Highlights
-
•
MV3-Exo inhibits CD4+ t cell immune function and induces M2-polarized macrophages.
-
•
TIM-3 is upregulated in MV3 cells-secreted exosomes.
-
•
TIM-3 inhibits CD4+ t cell immune function and induces M2-polarized macrophages.
-
•
TIM-3 shuttled by MV3-Exo promotes growth and metastasis of melanoma.
-
•
The study unveiled a protective target against metastasis of melanoma.
Keywords: Mv3 cells, Exosomes, Tim-3, Melanoma, Cd4+ t cell immune function, Macrophage m2 polarization, Metastasis
Abstract
This study is sought to determine the physiological mechanisms by which exosomes-encapsulated TIM-3 derived from melanoma cells might mediate CD4+ T cell immune function and macrophage M2 polarization in melanoma. Initially, exosomes were isolated from the human skin-derived melanoma cell line MV3for analysis of TIM-3 expression pattern. Next, the exosomes sourced from MV3 cells manipulated with sh-TIM-3 were co-incubated with CD4+ T cells to detect CD4+ T cell proliferation and MV3 cell migration and invasion, to observe the macrophage M2 polarization, and to determine levels of several EMT-related factors. Finally, melanoma nude mouse models were established to study the in vivo modulatory effects of TIM-3 from MV3 cells-derived exosomes. MV3 cells-derived exosomes inhibited CD4+ T cell immune function and promoted macrophage M2 polarization in melanoma. Our results revealed the abundance of TIM-3 in MV3 cells-derived exosomes. Of importance, silencing of TIM-3 shuttled by MV3 cells-derived exosomes improved CD4+ T cell immune function and inhibited macrophage M2 polarization to attenuate the growth and metastasis of melanoma cells. Collectively, MV3 cells-derived exosomes-loaded TIM-3 suppressed CD4+ T cell immune function and induced macrophage M2 polarization to improve occurrence and development of melanoma, therefore providing us with a potential therapeutic target for effectively combating melanoma.
Introduction
Melanoma is an invasive skin cancer associated with high incidence and mortality [1]. The high mortality rates of melanoma are prevalent due to its highly metastatic nature, while the depth of invasion is associated with poor prognosis and disease dissemination [2]. Statistically, the incidence of melanoma has been alarmingly increasing worldwide over the past 5 decades [3]. Recently, treatment with molecular targeted therapy and immunotherapy have elicited notable application as new effective therapies for the treatment of advanced melanoma [4]. However, the immunosuppressive microenvironment induced by regulatory T (Treg) cells has emerged as a major obstacle to the success of tumor immunotherapy [5]. A recent study reported that the reduction of CD4+T cells could result in almost complete elimination of the anti-tumor effect of the vaccine in melanoma, thus illustrating that CD4+T cells mediated the anti-tumor effect [6]. Moreover, as macrophages can facilitate tissue repair and immune response to pathogens, and its extensive functionality is apparent as it can adapt and regulate the requirements of the tissue microenvironment [7]. Moreover, depending on function, they can be classified as M1-polarized macrophages with anti-tumor functions and M2 tumor-associated macrophages (TAMs) that can radically induce tumor growth [8]. Currently, TAMs have been proposed as a promising therapeutic target for tumor immunotherapy, and targeted drug delivery of tumor promoted M2 like TAMs is challenging in the treatment of melanoma [9]. Therefore, the recovery of immune function of CD4+T cells and inhibition of M2-polarized macrophages will provide insight for the development of novel immunotherapies against melanoma.
Exosomes are small membrane vesicles (30 - 100 nm) originated from the luminal membranes of multivesicular bodies, and constitutively are secretions produced by fusion with the cell membrane [10]. Fundamentally, xxosomes can function are modulators of transport proteins for regulation of the intercellular communication within the tumor microenvironment [2]. Exosomes serve as a key mediator to affect cancer cell proliferating capability, epithelial-mesenchymal transition (EMT) process, and immune-evasion [11]. Exosomes can be derived from cancer cells, which can promote tumor progression while the melanoma-derived exosomes can mediate immune suppression in melanoma [12, 8]. Moreover, an existing study identified the ability of exosomes loaded with proteins, DNA, and coding or non-coding RNAs to function as intracellular cell-cell transmitters [13]. T-cell immunoglobulin and mucin domain 3 (TIM-3) mediated by exosomes could boost the migration, invasiveness, epithelial-mesenchymal transition (EMT), and metastatic potential of cancer cells [14]. TIM-3, encoded by the hepatitis A virus cellular receptor 2 gene (HAVCR2), is a trans-membrane receptor expressed by multiple cells including T lymphocytes and innate immune cells [15]. Principally, TIM-3 was discovered during identification of novel cell surface molecules that would mark interferon (IFN-γ) producing CD4+ T helper 1 (Th1) and CD8+ T cytotoxic 1 (Tc1) cells [16]. TIM-3, a member of the TIM family of genes has been genetically mapped in syntenic chromosomal regions in human (5q33.2) and mouse (11B1.1) involved in both allergy and autoimmune disease [17]. An existing study has implicated the dys-regulation ofTIM-3 in immune suppression in melanoma [18]. However, the mechanism by which MV3 cells-derived exosome communication influences the immune function of CD4+T cells and inhibition of M2-polarized macrophages in melanoma by interaction with TIM-3 remains elusive, highlighting a major gap in literature eliciting the significance of MV3 cells-derived exosomes against metastasis in the treatment of melanoma. Hence, we speculated that the transfer of TIM-3 via MV3 cells-derived exosomes might influence the growth and metastatic properties of melanoma cells.
Methods
Ethics statement
The current study was conducted with approval of the Ethic Committee of The First Affiliated Hospital of Soochow University. The animal experimentation was strictly designed and performed conforming to the Guide for the Care and Use of Laboratory Animal by the US National Institutes of Health. Adequate measures were taken to minimize the suffering of the enrolled animals.
Bioinformatics analysis
Melanoma-related RNA-seq data GSE98394 was obtained from the Gene Expression Omnibus (GEO) database. A total of 78 samples were identified in GSE98394, comprising of 51 tumor samples and 27 normal samples. The “edge” package of R language was employed to screen the differentially expressed genes (DEGs) with |logfc| > 2 and FDR < 0.01 as threshold. The top 100 immune related genes (IRG) were retrieved from the GeneCards database. Exosome proteome was obtained from the STRING database. The TCGA database (http://gepia2.cancer-pku.cn/#index) was used for comprehensive analysis of the differential expression genes of skin melanoma (SKCM) in TCGA (|Log2FC| Cutoff = 1 and p-value Cutoff = 0.01).
Cell culture
Human skin-derived melanoma cells MV3 and A375 (American Type Culture Collection [ATCC], VA, USA) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco, Carlsbad, California, USA) containing a combination of 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 U/mL streptomycin (Sigma-Aldrich, St Louis, MO, USA). Human monocytes (THP-1, ATCC) were cultured in Roswell Park Memorial Institute (RPMI)−1640 (Gibco) medium containing a combination of 10% FBS, 100 U/mL penicillin and 100 U/mL streptomycin, Normal human melanocytes (NHEM, PromoCell-China Co., Ltd., Nanjing, China) were cultured in medium 254 (Cascade Biologics®) containing human melanocyte growth supplement (HMGs) and 1% penicillin streptomycin. All cells were submitted to vigorous culture with 5% CO2 at 37 °C.
Mφ Macrophage induction as conducted as follows: the THP-1 cells in the logarithmic phase was centrifugally adjusted to attain a cell density of 2.5. × 105 cells/mL. THP-1 cells were supplemented with 100 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma). After incubation for 24 h in conditions devoid of light, the Mφ Macrophage was observed under the microscope. Next, flow cytometric analysis was conducted for detecting the expression of Mφ macrophage surface marker CD68. The cells were incubated with the FITC—CD6 antibody (562,117, 1 × 106 cells/5 μL, BD Biosciences, Franklin Lakes, NJ, USA) at 4 °C for 30 min prior to detection using the machine. The induced Mφ macrophages were co-cultured with CD4+ T cells, and the cell morphology was observed after 24 h.
MV3 cells were transduced with lentivirus-loaded sh-NC, sh-TIM-3–1 (5′-GAGCUAAGAAGCCUAGGGGTT), or sh-TIM-3–2 (5′-GCACCCAAGAAACGACAATTG) respectively. The sequence and plasmid were provided by the GenePharma Ltd. Company (Shanghai, China). After 48 h of treatments, the cells were reserved for subsequent experimentation.
Isolation and identification of exosomes
Exosomes were isolated from the culture supernatant of NHEM and MV3 cells by differential centrifugation. Briefly, the collected supernatant was subject to vigorous centrifugation at 300 × g for 10 min, 2000 × g for 10 min, and 10,000 × g for 30 min. Next, the supernatant was subject to continuous ultracentrifugation at 110,000 × g for 2 h, from which the collected precipitate was classified asexosome. The collected exosomes were rinsed with PBS and resuspended using 0.22 μm filter to eliminate smaller cell debris, the exosomes were rinsed with PBS and resuspended for another regimen of ultracentrifugation at 110,000 × g for 2 h to eliminate PBS. Next, the suspension was resuspended with phosphate buffer saline (PBS), followed by the second regimen ultracentrifugation under the same conditions, after which the precipitate was preserved at −80 °C for subsequent experimentation. The preceding centrifugation was conducted at 4 °C. Exosomes were incubated in exosome serum-free cell culture medium (C38010050, Viva Cell).
The exosomes were identified employing transmission electron microscopic (TEM) observation (H-7650, Hitachi, Tokyo, Japan). The exosomes (10 μL) were added to the sealing film, and the Formvar membrane of copper mesh was placed face-down on the suspension. Multiple copper meshes (2–3) were prepared for each exosome sample. The membrane was covered and the copper mesh was allowed to absorb for 20 min in a dry environment. Next, PBS (100 μL) was added to the sealing film. The copper mesh (Formvar film face down) was placed on the PBS drop with tweezers for 5 min prior to 2 rinses. Throughout the experiment, the film surface was kept wet while the other side was dry. Next, the copper mesh was immersed in 50 μL 1% glutaraldehyde for 5 min. The copper mesh was immersed into 100 μL of double distilled water for 2 min (8 rinses). Then copper mesh was immersed in 50 μL of uranium dioxygium oxalate for 5 min. Next, the copper mesh was immersed in 50 μL of Methyl cellulose UA solution for 10 min and operated on ice. The copper mesh was removed using a stainless steel ring. The redundant liquid on the filter paper was gently removed, and a thin layer of methyl cellulose membrane was leaved, which was air dried for 10 min. The dried copper mesh was placed at 100 keV for electron microscopy (Hitachi H-7650, Tokyo, Japan).
Several exosome surface marker proteins were assayed by Western blot analysis for identification. The exosomes were dissolved in Radio Immunoprecipitation Assay (RIPA) buffer containing Phenylmethylsulfonyl fluoride (PMSF) and quantified by the bicinchoninic acid (BCA) protein analysis kit (Thermo Fisher Scientific, Rockford, IL, Waltham, MA, USA). The antibodies used for western blot analysis of exosomes were as follows: CD81 (ab109201, 1: 5000, Rabbit, Abcam Inc., Cambridge, MA, USA), CD63 (ab134045, 1: 1000, Rabbit, Abcam), Calnexin (ab133615, 1: 2000, Rabbit, Abcam).
Nanoparticle tracking analysis (NTA) was performed for an extensive analysis of the size of exosome. The analysis was based on the principle of Brownian motion and diffusion coefficient for tracking and automatic analysis of the particle size. The exosomes were resuspended and mixed in 1 mL of PBS, with filtered PBS serving as control. Next, the size measurement of diluted exosomes was proceeded on NanoSight LM10 (NanoSight Ltd., Minton Park, UK) under the NTA measurement condition of 23.75 ± 5 °C for 60 s.
Flow cytometry
The spleens of male C57BL/6 nude mice (aging 6–8 weeks) were grounded applying a 200 mesh copper net to generate a single cell suspension. The supernatant was discarded after centrifugation at 800 × g for 5 min. Next, about 3 times the volume of red blood cell lysate (Beyotime, Shanghai, China) was added to the samples, allowed to stand for 5 min at ambient temperature. After centrifugation 800 × g for 5 min, the supernatant was discarded, and the samples were rinsed with PBS. The prepared suspension was blocked using Fc (anti-CD16/32, BD Biosciences) for incubation at 4 °C for 15 min. Next, the dead cells were excluded using the Live/Dead cell staining kit (Invitrogen, Life Technologies, Naerum, Denmark). Cells were incubated with the FITC—CD4 antibody (eBioscience, San Diego, CA, USA) at 4 °C for 30 min, and the binding antibody was removed with the addition of PBS. The isolated CD4+ T cells were identified with flow cytometric images and cultured in RPMI 1640 medium (Gibco) containing a combination of 2% FBS, 100 U/mL penicillin, and 100 U/mL streptomycin
The collected NHEM-derived exosomes and MV3-derived exosomes (30 μg) were co-incubated with the CD4+ T cells, respectively (2 × 104 cells/well) in 6-well plate (Costar®, Corning, Taipei, Taiwan, China). After 48 h, the expression pattern of TIM-3 on CD4+ T cells was detected. The CD4+ T cells after intervention were subjected to incubation with the PE-CF594-TIM-3 antibody (565,560, 1 × 106 cells/5 μL, BD Bioscience) at 4 °C for 30 min. Subsequently, we detected CD4+ T cells that secrete cytokines TNF-α,IFN-γ. After fixation and permeabilization with the Cytofix/Cytoperm kit (Invitrogen), the cells were stained with the anti-cytokine antibody bound with fluorescein (562,084, 0.2 mg/mL, APC-TNF-α, BD Bioscience; 559,326, 1 × 106 cells/20 μL, PE-IFN-γ, BD Bioscience). The carboxyfluorescein diacetate succinimide dilution protocol was adopted for assessing the proliferation of CD4+ T cells. The CD4+ T cells were stained with 1 μmol/L of Certified Functional Safety Expert (CFSE) (Sigma) at 37 °C for 10 min, after which the dead cells were excluded by the live/dead cell standing Kit (Biovision, Milpitas, CA, USA) staining and analyzed on flow cytometer (FACSverse, BD Bioscience).
Exosome uptake assay
The cell slides were placed on top of the culture dish, and the CD4+ T cells were placed into the culture dish for incubation. Upon reaching 50% cell density, the cell slides were removed for 3 rinses with PBS. Next, the cells were immersed in 4% paraformaldehyde (pH7.4) at 4 °C for 30 min. The ceells were then cleared with 2% Triton X-100 for 15 min. After 3 rinses with PBS, the cells were blocked with 2% bovine serum albumin (BSA) for 45 min. After removal of the blocking solution, MV3 cells-secreted exosomes were labeled with the PHK67 Cell Membrane Labeling Dye (Sigma) and incubated at room temperature for 24 h. The cell slides were rinsed with PBS for 3 times. Following that, the cells were dye employing antibody to CD4+T cell marker (ab133616, 1: 200, Abcam) and with 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, 2 μg/mL) individually in VECTASHIELD immobilizing matrix. After sealing, the uptake of exosomes was observed under confocal microscopy (LSM880, Carl Zeiss, Meditec, Oberkochen, Germany).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA content was extracted from the cells using Trizol (Article No. 16,096,020, Thermo Fisher Scientific, Waltham, MA, USA). The purity and concentration of the RNA were evaluated by spectrophotometry analysis of the absorbance of the solution at wavelengths of 260 and 280 nm. The ratio of A260/A280 of this sample was higher than or equal to 1.8. The sample RNA content was diluted to equivalent concentration, and the cDNA was synthesized by reverse transcription in strict accordance with the provided instructions of the reverse transcriptase kit (TaKaRa, Tokyo, Japan). The PCR was performed by LightCycle 480 SYBR Green I Master. The expression of target genes relative to GAPDH (loading control) was quantified based on the 2−ΔΔCt method. The primer sequences were presented in Supplementary Table 1.
Western blot analysis
The total protein content was extracted from the tissues and cells using the RIPA lysis buffer containing PMSF (P0013, Beyotime). The BCA Kit (P0028, Beyotime) was used to prepare the protein standard and BCA working solution in strict accordance with the instructions. The 2 μL protein sample and 18 μL distilled water were added in the sample well. A total of 200 μL of the BCA working solution was added for incubation in all standard wells and sample wells at 37 °C for 30 min. The optical density (OD) value of the sample at the wavelength of 560 nm was determined using the microplate reader. According to the protein standard curve, the concentration of the protein sample was calculated. After the addition and mixing of the loading buffer, the protein samples were boiled for 10 min for denaturation, and preserved at - 80 °C. According to the size of the target protein band, 8%−12% SDS gel was prepared, and 30 μg of protein samples were added for electrophoretic separation. The protein content from the gel was transferred onto the polyvinylidene fluoride (PVDF) membrane (1,620,177, BIO-RAD, Hercules, CA, USA) for blockade using 5% skim milk or 5% BSA at room temperature for 1 h. Subsequently, the membrane was subject to overnight incubation with the primary rabbit antibodies from Abcam: TIM-3 (ab241332, 1: 1000), E-cadherin (ab212059, 1: 1000), N-cadherin (ab76011, 1: 10,000), and Vimentin (ab92547, 1: 2000) at 4 °C. The following day, the membrane was rinsed with 1 × TBST at room temperature 3 times, 5 min per time, followed by incubation with the corresponding HRP-labeled secondary antibody goat anti-rabbit IgG (ab6721, at a dilution ratio of 1: 5000, Abcam) at room temperature for 1 h. After 3 rinses with 1 × TBST buffer solution at room temperature for 5 min per time, the membrane was immersed in ECL reaction solution (1,705,062, Bio-Rad) at room temperature for 1 min. Image Quant LAS 4000C gel imager (General Electric company, USA) was employed for exposure. The protein expression was summarized as the ratio of the gray value of protein for analysis relative to the expression of β-actin (ab5694, 1: 1000, rabbit, Abcam).
Scanning electron microscope
Aseptic slides were placed into the 12-well plate, which was supplemented with 2 mL of THP-1 Cell Suspension (2.5 × 105 cells/mL). After polarizaition into M0, M1 and M2 macrophages according to the aforementioned protocol, the supernatant was removed, and rinsed gently with PBS. The cells were fixed in 2.5% glutaraldehyde for 24 h and immersed into ethanol of variable concentrations (30%, 50%, 70%, and 90%) for 10 min, respectively. The cells were immersed in anhydrous ethanol for more than 10 min. The slide was dried in an ultra-vacuum for more than 4 h, plated with gold in vacuum for 40 s, and then observed with documentation of the findings under scanning electron microscopy.
Cell counting kit-8 (CCK-8) assay
The cells were seeded into a 96-well plate, 2 × 105 cells/well, and supplemented with 10 μL of CCK-8 (Sigma) after 24 h. The cells were incubated for another 2 h. The absorbance at the excitation wavelength of 450 nm was measured using the microplate reader (NYW-96 M, Beijing Noahway Instruments Co., Ltd., Beijing, China).
Transwell assay
Transwell chamber (8 mm; Corning, NY, USA) was adopted for assessing the migration of cells in a 24-well plate. The lower chamber was supplemented with 20% FBS DMEM (600 mL) and equilibrated at 37 °C for 1 h. After the intervention, MV3 was resuspended in DMEM without FBS. A total of 3 × 105 cells/mL were inoculated into the upper chamber for culture in 5% CO2 at 37 °C for 24 h. The cells were removed from Transwell chamber and fixed in 4% polyformaldehyde for 20 min. Subsequently, the cells were stained with 0.1% crystal violet for 10 min. The cells on the surface of chamber were removed using a cotton ball for observation under inverted fluorescence microscopy (TE2000, Nikon, Tokyo, Japan). Five fields were randomly selected for documenting the observations. The number of cells passing through the chamber was counted, and the average value was estimated. Transwell chamber with Matrigel was employed for cell invasion assay, while the other procedures were the same.
Animal experiment
Twelve male BALB/c nude mice (6–8 weeks old) were raised under standard environmental condition at 22–25 °C with relative humidity of 45–50% and 12 h light-dark cycle. The nude mice were randomly divided into two groups after one week of adaptive feeding.
Subcutaneous tumorigenesis experiment was as follows: Nude mice were subcutaneously injected with MV3 cells resuspended using PBS and transduced with sh-NC or sh-TIM-3 (1 × 106 cells/mouse) (n = 6). Subsequently, the macrophages treated with MV3-Exo-sh-NC—CD4+ T cells or MV3-Exo-sh-TIM-3-CD4+ T cells were injected into the tail vein of nude mice once a week. The tumor size of nude mice was measured 1 week after injection administration, while the tumor was surgically removed after 4 weeks. Tumor weight were calculated after isolation with the utility of the following formula: (length) × Width 2)/2.
Lung metastasis test was as follows: MV3 cells and macrophages were treated in compliance with the aforementioned protocols. MV3 cells and macrophages were simultaneously injected into the tail vein of nude mice. After two weeks, the lungs of nude mice were surgically harvested for quantification of the metastatic nodules.
Hematoxylin and eosin (HE) staining
The tumor or whole lung tissue of nude mice was fixed in 4% paraformaldehyde, dehydrated with gradient alcohol, transparent in xylene, and then embedded in paraffin. The paraffin-embedded tissues were sliced into 5 μM thick sections, which were conventionally prepared for use. After staining with hematoxylin (Beyotime) for 30 s, the sections were rinsed under tap water, followed by differentiation with 5% hydrochloric acid alcohol differentiation solution for 10 s and back to blue using the water anti-blue solution for 10 min. The sections were dyed utilizing eosin for 30 s, followed by dehydration, transparence, and sealing with neutral gum. The sections were observed under an optical microscope (XP-330, Shanghai Bingyu Optical Instrument Co., Ltd., Shanghai, China).
Immunohistochemistry
For Immunohistochemical analysis, the tissues were deparaffinized in xylene and rehydrated in gradient ethanol. After immersion in ethylenediaminetetraacetic acid (EDTA) antigenic retrieval solution (pH8.0), the tissue sections were incubated with the normal goat serum blocking buffer at room temperature for 1 h. The sections were subsequently probed to the anti-CD4 primary antibody (ab183685, 1: 200, Abcam) or anti-CD206 (ab64693, 1: 100, Abcam) at 4 °C overnight, and then incubated at room temperature for 1 h with peroxidase-conjugated secondary antibody (DS-0004, ZSGB-BIO, Beijing, China). The cell nuclei were subjected to staining with the hematoxylin (ZLI-9610, ZSGB-BIO).
Statistical analysis
Data analysis was performed using the SPSS 21.0 software (IBM-SPSS Inc., Chicago, IL, USA). All quantitative data were presented as mean ± standard deviation. An independent t-test was adopted for comparison of data between two groups. Data comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) with Tukey's post-hoc test. Data comparisons at different time points were performed by repeated measurement ANOVA with Tukey's post-hoc test. In all statistical references, a value of p < 0.05 were considered to be significant.
Results
MV3 cells-derived exosomes inhibit the immune function and proliferation of CD4+ T cells
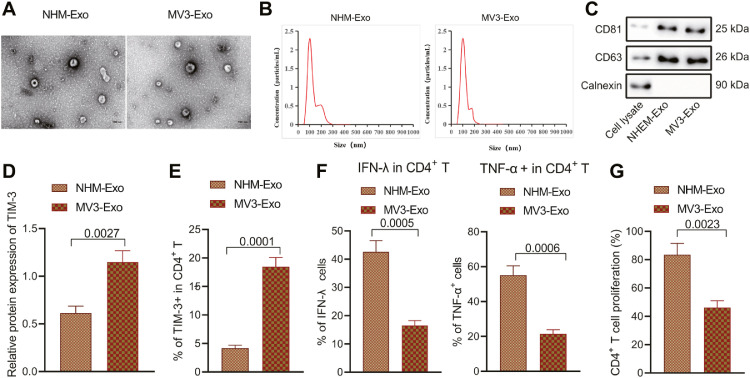
Tumor cells secrete exosomes under different stress conditions, and its function is reflective of passage of a variety of bioactive substances into host cells for d the regulation of host immune response [19]. In this study, ultracentrifugation was conducted to separate NHEM-Exo and MV3-Exo from the supernatant of culture cells for 48 h. The exosomes were observed under TEM (Fig. 1A), which revealed that the exosome was a round or oval membrane vesicle of the same shape, while the size of exosome was about 160 nm (Fig. 1B). Western blot analysis (Fig. 1C) illustrated that the surface markers of exosomes CD81 and CD63 were highly expressed, however calnexin were not expressed, which are illustrative of successful extraction of exosomes.
Fig. 1.
Effects of the exosomes from the NHEM cells and MV3 cells on the immune function and proliferation of CD4+ T cells. A, The exosomes from NHEM cells and MV3 cells observed under electron microscope. B, The diameter of exosomes determined by NTA. C, Protein levels of exosome surface markers determined by Western blot analysis. CD4+ T cells were co-cultured with NHEM-Exo and MV3-Exo, respectively. D, The protein level of TIM-3 in CD4+ T cells detected by Western blot. E, The expression pattern of TIM-3 in CD4+ T cell surface detected by flow cytometry. F, The proportion of CD4+ T cells producing cytokine IFN-γ and TNF-α detected by flow cytometry. G, The proliferation of CD4+ T cells detected by flow cytometry. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data between two groups were compared by the independent sample t-test.
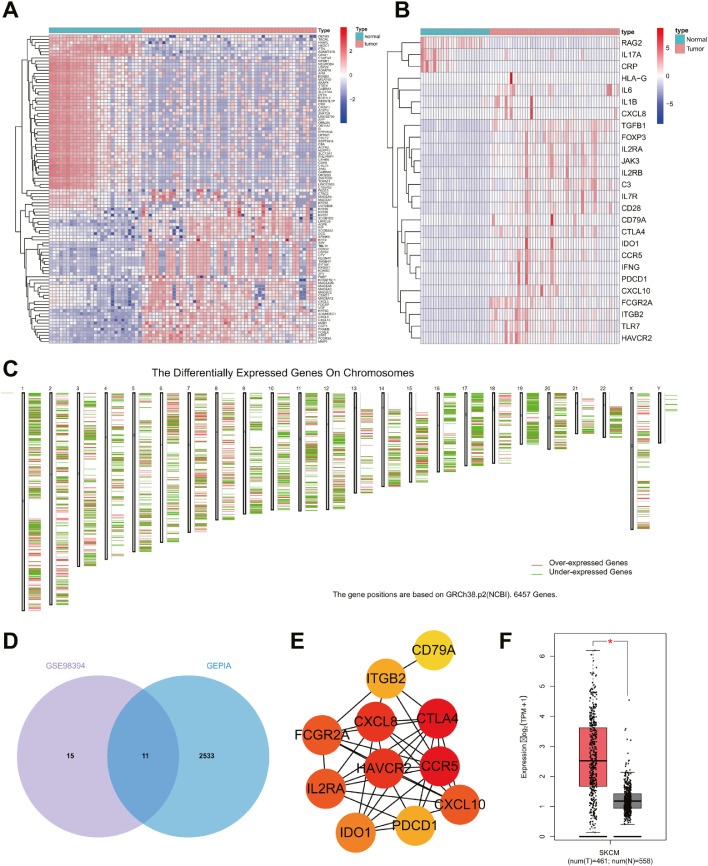
An existing study demonstrated that exosomes originating from tumor cells usually import immunosuppressive factors, which can interfere with the function of immune cells [20]. To further study the effect of melanoma cells-secreted exosomes on CD4+ T cells, 2254 DEGs were initially identified vfrom GSE98394, and 26 IRGs were identified, among which, 23 IRGs were upregulated (Fig. 2A, B). Moreover, 6457 DEGs were identified from the tumor samples of melanoma patients in the TCGA database, of which 2541 were upregulated (Fig. 2C). A total of 11 upregulated IRGs were identified by intersection between microarray data GSE98394 and TCGA database (Fig. 2D).
Fig. 2.
Bioinformatics prediction of TIM-3 expression pattern and biological function in melanoma cells-derived exosomes. A, The heat map of the top 50 DEGs in GSE98394, including 51 tumor samples and 27 normal samples. B, The heat map of differentially expressed IRG in GSE98394. C, DEGs in melanoma samples retrieved from TCGA database. D, Venn map of the intersection between the highly expressed IRG in GSE98394 and highly expressed genes in TCGA database. E, Protein interaction network of 11 IRGs. F, The expression pattern of HAVCR2 in melanoma samples from TCGA database.
Among the 11 upregulated IRGs, HAVCR2 (TIM-3) is a suppressor-molecule in T cell mediated immune response. Moreover, accumalting evidence has identified notable enrichment of HAVCR2 in the tumor cells-derived exosomes [14, 21]. Protein-protein interaction were further analyzed through the STRING database, which illustrated TIM-3 as one of the core genes (Fig. 2E). As shown in Fig. 2F, a high expression pattern of TIM-3 was evident among the samples of patients with cutaneous melanoma in the TCGA database.
Next, CD4+ T cells were isolated from the spleen of nude mice for co-culture with NHEM-Exo and MV3-Exo, respectively. Western blot and flow cytometric analyses were performed to characterize the expression patterns of TIM-3 and cytokine IFN-γ and TNF-α in CD4+ T cells. The results demonstrated notably higher protein level of TIM-3 and proportion of TIM-3+CD4+ T cells in CD4+ T cells co-incubated with MV3-Exo relative to the CD4+ T cells co-incubated with NHEM-Exo (Fig. 1D, E; Supplementary Fig. 1A). The proportion of cells producing cytokine IFN-γ and TNF-α in CD4+ T cells co-cultured with MV3-Exo was significantly lower relative ot the CD4+ T cells co-cultured with NHEM-Exo (Fig. 1F). It was, therefore, denoted that MV3-Exo could strikingly inhibit the immune function of CD4+ T cells. Flow cytometry revealed considerably inhibited proliferation of CD4+ T cells in CD4+ T cells co-cultured with MV3-Exo was significantly inhibited (Fig. 1G). The preceding findings validated that the MV3 cells-derived exosomes had effectively suppressed the immune function and proliferative ability of CD4+ T cells.
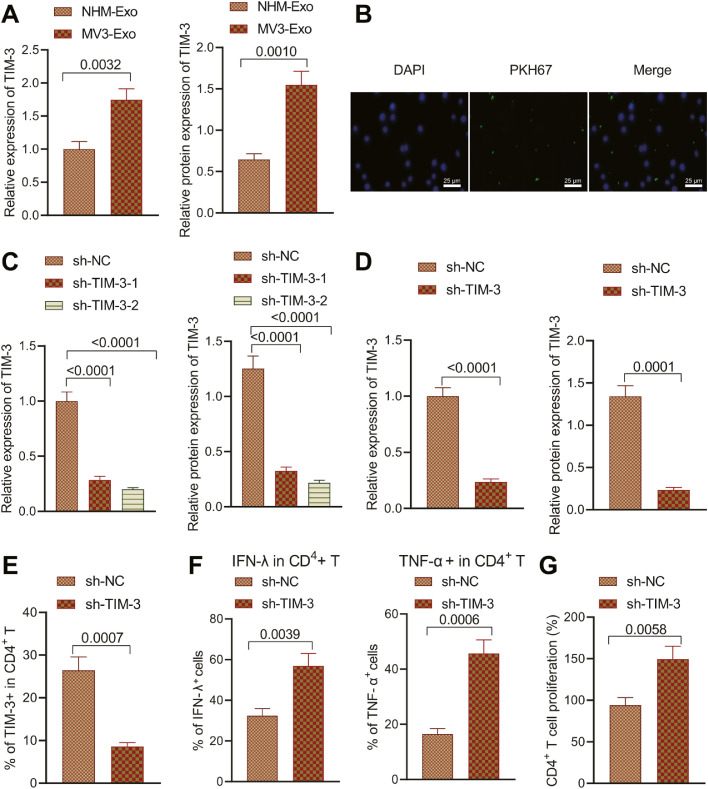
TIM-3 shuttled by MV3-cells derived exosomes inhibits the immune function and proliferative potential of CD4+T cells
The TIM-3 expression pattern in NHEM-Exo and MV3-Exo was detected, which elicited an elevated TIM-3 expression pattern in MV3-Exo (Fig. 3A; Supplementary Fig. 1B). Next, we sought to determine whether MV3-Exo could deliver TIM-3 into CD4+ T cells. MV3-Exo loaded with PKH26 (red) was co-cultured with CD4 + T cells (green) to observe the uptake of exosomes by CD4+ T cells under confocal fluorescence microscopy. The results are demonstrative of that red fluorescence in CD4+ T cells, suggesting that CD4+ T could capture a high proportion of MV3-Exo (Fig. 3B).
Fig. 3.
Effect of TIM-3 inhibition in MV3-Exo on immune function and proliferation of CD4+ T cells. A, TIM-3 expression pattern in NHEM-Exo and MV3-Exo determined by RT-qPCR and Western blot analysis. B, Uptake of exosomes by CD4+ T cells under confocal fluorescence microscopy. MV3 cells were transduced with sh-NC, sh-TIM-3–1, or sh-TIM-3–2. C, TIM-3 expression in MV3 cells measured by Western blot analysis. D, TIM-3 expression pattern in Exo derived from the MV3 cells transduced with sh-TIM-3 determined by RT-qPCR and Western blot analysis. CD4+ T cells were co-cultured with MV3-Exo-sh-NC and MV3-Exo-sh-TIM-3. E, Expression pattern of CD4+ T cell surface inhibitory receptor TIM-3 detected by flow cytometry. F, The proportion of CD4+ T cells producing IFN-γ and TNF-α detected by flow cytometry. G, CD4+ T cell proliferation detected by flow cytometry. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was carried out by the independent t-test. One-way ANOVA with Tukey's post-hoc test was applied for the comparison of data among multiple groups.
MV3 cells were transduced with sh-NC, sh-TIM-3–1, or sh-TIM-3–2 prior to analysis of the TIM-3 expression pattern, which revealed the best inhibitory effect on the expression pattern of TIM-3 with sh-TIM-32, thus was used for subsequent experimentation (Fig. 3C; Supplementary Fig. 1C).
After transduction with sh-TIM-3, the exosomes were isolated from the supernatant. RT-qPCR and Western blot analysis revealed a reduced TIM-3 expression pattern in MV3-Exo transduced with sh-TIM-3 (Fig. 3D; Supplementary Fig. 1D).
CD4+ T cells were co-cultured with MV3-Exo-sh-NC and MV3-Exo-sh-TIM-3 prior to detection of the expression patterns of CD4+ T cell surface inhibitory receptor, TIM-3 and cytokine IFN-γ and TNF-αe. The proportion of TIM-3+CD4+ T in CD4+ T cells co-cultured with MV3-Exo-sh-TIM-3 was lower relative to the CD4+ T cells co-cultured with MV3-Exo-sh-NC (Fig. 3E), while the proportion of IFN-γ+CD4+ T and TNF-α+CD4+ T cells in CD4+ T cells co-cultured with MV3-Exo-sh-TIM-3 was higher relative to the CD4+ T cells co-cultured with MV3-Exo-sh-NC (Fig. 3F). Thus, TIM-3 loss in MV3-Exo could appreciably limit the expression pattern of TIM-3 on the CD4+ T cell surface to facilitate the immune function of CD4+ T cells. Flow cytometric data illustrated elevated CD4+ T cell proliferation after incubation with MV3-Exo-sh-TIM-3 (Fig. 3G). The preceding findings elicited that the downregulation of TIM-3 in MV3 cells-derived exosomes could ameliorate CD4+ T cell immune function and enhance CD4+ T cell proliferation.
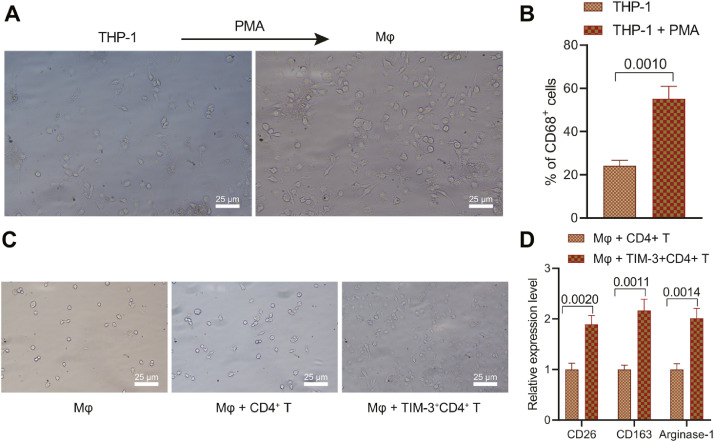
TIM-3+CD4+T cells induces macrophage M2 polarization
Immunotherapy, as one of the critical therapies for metastasis, aims to exert a chronic immunosurveillance effect to avoid recurrence by restoration of antitumor immunity in the tumor microenvironment, while macrophage M2 polarization induces tumor immunosuppression, leading to tumor deterioration and recurrence [9]. Therefore, the effect of MV3-Exo inducing CD4+ T cells in the transformation into TIM-3+CD4+T cells on macrophage polarization. Human monocytes THP-1 were stimulated with 100 ng/mL PMA for 24 h to prepare Mφ macrophages. THP-1 cells were observed to be round suspension cells under optical microscopy. After PMA stimulation, THP-1 cells became round and fusiform adherent cells, with extended pseudopodia Mφ macrophage (Fig. 4A). Flow cytometry demonstrated that after stimulation by PMA, a notably higher proportion of CD68+ cells in the control group after stimulation by PMA (Fig. 4B), thus suggesting that THP-1 cells was stimulated by PMA to isolate Mφ macrophages.
Fig. 4.
Overexpression of TIM-3 in CD4+ T cells promotes macrophage M2 polarization. A, Morphology of THP-1 and Mφ macrophages observed under optical microscopy. B, Expression pattern of the marker of macrophages CD68 detected by flow cytometry. C, Morphology of Mφ macrophages co-cultured with TIM-3+CD4+ T cells observed under the optical microscope and electron microscope. D, Expression patterns of several markers of M2-type macrophages (CD206, CD163, and Arginase-1) in Mφ macrophages co-cultured with TIM-3+CD4+ T cells. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.
The obtained Mφ macrophages were co-cultured with the CD4+ T cells and TIM-3+CD4+T cells for 24 h prior to the morphological observation of macrophages. Co-culture of Mφ macrophages with TIM-3+CD4+ T cells induced alterations in shape from round and spindle adherent cells to irregular adherent cells (Fig. 4C). RT-qPCR exhibited elevated expression patterns of markers of M2-type macrophages (CD206, CD163, and Arginase-1) in Mφ macrophages co-incubated with TIM-3+CD4+ T cells (Fig. 4D). The obtained findings indicated that the TIM-3+CD4+T cells promoted macrophage M2 polarization.
Macrophage M2 polarization promotes the growth and metastasis of melanoma cells
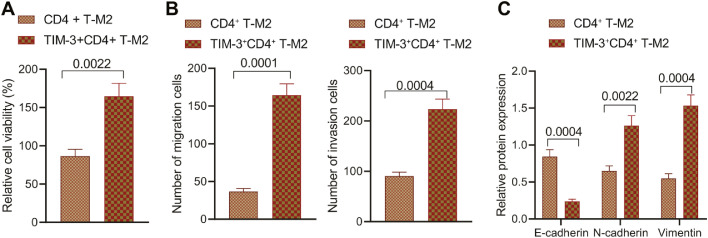
For purpose of additional validation, the macrophages induced by CD4+ T cells and then TIM-3+CD4+ T cells were submitted to co-culture with melanoma cells MV3 and A375. A combination of CCK-8 assay and Transwell assay exhibited elevated proliferation, migratory, and invasive properties of MV3 cells and A375 cells after incubation with TIM-3+CD4+ T-M2 (Fig. 5A, B; Supplementary Fig. 2A, B). Western blot analysis elected a reduced E-cadherin expression pattern along with increased expression patterns of N-cadherin and Vimentin in MV3 cells and A375 cells following co-culturing with TIM-3+CD4+ T-M2 (Fig. 5C; Supplementary Fig. 1E, 2C). The aforementioned findings illustrated that TIM-3+CD4+ T cells can promote the growth and metastasis of melanoma cells by inducing macrophage M2 polarization.
Fig. 5.
TIM-3+CD4+ T cells promote the growth and metastasis of MV3 cells by inducing macrophage M2 polarization. MV3 cells were co-cultured with TIM-3+CD4+ T-M2 macrophages. A, MV3 cell proliferation detected by CCK-8 assay. B, MV3 cell migration and invasion detected by Transwell assay. C, Expression pattern of several EMT-related factors in MV3 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was carried out by independent t-test.
Inhibition of TIM-3 in MV3-Exo inhibits the growth and metastasis of melanoma cells in vitro
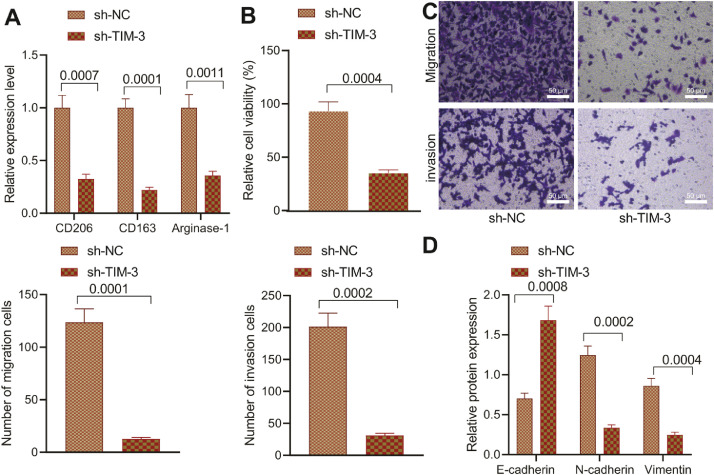
Co-culture system was adopted to detect the degree of M2 polarization of macrophages along with the growth and metastasis of MV3 cells and A375 cells after transduction with sh-NC and sh-TIM-3, respectively. RT-qPCR showed reduced expression patterns of several M2-type macrophage markers (CD206, CD163, and Arginase-1) in macrophages co-incubated with CD4+ T cells intervened with MV3-Exo-sh-TIM-3 (Fig. 6A). CCK-8 assay and Transwell assay exhibited suppressed the proliferating, migrating, and invading functions of MV3 cells and A375 cells after transduction with sh-TIM-3 and co-culturing with CD4+ T cells and macrophages (Fig. 6B, C; Supplementary Fig. 3A, B). Western blot analysis demonstrated an increased E-cadherin expression pattern along with decreased expression patterns of N-cadherin and Vimentin in MV3 cells and A375 cells transduced with sh-TIM-3 and co-cultured with CD4+ T cells and macrophages (Fig. 6D; Supplementary Fig. 1F, 3C). The preceding results suggested that downregulation of TIM-3 in MV3 cells-derived exosomes can facilitate inhibition of CD4+ T cells on macrophage M2 polarization, thus ultimately inhibiting the growth and metastatic activity of melanoma cells.
Fig. 6.
Inhibition of TIM-3 in MV3-Exo affects the anti-tumor effect of CD4+ T cells. CD4+ T cells were co-cultured with MV3-Exo-sh-TIM-3 and macrophages. A, Expression pattern of several markers of M2-type macrophages (CD206, CD163, and Arginase-1) in macrophages determined by RT-qPCR. B, MV3 cell proliferation detected by CCK-8 assay. C, MV3 cell migration and invasion detected by Transwell assay. D, Expression pattern of several EMT-related factors in MV3 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.
Inhibition of TIM-3 in MV3-Exo dampens the growth and metastasis of melanoma cells in vivo
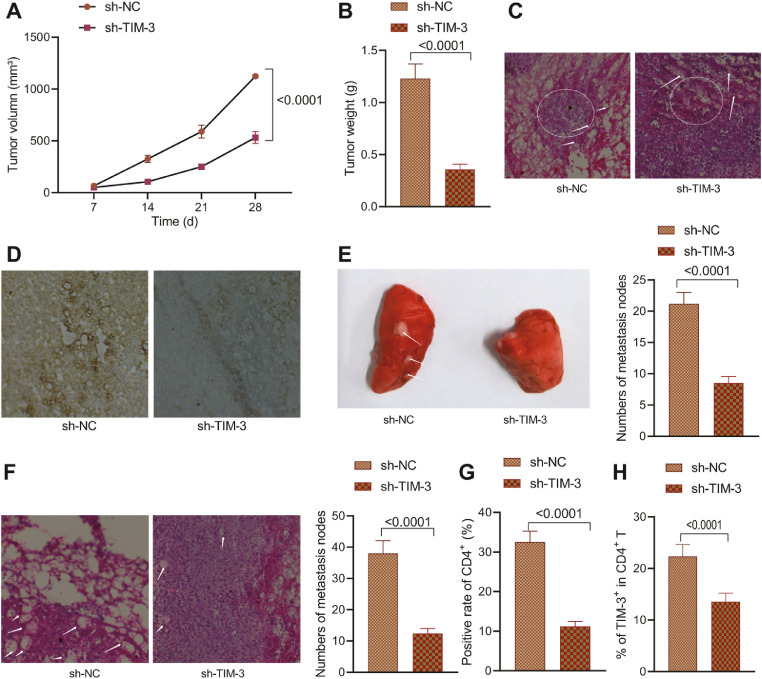
In vivo experiments were conducted in an attempt to explore the effects of MV3-Exo transduced with sh-TIM-3 on the growth and metastasis of melanoma cells. Nude mice were instilled with the MV3 cells via the armpit, while the MV3-Exo-sh-NC—CD4+ T cells- and MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages were instilled via the tail vein. After four weeks, an evaluation revealed lowered tumor volume in nude mice injected with the MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7A-C). HE staining displayed increased leukocyte infiltration in nude mice injected with MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7D). Besides, as suggested by CD206 immunohistochemistry staining, reduced M2 macrophage infiltration was evident in the nude mice injected with MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7E). Our findings elicited that inhibition of TIM-3 can restore the immune function of CD4+ T cells and inhibit the stimulative effect of M2 macrophages on melanoma growth in vivo.
Fig. 7.
Effect of depleted TIM-3 in MV3-Exo on the growth and metastasis of melanoma cells in nude mice. A, Tumor morphology in nude mice. B, Tumor volume in nude mice. C, Tumor weight in nude mice. D, Pathological changes of nude mice observed using HE staining. E, M2 macrophage infiltration in tumors of nude mice by CD206 immunohistochemistry staining. F, The number of lung metastatic nodes in nude mice. G, Population of infiltrated CD4+ T cells in tumors of nude mice by CD4+ immunohistochemistry staining. H, Percentage of TIM-3+CD4+T cells in lungs of nude mice by flow cytometry. p < 0.05. n = 6. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test, and repeated measurement ANOVA with Tukey's post-hoc test was applied for comparison of data at different time points.
Lung metastasis experiment illustrated a decreased number of metastatic lung nodules in nude mice injected with MV3 cells and MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7F). CD4+ immunohistochemistry staining revealed that the population of infiltrated CD4+ T cells had decreased in the lung tissues of nude mice injected with MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7G). Flow cytometry analysis showed a reduced percentage of TIM-3+CD4+ T cells in nude mice injected with MV3-Exo-sh-TIM-3-CD4+ T cells-treated macrophages (Fig. 7H). The obtained findings suggested that silencing of TIM-3 in MV3 cell-derived exosomes could facilitate the inhibition of macrophage M2 polarization by CD4+ T cells, thereby suppressing the malignancy and metastatic activity of melanoma cells in vivo.
Discussion
Melanoma is a fatal type of skin cancer, which is especially difficult to treat after its metastasis [22]. Immunotherapy, as one of the chief therapies for metastasis, aims for a chronic immunosurveillance effect to avoid recurrence of cancer by restoration of the antitumor immunity in tumor microenvironment, while macrophage M2 polarization can radically induce tumor immunosuppression, thus resulting in tumor deterioration and recurrence [9]. An existing study identified the significant impact of melanoma-released exosomes on the escape of melanoma cells from immune surveillance, however the crucial details of the mechanism remain elusive[23]. As the specific mechanisms that can influence the immune function of CD4+ T cells and macrophage M2 polarization in melanoma remain identified, the current study sought to investigate the effects of TIM-3 shuttled by MV3 cells-derived exosomes on the growth and metastasis in melanoma. Our findings demonstrated that TIM-3 shuttled by MV3 cells-derived exosomes could exert a suppressive effect on the immune function of CD4+ T cells and can further manipulate macrophage M2 polarization to facilitate the growth and metastasis of melanoma cells both in vitro and in vivo.
Our initial observations revealed the capacity of MV3 cells-derived exosomes to inhibit the immune function and proliferation of CD4+ T cells. Treg cells have been identified to be essential for the maintenance of peripheral immune tolerance and homeostasis [24]. CD4+T cells, fractions of white blood cells can exacerbate the degree of the cellular immune responses that provide protection against several forms of tumors, including melanoma [25]. Previous research has cited the vital functionality of EVs in intercellular communication and as vital components in tumor immunology [26]. Cancer cells by releasing exosomes can participate in the delivery of multiple biologically active substances into host cells and/or modulation of host immune responses [19]. Melanoma cells can essentially mediate various protumor processes with the production of exosomes, such as immune regulation and modification of tissue microenvironments [27]. Recent research has gained interest as exosomes isolated from melanoma cells elicit immunosuppressive properties with the functional suppression of primary human immune cells [28]. Meanwhile, melanoma cells-derived exosomes promote the melanoma growth by eliminating CD4+ T cells [23].
In addition, the current study demonstrated that macrophage M2 polarization enhanced the growth and metastasis of melanoma cells. An existing study identified an association between TAM infiltration and the facilitation of tumor growth, invasion and metastasis for exacerbated tumor progression [29]. Moreover, M2-type TAMs can precipitate to decreased survival in melanoma patients [30]. Our findings elicited that MV3 cells-derived exosomes induced macrophage M2 polarization by upregulating CD206, CD163, and Arginase-1, thus facilitating the growth and metastasis of melanoma. A n existing study elicited association of M2-skewed macrophage markers, including the scavenger receptors CD163, CD204, and CD206 with melanoma growth and progression [7]. Moreover, activation of M2-type macrophages characterized by typical M2 markers like arginase-1 has been identified in melanoma [31]. Exosomes can serve as terminal modulators of melanoma cell proliferation, epithelial-mesenchymal transition (EMT), and migration [11,32]. Gerlo et al. have verified the property of melanoma-sourced exosomes to elicit a tumor-promoting TAM phenotype in macrophages [13]. Morover, existing evidence verified that melanoma-secreted exosomes could effectively facilitate tumor survival and metastasis by M2 macrophage polarization [8], which is consistent with our findings.
Furthermore, our findings illustrated an abundance of TIM-3 in MV3 cells-derived exosomes, whereas inhibition of TIM-3 in MV3 cells-derived exosomes could induce CD4+ T cell immune function and suppress M2 macrophage polarization in melanoma. Additionally, a recent study identified a correlation between an increased TIM-3 expression with the inhibition of T cell responses and subseuqent T cell dysfunction, which is a definitive marker for gradual loss of T cell function in a hierarchical manner during tumor development [16]. As TIM-3 is an inhibitory receptor of CD4+ T cells, hereby targeting TIM-3 as a novel biomarker has been proposed in melanoma immunotherapy [33]. A recent study also exhibited the ability of that osteosarcoma cells-secreted exosomes to transport TIM-3 to induce macrophage M2 polarization, which consequently induce the invasiveness, metastasis, and EMT of tumors [14], while its effects on the progression of melanoma remain elusive. In the current study, either in vitro or in vivo experimentation validated that the silencing of TIM-3 shuttled by MV3 cells-secreted exosomes had definitively suppressed the growth and metastasis of melanoma cells.
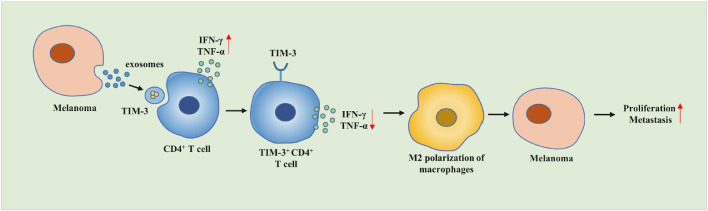
Conclusion
To conclude, our results demonstrated that the transfer of TIM-3 via MV3 cells could secrete exosomes to inhibit CD4+ T cell immune function and induce macrophage M2 polarization, which conjointly exacerbates the growth and metastasis of melanoma cells (Fig. 8). Our findings provide insight for the development of effective therapeutic strategies combating melanoma cells. However due to limited literature, the roles of TIM-3 shuttled by MV3 cells-derived exosomes in the CD4+ T cell immune function and macrophage M2 polarization in the progression of melanoma warrant extensive investigation.
Fig. 8.
Molecular mechanism of the effects of TIM-3 shuttled by MV3 cells-derived exosomes on the CD4+ T cell immune function and macrophage M2 polarization involved in the occurrence and development of melanoma.
CRediT authorship contribution statement
Xinghui Li: Conceptualization, Data curation, Formal analysis, Investigation. Yu Liu: Data curation, Formal analysis, Methodology, Supervision, Writing – original draft. Li Yang: Data curation, Formal analysis, Visualization, Writing – original draft. Yannan Jiang: Conceptualization, Software, Validation, Writing – review & editing. Qihong Qian: Conceptualization, Investigation, Methodology, Resources, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101334.
Appendix. Supplementary materials
Supplementary Fig. 1 Representative western blot images for the proteins measured in this study. A, Representative western blot images for TIM-3 protein in CD4+ T cells co-cultured with NHEM-Exo and MV3-Exo. B, Representative western blot images for TIM-3 protein in NHEM-Exo and MV3-Exo. C, Representative western blot images for TIM-3 protein in MV3 cells transduced with sh-NC, sh-TIM-3–1, or sh-TIM-3–2. D, Representative western blot images for TIM-3 protein in Exo derived from the MV3 cells transduced with sh-TIM-3. E, Representative western blot images for several EMT-related factors in MV3 cells co-cultured with TIM-3+CD4+ T-M2. F, Representative western blot images for several EMT-related factors in MV3 cells transduced with sh-TIM-3.
Supplementary Fig. 2 Validation experiments using A375 cells after being cocultured with TIM-3+CD4+ T-M2 macrophages. A, A375 cell proliferation detected by CCK-8 assay. B, A375 cell migration and invasion detected by Transwell assay. C, Expression pattern of several EMT-related factors in A375 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.
Supplementary Fig. 3 Validation experiments using A375 cells after transduction with sh-NC and sh-TIM-3. A, A375 cell proliferation detected by CCK-8 assay. B, A375 cell migration and invasion detected by Transwell assay. C, Expression pattern of several EMT-related factors in A375 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.
References
- 1.DiCaprio M.R., Abousayed M.M., Kambam M.L.R. Orthopaedic manifestations of melanoma and their management. J. Am. Acad. Orthop. Surg. 2020;28(13):e540–e549. doi: 10.5435/JAAOS-D-18-00757. [DOI] [PubMed] [Google Scholar]
- 2.Gowda R., Robertson B.M., Iyer S., Barry J., Dinavahi S.S., Robertson G.P. The role of exosomes in metastasis and progression of melanoma. Cancer Treat Rev. 2020;85 doi: 10.1016/j.ctrv.2020.101975. [DOI] [PubMed] [Google Scholar]
- 3.van der Weyden L., Brenn T., Patton E.E., Wood G.A., Adams D.J. Spontaneously occurring melanoma in animals and their relevance to human melanoma. J. Pathol. 2020;252(1):4–21. doi: 10.1002/path.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J., Carr M.J., Khushalani N.I. Principles of targeted therapy for melanoma. Surg. Clin. North Am. 2020;100(1):175–188. doi: 10.1016/j.suc.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Liu X., Sanders K.L., Edwards J.L., Ye J., Si F., Gao A., Huang L., Hsueh E.C., Ford D.A., Hoft D.F., Peng G. TLR8-mediated metabolic control of human treg function: a mechanistic target for cancer immunotherapy. Cell Metab. 2019;29(1):103–123. doi: 10.1016/j.cmet.2018.09.020. e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W., Zhang W., Wang X., Tan L., Li H., Wu J., Wu Q., Sun W., Chen J., Yin Y. HCA587 protein vaccine induces specific antitumor immunity mediated by CD4(+) T-cells expressing granzyme b in a mouse model of melanoma. Anticancer Agents Med. Chem. 2021;21(6):738–746. doi: 10.2174/1871520620666200728131951. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Janeiro A., Padilla-Ansala C., de Andrea C.E., Hardisson D., Melero I. Prognostic value of macrophage polarization markers in epithelial neoplasms and melanoma. A systematic review and meta-analysis. Mod. Pathol. 2020;33(8):1458–1465. doi: 10.1038/s41379-020-0534-z. [DOI] [PubMed] [Google Scholar]
- 8.Bardi G.T., Smith M.A., Hood J.L. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine. 2018;105:63–72. doi: 10.1016/j.cyto.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian Y., Qiao S., Dai Y., Xu G., Dai B., Lu L., Yu X., Luo Q., Zhang Z. Molecular-targeted immunotherapeutic strategy for melanoma via dual-targeting nanoparticles delivering small interfering RNA to tumor-associated macrophages. ACS Nano. 2017;11(9):9536–9549. doi: 10.1021/acsnano.7b05465. [DOI] [PubMed] [Google Scholar]
- 10.Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B.A., Callahan M.K., Yuan J., Martins V.R., Skog J., Kaplan R.N., Brady M.S., Wolchok J.D., Chapman P.B., Kang Y., Bromberg J., Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mannavola F., D’Oronzo S., Cives M., Stucci L.S., Ranieri G., Silvestris F., Tucci M. Extracellular vesicles and epigenetic modifications are hallmarks of melanoma progression. Int. J. Mol. Sci. 2019;21(1) doi: 10.3390/ijms21010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming V., Hu X., Weller C., Weber R., Groth C., Riester Z., Huser L., Sun Q., Nagibin V., Kirschning C., Bronte V., Utikal J., Altevogt P., Umansky V. Melanoma extracellular vesicles generate immunosuppressive myeloid cells by upregulating PD-L1 via TLR4 signaling. Cancer Res. 2019;79(18):4715–4728. doi: 10.1158/0008-5472.CAN-19-0053. [DOI] [PubMed] [Google Scholar]
- 13.Gerloff D., Lutzkendorf J., Moritz R.K.C., Wersig T., Mader K., Muller L.P., Sunderkotter C. Melanoma-derived exosomal miR-125b-5p educates tumor associated macrophages (TAMs) by targeting lysosomal acid lipase A (LIPA) Cancers (Basel) 2020;12(2) doi: 10.3390/cancers12020464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Z., Wang L., Wu C., Huang L., Ruan Y., Xue W. Tumor-derived exosomes induced M2 macrophage polarization and promoted the metastasis of osteosarcoma cells through tim-3. Arch. Med. Res. 2021;52(2):200–210. doi: 10.1016/j.arcmed.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Holderried T.A.W., de Vos L., Bawden E.G., Vogt T.J., Dietrich J., Zarbl R., Bootz F., Kristiansen G., Brossart P., Landsberg J., Dietrich D. Molecular and immune correlates of TIM-3 (HAVCR2) and galectin 9 (LGALS9) mRNA expression and DNA methylation in melanoma. Clin. Epigenetics. 2019;11(1):161. doi: 10.1186/s13148-019-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das M., Zhu C., Kuchroo V.K. Tim-3 and its role in regulating anti-tumor immunity. Immunol. Rev. 2017;276(1):97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acharya N., Sabatos-Peyton C., Anderson A.C. Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother. Cancer. 2020;8(1) doi: 10.1136/jitc-2020-000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J.J., Burger P., Taube J., Soni A., Chaichana K., Sheu M., Belcaid Z., Jackson C., Lim M. PD-L1, PD-1, LAG-3, and TIM-3 in melanoma: expression in brain metastases compared to corresponding extracranial tumors. Cureus. 2019;11(12):e6352. doi: 10.7759/cureus.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W.S., Choi D., Park J.M., Song H.Y., Seo H.S., Lee D.E., Byun E.B. Comparison of exosomes derived from non- and gamma-irradiated melanoma cancer cells as a potential antigenic and immunogenic source for dendritic cell-based immunotherapeutic vaccine. Vaccines (Basel) 2020;8(4) doi: 10.3390/vaccines8040699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig S., Floros T., Theodoraki M.N., Hong C.S., Jackson E.K., Lang S., Whiteside T.L. Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin. Cancer Res. 2017;23(16):4843–4854. doi: 10.1158/1078-0432.CCR-16-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng H., Zhang G.M., Li D., Zhang H., Yuan Y., Zhu H.G., Xiao H., Han L.F., Feng Z.H. Soluble form of T cell Ig mucin 3 is an inhibitory molecule in T cell-mediated immune response. J. Immunol. 2006;176(3):1411–1420. doi: 10.4049/jimmunol.176.3.1411. [DOI] [PubMed] [Google Scholar]
- 22.Yang K., Oak A.S.W., Slominski R.M., Brozyna A.A., Slominski A.T. Current molecular markers of melanoma and treatment targets. Int. J. Mol. Sci. 2020;21(10) doi: 10.3390/ijms21103535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Yang Y., Wang W., Zhang Y., Chen Z., Hao C., Zhang J. Melanoma-released exosomes directly activate the mitochondrial apoptotic pathway of CD4(+) T cells through their microRNA cargo. Exp. Cell Res. 2018;371(2):364–371. doi: 10.1016/j.yexcr.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Yu X., Lao Y., Teng X.L., Li S., Zhou Y., Wang F., Guo X., Deng S., Chang Y., Wu X., Liu Z., Chen L., Lu L.M., Cheng J., Li B., Su B., Jiang J., Li H.B., Huang C., Yi J., Zou Q. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat. Commun. 2018;9(1):3157. doi: 10.1038/s41467-018-05676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pour S.Rad, Morikawa H., Kiani N.A., Yang M., Azimi A., Shafi G., Shang M., Baumgartner R., Ketelhuth D.F.J., Kamleh M.A., Wheelock C.E., Lundqvist A., Hansson J., Tegner J. Exhaustion of CD4+ T-cells mediated by the kynurenine pathway in melanoma. Sci. Rep. 2019;9(1):12150. doi: 10.1038/s41598-019-48635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathew M., Zade M., Mezghani N., Patel R., Wang Y., Momen-Heravi F. Extracellular vesicles as biomarkers in cancer immunotherapy. Cancers (Basel) 2020;12(10) doi: 10.3390/cancers12102825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hood J.L. Natural melanoma-derived extracellular vesicles. Semin. Cancer Biol. 2019;59:251–265. doi: 10.1016/j.semcancer.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Sharma P., Diergaarde B., Ferrone S., Kirkwood J.M., Whiteside T.L. Melanoma cell-derived exosomes in plasma of melanoma patients suppress functions of immune effector cells. Sci. Rep. 2020;10(1):92. doi: 10.1038/s41598-019-56542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao M., Yan H., Han X., Weng L., Wei Q., Sun X., Lu W., Wei Q., Ye J., Cai X., Hu C., Yin X., Cao P. Ginseng-derived nanoparticles alter macrophage polarization to inhibit melanoma growth. J. Immunother. Cancer. 2019;7(1):326. doi: 10.1186/s40425-019-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lardone R.D., Chan A.A., Lee A.F., Foshag L.J., Faries M.B., Sieling P.A., Lee D.J. Mycobacterium bovis bacillus calmette-guerin alters melanoma microenvironment favoring antitumor T cell responses and improving M2 macrophage function. Front. Immunol. 2017;8:965. doi: 10.3389/fimmu.2017.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.W., Park S., Han H.K., Gye M.C., Moon E.Y. Di-(2-ethylhexyl) phthalate enhances melanoma tumor growth via differential effect on M1-and M2-polarized macrophages in mouse model. Environ. Pollut. 2018;233:833–843. doi: 10.1016/j.envpol.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 32.Bollard S.M., Casalou C., Goh C.Y., Tobin D.J., Kelly P., McCann A., Potter S.M. Circulating melanoma-derived extracellular vesicles: impact on melanoma diagnosis, progression monitoring, and treatment response. Pharmaceuticals (Basel) 2020;13(12) doi: 10.3390/ph13120475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Cai P., Wang N., Zhang Q., Chen F., Shi L., Zhang Y., Wang L., Hu L. Combined blockade of Tim-3 and MEK inhibitor enhances the efficacy against melanoma. Biochem. Biophys. Res. Commun. 2017;484(2):378–384. doi: 10.1016/j.bbrc.2017.01.128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Representative western blot images for the proteins measured in this study. A, Representative western blot images for TIM-3 protein in CD4+ T cells co-cultured with NHEM-Exo and MV3-Exo. B, Representative western blot images for TIM-3 protein in NHEM-Exo and MV3-Exo. C, Representative western blot images for TIM-3 protein in MV3 cells transduced with sh-NC, sh-TIM-3–1, or sh-TIM-3–2. D, Representative western blot images for TIM-3 protein in Exo derived from the MV3 cells transduced with sh-TIM-3. E, Representative western blot images for several EMT-related factors in MV3 cells co-cultured with TIM-3+CD4+ T-M2. F, Representative western blot images for several EMT-related factors in MV3 cells transduced with sh-TIM-3.
Supplementary Fig. 2 Validation experiments using A375 cells after being cocultured with TIM-3+CD4+ T-M2 macrophages. A, A375 cell proliferation detected by CCK-8 assay. B, A375 cell migration and invasion detected by Transwell assay. C, Expression pattern of several EMT-related factors in A375 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.
Supplementary Fig. 3 Validation experiments using A375 cells after transduction with sh-NC and sh-TIM-3. A, A375 cell proliferation detected by CCK-8 assay. B, A375 cell migration and invasion detected by Transwell assay. C, Expression pattern of several EMT-related factors in A375 cells determined by Western blot analysis. p < 0.05. Data are presented as the mean ± standard deviation of three technical replicates. Data comparison between two groups was conducted by the independent t-test.