Abstract
Ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP) family members (ENPP1–7) have been implicated in key biological and pathophysiological processes, including nucleotide and phospholipid signaling, bone mineralization, fibrotic diseases, and tumor-associated immune cell infiltration. ENPPs are single-pass transmembrane ecto-enzymes, with notable exceptions of ENPP2 (Autotaxin) and ENNP6, which are secreted and glycosylphosphatidylinositol (GPI)-anchored, respectively. ENNP1 and ENNP2 are the best characterized and functionally the most interesting members. Here, we review the structural features of ENPP1–7 to understand how they evolved to accommodate specific substrates and mediate different biological activities. ENPPs are defined by a conserved phosphodiesterase (PDE) domain. In ENPP1–3, the PDE domain is flanked by two N-terminal somatomedin B-like domains and a C-terminal inactive nuclease domain that confers structural stability, whereas ENPP4–7 only possess the PDE domain. Structural differences in the substrate-binding site endow each protein with unique characteristics. Thus, ENPP1, ENPP3, ENPP4, and ENPP5 hydrolyze nucleotides, whereas ENPP2, ENPP6, and ENNP7 evolved as phospholipases through adaptions in the catalytic domain. These adaptations explain the different biological and pathophysiological functions of individual members. Understanding the ENPP members as a whole advances our insights into common mechanisms, highlights their functional diversity, and helps to explore new biological roles.
Keywords: structure–function, phospholipase, phosphodiesterases, signaling, drug development, autotaxin, pyrophosphate, mineralization, cancer
Abbreviations: AP, alkaline phosphatase; ATX, autotaxin; cGAS, cyclic GMP-AMP synthase; CPPD, calcium pyrophosphate dehydrate; ENPP, ecto-nucleotide pyrophosphatase/phosphodiesterase; GPC, glycerophosphocholine; GPCR, G protein-coupled receptor; GPI, glycosylphosphatidylinositol; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; lysoPLD, lysophospholipase D; NAD, nicotinamide adenine dinucleotide; NUC, nuclease-like domain; PAF, platelet-activating factor; PDE, phosphodiesterase; PLC, phospholipase C; PPi, inorganic pyrophosphate; SM, sphingomyelin; STING, stimulator of interferon genes
A brief history of the ENPP family
The founding member of the family, ENPP1, was discovered more than 50 years ago as plasma cell-differentiation antigen PC-1 (1, 2) and subsequently characterized as NPP1 (3), along with ENPP2 and ENPP3, based on its enzymatic activity (4). The ENPP family initially comprised five members (ENPP1–5), at the time called NPPs. Subsequent identification of new NPPs showed that they are ecto-enzymes, thus the nomenclature was changed to ecto-nucleotide pyrophosphatase/phosphodiesterases (ENPPs). By the end of 2005, two additional members were added to the family: ENPP6 and 7.
A breakthrough was achieved some 20 years ago, when ENPP2 (better known as Autotaxin, ATX) was identified as the long-sought lysophospholipase D (lysoPLD) that produces the lipid mediator lysophosphatidic acid (LPA) from extracellular lysophosphatidylcholine (LPC) (5, 6). Thereafter, ENPP6 was characterized as a choline-specific phospholipase C (PLC) and ENPP7 as an alkaline sphingomyelinase that converts membrane sphingomyelin (SM) to ceramide and phosphocholine. The state of the art at that time is reviewed in (7).
All ENPP members share a homologous catalytic core, the phosphodiesterase (PDE) domain. They were grouped and named based on the nature of their substrates, their extracellular location, and their chronological discovery (4). From an evolutionary perspective, the family members are divided into two subgroups, ENPP1–3 and ENPP4–7 (8), which also reflect their structure and domain constitution.
Over the last decade, the three-dimensional structures of all ENPP members have been determined, shedding new light on their domain architecture and catalytic mechanisms. In this review, we will first discuss the family on the basis of their domain architecture that is crucial to endow them with specific biochemical characteristics. We then discuss the biochemical reactions mediated by the respective family members, considering the differences in their catalytic site and focusing on determinants of their substrate diversity. Finally, we will put the ENPPs in the context of their role in human (patho)physiology and their contribution to disease.
Domain architecture of ENPPs
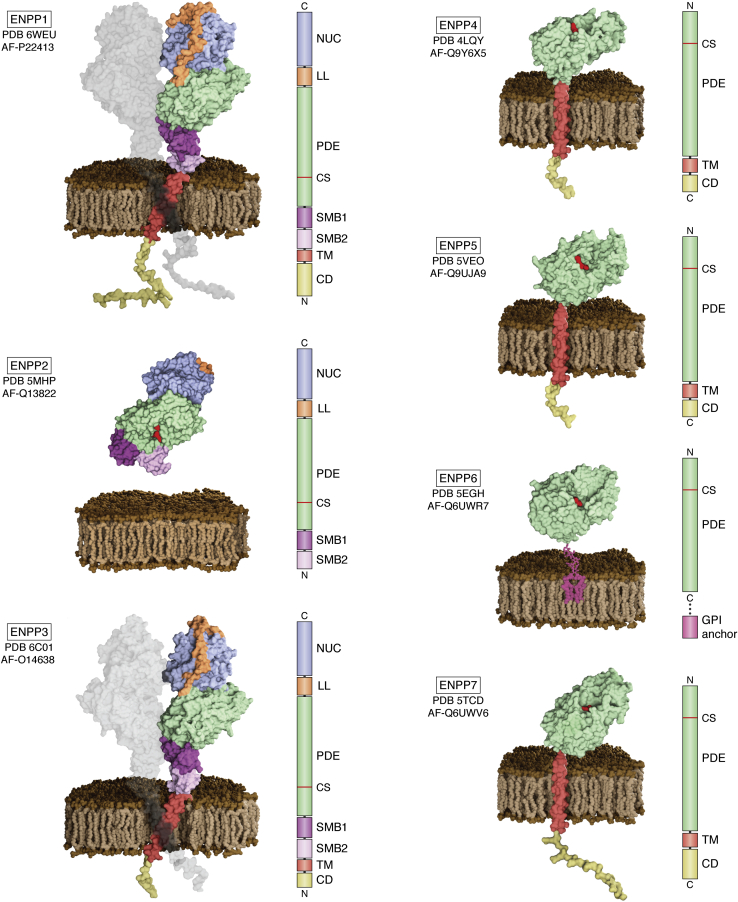
The ENPPs are classified on the basis of their conserved PDE domain, showing markedly different structures, as shown in Figure 1. ENPP1–3 are multidomain proteins, with two tandem N-terminal somatomedin B-like domains (SMB1–2), followed by the PDE domain, a “lasso” loop (LL), and a C-terminal nuclease-like domain (NUC). ENPP1 and ENPP3 are single-pass type II membrane proteins, whereas ENPP2 (ATX) is a secreted protein (9) processed by removal of its signal peptide (10). The last four ENPP members, ENPP4–7, only have the signature PDE domain. ENPP4, ENPP5, and ENPP7 are single-pass type I membrane proteins, while ENPP6 is GPI-anchored to the plasma membrane (11).
Figure 1.
Overview of ENPP1–7 structures and domain architecture. The structures are presented as surface models based on experimental findings and AlphaFold models (87). The ENPP1–3 multidomain subgroup is to the left, and the single-domain ENPP4–7 subgroup to the right. All family members are attached to the cell surface through a transmembrane domain, except for secreted ENPP2 and GPI-anchored ENPP6. Next to each structure, the colored bar represents the location of the different domains in the corresponding sequences. All pictures were generated in UCSF ChimeraX 1.2.5 (88), and the scenes were created and rendered using Blender 2.93.5 (https://www.blender.org/). PDE, Phosphodiesterase domain; CS, Catalytic site; SMB1/2, somatomedin like domain 1/2; NUC, Nuclease-like domain; LL, lasso loop; TM, transmembrane domain; CD, cytoplasmic domain; PDB files used: ENPP1: 6WEU, AF-P22413; ENPP2: 5MHP, AF-Q13822; ENPP3: 6C01, AF-O14638; ENPP4: 4LQY, AF-Q9Y6X5; ENPP5: 5VEO, AF-Q9UJA9; ENPP6: 5EGH, AF-Q6UWR7; and ENPP7: 5TCD, AF-Q6UWV6.
In the following paragraphs, we will discuss the domain structure and function of ENPP1–7.
Position and function of the SMB domains
The N-terminal SMB1/2 domains are tightly compacted by disulfide bridges and are structurally similar to the SMB domain of vitronectin (12). In ENPP1 and 3, the SMB domains do not show detectable intramolecular interactions, whereas the SMB1 domain of ENPP2 interacts tightly with the PDE domain to form an open tunnel or channel (13, 14), whose function will be discussed below. The conformational differences of the SMB domains among ENPP members originate from changes in the linkers connecting SMB1 and SMB2, or SMB2 and PDE (15).
The SMB domains of ENPP1 and ENPP3 were thought to contribute to homodimerization (16, 17, 18). ENPP1 dimerises in the crystals (18), and there is evidence that it is indeed also a dimer in cells (17, 18). The formation of the dimers in cells, however, appears to be mediated by cysteines through a zipper-like mechanism in the transmembrane domain, while the SMB domains contribute to trafficking to the plasma membrane (18). ENPP3 homodimerization has been observed both in solution and in crystals and is partially mediated by the SMB domains, but has no effect on its catalytic activity, and its physiological importance remains unclear (16).
The SMB2 domain of ENPP2 is unique in that it has an integrin-binding RGD motif, known to be associated with adhesion and immobilization on the cell surface (19, 20). However, structural studies led to the conclusion that integrin binding involves other residues (13) that may also modulate ENPP2 activity (21). Thus, the interaction mode of ENPP2 to integrins remains unclear to date.
The SMB domains of ENPP1 and ENPP2 are followed by the PDE domain—to be discussed below, and by a 50-amino acid so-called lasso loop. This loop tightly wraps around the structure, finally leading to the subsequent NUC domain, in the opposite side of the PDE domain (13, 16). This reinforcement from the LL is thought to maintain the tight interdomain interaction between the NUC and PDE domains (22).
The catalytic PDE domain
The PDE domain is the shared feature of the ENPP family, hosting the common pyrophosphatase/phosphodiesterase activity. The PDE fold was first established by the structure of the Xanthomonas axonapodis nucleotide pyrophosphatase/phosphodiesterase (XaNPP) (23). The fold is that of the alkaline phosphatase (AP) superfamily and has defined characteristics allowing it to specialize on diesters rather than monoesters, as observed for many members of the AP superfamily. The PDE domain is a monomer in which the active site is formed by residues residing on a central β-sheet structure and its flanking α-helices. The most characteristic adaptation of the PDE domain is observed in ENPP2, showing a deletion of some 20 residues compared with other ENPPs (e.g., 18 residues are deleted in human ENPP2 compared with human ENPP1) (13, 14). That deletion creates a unique hydrophobic pocket next to the catalytic site of ENPP2. Consistent with this, an ENPP1 mutant lacking the 18-aa "insertion loop" lost its nucleotide-hydrolyzing activity but instead gained the lysoPLD activity of ENPP2 (24).
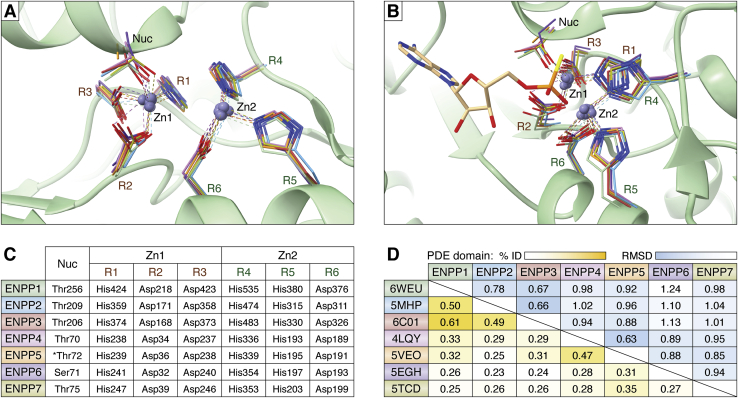
The catalytic site is characterized by two zinc ions essential for catalysis and located in a shallow groove, where the substrate binds. The two Zn2+ are coordinated by seven highly conserved residues: two histidines and an aspartate bind to Zn1, while two aspartates, a histidine and the catalytic nucleophile (a threonine for all ENPPs except for ENPP6 where it is a serine) hold Zn2, as depicted in Figure 2. The substrate-interacting residues vary between ENPP members, creating drastically different environments that determine substrate specificity. The PDE domains in ENPP1–7 are fairly well conserved. In humans, they share 23% to 61% identity (25), and the rms distance after structural superposition using flexible domains is 0.63 to 1.24 Å (26), see Figure 2D for pairwise comparisons.
Figure 2.
The conserved catalytic site of ENPP1–7.A, the zinc ions and the seven residues implicated in catalysis are shown for each ENPP with a different color; all structures have been superimposed in ChimeraX 1.2.5 (88), and the green ribbon model represents ENPP1; Nuc, nucleophile, R1–3, residues coordinating Zn1, R4–6, residues coordinating Zn2 (B) a different orientation of A, also showing binding of adenosine-5′-thio-monophosphate to better depict the substrate orientation. C, summary of the seven conserved residues involved in catalysis; ∗this residue was mutated to Alanine in the crystal structure. D, comparison of the catalytic PDE domains of all seven members for the human ENPPs: the sequence identity (yellow; saturation highlights highly conserved sequences) after multiple sequence alignment using ClustalW (25) is shown to the bottom half; the overall root mean square distance deviations (blue; saturation highlights more similar structures) after flexible structural superposition using RAPIDO (26) are shown; the mouse structure was used for ENNP6.
The role of the nuclease (NUC) domain
The NUC domain structure consists of a six-stranded antiparallel β-sheet surrounded by eight α-helices and is similar to bacterial nonspecific endonucleases, such as NucA (27). It appears to be catalytically dead as none of the residues required for catalysis are present in ENPP1–3 (7). It also encompasses an EF hand-like motif (EFL) that binds a calcium ion, but is not flanked by helices; this EFL motif appears to be important for ENPP1–3 activity and stability (4, 13). The NUC domain strongly interacts with the PDE domain contributing to its stability and enzymatic activity (28).
This interdomain interaction between the PDE and the NUC domains is conserved in ENPP1–3 and depends on a disulfide bridge, hydrogen bonds, and an N-linked glycan packed between both domains and a C-terminal α-helix and the lasso loop (13). The interface area between the NUC and the PDE domains varies among ENPP1–3 in the number of hydrogen bonds and salts bridges, but the disulfide bond is highly conserved in all three ENPP members (13, 18), and the PDE and NUC are firmly tethered. While many amino acids of the PDE domain participate in the interaction, only the C-terminal region of the NUC domain and an aspartate of the EFL motif are involved (13). The C-terminal region of the NUC domain has been shown to also be crucial for the protein’s secretion and catalytic activity (17, 22, 29).
Substrate specificity and catalytic mechanism
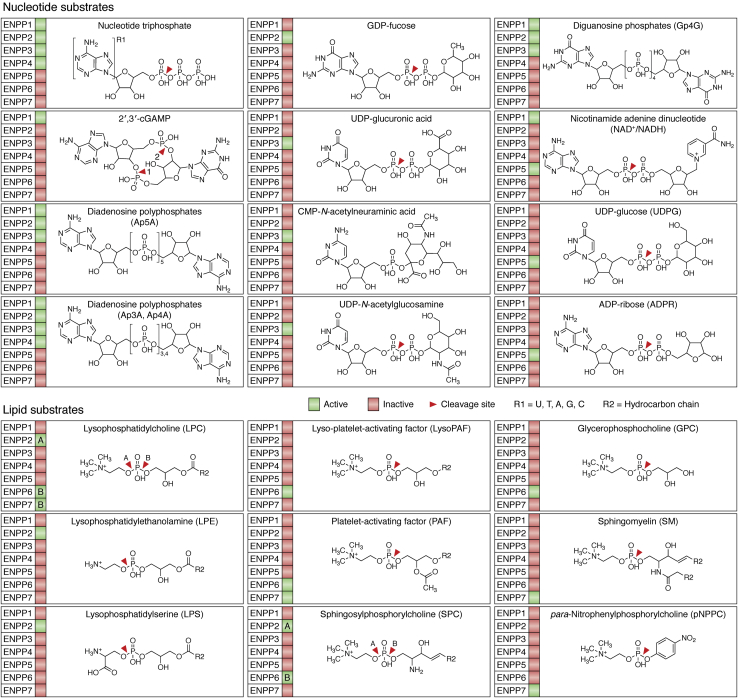
All ENPP members can hydrolyze extracellular nucleotides and, therefore, were originally assumed to be involved in purinergic signaling. Over the years, the preferred and physiologically relevant substrates for many family members turned out to be different, as summarized in Figure 3. ENPP1,3,4,5 specialize on nucleotide-base substrates (e.g., diadenosine triphosphate and cyclic nucleotides) but ENPP2,6,7 diverged in substrate preference, notably for lysophospholipids. A common feature is the action toward a phosphate diester bond, which remains the defining feature of the family.
Figure 3.
Substrates of ENPP1–7 divided into nucleotide-like and lipid-containing substrates. For each substrate, family members that have been shown to process these substrates in vitro or in vivo are highlighted in green; red suggests either that the substrate cannot be processed or data are not available. The red arrow-head indicates the scissile bond.
PDE domains have been suggested to support an associative two-step in-line displacement mechanism for catalysis, similar to what was originally described for alkaline phosphatases (30, 31). The latter mechanism was supported by crystallographic studies of transition state mimics for ENPP2 (29) and further backed by additional data for ENPP1 (32) and ENPP3 (33). In this model, the catalytic nucleophile is a threonine in all ENPPs (except for ENPP6, where a serine residue is used) and is activated by the Zn2 ion (see Fig. 2). Catalysis is initiated by a nucleophilic attack, cleaving the scissile phosphodiester bond and allowing formation of the first leaving group. A water-based attack on the formed enzyme–product intermediate completes the reaction and allows departure of the second leaving group. It must be noted, however, that recent mechanistic data for alkaline phosphatases argue that catalysis in these enzymes follows a concerted mechanism, where bond cleavage and nucleophilic attack occur in a single kinetic step (34). Similar studies of ENPP family members will be needed to fully understand the detailed mechanism of PDE-mediated catalysis.
Recognition and catalysis of nucleotide-based substrates
ENPP1,3–5 all recognize nucleotide-containing molecules as their primary substrates, yet show interesting differences in both substrate specificity and affinity. An overview of chemical structure of ENPP substrates is shown in Figure 3.
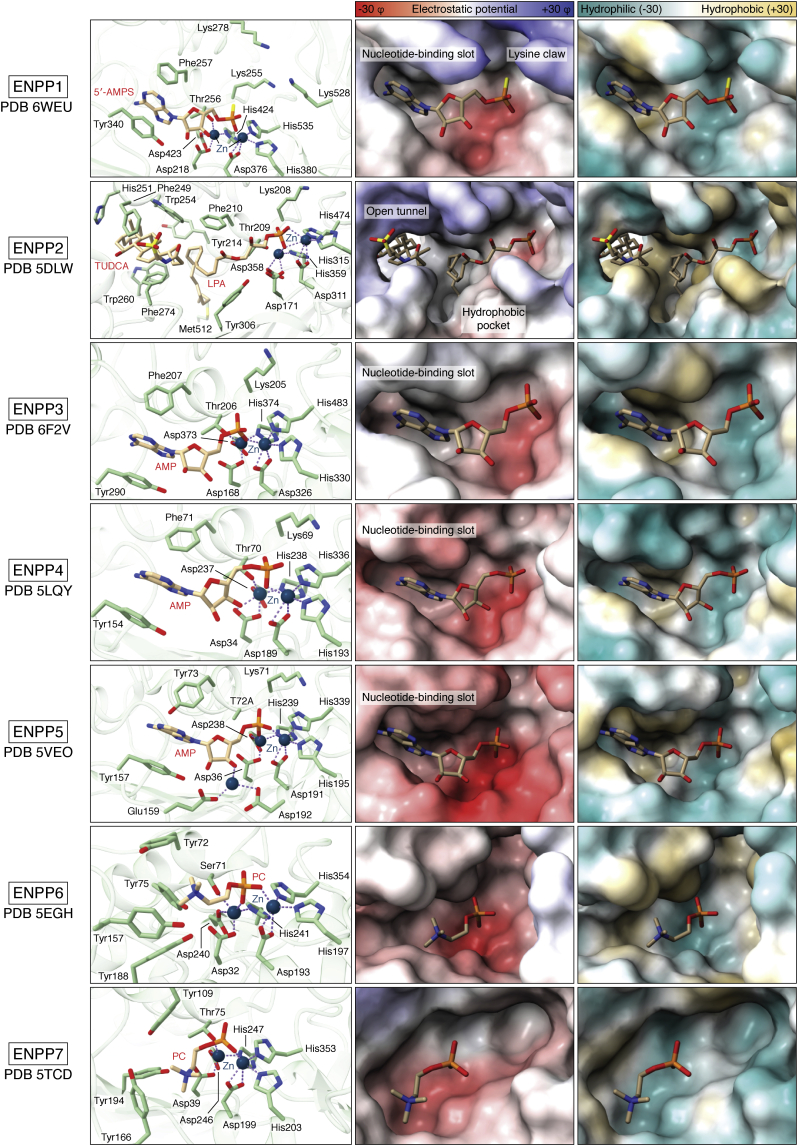
ENPP1, once known as plasma cell membrane glycoprotein (PC1), is arguably the best-studied family member that hydrolyzes ATP to generate AMP and inorganic pyrophosphate (PPi), thereby inhibiting mineralization processes (35, 36), as well as 2′,3′-cyclic GMP-AMP (cGAMP) to regulate the cyclic GMP-AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway (37, 38). Thus, ENPP1 participates in two distinct biochemical and biological processes. Interestingly, while ENPP1 has considerably higher affinity for ATP, as reflected by a Km of ∼10 to 50 μM (24, 39), compared with >1 mM for GTP, CTP and UTP, it displays the highest turnover rate (820 s−1) for GTP. Consequently, the catalytic efficiency (kcat/Km) for these nucleotide substrates is similar within one order of magnitude (0.017–0.34 s−1 μM−1) (24). The specificity of ENPP1 (18, 24) for nucleotides is determined by a well-defined nucleotide-binding slot, with π–π stacking interactions between tyrosine (Tyr340), phenylalanine (Phe257), and the nitrogenous base (Fig. 4). Another feature that favors the stability of the substrate-ENPP1 complex is the “lysine claw” (Lys255, Lys278, and Lys528). Upon substrate binding, the negative γ-phosphate of ATP attracts Lys278 and Lys528, along with Lys255, to enclose the phosphate with positive charges to provide electrostatic stability (40).
Figure 4.
Substrate recognition in the catalytic site. The first column shows the best characterized substrate for each family member as a stick model (carbons: creme; oxygens: red; nitrogen: blue; phosphate: orange) and a ribbon model of the corresponding structure (green) as well as the residues involved in substrate recognition and catalysis (green carbons; other atoms as for the substrate) and the zinc ions (blue). The second column shows the surface colored by electrostatic potential for each structure, and the same substrate as in the previous column as a stick model; the last column shows the surface colored by hydrophobicity. All pictures and calculations were generated with UCSF ChimeraX 1.2.5 (88) PDB files: 6WEU, 5DLW, 6F2V, 5LQY, 5VEO, 5EGH and 5TCD.
The PPi that gets released from all these reactions (and would presumably mostly originate from ATP, given its physiological abundance) is of particular physiological importance in the regulation of bone mineralization. This regulation depends on the balance between inorganic phosphate (Pi), the substrate, and PPi, which inhibits this process (24). These activities endow ENPP1 with particular pathophysiological interest in mineralization disorders and soft-tissue calcification, as it is the only ENPP family member showing various disease-associated loss-of-function mutations (18, 41, 42).
Of particular interest is the ability of ENPP1 to hydrolyze 2′,3′-cGAMP with a catalytic efficiency (0.57 s−1 μM−1) similar to that for ATP (0.27 s−1 μM−1) (39). 2′3′-cGAMP serves as an intracellular second messenger that can be secreted by tumor cells and thereby contributes to anticancer immunity and metastasis (37, 38). This ability of ENPP1 to hydrolyze 2′3′-cGAMP, but not 3′3′-cGAMP (32), brings into focus a new and exciting biological function for ENPP1, with a potential in cancer therapy (37, 42).
Other substrates for ENPP1 include the diadenosine polyphosphates diadenosine 5′,5′′′-P1,P3-triphosphate (Ap3A), diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A), and diadenosine 5′,5′′′-P1,P5-pentaphosphate, (Ap5A), the diguanosine polyphosphate diguanosine 5′,5′′′-P1,P4-tetraphosphate (Gp4G), the nicotinamide adenine dinucleotide (NAD+), and cyclic adenosine monophosphate (cAMP) (32, 43, 44), at least in vitro. It remains unclear whether any of these substrates is of functional relevance in vivo.
ENPP3 (also known as CD203c) and ENPP4 have low specificity for nucleotides with different nucleobase. ENPP3 prefers ATP to other NTPs by twofold (16). The low specificity of ENPP3 allows it to also degrade nucleotide sugars (UDP-N-acetylglucosamine, UDP-N-acetylgalactosamine, CMP-N-acetylneuraminic acid, GDP-fucose, and UDP-glucuronic acid), and diadenosine polyphosphates (Ap3A, Ap4A, and Ap5A), as well as Gp4G (16, 45), at least in vitro. For ENPP4, ATP and Ap3A have been identified as substrates in vitro, but the ATP hydrolysis rate by ENPP4 is negligible compared to that of ENPP1 (40). Ap3A hydrolysis is similar between ENPP1, ENPP2, and ENPP4 (40), but lower for ENPP3. In line with this promiscuity, ENPP3 and ENPP4 have a more open nucleotide-binding slot, and interactions with tyrosine (Tyr290 and Tyr154 respectively) and phenylalanine (Phe207 and Phe71 respectively) are weaker (Fig. 4). The catalytic site of ENPP4 is more solvent-exposed than others, while the substrate electrostatic environment is negatively charged, thereby decreasing its affinity for nucleotides (Fig. 4). Notably, the lysine claw is not present in ENPP3–4 as only the lysine closer to zinc ions is conserved (Lys205, Lys69, and Lys71, respectively). This clarifies why ENNP1 displays highly efficient ATP hydrolysis, when compared with other ENPPs, and explains why ENPP1 is a unique producer of PPi in vivo.
ENPP5 has unique features that distinguish it from the other ENPPs. It is the only ENPP containing three Zn2+ ions in the catalytic site (Fig. 2), where the third one is coordinated by an aspartate (Asp192) and a glutamate (Glu159). This additional Zn2+ ion interacts with the 2′ and 3′ oxygens of ribose substrates (46). Another unique feature of ENPP5 is the presence of a tyrosine (Tyr73) in the nucleotide-binding slot, rather than phenylalanine as in other family members. Reversal of this mutation, eliminating the hydroxyl group that would presumably clash with nucleotide substrates, enables NTP hydrolysis (46). Moreover, the catalytic site environment is negatively charged, similar to that of ENPP4 (Fig. 4). Although, ENPP5 can cleave substrates including UDP-glucose (UDPG) and ADP-ribose (ADPR). The highest catalytic rate is achieved on nicotinamide adenine dinucleotide (NAD+/NADH); however, the catalytic efficiency for NAD+ is low (0.00067 s−1 μM−1). It has been suggested that substrate recognition does not happen through the nucleotide-binding slot, at least in vitro, but instead involves other regions of the substrate and the protein where the third Zn2+ could be involved (46). Yet, the exact catalytic mechanism and substrate relevance remain to be resolved.
Recognition and catalysis of phospholipid substrates
Strikingly, the three remaining family members, ENPP2, ENPP6, and ENPP7, recognize phospholipids. ENPP2 evolved as lysoPLD, while ENPP6 is a choline-specific ENPP acting in a PLC-like manner. ENPP7, on the other hand, hydrolyzes SM as its preferred substrate.
ENPP2/ATX is the only secreted family member and acts as a lysoPLD that hydrolyzes extracellular lysophosphatidylcholine (LPC) species to the corresponding lysophosphatidic acid (LPA) products (5, 6, 47). LPAs are bioactive lipids that act on six distinct G protein-coupled receptors (GPCRs), thereby triggering various signaling pathways that lead to a variety of cellular responses in a cell- and receptor-dependent manner (48). ENPP2 recognizes monoacylglycerol fatty acid chain, but not the choline headgroup of LPCs. Accordingly, ENPP2 can also hydrolyze noncholine lysophospholipids, such as lysophosphatidylethanolamine (LPE) and lysophosphatidylserine (LPS), as well as sphingosylphosphorylcholine (SPC), at least in vitro. However, the physiological relevance of these substrates remains doubtful, as LPC is by far the most abundant lysophopholipid substrate in the extracellular milieu (49). It is worth noting that ENPP2 can also hydrolyze artificial nucleotide substrates.
From a structural point of view, ENPP2/ATX is the most interesting ENPP family member, as it has a unique tripartite active site formed by the catalytic site, common in all ENPPs and able to accommodate nucleotides, a hydrophobic pocket that binds the acyl chains of lipid substrates, and an open tunnel (or channel), whose function has long been elusive (13, 14). ENPP2 largely owes its ability to accommodate lipid substrates to a unique deletion of a stretch of about 20 residues (13, 14) (exactly 18 compared with ENPP1) in the PDE domain, which creates the hydrophobic pocket for acyl chain binding (Fig. 2). Although different acyl chain lengths can be accommodated in the hydrophobic pocket, there is a preference for short (14:0 > 16:0 > 18:0) and unsaturated (18:3 > 18:1) acyl chains (14, 50). Another unique structural and functional feature of ENPP2 is an open tunnel that is created between the PDE and SMB1 domains. The tunnel was originally described as an “exit channel” for the produced LPA molecules and may also play a role in delivering LPA to its cognate GPCRs (47, 50). Later, it was shown that the tunnel can also modulate ATX activity by binding certain compounds that act as “allosteric modulators.” Naturally occurring small molecules, such as bile salts (51), bind to this pocket and act as weak inhibitors. A kinetic analysis has shown that, at least in vitro, LPA binding in the tunnel can increase the catalytic rate for various LPC species (52). The tripartite site has provided an excellent template for the design of highly specific and potent ENPP2/ATX inhibitors (53). One inhibitor (GLPG1690) has reached phase 3 trials for idiopathic pulmonary fibrosis (https://www.businesswire.com/news/home/20210210005), which has now been discontinued as the benefit–risk profile no longer supported it. However, applications in liver disease (54, 55, 56) and pancreatic cancer (https://www.ionctura.com/admin/resources/ioa-289sitc-results-9-11final.pdf) are pursued. More recently, ATX has been shown to be chemorepulsive for tumor-infiltrating lymphocytes and circulating CD8+ T cells ex vivo and to suppress tumor infiltration of cytotoxic CD8+ T cells in an anticancer vaccination model, suggesting new therapeutic opportunities in cancer (57).
ENPP6 has been identified and characterized as glycerophosphocholine choline phosphodiesterase (GPC-Cpde). It was first isolated as a Ca2+-independent multimer of 62 kDa subunits from rat brain (11). ENPP6 is a PLC that produces phosphocholine from choline-containing lysophospholipids, including SPC, LPC, platelet-activating factor (PAF), lysoPAF, and glycerophosphocholine (GPC), but not others (58). In marked contrast to the case of ENPP2, ENPP6 recognizes its substrates through the choline headgroup, rather than by recognizing the acyl chains (58, 59). ENPP6 has tyrosine (Tyr72, Tyr75, Tyr157, and Tyr188) residues located close to the Zn2+ ions, surrounding the choline moiety (Fig. 2). These tyrosines electrostatically stabilize the positively charged choline though π–cation interactions (58). However, it is not yet clear how ENPP6 exactly binds its substrates. A “thumb loop” (337–350) near the catalytic site is unrelated to catalytic activity, but might have a role in catalysis given its size and position, interacting with the lipid part of the substrate.
ENPP7, also known as alkaline sphingomyelinase (Alk-SMase), hydrolyzes SM to ceramide and phosphocholine. Additionally, it can function as a lyso-PLC that hydrolyzes LPC and PAF to generate phosphocholine, similar to ENNP6, and the artificial substrate para-nitrophenylphosphorylcholine (pNPPC) (60). Its atomic structure was the last to be determined among the family, shedding new light on its substrate recognition (61). The ENPP7 catalytic site is more solvent-exposed compared with ENPP6 (Fig. 2). Moreover, there is no nucleotide-binding slot, and the choline moiety is surrounded and stabilized by tyrosines (Tyr109, Tyr166, and Tyr194), referred to as the cation-π box (61). In addition, a hydrophobic loop (342–351) and cation patches on the surface nearby the catalytic site may interact with bile salt micelles (61), although this remains to be clarified.
ENPPs in physiology and disease
The physiological functions of ENPP family members are largely defined by their different substrate preferences acquired through structural adaptations, as well as by their tissue-specific expression levels. In this section, we summarize the physiological roles of ENPP1–7 and their established or presumed roles in disease.
ENPP1 has long been known to regulate calcification in bone and other mineralization-competent tissues by converting extracellular nucleotides into PPi (18, 35, 36). PPi inhibits hydroxyapatite (HA) crystal formation and deposition and thereby prevents physiological overmineralization (18, 35). On the other hand, PPi is hydrolyzed by alkaline phosphatase activity into Pi and thereby promotes bone remodeling. A distorted Pi-PPi balance results in pathological calcification as is observed in aging, osteoarthritis, and other diseases. Most of our knowledge on ENPP1 in health and diseases comes from studies in KO mice and patients with ENPP1 mutations and calcification disorders (35). ENPP1-deficient mice are viable but suffer from reduced extracellular PPi levels causing pathological calcification of cartilage and soft tissues, leading to atherosclerosis and abnormal bone development (41, 42). ENPP1 has also been implicated in insulin resistance and type II diabetes (62), but these claims have raised severe doubts (63, 64).
Interestingly, loss-of-function mutations in ENPP1 are associated with several calcification disorders, including general arterial calcification of infancy (GACI), a life-threatening disease of early childhood (35). Of note, virtually all disease-associated mutations in ENPP1 map to the PDE and NUC domains, affecting ENPP1 protein stability but not the Thr256 nucleophile, the metal-binding site, or glycosylation sites (18, 24). Furthermore, reduced ENPP1 expression correlates with calcification in atherosclerotic plaques and has been implicated in Cole disease (41, 65). Conversely, upregulated ENPP1 expression can lead to pathological calcification in cartilage due to the formation of calcium pyrophosphate dihydrate (CPPD) crystals. Increased ENPP1 expression is detected in chondrocytes of aging persons and characterized by the accumulation of CPPD crystals in joints and may contribute to the pathogenesis of osteoarthritis. Therefore, ENPP1 has been investigated as a drug target for calcification disorders (66, 67).
ENPP1 also preferentially hydrolyzes 2′3′-cGAMP, but not its linkage isomer 3′3′-cGAMP, and negatively regulates the cGAS-STING pathway in the innate immune system. Of particular interest are recent studies linking ENPP1 to cancer and reduced immune cell infiltration through hydrolysis of 2′3′-cGAMP, which was shown to be exported by tumor cells to function as an anticancer “immunotransmitter” (37, 38, 42); the details of that mechanism are beyond the scope of this review. Briefly, ENPP1 loss suppresses metastasis, restores tumor immune infiltration, and potentiates response to immune checkpoint blockade in a manner dependent on tumor cGAS and host STING (37). Accordingly, ENPP1 is intensively investigated as a drug target not only for calcification disorders (66, 67), but also for its potential applications in cancer immunotherapy (38, 42, 68, 69, 70).
ENPP2 (ATX) is the only secreted family member and, as such, can easily diffuse in the extracellular environment. As a lysoPLD, ATX produces the bioactive lipid LPA (5, 6) that signals through six different GPCRs (LPAR1–6) (48, 71). ATX was identified as an “autocrine motility factor” for melanoma cells in the early 1990s (72), but it took another 10 years before it was uncovered as a lysoPLD present in serum and plasma. Recent studies on ATX-mediated T-cell migration and immune evasion (57) point to ATX as a dual-function protein that both produces and chaperones LPA for local delivery of LPA to its cognate receptors when ATX interacts with adhesive molecules (13, 73).
Enpp2-deficent mice are embryonic lethal due to defective vascular and neural development (74, 75). In the adult, ATX shows its highest expression in the brain, fibroblasts, and lymph nodes. Based on animal studies, the ATX-LPA signaling axis has been linked to T-cell migration, wound healing, inflammation, tissue fibrosis, tumor progression and immune evasion (57, 76, 77, 78, 79, 80). ATX is highly expressed in most solid tumors, in part attributable to ATX-expressing stromal cells such as cancer-associated fibroblasts (57); however, cancer-associated mutations or deletions inENPP2 have not been documented to date. In the nervous system, LPA signaling has been associated with neuropathic pain, cerebral ischemia, brain and spinal cord injury, as well as neurodegenerative diseases (81). Given its role in several pathologies, multiple ATX inhibitors are being developed, which can be classified into four types depending on their interaction with the tripartite site of the enzyme (53).
Finally, recent studies have revealed an unexpected role for ATX secreted by tumor cells, namely as a chemorepellent for CD8+ T cells, both in vitro and in tumor-bearing mice. ATX appears to repel T cells via the LPA6 receptor (LPAR6), notably independently of extracellular LPA levels (57). Consequently, pharmacological inhibition of ENPP2/ATX should stimulate T-cell infiltration into tumors, thereby enhancing the efficacy of immunotherapy, a scenario that warrants further studies.
ENPP3 was initially identified as a regulator of purinergic signaling, particularly in basophils and mast cells with a role in allergic responses (82) and serving as a potential biomarker for allergen sensitivity. In response to pathogens, basophils and mast cells release ATP to activate purinergic receptors. Enpp3 KO mice are highly sensitive to chronic allergic pathologies and show impaired ATP clearance and thereby suppress basophil and mast cell ATP-induced activities. As such, ENPP3 serves to avoid chronic allergic inflammation, at least in mice (82). To what extent these results extend to human pathology is currently unclear.
ENPP4 promotes platelet aggregation in human plasma through ADP production and activating of platelet P2Y1 and P2Y12 purinergic receptors. Receptor activation leads to calcium-mediated platelet activation, degranulation, and shape changes to extend the growing thrombus (40, 83). Therefore, ENNP4 may be involved in thrombotic disorders, which needs to be confirmed using Enpp4 knockout mice.
ENPP5 is among the least characterized ENPP family members. It shows no activity toward many known ENPP substrates but, instead, cleaves nicotinamide adenine dinucleotide (NAD) (46). This suggests a role for ENPP5 in NAD-based neurotransmission, but this hypothesis awaits validation using Enpp5 KO mice.
ENPP6 is the only GPI-anchored ENPP family member; whether its GPI anchor is susceptible to phospholipase attack, generating soluble ENPP6, remains to be examined. ENPP6 is a choline-specific ENPP (84) that functions as a lysoPLC hydrolyzing classical PLC substrates as well as LPC to form monoacylglycerol and phosphorylcholine. Through the latter activity, ENPP6 regulates choline metabolism in vivo as revealed from analysis of Enpp6 knockout mice. These mice exhibit known choline-deficient phenotypes, such as fatty liver and hypomyelination, of potential relevance for human choline-metabolism disorders (58, 85).
ENPP7 stands out as an alkaline sphingomyelinase that functions in the intestine to cleave SM into ceramide and phosphocholine in a PLC-like manner and has been associated with hepatobiliary diseases. It can be cleaved by pancreatic trypsin from the mucosa and is released to the lumen, which can explain some aspects of its role in SM to ceramide digestion (86), but further research is clearly needed (84). Enpp7 knockout mice show altered lipid digestion and absorption, resulting in the accumulation of 90% of ingested sphingomyelin in colon (84). Reduced ENPP7 activity is detected in colorectal cancer and in familial adenomatous polyposis, suggesting a link between decreased SM metabolism and colon tumorigenesis (60, 61).
Conclusion and outlook
ENPP family members have long been implicated in purinergic signaling, but it is now clear that they are of much wider biological importance, as revealed by their broad substrate repertoire, their structural divergence, and by animal and human data. Structural adaptations in the respective PDE domains underlie the unique substrate specificity of each ENPP; but still the physiological substrates for ENPP3 and ENPP4 remain unclear. While our knowledge on the biochemistry of ENPPs has made major leaps in the last decade, our insights into the ENPP family remain incomplete, not least because some members await generation and analysis of KO mice. Hence, new studies of immediate relevance for human disease will undoubtedly be initiated on this interesting but still incompletely understood phosphodiesterase family in the years to come.
Structural analysis has greatly helped to tidy up the divergence between ENPP family members, highlighting common architectures and mechanisms, and aiding our understanding of how disease-causing mutations affect ENPP1 function. More importantly, the structures have created excellent opportunities to aid drug development, which will continue to evolve. ENPP1 and ENPP2/ATX are currently the most interesting targets for pharmaceutical intervention and many specific and potent inhibitors exist, especially for ENPP2. While the ENPP2 inhibitors have historically attracted more interest, a major opportunity for cancer immunotherapy lies in the recent finding that ENPP1 hydrolyzes cGAMP and thereby can modulate immune cell recruitment. Since ENPP2/ATX has also been implicated in creating an immune-evasive tumor microenvironment, this suggests opportunities for examining these two enzymes jointly as targets to boost the efficacy of cancer immunotherapies.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
R. B. and F. S.-P. investigation; A. P. supervision; R. B. visualization; R. B. writing—original draft; F. S.-P., W. H. M., and A. P. writing—reviewing and editing.
Edited by Wolfgang Peti
References
- 1.Takahashi T., Old L.J., Boyse E.A. Surface alloantigens of plasma cells. J. Exp. Med. 1970;131:1325–1341. doi: 10.1084/jem.131.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Driel I.R., Goding J.W. Plasma cell membrane glycoprotein PC-1. Primary structure deduced from cDNA clones. J. Biol. Chem. 1987;262:4882–4887. [PubMed] [Google Scholar]
- 3.Goding J.W., Terkeltaub R., Maurice M., Deterre P., Sali A., Belli S.I. Ecto-phosphodiesterase/pyrophosphatase of lymphocytes and non-lymphoid cells: Structure and function of the PC-1 family. Immunol. Rev. 1998;161:11–26. doi: 10.1111/j.1600-065x.1998.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 4.Bollen M., Gijsbers R., Ceulemans H., Stalmans W., Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit. Rev. Biochem. Mol. Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
- 5.Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 6.Umezu-Goto M., Kishi Y., Taira A., Hama K., Dohmae N., Takio K., Yamori T., Mills G.B., Inoue K., Aoki J., Arai H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefan C., Jansen S., Bollen M. NPP-type ectophosphodiesterases: Unity in diversity. Trends Biochem. Sci. 2005;30:542–550. doi: 10.1016/j.tibs.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Hausmann J., Perrakis A., Moolenaar W.H. Structure-function relationships of autotaxin, a secreted lysophospholipase D. Adv. Biol. Regul. 2013;53:112–117. doi: 10.1016/j.jbior.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Moolenaar W.H. Lysophospholipids in the limelight: Autotaxin takes center stage. J. Cell Biol. 2002;158:197–199. doi: 10.1083/jcb.200206094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen S., Stefan C., Creemers J.W.M., Waelkens E., Van Eynde A., Stalmans W., Bollen M. Proteolytic maturation and activation of autotaxin (NPP2), a secreted metastasis-enhancing lysophospholipase D. J. Cell Sci. 2005;118:3081–3089. doi: 10.1242/jcs.02438. [DOI] [PubMed] [Google Scholar]
- 11.Greiner-Tollersrud L., Berg T., Stensland H.M.F.R., Evjen G., Greiner-Tollersrud O.K. Bovine brain myelin glycerophosphocholine choline phosphodiesterase is an alkaline lysosphingomyelinase of the eNPP-family, regulated by lysosomal sorting. Neurochem. Res. 2013;38:300–310. doi: 10.1007/s11064-012-0921-z. [DOI] [PubMed] [Google Scholar]
- 12.Zhou A., Huntington J.A., Pannu N.S., Carrell R.W., Read R.J. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 13.Hausmann J., Kamtekar S., Christodoulou E., Day J.E., Wu T., Fulkerson Z., Albers H.M.H.G., Van Meeteren L.A., Houben A.J.S., Van Zeijl L., Jansen S., Andries M., Hall T., Pegg L.E., Benson T.E., et al. Structural basis of substrate discrimination and integrin binding by autotaxin. Nat. Struct. Mol. Biol. 2011;18:198–205. doi: 10.1038/nsmb.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimasu H., Okudaira S., Hama K., Mihara E., Dohmae N., Inoue A., Ishitani R., Takagi J., Aoki J., Nureki O. Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat. Struct. Mol. Biol. 2011;18:205–213. doi: 10.1038/nsmb.1998. [DOI] [PubMed] [Google Scholar]
- 15.Döhler C., Zebisch M., Krinke D., Robitzki A., Sträter N. Crystallization of ectonucleotide phosphodiesterase/pyrophosphatase-3 and orientation of the SMB domains in the full-length ectodomain. Acta Crystallogr. F Struct. Biol. Commun. 2018;74:696–703. doi: 10.1107/S2053230X18011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelik A., Randriamihaja A., Illes K., Nagar B. Structural basis for nucleotide recognition by the ectoenzyme CD203c. FEBS J. 2018;285:2481–2494. doi: 10.1111/febs.14489. [DOI] [PubMed] [Google Scholar]
- 17.Gijsbers R., Ceulemans H., Bollen M. Functional characterization of the non-catalytic ectodomains of the nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem. J. 2003;371:321–330. doi: 10.1042/BJ20021943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen S., Perrakis A., Ulens C., Winkler C., Andries M., Joosten R.P., Van Acker M., Luyten F.P., Moolenaar W.H., Bollen M. Structure of NPP1, an ectonucleotide pyrophosphatase/phosphodiesterase involved in tissue calcification. Structure. 2012;20:1948–1959. doi: 10.1016/j.str.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Pamuklar Z., Federico L., Liu S., Umezu-Goto M., Dong A., Panchatcharam M., Fulerson Z., Berdyshev E., Natarajan V., Fang X., van Meeteren L.A., Moolenaar W.H., Mills G.B., Morris A.J., Smyth S.S. Autotaxin/lysopholipase D and lysophosphatidic acid regulate murine hemostasis and thrombosis. J. Biol. Chem. 2009;284:7385–7394. doi: 10.1074/jbc.M807820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulkerson Z., Wu T., Sunkara M., Kooi C.V., Morris A.J., Smyth S.S. Binding of autotaxin to integrins localizes lysophosphatidic acid production to platelets and mammalian cells. J. Biol. Chem. 2011;286:34654–34663. doi: 10.1074/jbc.M111.276725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyruchaud O., Saier L., Leblanc R. Autotaxin implication in cancer metastasis and autoimunne disorders: Functional implication of binding autotaxin to the cell surface. Cancers (Basel) 2020;12:1–15. doi: 10.3390/cancers12010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen S., Callewaert N., Dewerte I., Andries M., Ceulemans H., Bollen M. An essential oligomannosidic glycan chain in the catalytic domain of autotaxin, a secreted lysophospholipase-D. J. Biol. Chem. 2007;282:11084–11091. doi: 10.1074/jbc.M611503200. [DOI] [PubMed] [Google Scholar]
- 23.Zalatan J.G., Fenn T.D., Brunger A.T., Herschlag D. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: Implications for mechanism and evolution. Biochemistry. 2006;45:9788–9803. doi: 10.1021/bi060847t. [DOI] [PubMed] [Google Scholar]
- 24.Kato K., Nishimasu H., Okudaira S., Mihara E., Ishitani R., Takagi J., Aoki J., Nureki O. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16876–16881. doi: 10.1073/pnas.1208017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosca R., Brannetti B., Schneider T.R. Alignment of protein structures in the presence of domain motions. BMC Bioinformatics. 2008;9:352. doi: 10.1186/1471-2105-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh M., Meiss G., Pingoud A., London R.E., Pedersen L.C. Structural insights into the mechanism of nuclease A, a ββα metal nuclease from Anabaena. J. Biol. Chem. 2005;280:27990–27997. doi: 10.1074/jbc.M501798200. [DOI] [PubMed] [Google Scholar]
- 28.Koyama M., Nishimasu H., Ishitani R., Nureki O. Molecular dynamics simulation of autotaxin: Roles of the nuclease-like domain and the glycan modification. J. Phys. Chem. B. 2012;116:11798–11808. doi: 10.1021/jp303198u. [DOI] [PubMed] [Google Scholar]
- 29.Jansen S., Andries M., Derua R., Waelkens E., Bollen M. Domain interplay mediated by an essential disulfide linkage is critical for the activity and secretion of the metastasis-promoting enzyme autotaxin. J. Biol. Chem. 2009;284:14296–14302. doi: 10.1074/jbc.M900790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E.E., Wyckoff H.W. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 31.Holtz K.M., Kantrowitz E.R. The mechanism of the alkaline phosphatase reaction: Insights from NMR, crystallography and site-specific mutagenesis. FEBS Lett. 1999;462:7–11. doi: 10.1016/s0014-5793(99)01448-9. [DOI] [PubMed] [Google Scholar]
- 32.Kato K., Nishimasu H., Oikawa D., Hirano S., Hirano H., Kasuya G., Ishitani R., Tokunaga F., Nureki O. Structural insights into cGAMP degradation by ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nat. Commun. 2018;9:1–4. doi: 10.1038/s41467-018-06922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Döhler C., Zebisch M., Sträter N. Crystal structure and substrate binding mode of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) Sci. Rep. 2018;8:10874. doi: 10.1038/s41598-018-28814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roston D., Cui Q. Substrate and transition state binding in alkaline phosphatase analyzed by computation of oxygen isotope effects. J. Am. Chem. Soc. 2016;138:11946–11957. doi: 10.1021/jacs.6b07347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts F., Zhu D., Farquharson C., Macrae V.E. ENPP1 in the regulation of mineralization and beyond. Trends Biochem. Sci. 2019;44:616–628. doi: 10.1016/j.tibs.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Terkeltaub R. Physiologic and pathologic functions of the NPP nucleotide pyrophosphatase/phosphodiesterase family focusing on NPP1 in calcification. Purinergic Signal. 2006;2:371–377. doi: 10.1007/s11302-005-5304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Duran M.A., Dhanota N., Chatila W.K., Bettigole S.E., Kwon J., Sriram R.K., Humphries M.P., Salto-Tellez M., James J.A., Hanna M.G., Melms J.C., Vallabhaneni S., Litchfield K., Usaite I., et al. Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 2021;11:1212–1227. doi: 10.1158/2159-8290.CD-20-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carozza J.A., Böhnert V., Nguyen K.C., Skariah G., Shaw K.E., Brown J.A., Rafat M., von Eyben R., Graves E.E., Glenn J.S., Smith M., Li L. Extracellular cGAMP is a cancer-cell-produced immunotransmitter involved in radiation-induced anticancer immunity. Nat. Cancer. 2020;1:184–196. doi: 10.1038/s43018-020-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L., Yin Q., Kuss P., Maliga Z., Millán J.L., Wu H., Mitchison T.J. Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albright R.A., Ornstein D.L., Cao W., Chang W.C., Robert D., Tehan M., Hoyer D., Liu L., Stabach P., Yang G., De La Cruz E.M., Braddock D.T. Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke. J. Biol. Chem. 2014;289:3294–3306. doi: 10.1074/jbc.M113.505867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nitschke Y., Weissen-Plenz G., Terkeltaub R., Rutsch F. Npp1 promotes atherosclerosis in ApoE knockout mice. J. Cell. Mol. Med. 2011;15:2273–2283. doi: 10.1111/j.1582-4934.2011.01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onyedibe K.I., Wang M., Sintim H.O. ENPP1, an old enzyme with new functions, and small molecule inhibitors—a STING in the tale of ENPP1. Molecules. 2019;24:4192. doi: 10.3390/molecules24224192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namasivayam V., Lee S.Y., Müller C.E. The promiscuous ectonucleotidase NPP1: Molecular insights into substrate binding and hydrolysis. Biochim. Biophys. Acta Gen. Subj. 2017;1861:603–614. doi: 10.1016/j.bbagen.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Durnin L., Kurahashi M., Sanders K.M., Mutafova-Yambolieva V.N. Extracellular metabolism of the enteric inhibitory neurotransmitter β-nicotinamide adenine dinucleotide (β-NAD) in the murine colon. J. Physiol. 2020;598:4509–4521. doi: 10.1113/JP280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vollmayer P., Clair T., Goding J.W., Sano K., Servos J., Zimmermann H. Hydrolysis of diadenosine polyphosphates by nucleotide pyrophosphatases/phosphodiesterases. Eur. J. Biochem. 2003;270:2971–2978. doi: 10.1046/j.1432-1033.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 46.Gorelik A., Randriamihaja A., Illes K., Nagar B. A key tyrosine substitution restricts nucleotide hydrolysis by the ectoenzyme NPP5. FEBS J. 2017;284:3718–3726. doi: 10.1111/febs.14266. [DOI] [PubMed] [Google Scholar]
- 47.Perrakis A., Moolenaar W.H. Thematic review series: Lysophospholipids and their receptors: Autotaxin: Structure-function and signaling. J. Lipid Res. 2014;55:1010–1018. doi: 10.1194/jlr.R046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yung Y.C., Stoddard N.C., Chun J. LPA receptor signaling: Pharmacology, physiology, and pathophysiology. J. Lipid Res. 2014;55:1192–1214. doi: 10.1194/jlr.R046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Meeteren L.A., Ruurs P., Christodoulou E., Goding J.W., Takakusa H., Kikuchi K., Perrakis A., Nagano T., Moolenaar W.H. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J. Biol. Chem. 2005;280:21155–21161. doi: 10.1074/jbc.M413183200. [DOI] [PubMed] [Google Scholar]
- 50.Nishimasu H., Ishitani R., Aoki J., Nureki O. A 3D view of autotaxin. Trends Pharmacol. Sci. 2012;33:138–145. doi: 10.1016/j.tips.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Keune W.-J., Hausmann J., Bolier R., Tolenaars D., Kremer A., Heidebrecht T., Joosten R.P., Sunkara M., Morris A.J., Matas-Rico E., Moolenaar W.H., Oude Elferink R.P., Perrakis A. Steroid binding to autotaxin links bile salts and lysophosphatidic acid signalling. Nat. Commun. 2016;7:11248. doi: 10.1038/ncomms11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salgado-Polo F., Fish A., Matsoukas M.T., Heidebrecht T., Keune W.J., Perrakis A. Lysophosphatidic acid produced by autotaxin acts as an allosteric modulator of its catalytic efficiency. J. Biol. Chem. 2018;293:14312–14327. doi: 10.1074/jbc.RA118.004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salgado-Polo F., Perrakis A. The structural binding mode of the four autotaxin inhibitor types that differentially affect catalytic and non-catalytic functions. Cancers (Basel) 2019;11:1577. doi: 10.3390/cancers11101577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaffe E., Katsifa A., Xylourgidis N., Ninou I., Zannikou M., Harokopos V., Foka P., Dimitriadis A., Evangelou K., Moulas A.N., Georgopoulou U., Gorgoulis V.G., Dalekos G.N., Aidinis V. Hepatocyte autotaxin expression promotes liver fibrosis and cancer. Hepatology. 2017;65:1369–1383. doi: 10.1002/hep.28973. [DOI] [PubMed] [Google Scholar]
- 55.Baader M., Bretschneider T., Broermann A., Rippmann J.F., Stierstorfer B., Kuttruff C.A., Mark M. Characterization of the properties of a selective, orally bioavailable autotaxin inhibitor in preclinical models of advanced stages of liver fibrosis. Br. J. Pharmacol. 2018;175:693–707. doi: 10.1111/bph.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bain G., Shannon K.E., Huang F., Darlington J., Goulet L., Prodanovich P., Ma G.L., Santini A.M., Stein A.J., Lonergan D., King C.D., Calderon I., Lai A., Hutchinson J.H., Evans J.F. Selective inhibition of autotaxin is efficacious in mouse models of liver fibrosis. J. Pharmacol. Exp. Ther. 2017;360:1–13. doi: 10.1124/jpet.116.237156. [DOI] [PubMed] [Google Scholar]
- 57.Matas-Rico E., Frijlink E., van der Haar Àvila I., Menegakis A., van Zon M., Morris A.J., Koster J., Salgado-Polo F., de Kivit S., Lança T., Mazzocca A., Johnson Z., Haanen J., Schumacher T.N., Perrakis A., et al. Autotaxin impedes anti-tumor immunity by suppressing chemotaxis and tumor infiltration of CD8+ T cells. Cell Rep. 2021;37:110013. doi: 10.1016/j.celrep.2021.110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morita J., Kano K., Kato K., Takita H., Sakagami H., Yamamoto Y., Mihara E., Ueda H., Sato T., Tokuyama H., Arai H., Asou H., Takagi J., Ishitani R., Nishimasu H., et al. Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase. Sci. Rep. 2016;6:1–7. doi: 10.1038/srep20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morita J., Kato K., Mihara E., Ishitani R., Takagi J., Nishimasu H., Aoki J., Nureki O. Expression, purification, crystallization and preliminary X-ray crystallographic analysis of Enpp6. Acta Crystallogr. F Struct. Biol. Commun. 2014;70:794–799. doi: 10.1107/S2053230X14008929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parrill A.L., Wanjala I.W., Pham T., Baker D.L. Computational identification and experimental characterization of substrate binding determinants of nucleotide pyrophosphatase/phosphodiesterase 7. BMC Biochem. 2011;12:65. doi: 10.1186/1471-2091-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorelik A., Liu F., Illes K., Nagar B. Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition. J. Biol. Chem. 2017;292:7087–7094. doi: 10.1074/jbc.M116.769273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldfine I.D., Maddux B.A., Youngren J.F., Reaven G., Accili D., Trischitta V., Vigneri R., Frittitta L. The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities. Endocr. Rev. 2008;29:62–75. doi: 10.1210/er.2007-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefan C., Wera S., Stalmans W., Bollen M. The inhibition of the insulin receptor by the receptor protein PC-1 is not specific and results from the hydrolysis of ATP. Diabetes. 1996;45:980–983. doi: 10.2337/diab.45.7.980. [DOI] [PubMed] [Google Scholar]
- 64.Zhao T., Liu Z., Zhang D., Liu Y., Yang Y., Zhou D., Chen Z., Yu L., Zhang Z., Feng G., He L., Xu H. The ENPP1 K121Q polymorphism is not associated with type 2 diabetes or obesity in the Chinese Han population. J. Hum. Genet. 2011;56:12–16. doi: 10.1038/jhg.2010.124. [DOI] [PubMed] [Google Scholar]
- 65.Schlipf N.A., Traupe H., Gilaberte Y., Peitsch W.K., Hausser I., Oji V., Schmieder A., Schneider S.W., Demmer P., Rösler B., Fischer J. Association of Cole disease with novel heterozygous mutations in the somatomedin-B domains of the ENPP1 gene: Necessary, but not always sufficient. Br. J. Dermatol. 2016;174:1152–1156. doi: 10.1111/bjd.14328. [DOI] [PubMed] [Google Scholar]
- 66.Albright R.A., Stabach P., Cao W., Kavanagh D., Mullen I., Braddock A.A., Covo M.S., Tehan M., Yang G., Cheng Z., Bouchard K., Yu Z.-X., Thorn S., Wang X., Folta-Stogniew E.J., et al. ENPP1-Fc prevents mortality and vascular calcifications in rodent model of generalized arterial calcification of infancy. Nat. Commun. 2015;6:10006. doi: 10.1038/ncomms10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stabach P.R., Zimmerman K., Adame A., Kavanagh D., Saeui C.T., Agatemor C., Gray S., Cao W., De La Cruz E.M., Yarema K.J., Braddock D.T. Improving the pharmacodynamics and in vivo activity of ENPP1-Fc through protein and glycosylation engineering. Clin. Transl. Sci. 2021;14:362–372. doi: 10.1111/cts.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar M., Lowery R.G. Development of a high-throughput assay to identify inhibitors of ENPP1. SLAS Discov. 2021;26:740–746. doi: 10.1177/2472555220982321. [DOI] [PubMed] [Google Scholar]
- 69.Cogan D., Bakhoum S.F. Re-awakening innate immune signaling in cancer: The development of highly potent ENPP1 inhibitors. Cell Chem. Biol. 2020;27:1327–1328. doi: 10.1016/j.chembiol.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Gamal M.I., Ullah S., Zaraei S.-O., Jalil S., Zaib S., Zaher D.M., Omar H.A., Anbar H.S., Pelletier J., Sévigny J., Iqbal J. Synthesis, biological evaluation, and docking studies of new raloxifene sulfonate or sulfamate derivatives as inhibitors of nucleotide pyrophosphatase/phosphodiesterase. Eur. J. Med. Chem. 2019;181:111560. doi: 10.1016/j.ejmech.2019.07.063. [DOI] [PubMed] [Google Scholar]
- 71.Noguchi K., Herr D., Mutoh T., Chun J. Lysophosphatidic acid (LPA) and its receptors. Curr. Opin. Pharmacol. 2009;9:15–23. doi: 10.1016/j.coph.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Stracke M.L., Krutzsch H.C., Unsworth E.J., Arestad A., Cioce V., Schiffmann E., Liotta L.A. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 1992;267:2524–2529. [PubMed] [Google Scholar]
- 73.Houben A.J.S., Van Wijk X.M.R., Van Meeteren L.A., Van Zeijl L., Van De Westerlo E.M.A., Hausmann J., Fish A., Perrakis A., Van Kuppevelt T.H., Moolenaar W.H. The polybasic insertion in autotaxin α confers specific binding to heparin and cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2013;288:510–519. doi: 10.1074/jbc.M112.358416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moolenaar W.H., Houben A.J.S., Lee S.-J., van Meeteren L.A. Autotaxin in embryonic development. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2013;1831:13–19. doi: 10.1016/j.bbalip.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 75.van Meeteren L.A., Ruurs P., Stortelers C., Bouwman P., van Rooijen M.A., Pradère J.P., Pettit T.R., Wakelam M.J.O., Saulnier-Blache J.S., Mummery C.L., Moolenaar W.H., Jonkers J. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol. Cell. Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ninou I., Magkrioti C., Aidinis V. Autotaxin in pathophysiology and pulmonary fibrosis. Front. Med. 2018;5:1–11. doi: 10.3389/fmed.2018.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamazaki T., Joshita S., Umemura T., Usami Y., Sugiura A., Fujimori N., Shibata S., Ichikawa Y., Komatsu M., Matsumoto A., Igarashi K., Tanaka E. Association of serum autotaxin levels with liver fibrosis in patients with chronic hepatitis C. Sci. Rep. 2017;7:1–10. doi: 10.1038/srep46705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Magkrioti C., Galaris A., Kanellopoulou P., Stylianaki E.A., Kaffe E., Aidinis V. Autotaxin and chronic inflammatory diseases. J. Autoimmun. 2019;104:102327. doi: 10.1016/j.jaut.2019.102327. [DOI] [PubMed] [Google Scholar]
- 79.Birgbauer E. Lysophosphatidic acid signalling in nervous system development and function. Neuromolecular Med. 2021;23:68–85. doi: 10.1007/s12017-020-08630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Auciello F.R., Bulusu V., Oon C., Tait-Mulder J., Berry M., Bhattacharyya S., Tumanov S., Allen-Petersen B.L., Link J., Kendsersky N.D., Vringer E., Schug M., Novo D., Hwang R.F., Evans R.M., et al. A Stromal Lysolipid-Autotaxin Signaling Axis Promotes Pancreatic Tumor Progression. Cancer Discov. 2019;9:617–627. doi: 10.1158/2159-8290.CD-18-1212. PMID: 30837243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yung Y.C., Stoddard N.C., Mirendil H., Chun J. Lysophosphatidic acid signaling in the nervous system. Neuron. 2015;85:669–682. doi: 10.1016/j.neuron.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsai S.H., Kinoshita M., Kusu T., Kayama H., Okumura R., Ikeda K., Shimada Y., Takeda A., Yoshikawa S., Obata-Ninomiya K., Kurashima Y., Sato S., Umemoto E., Kiyono H., Karasuyama H., et al. The ectoenzyme E-NPP3 negatively regulates ATP-dependent chronic allergic responses by basophils and mast cells. Immunity. 2015;42:279–293. doi: 10.1016/j.immuni.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 83.Gachet C. P2Y12 receptors in platelets and other hematopoietic and non-hematopoietic cells. Purinergic Signal. 2012;8:609–619. doi: 10.1007/s11302-012-9303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duan R.D. Alkaline sphingomyelinase (NPP7) in hepatobiliary diseases: A field that needs to be closely studied. World J. Hepatol. 2018;10:246–253. doi: 10.4254/wjh.v10.i2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stewart A.J., Leong D.T.K., Farquharson C. PLA2 and ENPP6 may act in concert to generate phosphocholine from the matrix vesicle membrane during skeletal mineralization. FASEB J. 2018;32:20–25. doi: 10.1096/fj.201700521R. [DOI] [PubMed] [Google Scholar]
- 86.Wu J., Liu F., Nilsson Å., Duan R.D. Pancreatic trypsin cleaves intestinal alkaline sphingomyelinase from mucosa and enhances the sphingomyelinase activity. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:967–973. doi: 10.1152/ajpgi.00190.2004. [DOI] [PubMed] [Google Scholar]
- 87.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]