in lung cancer. J Thorac Oncol 2012; 7: 459–467. [DOI] [PubMed] [Google Scholar]
- 216. Eder JP, Vande Woude GF, Boerner SA, et al. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res 2009; 15: 2207–2214. [DOI] [PubMed] [Google Scholar]
- 217. Van Der Steen N, Giovannetti E, Pauwels P, et al. CMET exon 14 skipping: from the structure to the clinic. J Thorac Oncol 2016; 11: 1423–1432. [DOI] [PubMed] [Google Scholar]
- 218. Fujino T, Kobayashi Y, Suda K, et al. Sensitivity and resistance of MET exon 14 mutations in lung cancer to eight MET tyrosine kinase inhibitors in vitro. J Thorac Oncol 2019; 14: 1753–1765. [DOI] [PubMed] [Google Scholar]