Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common type of malignant lymphoma with biologically and clinically heterogeneous features. Recently, the tumor microenvironment of this disease has been recognized as an important biological aspect of tumor development and therapeutic targets. Recurrent genetic alterations play significant roles in immune recognition of lymphoma cells. In particular, novel genetic alterations promoting phagocytosis were identified, suggesting a potential therapeutic strategy targeting the “don’t eat me” signal.
Keywords: microenvironment, phagocytosis, TMEM30A, Diffuse large B-cell lymphoma
INTRODUCTION
The molecular genetic characteristics of mature B-cell lymphoma were recently elucidated, leading the way to personalized medicine based on the identified gene mutations and molecular genetic classification.1-3 Diffuse large B-cell lymphoma (DLBCL) is the most common type of malignant lymphoma, accounting for approximately 40% of all malignant lymphomas with biologically and clinically heterogeneous features.4 Biologically, DLBCL is subdivided into an activated B-cell-like type (ABC-DLBCL) and a germinal center B-cell-like type (GCB-DLBCL) depending on the cell of origin (COO) of the tumor.5-9 The COO is classified by its gene expression profile, and a recent large-scale gene mutation analysis proposed new genetic classifications and identified drug target genes.10,11 In addition, the importance of the tumor microenvironment (TME) of DLBCL has increased with the development and employment of novel immunotherapeutic strategies, such as CAR-T therapy, in the current standard of care for this disease.12 A recent comprehensive study identified several driver genetic alterations impacting the checkpoint of immune recognition.13,14 In particular, the discovery of novel genetic alterations affecting the macrophage engulfment (“eat me” and “don’t eat me” signal) suggests a potential therapeutic target focusing on the cross-talk between macrophages and lymphoma cells.15
In this review, we describe the recent advances in the understanding of the biology of TME in aggressive B-cell lymphomas. We highlight the clinical and biological significance of TMEM30A genetic alterations, and discuss the potential development of predictive and prognostic biomarkers for next-generation immune-checkpoint inhibitors targeting phagocytosis.
GENETIC LANDSCAPE OF DLBCL
Recent advances in genetic technologies using next-generation sequencing revealed several recurrent genetic abnormalities in DLBCL.1-3 Large-scale genetic analyses with clinical information have been reported by two independent groups. These studies established new molecular classifications based on multiple genetic abnormalities correlated with the prognosis of DLBCL.10,11 Chapuy et al. performed a multi-omics analysis of more than 300 DLBCL cases that demonstrated that DLBCL can be divided into five groups (C1, C2, C3, C4, C5) based on the combination of recurrent genetic abnormalities. This enabled the risk stratification of DLBCL patients and promoted targeted therapy. For example, cases classified as C5 frequently have CD79B and MYD88-L265P mutations, poor outcomes are observed in cases of ABC-DLBCL, and sensitivity to BTK inhibitors or lenalidomide is noted. Furthermore, C1 and C5 were significantly enriched in ABC-DLBCL, and C3 and C4 were enriched in GCB-DLBCL, further dividing COO into two groups with different prognoses (e.g., C5 and C3 as the unfavorable group, and C1 and C4 as the favorable group).10 Schmitz et al. also reported that approximately half of DLBCL cases can be divided into four classifications (MCD, BN2, N1, and EZB) based on the pattern of genetic abnormality.16 Importantly, these two groups can be consistently genetically classified by clinical outcomes (e.g., C5 and MCD are characterized by CD79B and MYD88 mutations, ABC-DLBCL enrichment, and poor outcome), suggesting highly reproducible genetic subtypes that can serve as a foothold for personalized medicine for DLBCL. However, the correlation between the genetic subtypes and TME composition remains unclear, which may limit the clinical implications of subtyping for future immune-therapeutic strategies.
BIOLOGY OF TME OF DLBCL
DLBCL is less dependent on its microenvironment, consistent with the complete disorganization of normal lymphoid structure. However, increasing evidence suggests that the immune system is essential for disease development and outcome of DLBCL, similar to other B-cell lymphomas and solid cancers. The features of the TME differ between different lymphoma types.13,17 In DLBCL, disrupted cross-talk between lymphoma cells and the microenvironment plays a role in the ability of lymphoma cells to escape immune surveillance of the host. The mechanisms of immune escape include i) hiding from the immune system by losing or reducing recognition molecules (altering immune recognition), ii) suppressing antitumor immune function, and iii) creating a lymphoma-supportive microenvironment.18 Altering immune recognition, in particular, is deeply involved in tumor development and progression in DLBCL, and its molecular basis has been actively investigated.
Attenuated expression of MHC systems plays a key role in the immune escape of DLBCL.19,20 MHC class I (MHC-I) proteins, which are present on most nucleated cells, mediate the presentation of self, non-self, and neo-peptides to cytotoxic CD8+T cells.21,22 Frequent deficiency of MHC-I expression on the surface of DLBCL cells was observed in DLBCL based on genetic mechanisms such as inactivation of beta-2-microglobulin (B2M).23 In contrast, the COO of B-cell lymphomas is a professional antigen-presenting cell. Thus, MHC class II (MHC-II) is normally expressed, and selection in the light zone (LZ) of the germinal center (GC) involves antigen presentation via MHC-II to T follicular helper (TFH) cells and follicular dendritic cells (FDCs).24,25 Thus, antigen presentation must be concealed for these cells to escape death in GC-driven B-cell lymphomas. Accordingly, MHC-II expression is often lost in GC-derived neoplasms. In the COO-specific context, loss of MHC-II expression occurs more often in ABC-DLBCL than in GCB-DLBCL.26 As GC B-cells transition to plasma cells, the expression of transactivators of MHC-II is silenced, resulting in the subsequent loss of MHC-II expression. Of note, a recent study demonstrated that loss of MHC-II expression also defines tumors originating from the dark zone (DZ) of GC, which is associated with an inferior prognosis and immune “cold” microenvironment.27 Moreover, the impact of MHC-II deficiency on the immune microenvironment and outcome was much stronger in GCB-DLBCL than in ABC-DLBCL, reflecting a substantial degree of dependence on microenvironmental cells for survival and proliferation signals in the DLBCL subtype.27 Clinically, loss of MHC class II expression in DLBCL tumor cells correlates with poor patient survival and lower numbers of TILs,28-30 mainly due to reduced immune reactivity against tumors by immune chemotherapies.
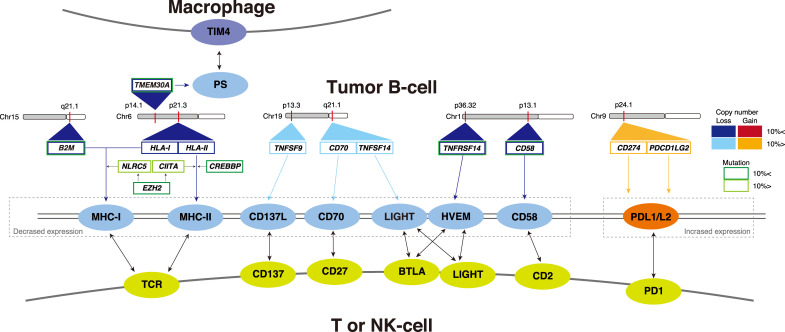
Mutational landscape studies highlighted the recurrent mutations involved in immune recognition in DLBCL (Figure 1). Inactivating mutations and deletions in the B2M gene impair MHC-I assembly and cell surface expression. These events occur in 30% of DLBCL cases. Copy number loss of HLA-I loci at chromosome 6p21 is also a recurrent genetic event associated with reduced MHC-I expression in lymphoma cells.31 The genetic mechanism of the loss of MHC-II proteins in DLBCL is more complicated. In addition to deleted HLA-II loci, they inactivate CIITA, an essential activator of the MHC class II gene, by inactivating somatic mutations, whereas CIITA translocations are recurrent genetic events in cHL and PMBCL.2,32,33 Of note, recent analyses revealed that MHC-II is downregulated by epigenetic aberrations. For example, CREBBP mutations downregulate MHC-II expression, which results in reduced T-cell infiltration in follicular lymphoma (FL) and GCB-DLBCL.34,35 CREBBP binds and acetylates the regulatory sequences of several genes involved in antigen presentation/processing, including the CIITA transactivator and multiple MHC class II loci, thus functioning in tumor immune escape. Moreover, HDAC3-specific inhibitors can rescue the expression of MHC-II in CREBBP-mutant lymphoma cells and restore the ability of TILs to kill DLBCL cells in an MHC class II-dependent manner.35 EZH2 Y641 mutation is also associated with marked silencing of both MHC-I and MHC-II genes, and EZH2-mutant lymphomas in both mice and humans exhibit reduced expression of these genes, and a reduction in lymphoma-infiltrating CD4+ and CD8+ T cells. EZH2 inhibitors also restored MHC-I and MHC-II expression in in vitro models.27 Collectively, these studies strongly suggest the potential of epigenetic reprogramming to prime the host immune system, providing an attractive rationale for the combination treatment strategy of epigenetic modifiers with immune-checkpoint inhibitors for a subset of DLBCL.
Fig. 1.
Recurrent genetic alterations impacting the tumor microenvironment of DLBCL.
Conversely and consistent with the low expression of programmed death-ligand 1(PD-L1) in DLBCL, gain-of-function genetic changes in the PD1/PD-L1 axis are rare in DLBCL patients (~10%).1,31,36 In particular, the C1 genetic subtype harbors gains, amplifications, and translocations of the PD-L1/PD-L2 locus associated with increased expression, but its frequency is low (20% of this subtype).10 This suggests that the PD1/PD-L1 axis does not play a major role in the immune architecture of tumors, which explains the lower activity of immune-checkpoint inhibitors in DLBCL patients than in those with other cancers.37
DISCOVERY OF NOVEL GENETIC ALTERATIONS OF TMEM30A IN DLBCL
In addition to the aforementioned recurrent genetic alterations that induce immune escape, our previous mutation landscape study uncovered recurrent mutations of TMEM30A with a novel biological mechanism impacting the macrophage checkpoint.14 TMEM30A mutations have several biologically and clinically distinct features. First, although the TMEM30A mutation is a gene mutation found in DLBCL with a frequency of approximately 5%–10%, 80% of TMEM30A mutations have loss-of-function mutations (nonsense, frameshift, and splice-site mutations), which is the highest proportion of loss-of-function mutations among the 57 driver genes examined in this study. Furthermore, copy number analysis revealed that TMEM30A is located in a gene region (6q14.1) that is prone to copy number attenuation. Indeed, more than 80% of cases with loss-of-function TMEM30A mutation have copy number attenuation resulting in the deficiency of both alleles, reflecting tumor suppressor features based on a double-hit theory. Furthermore, gene expression analysis confirmed that the TMEM30A mutation also reduced its own gene expression.
Another major feature of the TMEM30A mutation is that it appears specifically in aggressive B-cell lymphoma (BCL). Search of a public database confirmed the presence of the mutation only in aggressive BCL, including DLBCL, primary central nervous system lymphoma, and transformed FL, although almost no mutations were identified in other hematological malignancies or solid cancers. Of note, in six cases with transformed FL, the TMEM30A mutation appeared only when DLBCL was diagnosed at the time of recurrence in all cases, whereas biopsy material obtained at FL diagnosis did not have the TMEM30A mutation. This supports the specific appearance of the TMEM30A mutation in aggressive BCL.
The clinical importance of the TMEM30A mutation has also been reported. The prognostic significance of all gene mutations (mutations, copy number abnormalities, and fusion genes) in a comprehensive analysis revealed that the TMEM30A mutation is a strong favorable prognostic factor in R-CHOP therapy, especially in cases with bi-allelic alterations of TMEM30A. The prognostic effects were independent of the clinical prognostic factors and COO. Importantly, favorable prognostic effects of the TMEM30A mutation were confirmed in the reanalysis of another study dataset.10,11 The TMEM30A mutation is a constituent gene mutation of the C1 and BN2 groups, which have a good prognosis in the genetic classification proposed by previous studies, suggesting the reproducibility of the prognostic significance of the TMEM30A mutation. TMEM30A loss-of-function drives lymphomagenesis by increasing BCR-dependent signaling. Thus, the TMEM30A mutation is a tumor suppressor gene that appears specifically in DLBCL and is a prognostic factor in DLBCL patients who received R-CHOP therapy.
Concerning the biological insight underlying the clinical and genetic significance of the TMEM30A mutation, previous studies demonstrated that TMEM30A is one of the main players regulating the “eat me” signal that promotes phagocytosis of macrophages.38-40 Based on these studies, we examined the relationship between phosphatidylserine (PS) exposure and phagocytosis using primary samples, cell lines, and animal models. The study confirmed that TMEM30A knockout DLBCL promotes PS exposure and phagocytosis. Furthermore, in an experiment using an inhibitor of signal regulatory protein alpha (SIRPα)—another “don’t eat me” signal that suppresses phagocytosis—the tumor suppressive effects increased and the survival time was significantly prolonged in knockout mice compared with normal TMEM30A mice. Thus, TMEM30A has the potential to predict the therapeutic response to macrophage checkpoint inhibitors. Moreover, the increased phagocytosis caused by TMEM30A mutation may be related to the biological background of the good prognosis of TMEM30A mutation-positive DLBCL ( Figure 1).
Fig. 2.
Biological significance of TMEM30A in BCL.
CLINICAL IMPACT OF “EAT ME” AND “DON’T EAT ME” SIGNALS IN BCL
Inhibition of macrophage-mediated phagocytosis has emerged as an essential mechanism for tumor immune evasion and is an attractive therapeutic target as a next-generation immune-checkpoint inhibitor. During apoptosis, phagocytosis is mainly induced by exposure to PS on the cell membrane surface.41-43 PS is predominantly confined to the inner leaflet of the plasma membrane in cells, but it is externalized on the cell surface during apoptosis. This externalized PS is required for the effective phagocytosis of apoptotic cells by macrophages. In 2014, Segawa et al. discovered that CDC50A (TMEM30A) plays an essential role in flipping PS inside the plasma membrane to avoid engulfment of living cells. Importantly, they knocked out TMEM30A in a mouse model and demonstrated that the deficiency of TMEM30A promoted PS exposure on the cell membrane surface, resulting in the elimination of cancer cells by macrophage engulfment.38
The “don’t eat me” signal based on the phagocytosis checkpoint axis, CD47- SIRPα, was identified in the 1990s.44 CD47 was first identified as a “marker of self” in red blood cells, and was highly upregulated in malignant hematopoietic and non-hematopoietic cells.45 CD47 inhibits cellular phagocytosis through its interaction with SIRPα expressed on phagocytic cells, which include macrophages and dendritic cells. Due to the inhibitory signal of phagocytosis, CD47 overexpression confers an unfavorable prognostic effect in several cancer types.46,47 Chao et al. reported that CD47 expression correlates with an aggressive phenotype in non-Hodgkin lymphoma and an overall poor clinical prognosis following immune chemotherapies in DLBCL.48 The indicated biological and clinical significance of “don’t eat me” signal prompted the idea of drug-related inhibition of the CD47 signal to increase macrophage phagocytic activity (Table 1). Indeed, several CD47 inhibitors impaired tumor growth, inhibited metastatic spread, and inhibited tumor regression in a preclinical model.49,50 Moreover, this activity reportedly increased when combined with cancer targeting antibodies that provide exogenous prophagocytic signals. For example, synergistic suppression of tumor growth was observed when CD47-targeting agents were combined with monoclonal antibodies such as rituximab.51 These antibodies have active Fc domains that bind to Fc-gamma receptors on macrophages, resulting in the stimulation of phagocytosis. This mechanism provides rationale for exploring combinations of antitumor antibodies with CD47 targeting agents in clinical trials.
Table 1. Development of novel agents targeting “don’t eat me” signal.
| Name | Target | Clinical | Disease |
|---|---|---|---|
| Hu5F9-G4 | CD47 | Phase I/II | lymphomas |
| TI-061 | CD47 | Phase I/II | solid cancers |
| TTI-622 | SIRPα | Phase I | lymphomas |
| TTI-621 | SIRPα | Phase I | solid and hematologic malignancies |
| SRF231 | CD47 | Phase I | solid and hematologic malignancies |
| SHR-1603 | CD47 | Phase I | solid and hematologic malignancies |
| OSE-172 | SIRPα | Phase I | solid cancers |
| NI-1701 | CD47 | Phase I | hematologic malignancies |
| IBI188 | CD47 | Phase I | solid and hematologic malignancies |
Among candidate CD47 blockades, Hu5F9-G4 (5F9) is a humanized IgG4 monoclonal antibody undergoing clinical trials.50 5F9 was engineered with a human IgG4 isotype to minimize potential off-target effects on normal tissues. 5F9 binds to CD47 on tumor tissues and its antitumor effects are primarily dependent on the inhibition of CD47 signaling, although for optimal activity, an IgG4 Fc domain is required. Indeed, a recent clinical trial demonstrated marked anti-lymphoma effects with CD47 blockade in relapsed/refractory DLBCL. Advani et al. first reported the significant efficacy of 5F9 in combination with rituximab, without severe toxicities, in 15 patients with relapsed and refractory DLBCL, and seven FL patients exhibiting resistance to immune chemotherapies. Overall, 11 patients responded to the agent, including five complete responses of DLBCL. In addition, 91% of patients remained in remission at a median follow-up of 6.2 months in the DLBCL group.52 An ongoing phase II trial enrolling more patients is seeking to validate these findings.
CONCLUSION
DLBCL is a disease with a complicated pathogenesis that is based on genetic alterations of tumor cells, composition of the TME, and the escape from attack by tumor-associated immune cells. As a mechanism of immune escape, several genetic alterations that affect immune recognition and elimination have been identified. These recent biological insights regarding immune evasion by lymphomas have enabled the development of multiple promising immunotherapeutic strategies. Among them, CD47 blockades are less toxic and induce stable responses in patients with relapsed or refractory DLBCL. TMEM30A is a potentially novel biomarker for next-generation checkpoint inhibitors.
Footnotes
CONFLICT OF INTEREST
DE received honoraria from Kyowa Kirin Co., Ltd., Eisai Co., Ltd. and research fund from Chugai Pharmaceutical Co., Ltd. and Nipponshinyaku Pharmaceutical Co., Ltd.
REFERENCES
- 1.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011; 43: 830-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011; 476: 298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012; 109: 3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016; 127: 2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403: 503-511. [DOI] [PubMed] [Google Scholar]
- 6.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002; 346: 1937-1947. [DOI] [PubMed] [Google Scholar]
- 7.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008; 359: 2313-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013; 502: 333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur SE, Jiang A, Grande BM, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun. 2018; 9: 4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018; 24: 679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018; 378: 1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brudno JN, Kochenderfer JN. Chimeric antigen receptor T-cell therapies for lymphoma. Nat Rev Clin Oncol. 2018; 15: 31-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014; 14: 517-534. [DOI] [PubMed] [Google Scholar]
- 14.Ennishi D, Hsi ED, Steidl C, Scott DW. Toward a new molecular taxonomy of diffuse large B-cell lymphoma. Cancer Discov. 2020; 10: 1267-1281. [DOI] [PubMed] [Google Scholar]
- 15.Ennishi D, Healy S, Bashashati A, et al. TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma. Nat Med. 2020; 26: 577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott DW, King RL, Staiger AM, et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood. 2018; 131: 2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarte K. Role of the microenvironment across histological subtypes of NHL. Hematology (Am Soc Hematol Educ Program). 2017; 2017: 610-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144: 646-674. [DOI] [PubMed] [Google Scholar]
- 19.Drénou B, Le Friec G, Bernard M, et al. Major histocompatibility complex abnormalities in non-Hodgkin lymphomas. Br J Haematol. 2002; 119: 417-424. [DOI] [PubMed] [Google Scholar]
- 20.Wargo JA, Reddy SM, Reuben A, Sharma P. Monitoring immune responses in the tumor microenvironment. Curr Opin Immunol. 2016; 41: 23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015; 35: S185-S198. [DOI] [PubMed] [Google Scholar]
- 22.Ardolino M, Azimi CS, Iannello A, et al. Cytokine therapy reverses NK cell anergy in MHC-deficient tumors. J Clin Invest. 2014; 124: 4781-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of β2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011; 20: 728-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victora GD, Dominguez-Sola D, Holmes AB, et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012; 120: 2240-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannard O, McGowan SJ, Ersching J, et al. Ubiquitin-mediated fluctuations in MHC class II facilitate efficient germinal center B cell responses. J Exp Med. 2016; 213: 993-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson ST, Vanpatten KA, Fernandez DR, et al. Partial plasma cell differentiation as a mechanism of lost major histocompatibility complex class II expression in diffuse large B-cell lymphoma. Blood. 2012; 119: 1459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ennishi D, Takata K, Béguelin W, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 2019; 9: 546-563. [DOI] [PubMed] [Google Scholar]
- 28.Rimsza LM, Roberts RA, Miller TP, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004; 103: 4251-4258. [DOI] [PubMed] [Google Scholar]
- 29.Rimsza LM, Roberts RA, Campo E, et al. Loss of major histocompatibility class II expression in non-immune-privileged site diffuse large B-cell lymphoma is highly coordinated and not due to chromosomal deletions. Blood. 2006; 107: 1101-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rimsza LM, LeBlanc ML, Unger JM, et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2008; 112: 3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monti S, Chapuy B, Takeyama K, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012; 22: 359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimsza LM, Chan WC, Gascoyne RD, et al. CIITA or RFX coding region loss of function mutations occur rarely in diffuse large B-cell lymphoma cases and cell lines with low levels of major histocompatibility complex class II expression. Haematologica. 2009; 94: 596-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steidl C, Shah SP, Woolcock BW, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011; 471: 377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci USA. 2015; 112: E1116-E1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Vlasevska S, Wells VA, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017; 7: 322-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgiou K, Chen L, Berglund M, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016; 127: 3026-3034. [DOI] [PubMed] [Google Scholar]
- 37.Lesokhin AM, Ansell SM, Armand P, et al. Nivolumab in patients with relapsed or refractory hematologic malignancy: preliminary results of a Phase Ib Study. J Clin Oncol. 2016; 34: 2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segawa K, Kurata S, Yanagihashi Y, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014; 344: 1164-1168. [DOI] [PubMed] [Google Scholar]
- 39.Segawa K, Kurata S, Nagata S. The CDC50A extracellular domain is required for forming a functional complex with and chaperoning phospholipid flippases to the plasma membrane. J Biol Chem. 2018; 293: 2172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen JP, Vestergaard AL, Mikkelsen SA, et al. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front Physiol. 2016; 7: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J Biol Chem. 2001; 276: 1071-1077. [DOI] [PubMed] [Google Scholar]
- 42.Elliott JI, Surprenant A, Marelli-Berg FM, et al. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat Cell Biol. 2005; 7: 808-816. [DOI] [PubMed] [Google Scholar]
- 43.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010; 39: 407-427. [DOI] [PubMed] [Google Scholar]
- 44.Barclay AN, van den Berg TK. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu Rev Immunol. 2014; 32: 25-50. [DOI] [PubMed] [Google Scholar]
- 45.Chao MP, Weissman IL, Majeti R. The CD47–SIRPα pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol. 2012; 24: 225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009; 138: 286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willingham SB, Volkmer JP, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA. 2012; 109: 6662-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao MP, Alizadeh AA, Tang C, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010; 142: 699-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russ A, Hua AB, Montfort WR, et al. Blocking “don’t eat me” signal of CD47-SIRPα in hematological malignancies, an in-depth review. Blood Rev. 2018; 32: 480-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Wang L, Zhao F, et al. Pre-clinical development of a humanized anti-CD47 antibody with anti-cancer therapeutic potential. PLoS One. 2015; 10: e0137345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sockolosky JT, Dougan M, Ingram JR, et al. Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc Natl Acad Sci USA. 2016; 113: E2646-E2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Advani R, Flinn I, Popplewell L, et al. CD47 blockade by Hu5F9-G4 and rituximab in non-Hodgkin’s lymphoma. N Engl J Med. 2018; 379: 1711-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]