Summary
Background
Major challenges in large scale genetic association studies include not only the identification of causative single nucleotide polymorphisms (SNPs), but also accounting for SNP-SNP interactions. This study thus proposes a novel feature engineering approach integrating potentially functional coding haplotypes (pfcHap) with machine-learning (ML) feature selection to identify biologically meaningful, possibly causative genetic factors, that take into consideration potential SNP-SNP interactions within the pfcHap, to best predict for methotrexate (MTX) response in rheumatoid arthritis (RA) patients.
Methods
Exome sequencing from 349 RA patients were analysed, of which they were split into training and unseen test set. Inferred pfcHaps were combined with 30 non-genetic features to undergo ML recursive feature elimination with cross-validation using the training set. Predictive capacity and robustness of the selected features were assessed using six popular machine learning models through a train set cross-validation and evaluated in an unseen test set.
Findings
Significantly, 100 features (95 pfcHaps, 5 non-genetic factors) were identified to have good predictive performance (AUC: 0.776-0.828; Sensitivity: 0.656-0.813; Specificity: 0.684-0.868) across all six ML models in an unseen test dataset for the prediction of MTX response in RA patients.
Interpretation
Majority of the predictive pfcHap SNPs were predicted to be potentially functional and some of the genes in which the pfcHap resides in were identified to be associated with previously reported MTX/RA pathways.
Funding
Singapore Ministry of Health's National Medical Research Council (NMRC) [NMRC/CBRG/0095/2015; CG12Aug17; CGAug16M012; NMRC/CG/017/2013]; National Cancer Center Research Fund and block funding Duke-NUS Medical School.; Singapore Ministry of Education Academic Research Fund Tier 2 grant MOE2019-T2-1-138.
Keywords: Rheumatoid Arthritis, Methotrexate, Genetic polymorphism, Machine learning, Feature selection, Haplotypes
Research in Context.
Evidence before this study
Methotrexate (MTX) is a first-line medication for rheumatoid arthritis (RA) patients despite low monotherapy response of only 25 to 45 percent. Identification of non-responders is necessary to help mitigate disease progression. However, evaluation of treatment response often takes three to six months following MTX administration. Current research on MTX response in RA are primarily focused on genetic variation in specific genes involved in MTX-related pathways and RA susceptibility with few interrogating the entire genome using genome-wide association studies (GWAS). Nonetheless, several challenges are associated with current GWAS including high data dimensionality and the identification of causative variations.
Added value of this study
In this study, we employed a cost-effective exome sequencing approach to interrogate haplotype of single nucleotide polymorphisms (SNPs) in the coding region of genes which is deemed as one of the most informative functional regions of the genome. To reduce the high dimensionality data, and capitalize on the property that SNPs within a functional unit (e.g. coding region) interact to modulate structure/function of the target protein, we inferred SNP haplotypes in the coding region and employed machine-learning (ML) to identify potentially functional coding haplotypes (pfcHaps) that best predicts MTX response in RA patients. Notably, the predictive pfcHap SNPs and genes were predicted to be functional and associated with previously reported MTX/RA pathways, respectively, highlighting the promise of this approach.
Implications of all the available evidence
Taken together, we envision that the best predictors identified will be effective in aiding decision-making for the treatment of RA patients after further validation in larger multi-institutional studies. Furthermore, we believe that our analysis pipeline for handling and interpretation of genetic data will also be applicable in other contexts beyond MTX response in RA patients.
Alt-text: Unlabelled box
Introduction
Of the diverse factors influencing drug response, elucidating the genetic basis that underlie differences in drug response may facilitate the development of tools to predict drug response even before the drug is administered. Although there is growing interest in pharmacogenomics, the focus has thus far been primarily on the identification of common gene variants with large effect size on known pharmacokinetic, pharmacodynamics and/or immuno-pharmacogenomics candidate genes.1, 2, 3 However, polymorphisms in other genes or rare gene variants, either alone or in combination, may, also modulate drug response but have yet to be thoroughly investigated.3 With the advent of high throughput genomic tools, it is now possible to explore the association of other genes with drug response using GWAS (Genome Wide Association Study), exome sequencing or even WGS (Whole Genome Sequencing).3 Current genomic approaches which mainly focused on tag-SNPs in GWAS only represent a very small proportion of all potentially functional SNPs (pfSNPs) in the human genome with likelihood to be causative. While WGS would be the most ideal approach to examine all SNPs including pfSNPs, exome sequencing is a cost-effective way to facilitate the interrogation of all pfSNPs in the most informative functional region of the genome, namely, the coding region.
Thus far, traditional statistical methods have been the primary tool to associate genetic and other features with drug response and/or disease susceptibility. However, these statistical approaches have some limitations.4, 5, 6 This includes the requirement for large sample sizes, which can potentially be mitigated by machine learning (ML) which are suited for high dimensionality complex problems, including the consideration for non-linear interactions between features.7,8 Nonetheless, a major limitation of biomedical datasets, even for ML, is that their high dimensionality is often coupled with limited labelled sample size, which pose a challenge for learning models to predict individualized response to drugs. ML-based dimensionality reduction and feature selection strategies can help reduce the feature space and select the most informative features that can accurately predict an outcome. Nonetheless, to achieve acceptable accuracy in pharmacogenomics, careful data pre-processing and feature handcrafting with strong domain knowledge9 is necessary.
Here, we introduce a novel biologically meaningful, feature pre-processing/engineering strategy focused on haplotypes of SNPs in the coding regions of genes with the potential to be functional (pfcHap). By integrating pfcHap together with ML feature elimination and selection, the strategy identifies a signature of potentially causative genetic and non-genetic factors that can robustly predict response to the methotrexate (MTX) drug in Rheumatoid Arthritis (RA) patients across diverse ML models.
RA was proposed to be particularly appropriate for personalized therapy because of the costly therapy due to prolonged disease duration, low response to the conventional therapy, trial-and-error nature of therapy prescription, and the risk of serious drug-induced side effects.10 The disability brought on by comorbidities such as coronary artery disease and hyperlipidaemia11 in combination with poorly controlled RA adds to the healthcare costs and further strains the health care system.
RA is a chronic inflammatory disease involving primarily the joints with a prevalence of an average of ∼5 per 1000 people12 that varies across different populations.13 It imposes huge socioeconomic burden on both the patient and society as it commonly affects middle-aged adults at their economic peak.14, 15, 16, 17, 18, 19, 20, 21, 22 Inadequate treatment of RA leads to irreversible joint damage resulting in potential disabilities that affects the patient's quality of life and work productivity, and even premature death.23, 24, 25 Hence, timely and appropriate control of this condition is critical to minimize the morbidity and mortality.24 Amongst the disease-modifying antirheumatic drugs (DMARDs) in RA, MTX is the anchor agent and the recommended first-line choice for the majority of RA cases.26,27 Approximately 25 to 40 percent of patients improve with MTX monotherapy, which is further increased to 50 percent for patients receiving combination therapy with glucocorticoids.28 Patients with inadequate response to MTX monotherapy are offered alternative biologic and targeted synthetic DMARDs.12 The current state-of-the-art management of RA is still primarily based on trial-and-error with recommendations from the European League Against Rheumatism (EULAR) being to assess the effectiveness of MTX therapy between three to six months of administering the drug and re-evaluating the treatment approach of poor responders thereafter.29 This suggests that rheumatologists only know the effectiveness of MTX after the patient is already on the drug for 3-6 months. Earlier identification of poor responders of MTX, preferably even before drug administration, will enable prompt initiation of alternative treatment which could help mitigate disease progression.
To date, the study of MTX response in RA have been focused on the genetic variability in specific genes, often involving those in MTX-related pathways or RA susceptibility or Genome wide association study (GWAS) interrogating individual SNPs.30 A recent review summarised 125 SNPs from 34 genes involved with MTX metabolism, transport or RA progression/pathogenesis were previously evaluated for associations with MTX response.31 However, some of these studies have reported contradictory results, including conflicting reports of associations of polymorphism rs1045642 (3435C > T) in the ATP-binding cassette B1 (ABCB1) transporter gene, with MTX efficacy in two separate Japanese cohort studies.32,33
While there is recent increasing interest to employ ML for electronic diagnosis, prediction of disease progression and drug response of RA patients,34 these methods remain at its infancy, with few studies exploring predictive models to evaluate MTX drug response.34 They mainly focus on specific subsets of SNPs with non-genetic factors35,36 or other molecular signatures (e.g. transcription/epigenetic-based signatures).37 Most of the methotrexate predictive models employed simple machine learning models such as logistic regression and mainly focused on electronic medical records, or juvenile RA. Thus far, more complex ML models with careful domain-based, biologically meaningful feature handcrafting have yet to be applied comprehensively to improve predictive performance of response to RA drugs.
Previous haplotype-based studies for other diseases mainly examined haplotype of SNPs within specific window sizes38 or employed to account for familial correlation for association between rare haplotypes and complex disease.39 Here, we report a novel, cost-effective approach through exome sequencing to interrogate haplotypes of SNPs in the coding regions of genes (pfcHap) of the entire human genome, since coding regions, which are translated into proteins represent one of the most functional regions of genes. As complex phenotypes are likely due to the interaction of multiple SNPs in a functional unit (e.g. coding region) rather than single SNP acting in isolation, and SNPs altering amino acids in the same protein may interact with each other to alter the folding or function of protein (e.g. binding to substrates, etc), this approach of focusing on pfcHap has the potential for identifying signatures of pfcHap that not only account for SNP-SNP interactions but also a higher likelihood of being the causative combination of variants.
Methods
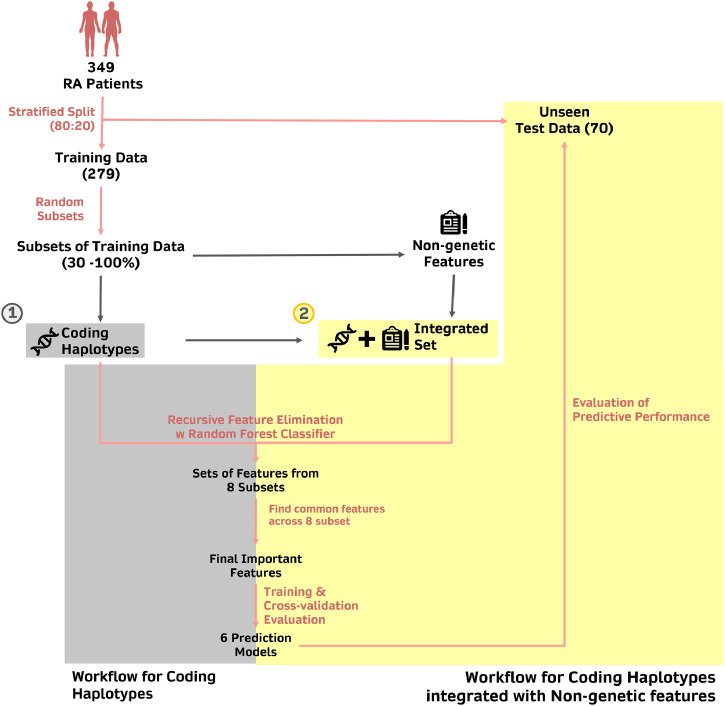
Fig. 1 illustrates our strategy to identify biologically meaningful features with good prediction performance for MTX response in RA patients.
Figure 1.
Pipeline employed to identify the predictors of MTX response.
349 patient samples were first divided into training (n=279, 70%) and test (n=70, 30%) sets using a stratified split, such that datasets consist of the proportion of responders and non-responders that is representative of the original dataset. The training set was then further split into eight subsets consisting of different sample size ranging from 30% to 100% of the samples in the training set. Within each subset, the important features (coding haplotypes or integration of coding haplotypes with non-genetic features) were selected using recursive feature elimination with cross-validation (RFECV), applied with Random Forest Classifier as the estimator of choice. The important features that were commonly identified in all eight subsets were then shortlisted and identified as the set of important features that are predictive of MTX response. The predictive performance of these features was assessed in six different machine learning models, using cross-validation within the training set and the unseen test dataset.
Study cohort
This study examined 349 subjects of Chinese ethnicity receiving MTX treatment for RA. Patients were at least 18 years old and satisfy the 1987 American College of Rheumatology revised criteria or the 2010 American College of Rheumatology/European League Against Rheumatism criteria for RA. This study was endorsed by the National Healthcare Group Domain Specific Review Board (DSRB 2015/00582). All protocols were carried out according to the Declaration of Helsinki and informed consent was collected from all patients. All patients received at least 3 months of MTX treatment at 15 mg per week and >90% of the patients completed 2 years of treatment. MTX drug response is defined as remission within/at two years post-treatment, which is determined by evaluation of Disease Activity Score in 28 joints (DAS28). DAS28 is a composite score representative of RA activity that includes the number of tender joints and swollen joints, erythrocyte sedimentation rate, and a global assessment of health.40 Responders to MTX were classified as patients with DAS28<2.6 following MTX treatment. Various non-genetic features (including demographic/clinical characteristics and medication status) of patients were also recorded and summarised in Table S1.
Exome sequencing, sequence alignment, and single variant analysis
Enrichment of the exome region of genomic DNA was performed with the Nimblegen SeqCap EZ kit (Roche) and with Agilent SureSelect Human All Exon kit (Agilent Technologies, CA). Products were then purified using AMPure XP system (Beckman Coulter, Beverly, USA) and quantified using the Agilent high sensitivity DNA assay on the Agilent Bioanalyzer 2100 system. The exome sequencing was performed by commercial providers using the IlluminaHiSeq2000 100PE platform.
Using the BWA-MEM algorithm,41 the sequenced data was aligned to the hs37d5 human reference genome. Following, PICARD was used to remove duplicated reads. Each sample was processed separately using the base recalibration and haplotypecaller modules of GATK v3. Thereafter, using genotypeGVCF, variants were called on all the samples together.42 As per GATK best practice for hard filtering,43 SNP filtering was performed with "QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < -8.0" as the criteria and indel using "QD < 2.0 || FS > 200.0 || ReadPosRankSum < -20.0".
Training and test data
The total dataset from 349 subjects were split into a training (80%, N = 279) and an unseen test set (20%, N = 70) in a stratified manner to maintain the ratio between MTX responders and non-responders. Splitting of dataset was performed within the Python (3.8.3) environment using the Scikit-learn module.44 To avoid data leakage, subsequent processing steps were performed on the training and test datasets separately. The training dataset (80%) is further processed into 8 subsets of variable sample sizes through stratified random sampling with replacement.
Genetic/Biological-based feature engineering: Data processing and defining haplotypes
Biallelic variants with >10% genotype missingness or deviate from Hardy-Weinberg equilibrium (p-value > 0.001) were removed before downstream analyses. Thereafter, using the UCSC table browser45 as reference, remaining variants within coding regions were further selected using BEDTools Suite.46 These variants were annotated using ANNOVAR based on the hg19 reference genome.47 Genotype phasing was then performed using the BEAGLE 5.1 software together with HapMap Phase II recombination maps for each respective chromosome48,49 Using PLINK v1.07 software,50 all variants within coding regions of a gene were used for haplotype construction. Only haplotypes with a minor haplotype frequency > 0.01 were further analysed. Haplotypes carrying all reference SNPs were removed. The final total of ∼39,000 haplotypes were combined with 30 non-genetic features for downstream analyses.
Selecting features/predictors that are predictive for MTX responses in RA patients
Within each training subset, features with near zero variance (i.e., features which are almost constant across all samples) or display > 95% correlation with other features were excluded. To identify a signature of robust features that are important for prediction, we utilised recursive feature elimination with cross-validation (RFECV), as implemented in the Scikit-learn module,44 incorporating a Random Forest classifier as the estimator. The process was performed using a 5-fold cross-validation until an optimal number of features was selected. To obtain features with high stability of importance,51 features that are common across all 8 training subsets were selected for further evaluation of their predictive performance.
Evaluating predictive performances of selected features
Selected features were evaluated across six ML diverse algorithms: Neural Networks, Support Vector Machines, two regression-based algorithms (Logistic regression and Elastic nets), two tree-based algorithms (Random Forests and Boosted Trees) for a broad representation, as there is currently no consensus as to which ML algorithm is the most appropriate for genomic data. Predictions were performed using the Python Scikit-learn44 module with default parameters. Performance evaluation of each classifier was conducted using a 5-fold cross-validation of the training set (n=279), and the area under the curve (AUC) of a receiver operating characteristic (ROC) curve was generated. Since a smaller number of features is generally preferable for facilitating clinical implementation,35 the minimum number of features that achieve good predictive performance in the training set was selected as the best predictors. These predictors were then tested in the unseen test set using the six ML models, and the AUC of a ROC curve was generated.
Analysis for potential functions of SNPs within selected haplotypes
To explore for possible functional mechanisms underlying the differences in MTX responses associated with selected haplotypes, SNPs within selected haplotypes were evaluated for their potential or predicted functionality, through interrogation of an updated potentially functional SNP (pfSNP) resource developed by our laboratory52 which has included data from more recently published prediction databases53 including expression-associated SNPs or (expression quantitative trait loci (eQTLs)).54,55 The functionality of SNPs was assessed for their potential to alter important functional regions (e.g., transcription factor binding sites, miRNA binding sites, exonic splice enhancer/silencer (ESE/ESS), etc) or induce nonsense-mediated decay (NMD). Additionally, non-synonymous SNPs were also evaluated for their potential deleterious effects while synonymous SNPs were identified for possible codon usage bias.52
To further explore the functions of these haplotypes, pathway enrichment analyses were performed on the genes corresponding to the most predictive haplotypes using ConsensusPathDB resource.56
Ethics
This study was endorsed by the National Healthcare Group Domain Specific Review Board (DSRB 2015/00582). All protocols were carried out according to the Declaration of Helsinki and informed consent was collected from all patients.
Role of funders
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Any opinions, findings, or recommendations expressed in this material are those of the authors and do not reflect the views of the funders.
Results
Combination of biological and ML approaches reduces complexity of dataset and identifies a small set of features predictive for MTX responses
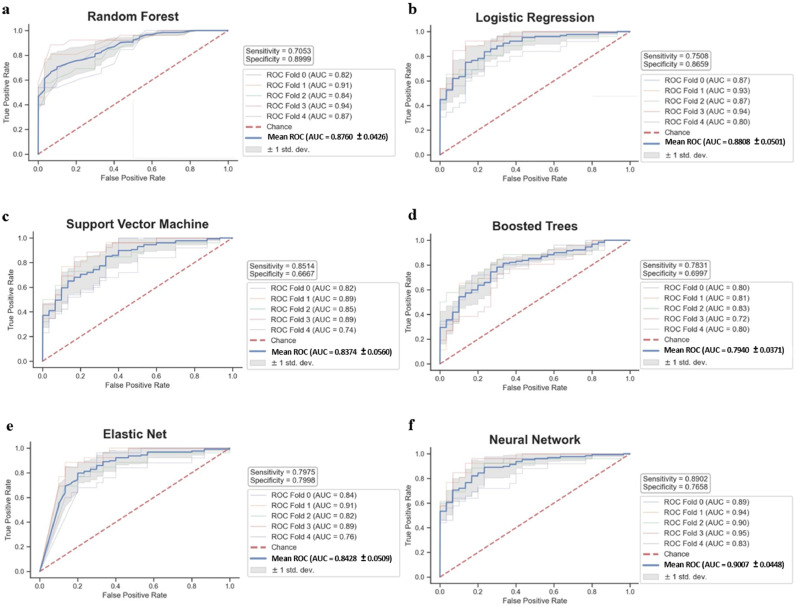
A total of 114,000 SNPs were identified from exome sequencing of blood DNA. With such high-dimensional data where the potential predictors far outnumber the number of samples, the identification of features with good predictive performance in unseen datasets becomes very challenging. Here, two consecutive steps were employed to mitigate this ‘curse of dimensionality’. The first step was a biologically meaningful, feature handcrafting, serving as a genetic approach of dimensionality reduction. This involved the derivation of haplotypes from SNPs in the coding regions of genes (pfcSNPs) as these are functional regions and are more likely to be causative, potentially altering protein structure and/or function. A total of 52,331 pfcHaps were derived from ∼13,000 genes, which represented a ∼54.1% reduction in the number of potential predictors. The exclusion of pfcHaps that comprises only reference alleles further reduced the number to 39,160 leading to a ∼65.6% reduction in potential predictors. The second step utilised ML recursive feature elimination with cross validation (RFECV) with Random Forest classifiers. Repeated eight times, RFECV on each training subset reduced the set of 39,160 pfcHaps, and identified between 411 to 2602 important pfcHaps, of which 120 were commonly identified in all eight subsets and used for training with cross-validation of the six ML models. These 120 pfcSHaps displayed reasonable cross-validation AUCs of between 0.794 and 0.901 with sensitivity of between 0.705 and 0.890 and specificity of 0.667 and 0.900 (Fig. 2).
Figure 2.
Predictive performance of 120 haplotypes (from Haplotype-only analysis) in the training set using 5-fold cross-validation.
ROC curves of 120 haplotypes using (a) Random Forest, (b) Logistic Regression, (c) Support Vector Machine, (d) Boosted Trees, (e) Elastic Net, and (f) Neural Network.
Integrating coding haplotypes with non-genetic factors identifies a smaller set of features with similar predictive capacity for MTX response
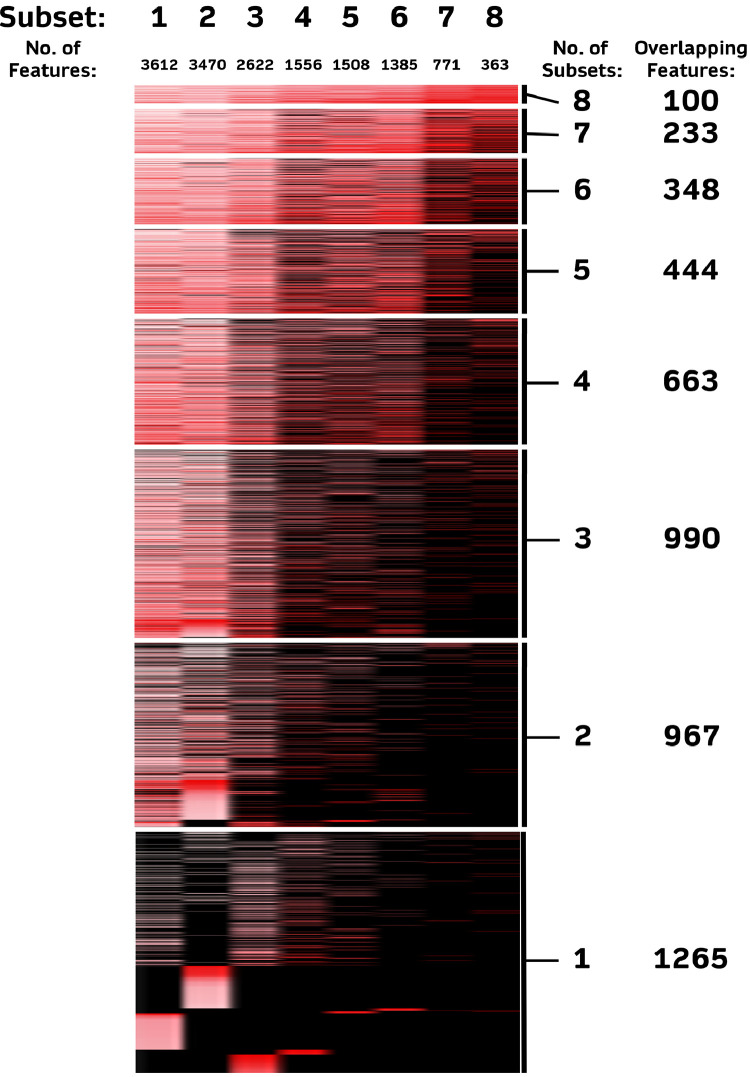
To determine whether the integration of pfcHaps with non-genetic features from medical records can improve classifier accuracy and/or reduce the number of features required for accurate prediction, we combined the 39,160 pfcHaps with 30 non-genetic factors (Table S1) and re-performed the same RFECV with Random Forest classifier in the eight training subsets. Between 363 to 3612 important features were identified in the eight training subsets (Fig. 3), of which 100 (95 pcfHaps and 5 non-genetic features) were common across all eight training subsets. features. The 5 non-genetic features were platelet count, haemoglobin levels, duration of morning stiffness, age, and presence of anti-cyclic citrullinated peptides antibody (anti-CCP). These 100 features were then trained on the six ML models, and exhibited improved predictive performance as compared to using haplotypes alone, with cross-validation AUCs between 0.822 and 0.906, sensitivity between 0.744 and 0.837 and specificity between 0.766 and 0.866 (Fig. 4). In addition to the improvements in predictive performances, there was a 16.7% reduction in the number of features, from 120 to 100, compared to the analyses focused on pfcHaps only. As such, these 100 features were noted to be the best predictors and were tested in an unseen test set. Significantly, the robustness of these 100 features to predict MTX in the unseen dataset was evident from the good predictive performance with AUCs between 0.775 and 0.828, sensitivity between 0.656 and 0.813 and specificity between 0.684 and 0.868 (Fig. 5) across all six ML models.
Figure 3.
Number of important features (coding haplotypes and non-genetic factors) identified in eight training subsets of variable sample sizes.
Columns represent the different training subsets and each row represent the features. Intensity of red represent the importance of the feature in each subset (i.e., Greater intensity represent features of greater importance and vice versa); Black represents features that are not found to be important in the respective subset.
Figure 4.
Predictive performance of 95 haplotypes and 5 non-genetic factors in the training set using 5-fold cross-validation.
ROC curves of 95 haplotypes and 5 non-genetic factors using (a) Random Forest, (b) Logistic Regression, (c) Support Vector Machine, (d) Boosted Trees, (e) Elastic Net, and (f) Neural Network.
Figure 5.
Predictive performance of 95 haplotypes and 5 non-genetic factors in the unseen test set.
ROC curves of 95 haplotypes and 5 non-genetic factors using (a) Random Forest, (b) Logistic Regression, (c) Support Vector Machine, (d) Boosted Trees, (e) Elastic Net, and (f) Neural Network.
Majority of SNPs within selected haplotypes are associated with changes in gene expression
Of the 100 features determined to be the best predictors, 95 were pfcHaps derived from 142 unique SNPs. 93.0% [132] of these SNPs are eQTL SNPs[54,55] (Table 1, black box). Approximately 40.8% (58) are non-synonymous while 59.2%(84) are synonymous SNPs. Majority(45) of the non-synonymous SNPs are predicted to be benign, while 5 and 8 SNPs are predicted to be possibly damaging and deleterious, respectively. 18.3%(26) of the 142 SNPs are also predicted to potentially alter transcription factor binding sites while 2.1%(3) can potentially affect miRNA binding sites and 52.1%(74) can potentially modify ESE/ESS sites. The SNPs and their potential functions are summarised in Table 1.
Table 1.
Potential function of 142 SNPs in 95 coding haplotypes which are identified as potential predictors of methotrexate response.
 |
 |
 |
Genes of the 95 predictive haplotypes are involved in a variety of pathways
To gain mechanistic insights into these 95 predictive pfcHaps, a pathway enrichment analysis was performed. Genes of these pfcHaps are enriched in diverse pathways, including Rho activation of PAKs, ROCKs, and CITs, complement and coagulation cascade, beta-2 cell surface interactions, ion channel transport, transcriptional activation of mitochondrial biogenesis and rheumatoid arthritis (Figure S1).
Discussion
The current study aims to identify biologically meaningful features that are predictive for MTX response in RA patients. Functional coding haplotypes (pfcHaps) present as biologically meaningful predictors and provide us with an opportunity to better understand the combined role that multiple SNPs within a single functional unit have in influencing the function of genes to explain the variation in phenotypes between individuals. Interrogation at a haplotype level has the potential to alleviate problems commonly faced in current genome wide association studies (GWAS) involving large number of SNPs which probe tag-SNPs that are in linkage disequilibrium with the causal SNP but are not necessarily the causal SNPs.57 More importantly, by focusing on haplotypes we have greatly reduced the complexity of the study and partially mitigated the curse of dimensionality. As such, this approach of feature handcrafting can be viewed as a biologically meaningful, genetic dimensionality reduction strategy. We supplemented the genetic dimensionality reduction strategy with ML feature elimination/selection using RFECV with Random Forest classifiers. Random Forest classifiers were chosen for our study due to its robustness to over-fitting, its computational efficiency, and the provision of feature importance scores.58 When coupled with RFECV, Random Forest is able to identify a small subset of features that produces the highest accuracy in the specific classifier model.58,59 To ensure that the selected features are stable and robust to differences in sample size,51,60 RFECV feature selection was performed on eight random subsets of training data with variable sample sizes. Only features that are common across all eight random, variable sized training subsets were selected for further evaluation of their predictive performance.
Overall, our proposed strategy allowed us to overcome issues of redundancy and irrelevancy of information that are commonly faced when handling high dimensional data which would have led to reduced efficiency and accuracy of ML models trained.61
Our strategy and the incorporation of non-genetic factors identified 95 coding haplotypes and 5 non-genetic factors for training. Notably, the identified features displayed good predictive performance classifying the MTX response of 70 patients whose data had been placed aside as the unseen test dataset. The overall performance (AUC, sensitivity, specificity) of the unseen test set suggested that the performance obtained during classifier training was not simply a case of overfitting and must have had some genuine ability to distinguish MTX response. Furthermore, the observed robustness of our features across diverse ML models indicated a non-dependency of ML strategy for effective classification prediction.
The 5 non-genetic predictive features were previously reported to be linked to RA/MTX. Platelet count, haemoglobin value, duration of morning stiffness and presence of anti-CCP are well-known markers for both the diagnosis of RA and the evaluation of disease activity.62, 63, 64, 65 Conversely, a patient's age is also generally an underlying factors influencing their response to drugs due to changes in pharmacokinetics and pharmacodynamics with age.66
We further investigated the significance of the 142 non-reference SNPs within these 95 predictive coding haplotypes (Table 1). Curiously, although coding regions of the genes, which should affect structure and function, were interrogated, majority (93.0%) of these SNPs were previously predicted as eQTLs (i.e., associated with changes in gene expression) suggesting that perhaps even polymorphisms within coding regions may influence gene expression. Of note, among the non-synonymous SNPs, most are predicted to be benign with only a few predicted to be possibly damaging or have deleterious effect. Several SNPs were also predicted to alter consensus binding sites including transcription factor/miRNA binding sites as well as ESE/ESS. Taken together, these SNPs in the predictive pfcHap may alter the expression, structure and/or activity/function of the gene/gene product, either individually or through interaction with other SNPs within the same coding haplotype. Overall, the enrichment of potentially functional SNPs within the predictive pfcHaps highlights the ability of our approach to select haplotype of SNPs that are likely to be functional.
Identifying predictive pfcHap as haplotype of SNPs in functional coding region offers us an opportunity to examine how multiple SNPs altering amino acids in the same protein may alter the structure or function (e.g. binding to substrates) of the protein differently from isolated SNPs. An example is the predictive pfcHap STING1_1.0 (HAQ) (Table 1) which comprises 3 non-synonymous SNPs (rs7380824 (R71H), rs78233829 (G230A), rs11554776 (R293Q)) within the STING1 (Stimulator of Interferon Gene 1) gene that was involved in both MTX and RA (Figure S2). Specifically, STING1 is implicated in the cGAS-STING pathway that contributes to inflammatory response in RA,67 and MTX has been reported to inhibit the activation of STING and downstream effects.68 Individuals harbouring the HAQ haplotype of the predictive pfcHap STING1_1.0 were previously reported to be more susceptible to viral infection and less responsive to DNA vaccines.69 An incomplete STING1 crystal structure70 (4KSY) encompassing the region of only two of the 3 polymorphisms, namely G230A (rs78233829) and R293Q (rs11554776) is available on RCSB PDB71 (www.rcsb.org) and the PyMol72 software (www.pymol.org/pymol) was employed to predict the potential changes in STING1 protein structure associated with these 2 aa changes (G230A and R293Q), either alone or together. As shown in Figure S3 and Video S1 the G230A polymorphism not only potentially altered the beta strand at the site of the polymorphism (Region i) but it was also predicted to alter the alpha helix structure at a distant site (∼aa263; Region ii) that is closer to the R293Q (Region ii). As these 2 polymorphisms reside within the cyclic dinucleotide binding region (aa153-aa340) of the STING1 protein,73 it is thus likely that the AQ haplotype will modulate the binding to cyclic dinucleotide differently from either of the single polymorphisms (either G230A or R293Q). Apart from this, the positive charge contributed by R293 has additionally been proposed to be important for disulfide bond formation or related modifications in relation to the neighbouring conserved C292.69 Thus, the absence of a positive charge from Q293 may possibly hinder functions or interactions associated with C292 that are needed for gene function. Hence, identifying predictive pfcHaps rather than isolated SNPs provides opportunities to explore interactions between SNPs within the gene (pfcHaps) and whether these interactions may modulate protein folding/structure and/or function.
To our knowledge, none of the SNPs within the pfcHaps were previously reported to be associated with MTX response in RA patients. Hence, we probed for previously reported associations with the genes of these predictive pfcHaps. Using the Python PyMed library, we performed a batch query interrogating the PubMed database using names of genes of these predictive haplotypes together with key terms including “Rheumatoid Arthritis”, “Methotrexate, and “MTX”. Our search identified several publications where these genes were mentioned together with the key terms (Figure S2). Ten genes have been reported to be associated with either MTX or RA with 4 of them (CR1, ICAM1, MMP3, MTR) being mentioned in numerous publications (28 – 63 publications) (Figure S2). Although majority (84) of the genes of the predictive pfcHaps had not been previously reported to be associated with RA and/or MTX, our pathway enrichment analyses (Figure S1) highlighted the relevance of the predicted pfcHaps to those pathways reported to be associated with MTX and/or RA (summarized in Table S2).
Thus far, studies that examined MTX response in RA patients mainly focussed on statistical association of non-genetic factors74 and/or genetic variants employing either the very popular genome-wide association studies (GWAS)30 or gene specific association analyses interrogating specific genes in the MTX/RA pathways31,32 with MTX response. This popular classical statistical approach employs Raw P-Value Thresholding (RPVT),75 where a P-value is assigned to each SNP; and the inferred confidence of a variant in accounting for the phenotype in the dataset is assessed by its statistical significance via comparison to a predefined threshold that considers a balance between Type I and Type 2 errors. However, since statistics merely derive population inference of a relationship between the data and the outcome variable from a sample76; and its main purpose is not to make prediction of a future dataset, statistically significant association in one dataset are not necessarily predictive of the outcome in a future dataset.4, 5, 6 Furthermore, as statistical approach evaluates individual SNP independently and in parallel, it does not consider potential higher order interactions amongst SNPs77,78 and is less able to identify variants with small effects because of statistical power constraints due to excessive multiple testing.75 Additionally, as classical statistical approach was originally designed for datasets with limited dependent and independent variables, statistical inferences are less precise with large number of variables as observed in GWAS studies since the possible associations among the many variables also increase drastically leading to more complex relationships.76
On the other hand, the Machine Learning (ML) approach employed in this study is particularly suited for dealing with rich, unwieldy, ‘wide’ data where the independent variables (e.g. SNPs) exceeds the number of samples,76 since it ‘makes minimal assumptions about the data-generating systems’ and is effective even with data from less well-controlled experimental design or with ‘complicated nonlinear interactions’.76 Being a ‘statistic-free’79 approach, type 1 error no longer poses an issue as there is no necessity to determine the population distribution or evaluate the P-value / confidence intervals or test the null hypothesis.79 As such, ML approach was reported to be more robust in identifying SNPs with small effects80 or SNP sets with more complex epistasis.81, 82, 83 While statistical approach concentrates on inference of relationship, ML focuses on making generalizable predictions using general algorithms to identify patterns in complex data76 based on its empirical capabilities. It does so by training different models to achieve the best predictive performance on an unseen test set which is required to minimize overfitting to the training dataset.
Nonetheless, ML approaches suffer from 2 major limitations. Similar to the statistical approach, the first limitation of the ML approach is the ‘curse of dimensionality’ where there are too many features in the dataset complicating the training of ML models.84,85 When models are trained with too many variables, they may not capture all possible combinations leading to high-variance and overfitting of the training model, resulting in the model being unable to predict accurately when less frequently occurring combinations in the test set are fed into the model. Another aspect of the ‘curse of dimensionality’, which greatly affects clustering or nearest neighbours-based ML methods, is ‘distance concentration’ with convergence of all pairwise distances between different samples/points as the number of variables increases, leading to difficulty in clustering high dimension data. Several machine learning tools of feature selection / reduction / extraction have been developed to mitigate this ‘curse of dimensionality’. In this study, we included a genetic approach to dimensionality reduction by focusing on haplotypes of coding SNPs, which greatly reduced the complexity from 114,000 SNPs to 52,331 pfcHaps, partially mitigating the curse of dimensionality, before ML feature selection approach was employed to further reduce the dimensionality and identify features for model training. Furthermore, focusing on haplotypes of coding SNPs (pfcHaps) partially addresses the clustering issue of ML by clustering SNPs in the coding regions of genes in a biologically meaningful way. The second limitation of the ML approach is its low interpretability where the underlying mechanism is less clear, since ML sacrifice interpretability for predictive power, leading to less acceptance of these approaches by many biomedical scientists.79 In this study, by focusing on pfcHaps, the interpretability issue is also partially addressed, since the coding region of genes represents one of the most functional regions of genes that is translated into protein and are thus most likely to be biologically relevant. In fact, majority of the SNPs within the predictive pfcHaps were found to be potentially functional (e.g., altering important functional sites including transcription factor binding, splicing, microRNA, etc and/or associated with changes in gene expression and/or predicted to have deleterious effect on the protein) (Table 1). The interpretability and biological significance of the features selected through our algorithm is further highlighted by previous reports of the association of some of the genes of the predictive pfcHaps with either MTX and/or RA (Figure S2) as well as the inferred enriched pathways of some other predicted pfc genes being associated with MTX/RA (Figure S1, Table S2).
In summary, our approach not only uncovered known non-genetic factors that were previously associated with MTX/RA, but also novel genetic features, namely haplotype of coding SNPs (pfcHap) that can predict MTX response in RA patients. Some of these predictive pfcHap resides in genes that were previously reported to be associated with MTX/RA or in pathways associated with MTX/RA, highlighting novel connections that have yet to be investigated, providing us with an opportunity to gain new mechanistic insights into the contributing factors that may account for differences in MTX response in RA patients. Our findings thus serve to complement currently reported non-genetic and genetic features that are associated with MTX response by providing a totally different set of effective genetic features, pfcHap, that can predict MTX response. It would be worthwhile to evaluate if these MTX predictive features that we identified will also be able to predict response to other small molecule anti-arthritic drugs and/or biological agents used for the treatment of RA.
Further optimization and sensitivity analyses of the current model can be performed to fine tune the model as well as assess alternative models in the unseen test dataset. Another worthwhile future direction is to focus on examining whole genome sequencing to identify more predictive haplotypes in all functional regions, including the promoter, intron, 3’ and 5’UTRs, non-coding regions, etc, to further improve the predictive performance in unseen data, not only in other patients with similar profiles but also in patients of different profiles (e.g., different ethnicity, etc) so that the predictive features identified can be generalizable. The effectiveness of other ML feature engineering algorithms that have been successfully applied in biomedical studies86, 87, 88, 89, 90 could also be explored.
While further validation and refinement of these predictors will be necessary, we can remain hopeful that these predictors have the potential to be used in clinical practice to facilitate decision-making for the treatment of RA patients. This approach may also be applicable for the identification of predictive features associated with response to other drugs and even disease susceptibility or traits.
Declaration of Competing Interest
CGL, KPL, CCK, SSC, AJWL, and LJL declare that they have a pending Coversheet IP application.
Acknowledgments
Contributors
CGL and KPL conceived the study, directed the research, and edited the manuscript. AJWL, LJL, and BNSO designed and performed the bioinformatics analysis. AJWL drafted the manuscript. ETK, JWLT, TRASG, KPL provided the clinical samples and clinical insights. CCK analysed the sequencing data and provided scientific insights. SSC and LTK provided scientific insights and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by grants from the Singapore Ministry of Health's National Medical Research Council (NMRC) [NMRC/CBRG/0095/2015] (to NCC), CG12Aug17 (to TTSH), CGAug16M012 (to TTSH) and NMRC/CG/017/2013 (to TTSH); National Cancer Center Research Fund and block funding Duke-NUS Medical School to A/P Caroline G.L. LEE.; Singapore Ministry of Education Academic Research Fund Tier 2 grant MOE2019-T2-1-138 to LTK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Any opinions, findings, or recommendations expressed in this material are those of the authors and do not reflect the views of the funders.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author, but restrictions apply to the availability of these data, which were used under the license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the corresponding author.
The TTSH Rheumatoid Arthritis Study Group consists of Andrea Ee Ling Ang, Grace Yin Lai Chan, Madelynn Tsu-Li Chan, Faith Li-Ann Chia, Hiok Hee Chng, Choon Guan Chua, Hwee Siew Howe, Ee Tzun Koh, Li Wearn Koh, Kok Ooi Kong, Weng Giap Law, Samuel Shang Ming Lee, Khai Pang Leong, Tsui Yee Lian, Xin Rong Lim, Jess Mung Ee Loh, Mona Manghani, Justina Wei Lynn Tan, Sze-Chin Tan, Claire Min-Li Teo, Bernard Yu-Hor Thong, Paula Permatasari Tjokrosaputro, Chuanhui Xu.
Funding
This work was supported by grants from the Singapore Ministry of Health's National Medical Research Council (NMRC) [NMRC/CBRG/0095/2015 (to NCC), CG12Aug17 (to TTSH), CGAug16M012 (to TTSH) and NMRC/CG/017/2013]; National Cancer Center Research Fund and block funding Duke-NUS Medical School to A/P Caroline G.L. LEE. Singapore Ministry of Education Academic Research Fund Tier 2 grant MOE2019-T2-1-138 to LTK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103800.
Contributor Information
Khai Pang Leong, Email: khai_pang_leong@ttsh.com.sg.
Caroline G. Lee, Email: bchleec@nus.edu.sg.
Appendix. Supplementary materials
References
- 1.Relling MV., Klein TE. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relling MV., Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The clinical pharmacogenetics implementation consortium: 10 years later. Clin Pharmacol Ther. 2020;107:171–175. doi: 10.1002/cpt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roden DM, Mcleod HL, Relling MV, Williams MS, Mensah GA, Peterson JF, et al. Pharmacogenomics HHS public access. Lancet. 2019;394:521–532. doi: 10.1016/S0140-6736(19)31276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poldrack RA, Huckins G, Varoquaux G. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry. 2020;77:534–540. doi: 10.1001/jamapsychiatry.2019.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varga TV., Niss K, Estampador AC, Collin CB, Moseley PL. Association is not prediction: A landscape of confused reporting in diabetes – A systematic review. Diabetes Res Clin Pract. 2020;170 doi: 10.1016/j.diabres.2020.108497. [DOI] [PubMed] [Google Scholar]
- 6.Goh WW, Bin, Wong L. Dealing with confounders in omics analysis. Trends Biotechnol. 2018;36:488–498. doi: 10.1016/j.tibtech.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyay A, Lu T-P. Gene-gene interaction: the curse of dimensionality. Ann Transl Med. 2019;7:813. doi: 10.21037/atm.2019.12.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botta V, Louppe G, Geurts P, Wehenkel L. Exploiting SNP correlations within random forest for genome-wide association studies. PLoS One. 2014;9:e93379. doi: 10.1371/JOURNAL.PONE.0093379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15 doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karsdal MA, Bay-Jensen AC, Henriksen K, Christiansen C, Genant HK, Chamberlain C, et al. Rheumatoid arthritis: A case for personalized health care? Arthritis Care Res. 2014;66:1273–1280. doi: 10.1002/acr.22289. [DOI] [PubMed] [Google Scholar]
- 11.Dougados M, Soubrier M, Antunez A, Balint P, Balsa A, Buch MH, et al. Prevalence of comorbidities in rheumatoid arthritis and evaluation of their monitoring: results of an international, cross-sectional study (COMORA) Ann Rheum Dis. 2014;73:62–68. doi: 10.1136/ANNRHEUMDIS-2013-204223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA - J Am Med Assoc. 2018;320:1360–1372. doi: 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- 13.Silman AJ, Pearson JE. Epidemiology and genetics of rheumatoid arthritis. Arthritis Res. 2002;4:S265–S272. doi: 10.1186/ar578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikiphorou E, Guh D, Bansback N, Zhang W, Dixey J, Williams P, et al. Work disability rates in RA. Results from an inception cohort with 24 years follow-up. Rheumatology. 2012;51:385–392. doi: 10.1093/rheumatology/ker401. [DOI] [PubMed] [Google Scholar]
- 15.Young A, Dixey J, Kulinskaya E, Cox N, Davies P, Devlin J, et al. Which patients stop working because of rheumatoid arthritis? Results of five years’ follow up in 732 patients from the early RA study (ERAS) Ann Rheum Dis. 2002;61:335–340. doi: 10.1136/ard.61.4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albers JMC, Kuper1 HH, Van Riel PLCM, Prevoo MLL, Van ’t Hof2 MA, Van Gestel AM, et al. Socio-economic consequences of rheumatoid arthritis in the first years of the disease. 1999;38 doi: 10.1093/rheumatology/38.5.423. [DOI] [PubMed] [Google Scholar]
- 17.Verstappen SMM, Boonen A, Bijlsma JWJ, Buskens E, Verkleij H, Schenk Y, et al. Working status among Dutch patients with rheumatoid arthritis: Work disability and working conditions. Rheumatology. 2005;44:202–206. doi: 10.1093/rheumatology/keh400. [DOI] [PubMed] [Google Scholar]
- 18.Kwon J-M, Rhee J, Ku H, Lee E-K. Socioeconomic and employment status of patients with rheumatoid arthritis in Korea. Epidemiol Health. 2012;34 doi: 10.4178/epih/e2012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh ET, Leong KP, Tsou IYY, Lim VH, Pong LY, Chong SY, et al. The reliability, validity and sensitivity to change of the Chinese version of SF-36 in oriental patients with rheumatoid arthritis. Rheumatology. 2006;45:1023–1028. doi: 10.1093/rheumatology/kel051. [DOI] [PubMed] [Google Scholar]
- 20.Lim XR, Xiang W, Tan JWL, Koh LW, Lian TY, Leong KP, et al. Incidence and patterns of malignancies in a multi-ethnic cohort of rheumatoid arthritis patients. Int J Rheum Dis. 2019;22:1679–1685. doi: 10.1111/1756-185X.13655. [DOI] [PubMed] [Google Scholar]
- 21.Koh ET, Tan JWL, Thong BYH, Teh CL, Lian TY, Law WG, et al. Major trends in the manifestations and treatment of rheumatoid arthritis in a multiethnic cohort in Singapore. Rheumatol Int. 2013;33:1693–1703. doi: 10.1007/s00296-012-2602-2. [DOI] [PubMed] [Google Scholar]
- 22.Wong SH. Annual costs of rheumatoid arthritis in Singapore: a pilot study. BSc Thesis 2011.
- 23.Radner H, Smolen JS, Aletaha D. Comorbidity affects all domains of physical function and quality of life in patients with rheumatoid arthritis. Rheumatology. 2011;50:381–388. doi: 10.1093/rheumatology/keq334. [DOI] [PubMed] [Google Scholar]
- 24.Aletaha D, Strand V, Smolen JS, Ward MM. Treatment-related improvement in physical function varies with duration of rheumatoid arthritis: A pooled analysis of clinical trial results. Ann Rheum Dis. 2008;67:238–243. doi: 10.1136/ard.2007.071415. [DOI] [PubMed] [Google Scholar]
- 25.Kwan YH, Koh ET, Leong KP, Wee HL. Association between helplessness, disability, and disease activity with health-related quality of life among rheumatoid arthritis patients in a multiethnic Asian population. Rheumatol Int. 2014;34:1085–1093. doi: 10.1007/s00296-013-2938-2. [DOI] [PubMed] [Google Scholar]
- 26.Lau CS, Chia F, Dans L, Harrison A, Hsieh TY, Jain R, et al. 2018 update of the APLAR recommendations for treatment of rheumatoid arthritis. Int J Rheum Dis. 2019;22:357–375. doi: 10.1111/1756-185X.13513. [DOI] [PubMed] [Google Scholar]
- 27.Weinblatt ME. Methotrexate in rheumatoid arthritis: a quarter century of development. Trans Am Clin Climatol Assoc. 2013;124:16–25. [PMC free article] [PubMed] [Google Scholar]
- 28.Nam JL, Villeneuve E, Hensor EMA, Conaghan PG, Keen HI, Buch MH, et al. Remission induction comparing infliximab and high-dose intravenous steroid, followed by treat-to-target: A double-blind, randomised, controlled trial in new-onset, treatment-naive, rheumatoid arthritis (the IDEA study) Ann Rheum Dis. 2014;73:75–85. doi: 10.1136/annrheumdis-2013-203440. [DOI] [PubMed] [Google Scholar]
- 29.Smolen JS, Landewé R, Bijlsma J, Burmester G, Chatzidionysiou K, Dougados M, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann Rheum Dis. 2017;76:960–977. doi: 10.1136/annrheumdis-2016-210715. [DOI] [PubMed] [Google Scholar]
- 30.Taylor JC, Bongartz T, Massey J, Mifsud B, Spiliopoulou A, Scott IC, et al. Genome-wide association study of response to methotrexate in early rheumatoid arthritis patients. Pharmacogenomics J. 2018;18:528–538. doi: 10.1038/s41397-018-0025-5. 2018 184. [DOI] [PubMed] [Google Scholar]
- 31.Qiu Q, Huang J, Shu X, Fan H, Zhou Y, Xiao C. Polymorphisms and pharmacogenomics for the clinical efficacy of methotrexate in patients with rheumatoid arthritis: a systematic review and meta-analysis. Sci Rep. 2017;7 doi: 10.1038/srep44015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato T, Hamada A, Mori S, Saito H. Genetic polymorphisms in metabolic and cellular transport pathway of methotrexate impact clinical outcome of methotrexate monotherapy in japanese patients with rheumatoid arthritis. Drug Metab Pharmacokinet. 2012;27:192–199. doi: 10.2133/dmpk.DMPK-11-RG-066. [DOI] [PubMed] [Google Scholar]
- 33.Takatori R, Takahashi KA, Tokunaga D, Hojo T, Fujioka M, Asano T, et al. ABCB1 C3435T polymorphism influences methotrexate sensitivity in rheumatoid arthritis patients. Clin Exp Rheumatol. 2006;24:546–554. [PubMed] [Google Scholar]
- 34.Hü Gle M, Omoumi P, Van Laar JM, Boedecker J, Hü Gle T. Applied machine learning and artificial intelligence in rheumatology. Rheumatol Adv Pract. 2020;20:1–10. doi: 10.1093/rap/rkaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosselt HR, Verhoeven MMA, Bulatović-ćalasan M, Welsing PM, de Rotte MCFJ, Hazes JMW, et al. Complex machine-learning algorithms and multivariable logistic regression on par in the prediction of insufficient clinical response to methotrexate in rheumatoid arthritis. J Pers Med. 2021;11:1–12. doi: 10.3390/jpm11010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan Y, Zhang H, Quang D, Wang Z, Parker SCJ, Pappas DA, et al. Machine learning to predict anti–tumor necrosis factor drug responses of rheumatoid arthritis patients by integrating clinical and genetic markers. Arthritis Rheumatol. 2019;71:1987–1996. doi: 10.1002/art.41056. [DOI] [PubMed] [Google Scholar]
- 37.Tao W, Concepcion AN, Vianen M, Marijnissen ACA, Lafeber FPGJ, Radstake TRDJ, et al. Multiomics and machine learning accurately predict clinical response to adalimumab and etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73:212–222. doi: 10.1002/art.41516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howard DM, Adams MJ, Clarke TK, Wigmore EM, Zeng Y, Hagenaars SP, et al. Haplotype-based association analysis of general cognitive ability in Generation Scotland, the English Longitudinal Study of Ageing, and UK Biobank. Wellcome Open Res. 2017;2:61. doi: 10.12688/wellcomeopenres.12171.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Wang M, Zhang H, Stewart WCL, Lin S. Logistic Bayesian LASSO for detecting association combining family and case-control data 06 Biological Sciences 0604 Genetics. BMC Proc. 2018;12:163–167. doi: 10.1186/s12919-018-0139-4. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prevoo MLL, Van'T Hof MA, Kuper HH, Van Leeuwen MA, Van De Putte LBA, Van Riel PLCM. Modified disease activity scores that include twenty-eight-joint counts development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Depristo MA, Banks E, Poplin R, Garimella KV., Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–501. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poplin R, Ruano-Rubio V, DePristo M, Fennell T, Carneiro M, Van der Auwera G, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv. 2017 doi: 10.1101/201178. [DOI] [Google Scholar]
- 44.Pedregosa Fabianpedregosaf, Michel V, Grisel OLIVIERGRISELO, Blondel M, Prettenhofer P, Weiss R, et al. Scikit-learn: Machine Learning in Python. 2011;12 [Google Scholar]
- 45.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool n.d. https://doi.org/10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed]
- 46.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data n.d. https://doi.org/10.1093/nar/gkq603 2021. [DOI] [PMC free article] [PubMed]
- 48.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skipper M. Genomics: HapMap Phase II unveiled. Nat Rev Genet. 2007;8:826–827. doi: 10.1038/nrg2235. [DOI] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nogueira S, Sechidis K, Brown G. On the Stability of Feature Selection Algorithms. 2018;18 [Google Scholar]
- 52.Wang J, Ronaghi M, Chong SS, Lee CGL. pfSNP: An integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses 2010. https://doi.org/10.1002/humu.21331. [DOI] [PubMed]
- 53.Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018;46:109–113. doi: 10.1093/nar/gky399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. BioRxiv. 2018 doi: 10.1101/447367. [DOI] [Google Scholar]
- 55.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 58.Poona NK, Van Niekerk A, Nadel RL, Ismail R. Random Forest (RF) wrappers for waveband selection and classification of hyperspectral data. Appl Spectrosc. 2016;70:322–333. doi: 10.1177/0003702815620545. [DOI] [PubMed] [Google Scholar]
- 59.Chen Q, Meng Z, Liu X, Jin Q, Su R. Decision variants for the automatic determination of optimal feature subset in RF-RFE. Genes (Basel) 2018;9 doi: 10.3390/genes9060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loke SY, Munusamy P, Koh GL, Chan CHT, Madhukumar P, Thung JL, et al. A circulating miRNA signature for stratification of breast lesions among women with abnormal screening mammograms. Cancers (Basel) 2019;11 doi: 10.3390/cancers11121872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Zhang A. Feature selection for classifying high-dimensional numerical data. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2004;2 doi: 10.1109/cvpr.2004.1315171. [DOI] [Google Scholar]
- 62.Yazici S, Yazici M, Erer B, Erer B, Calik Y, Ozhan H, et al. The platelet indices in patients with rheumatoid arthritis: Mean platelet volume reflects disease activity. Platelets. 2010;21:122–125. doi: 10.3109/09537100903474373. [DOI] [PubMed] [Google Scholar]
- 63.Abdul Wahab A, Mohammad M, Rahman MM, Mohamed Said MS. Anti-cyclic citrullinated peptide antibody is a good indicator for the diagnosis of rheumatoid arthritis. Pakistan J Med Sci. 2012;29:773. doi: 10.12669/pjms.293.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padjen I, Öhler L, Studenic P, Woodworth T, Smolen J, Aletaha D. Clinical meaning and implications of serum hemoglobin levels in patients with rheumatoid arthritis. Semin Arthritis Rheum. 2017;47:193–198. doi: 10.1016/j.semarthrit.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Khan NA, Yazici Y, Calvo-Alen J, Dadoniene J, Gossec L, Hansen TM, et al. Reevaluation of the role of duration of morning stiffness in the assessment of rheumatoid arthritis activity. J Rheumatol. 2009;36:2435–2442. doi: 10.3899/jrheum.081175. [DOI] [PubMed] [Google Scholar]
- 66.Mangoni AA, Jackson SHD. Age-related changes in pharmacokinetics and pharmacodynamics: Basic principles and practical applications. Br J Clin Pharmacol. 2004;57:6–14. doi: 10.1046/j.1365-2125.2003.02007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Li R, Lin H, Qiu Q, Lao M, Zeng S, et al. Accumulation of cytosolic dsDNA contributes to fibroblast-like synoviocytes-mediated rheumatoid arthritis synovial inflammation. Int Immunopharmacol. 2019;76 doi: 10.1016/j.intimp.2019.105791. [DOI] [PubMed] [Google Scholar]
- 68.Luteijn RD, Zaver SA, Gowen BG, Wyman SK, Garelis NE, Onia L, et al. SLC19A1 transports immunoreactive cyclic dinucleotides. Nature. 2019;573:434–438. doi: 10.1038/s41586-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, et al. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP containing mixed Phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV., et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8 2015.
- 73.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, et al. Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING n.d. https://doi.org/10.1016/j.molcel.2012.05.029. 2021 [DOI] [PMC free article] [PubMed]
- 74.Sergeant JC, Hyrich KL, Anderson J, Kopec-Harding K, Hope HF, Symmons DPM, et al. Prediction of primary non-response to methotrexate therapy using demographic, clinical and psychosocial variables: results from the UK Rheumatoid Arthritis Medication Study (RAMS) Arthritis Res Ther. 2018;20:147. doi: 10.1186/s13075-018-1645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mieth B, Rozier A, Rodriguez JA, Höhne MMC, Görnitz N, Müller K-R. DeepCOMBI: explainable artificial intelligence for the analysis and discovery in genome-wide association studies. NAR Genomics Bioinforma. 2021;3 doi: 10.1093/NARGAB/LQAB065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bzdok D, Altman N, Krzywinski M. Points of Significance: Statistics versus machine learning. Nat Methods. 2018;15:233–234. doi: 10.1038/NMETH.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edwards SL, Beesley J, French JD, Dunning M. Beyond GWASs: Illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–515. doi: 10.1038/NRG3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun S, Dong B, Zou Q. Revisiting genome-wide association studies from statistical modelling to machine learning. Brief Bioinform. 2021;22:1–10. doi: 10.1093/BIB/BBAA263. [DOI] [PubMed] [Google Scholar]
- 80.Romagnoni A, Jégou S, Van Steen K, Wainrib G, Hugot JP, Peyrin-Biroulet L, et al. Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Rep. 2019;9 doi: 10.1038/S41598-019-46649-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leem S, Jeong HH, Lee J, Wee K, Sohn KA. Fast detection of high-order epistatic interactions in genome-wide association studies using information theoretic measure. Comput Biol Chem. 2014;50:19–28. doi: 10.1016/J.COMPBIOLCHEM.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Wei WH, Hemani G, Haley CS. Detecting epistasis in human complex traits. Nat Rev Genet. 2014;15:722–733. doi: 10.1038/nrg3747. 2014 1511. [DOI] [PubMed] [Google Scholar]
- 83.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun. 2018;9:1–9. doi: 10.1038/s41467-018-06634-y. 2018 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The curse of dimensionality. Why high dimensional data can be so… | by Tony Yiu | Towards Data Science n.d. 2021
- 85.What is curse of dimensionality in machine learning? n.d. 2021
- 86.Yoon S, Kim S. Proc. - 2008 IEEE Int. Conf. Bioinforma. Biomed. Work. BIBMW. 2008. AdaBoost-based multiple SVM-RFE for classification of mammograms in DDSM; pp. 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim S. Margin-maximised redundancy-minimised SVM-RFE for diagnostic classification of mammograms. Int J Data Min Bioinform. 2014;10:374–390. doi: 10.1504/IJDMB.2014.064889. [DOI] [PubMed] [Google Scholar]
- 88.Su R, Xiong S, Zink D, Loo L-H. High-throughput imaging-based nephrotoxicity prediction for xenobiotics with diverse chemical structures. Arch Toxicol. 2016;90:2793–2808. doi: 10.1007/s00204-015-1638-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang F, Mao KZ. Robust feature selection for microarray data based on multicriterion fusion. IEEE/ACM Trans Comput Biol Bioinforma. 2011;8:1080–1092. doi: 10.1109/TCBB.2010.103. [DOI] [PubMed] [Google Scholar]
- 90.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422. doi: 10.1023/A:1012487302797. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.