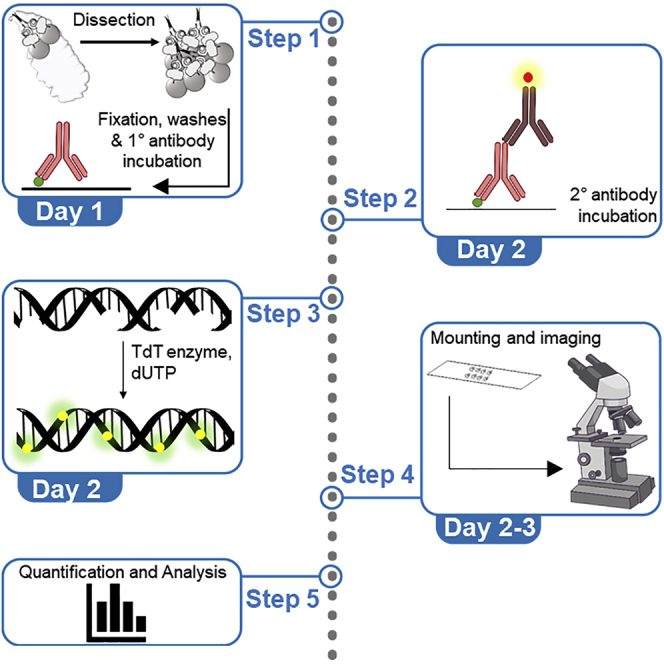
Summary
Cell death maintains tissue homeostasis by eliminating dispensable cells. Misregulation of cell death is seen in diseases like cancer, neurodegeneration, etc. Therefore, cell death assays like TUNEL have become reliable tools, where fragmented DNA of dying cells gets fluorescently labeled and can be detected under microscope. We used TUNEL assay in Drosophila melanogaster third-instar larval eye-antennal imaginal discs to label and quantify cell death. This assay is sensitive to detect DNA fragmentation, an important event, during apoptosis in retinal neurons.
For complete details on the use and execution of this profile, please refer to Wang et al. (1999), Tare et al. (2011), and Mehta et al. (2021).
Subject areas: Cell Biology, Microscopy, Model Organisms, Neuroscience, Molecular/Chemical Probes
Graphical abstract
Highlights
-
•
Combines immunohistochemistry and TUNEL assay to measure cell death
-
•
Marks the fragmented DNA to quantify dying cells in specific regions of the tissue
-
•
This protocol can be adapted to assay cell death in other tissues as well
Cell death maintains tissue homeostasis by eliminating dispensable cells. Misregulation of cell death is seen in diseases like cancer, neurodegeneration, etc. Therefore, cell death assays like TUNEL have become reliable tools, where fragmented DNA of dying cells gets fluorescently labeled and can be detected under microscope. We used TUNEL assay in Drosophila melanogaster third-instar larval eye-antennal imaginal discs to label and quantify cell death. This assay is sensitive to detect DNA fragmentation, an important event, during apoptosis in retinal neurons.
Before you begin
-
1.
Prepare fresh buffers, fixatives and antibody solutions that would be required for the protocol.
-
2.
Set up all experiments synchronously to ensure enough number of larvae are available for all experimental groups. This is to ensure least variability across groups.
Note: This protocol describes the use of Terminal deoxynucleotidyl transferase (TdT) deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) assay. Here we used Drosophila melanogaster’s third instar larvae eye-antennal imaginal discs for TUNEL staining (Tare et al., 2011, 2016; Raj et al., 2020; Irwin et al., 2020; Gogia et al., 2020; Sarkar et al., 2018; Steffensmeier et al., 2013; Woodfield et al., 2013; Zhu et al., 2017). TUNEL staining can be used for other imaginal discs and tissue as well.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat-Elav-7E8A10 anti-elav (dilution 1:100) | Developmental Studies Hybridoma Bank | Antibody Registry ID: AB_528218 |
| Goat anti-rat IgG conjugated with Cy5 (dilution 1:250)s | The Jackson Laboratory | Cat# 112-175-143 |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium Chloride | Sigma-Aldrich | Product# S7653 |
| Potassium Chloride | Sigma-Aldrich | Product# P9333 |
| Sodium Phosphate, Dibasic | Sigma-Aldrich | Product# S7907 |
| Potassium Phosphate, Monobasic | Sigma-Aldrich | Product# P5655 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| Vectashield | Vector labs | Cat# H1000 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| Normal Donkey Serum | Fisher scientific | Cat# 5058837 |
| Sodium citrate | Thermo Fisher Scientific | Cat# S279 |
| Critical commercial assays | ||
| TUNEL dilution buffer | Roche Diagnostics | Cat# 11966006001 |
| In Situ Cell Death Detection Kit, TMR red | Roche Diagnostics | Cat# 12156792910 |
| Experimental models: Organisms/strains | ||
|
D. melanogaster Canton-S |
Bloomington Drosophila Stock Center | BDSC: 64349; FlyBase: FBsn0000274 |
|
D. melanogaster ey-Gal4 (III) |
(Hazelett et al., 1998) | N/A |
|
D. melanogaster L2/CyO; ey-Gal4/ey-Gal4 |
(Singh et al., 2005, 2006; Singh and Choi, 2003) | N/A |
|
D. melanogaster GMR-Gal4 w[∗]; P{w[+mC]=longGMR-Gal4}2 |
Bloomington Drosophila Stock Center | BDSC: 8605; FlyBase: FBti0058798 |
|
D. melanogaster GMR-hid, GMR-Gal4 |
A gift from Dr. Andreas Bergmann | N/A |
|
D. melanogaster Viropana 4 (III) |
(Mehta et al., 2021) | N/A |
| Software and algorithms | ||
| ImageJ software | National Institutes of Health | http://rsb.info.nih.gov/ij/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Materials and equipment
1× Phosphate Buffered Saline (PBS)
| Reagent | Final concentration |
|---|---|
| NaCl | 137 mM |
| KCl | 2.7 mM |
| Na2HPO4 | 4.3 mM |
| KH2PO4 | 1.5 mM |
Note: pH: 7.4, store in 4°C for up to a month. See troubleshooting 1.
Fixative solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 16% Paraformaldehyde | 4% | 50 μL |
| 1× PBS | n/a | 150 μL |
| Total | n/a | 200 μL |
Note: Prepare fresh, store in 4°C. See troubleshooting 1.
1× PBS-Triton-X-100
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× PBS | n/a | 500 mL |
| Triton X 100 | 0.2% | 1 mL |
Note: Store in 4°C for up to a month.
1× PBST-NDS
| Reagent | Final concentration | Amount |
|---|---|---|
| Normal Donkey Serum (NDS) | 10% | 1 mL |
| 1× PBST | n/a | 9 mL |
| Total | n/a | 10 mL |
Note: Store in −20°C in small aliquots to avoid repeated freeze-thaw cycles.
100 mM Sodium Citrate Stock Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium citrate | 100 mM | 0.294 g |
| ddH2O | n/a | 10 mL |
Note: Prepare fresh when needed.
10% Triton X-100 Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| TritonX-100 | 10% | 100 μL |
| ddH2O | n/a | 900 μL |
Note: See troubleshooting 2.
TUNEL labeling solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Labeling Solution (in situ cell death detection TMR red kit, Roche) | n/a | 25 μL |
| Dilution buffer | n/a | 25 μL |
| Enzyme solution | n/a | 5 μL |
| Total | n/a | 55 μL |
Note: Prepare fresh when needed. See troubleshooting 2.
Step-by-step method details
Dissection, fixation and primary antibody incubation – Day 1
Timing: 1–2 h
-
1.Dissection
-
a.Dissect eye-antennal imaginal discs along with the brain lobes (Figure 1) from wandering third instar larvae (Singh et al., 2002).
-
b.Dissection is performed in cold 1× Phosphate Buffered Saline (PBS) for no longer than 20 min at room temperature (RT) (21°C–23°C).
-
a.
Note: Tissue is prone to disintegration if left for longer periods without fixation. It is crucial to dissect as quickly as possible and transfer the tissue into the fixative solution. See troubleshooting 1.
-
2.
Fix tissue in freshly prepared 4% Paraformaldehyde (diluted in 1× PBS) for 20 min at RT.
Note: Fixation is a critical step to maintain the morphology and antigenicity within the cells. Longer exposures to fixatives can lead to improper or no staining. See troubleshooting 1.
-
3.
Wash the tissue in 1× PBS once followed by washes in PBST (1×PBS+0.2% Triton-X-100) three times for 10 min each.
-
4.
Prepare primary antibody (Rat anti-Elav) at a 1:100 dilution in 10% Normal Donkey Serum (NDS) in 1XPBST (PBST-NDS) solution.
-
5.
Incubate in primary antibody overnight (∼8–16 h) at 4°C (Figure 1).
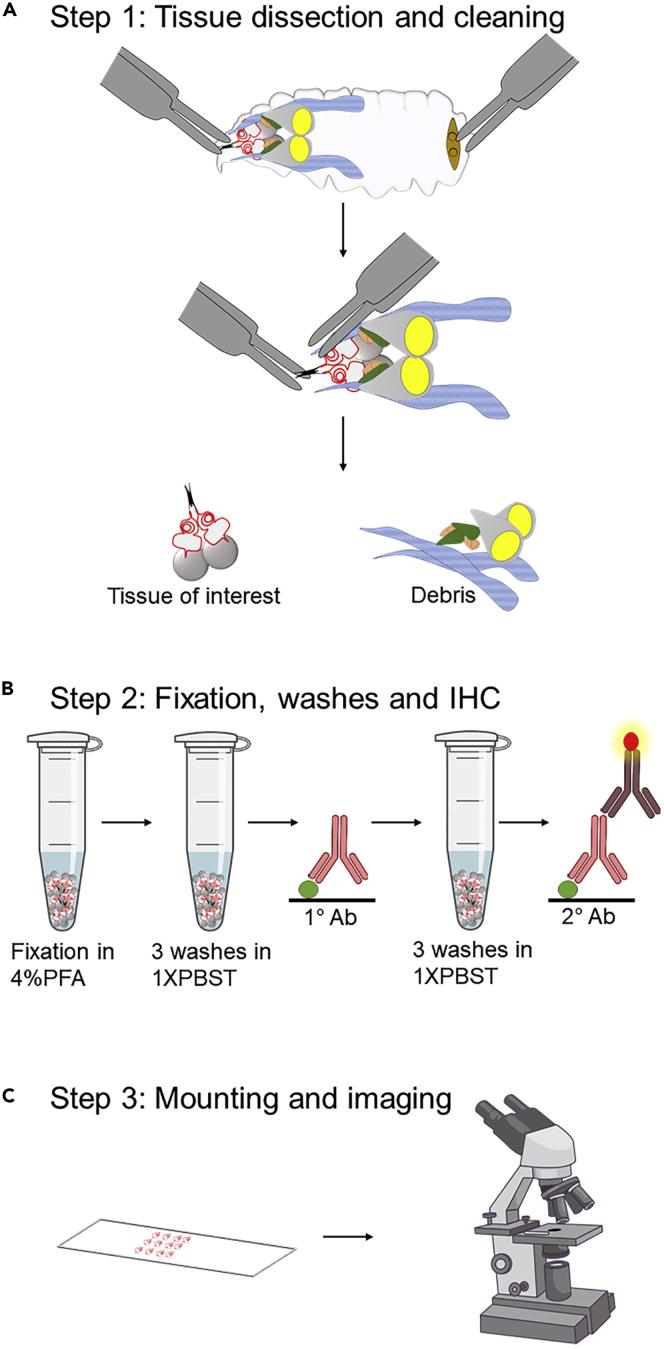
Figure 1.
Step-by-step schematic representation of dissection and mounting
(A) Desired tissue is dissected and cleaned in 1× PBS.
(B) Fixation is done in 4% paraformaldehyde for 20 min and the tissue is washed 3 times for 10 min each before incubating overnight (∼8–16 h) in primary antibody. The tissue is then washed 3 times for 10 min each and incubated in secondary antibody for 2 h followed by washes. In this study we processed the tissue after secondary for TUNEL assay.
(C) The washed tissue was then mounted onto a slide and visualized under confocal microscope.
Secondary antibody incubation, TUNEL labeling – Day 2
Timing: 6–8 h
-
6.
Wash the tissue in 1× PBST, thrice for 10 min each.
-
7.
Prepare secondary antibody (anti-Rat Cy5) at a dilution of 1:300 in 1× PBST-NDS.
Note: Do not use Cy3 secondary antibody as that will overlap with the TUNEL signal in the same channel.
-
8.
Incubate the tissue in a secondary antibody for 2 h in the dark at RT.
-
9.
Wash the tissue in 1× PBST thrice for 10 min each.
-
10.
Incubate in freshly prepared 495 μL of 100 mM Sodium Citrate stock solution + 5 μL of 10% Triton X-100 for 30 min in the dark at 65°C.
Note: Prepare freshly the Sodium Citrate and Triton X-100 solution with 500 μL per tube every time.
-
11.
Wash the tissue in 1× PBST, thrice for 10 min each.
-
12.
Incubate the tissue in ∼30 μL of TUNEL-Dilution Buffer (Roche) for 10 min in the dark at RT.
-
13.
Incubate the tissue in a TUNEL labeling solution for 2 h at 37°C.
Note: For our experiments, we have used the In Situ Cell Death Detection Kit, TMR red. Alternatively, the fluorescein kits can also be used. In case of using GFP or mCherry reporters in the crosses, only TMR or fluorescein kits can be used respectively. See troubleshooting 2.
-
14.
Wash the tissue in 1× PBST, thrice for 10 min each.
Note: 1XPBST helps in effective washing and removal of excess antibodies. See troubleshooting 3.
Mounting
Timing: 20–30 min
-
15.
Transfer the tissue onto a clean glass slide and carefully remove the brain lobes and detach the eye-antennal imaginal discs from the mouth hooks (Figure 1).
-
16.
Arrange the eye-antennal discs on the slide and remove excess PBST.
-
17.
Mount in Vectashield mounting medium. Store the slides in a slide holder that can be stored at −20°C.
Imaging and analysis
Timing: 2–3 h
-
18.
Image the slides under a confocal microscope using appropriate channels.
-
19.
All images were taken in 20 X magnification. The confocal parameters were set to gain: 1, offset: 3%. The intensities of all the lasers and photomultiplier tube (PMT) detectors were optimized and set to avoid excessive or weak signal. The Hi–Lo feature in the software was used to assess the intensity saturation of the lasers hitting the sample.
Note: See troubleshooting 4 and troubleshooting 5
-
20.
TUNEL labeling will be visible in the Cy3 channel if you use this In Situ Cell Death Detection, TMR red Kit. See troubleshooting 4.
-
21.
Save all images in the original and other appropriate formats for later analysis.
-
22.
The .tif images are used for quantification of TUNEL positive nuclei.
-
23.
Perform statistical analysis using MS Excel or GraphPad Prism and plot a graph.
Expected outcomes
TUNEL assay is useful in labeling and detecting fragmented DNA that is one of the events during apoptosis, a type of programmed cell death (White et al., 1994, Mccall and Peterson, 2004). In this assay, a deoxynucleotidyl transferase (TdT) is used to transfer fluorescently labeled nucleotides to the free 3′-OH DNA ends. These nucleotides are then detected by fluorescence microscopy or confocal microscopy (Singh and Gopinathan, 1998). We have optimized the volume of TUNEL labeling solution and the concentration of Triton-X-100 for permeabilization. We have also used this assay in combination with immunohistochemistry (IHC) to study protein localization in Drosophila eye-antennal imaginal discs as it allows simultaneous visualization of other markers along with assaying cell death. We have used a pan-neuronal marker, Elav to stain the nuclei of the photoreceptor neurons in the eye-antennal imaginal discs and performed TUNEL labeling to mark the dying cell nuclei (Tare et al., 2011, 2016; Steffensmeier et al., 2013; Singh et al., 2006; Sarkar et al., 2018; Raj et al., 2020; Moran et al., 2013; Irwin et al., 2020; Gogia et al., 2020; Yeates et al., 2020). Imaging these discs will show signals / puncta that indicate cell death i.e., exposed DNA or fragmented DNA.
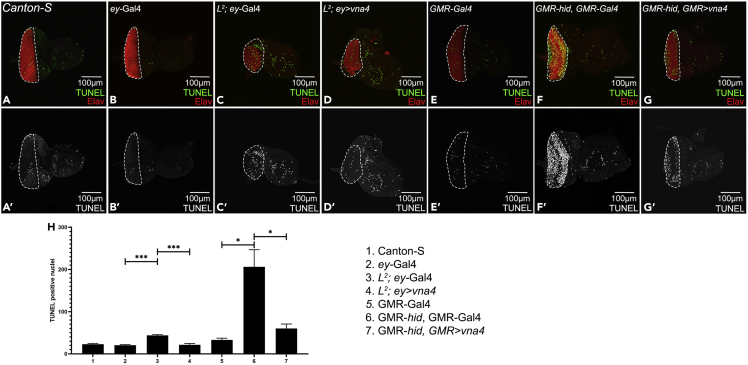
In wild-type Canton-S discs, TUNEL staining marks a few nuclei indicating some basal level of developmental cell death that might be happening (Figures 2A, A′). The ey-Gal4 and GMR-Gal4 controls used in this study also exhibit minimal cell death (Figures 2C, C′, F, F′, H). In contrast, the L2 mutant eye discs exhibit the loss of ventral eye phenotype (Singh and Choi, 2003; Singh et al., 2005) due to increased cell death (Singh et al., 2006) as evident by increased TUNEL labeling (Figures 2C, C′, H) when compared to ey-Gal4 control (Figures 2C, C′, H). In the positive genetic control, GMR-hid, GMR-Gal4, misexpression of hid initiates apoptosis (Grether et al., 1995) and thus exhibits significantly higher number of TUNEL positive nuclei (Figures 2E, E′, H) compared to GMR-Gal4 control (Figures 2F, F′, H). We have recently shown that misexpression of viropana 4, a novel newt gene, rescues the GMR-hid, GMR-Gal4 and L2 mutant phenotype by downregulating the cell death as evident from TUNEL assay (Figures 2D, D′, G, G′, H) and Dcp1 (Death caspase-1) staining (Mehta et al., 2021). We have used the same example to explain the TUNEL protocol and quantification (Figure 2).
Figure 2.
TUNEL assay labels free 3′-OH DNA ends
(A–G) Eye-antennal imaginal discs stained for Elav (red) and showing TUNEL positive nuclei (green), and (A′–G′) split channel for TUNEL in (A, A′) Canton-S, (B, B′) ey-Gal4, (C, C′) L2; ey-Gal4, (D, D′) L2; ey>vna4, (E, E′) GMR-Gal4, (F, F′) GMR-hid, GMR-Gal4, and (G, G′) GMR-hid, GMR>vna4. (H) Quantification of the TUNEL positive nuclei (green) in the fly retina (marked by dotted line) shows that the average number of dying cells were significantly reduced when vna4, a newt gene is misexpressed in the GMR-hid, GMR-Gal4 or L2-mutant background as compared to GMR-hid, GMR-Gal4 or L2-mutant only. The orientation of all imaginal discs is dorsal up and posterior left. The magnification of all discs is 20×. A total of five eye-antennal imaginal discs (n = 5) was analyzed for each genotype. Error bars show standard deviation (mean ± SEM), and symbols above the error bar signify as ∗∗∗ p-value <0.001, ∗∗ p-value <0.01, ∗ p-value <0.05, and ns, not significant, p-value >0.05 respectively.
Quantification and statistical analysis
The TUNEL assay marks damaged and exposed 3′-OH ends of DNA in cells. Imaging the nuclei of such cells reveals clear signals and makes it possible to accurately quantify them. For quantification of TUNEL positive nuclei, the .tif image was opened in the Fiji/ImageJ software and a region of interest was drawn. Our interest was to observe and quantify the cell death occurring in the eye region. The posterior region of the third instar eye-antennal imaginal disc consists of photoreceptors that form the adult compound eye. During development, there is a basal level of cell death to remove excess cells. Therefore, the range of cell death varies in the eyes of different genotypes.
Note: The L2 mutant flies used in this study shows a loss of ventral eye phenotype (smaller area) and the GMR-hid, GMR-Gal4 flies that serve as a positive control have a much larger eye area. We can also normalize the data by dividing the TUNEL positive nuclei by area of the region of interest. This helps in considering how much cell death is happening (per unit area) in eyes of each genotype. Alternatively, the number of dead nuclei can be normalized by dividing with the total number of nuclei as well.
For quantification of TUNEL positive nuclei, the .tif image was opened in the Fiji/ImageJ software and a region of interest was drawn. The split channel function was used and the number of TUNEL positive nuclei were counted manually in at least 5 discs of each genotype (Singh et al., 2005; Gogia et al., 2020; Irwin et al., 2020; Raj et al., 2020; Yeates et al., 2020). Average was calculated and used to plot graphs. Statistical analysis was performed using Student’s t-test since we have compared L2; ey-Gal4 and L2; ey>vna4 with ey-Gal4 and GMR-hid, GMR-Gal4 and GMR-hid, GMR>vna4 with GMR-Gal4. p-value<0.05 was considered statistically significant.
Limitations
TUNEL assay labels all free 3′-OH ends on the DNA which could also be due to other kinds of cellular insults or cellular processes. TUNEL assay cannot differentiate between different modes of cell death. Finally, this protocol shows the number of TUNEL positive nuclei and not the extent of DNA damage. Another limitation of this protocol is that the incubation steps at higher temperatures can also damage some cells.
Troubleshooting
Problem 1
Degradation of tissue or soft tissue during mounting (steps 1 and 2)
Potential solution
This could be due to improper buffer pH or even insufficient fixing.
Make sure that the pH of 1× PBS is maintained at 7.4.
Fix the tissue properly by 4% paraformaldehyde. Make sure the tissue is submerged in the fixative.
Problem 2
Weaker signal or no staining (step 13).
Potential solution
This problem could be due to impermeability of the TUNEL labeling solution in the cell. Permeabilize cells with 10% Triton X-100 and incubate at 65°C. Always ensure there are no pipetting errors while dispensing TritonX-100 for making PBST solution. TritonX-100 is viscous in nature.
Make sure TritonX-100 is fully dissolved in PBST.
The other possible reason could be not enough labeling solution and enzyme solution in the sample. Optimize their concentration as per the tissue sample used.
Incubate the samples with labeling mix for a longer time or incubate at higher temperature. Note that during the staining process, samples should not dry out.
TUNEL signal fades within a short period after mounting, so the slide must be imaged immediately. Also, the slides should be stored in dark in the slide folder.
Always make sure to run the positive sample to avoid the confusion if the staining didn’t work in the experimental sample.
Problem 3
Higher background, reduced signal to noise ratio (step 14)
Potential solution
This problem could be due to excessive staining of the sample.
Washing step is critical. Please make sure to give proper washes in between every step as per the protocol.
Concentration of the labeling mix is reduced by diluting with TUNEL dilution buffer. This step needs to be optimized by changing the concentration of the labeling mix.
Problem 4
Overlap of dye and fluorophore (step 20)
Potential solution
In order to study protein localization, make sure not to use Cy3 fluorophore in the secondary antibody. Otherwise, it will overlap with TMR red dye wavelength. The Roche cell death kit used in the study has TMR-red dye for TUNEL detection.
Problem 5
Nonspecific labeling (step 19).
Potential solution
Concentration of enzyme solution in the TUNEL labeling solution can be optimized or diluted further with TUNEL dilution buffer.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Amit Singh, Email ID: asingh1@udayton.edu .
Materials availability
This protocol does not report any newly generated material.
Acknowledgments
We thank Bloomington Drosophila Stock Center (BDSC) for Drosophila strains and the Developmental Studies Hybridoma Bank (DSHB) for antibodies. Confocal microscopy was supported by the core facility at University of Dayton. A.S. is supported by 1R15GM124654-01, 1RO1EY032959-01 from NIH, Schuellein Chair Endowment Fund, and STEM Catalyst Grant from the University of Dayton.
Author contributions
A.V.C. and P.D. performed the experiments; A.S.M. and A.S. provided inputs in data analysis; and A.V.C. and A.S. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
All datasets generated or analyzed during this study are included in the protocol. This paper does not report any original code.
References
- Gogia N., Sarkar A., Mehta A.S., Ramesh N., Deshpande P., Kango-Singh M., Pandey U.B., Singh A. Inactivation of Hippo and cJun-N-terminal Kinase (JNK) signaling mitigate FUS mediated neurodegeneration in vivo. Neurobiol. Dis. 2020;140:104837. doi: 10.1016/j.nbd.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M.E., Abrams J.M., Agapite J., White K., Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hazelett D.J., Bourouis M., Walldorf U., Treisman J.E. Decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Irwin M., Tare M., Singh A., Puli O.R., Gogia N., Riccetti M., Deshpande P., Kango-Singh M. A positive feedback loop of hippo- and c-jun-amino-terminal kinase signaling pathways regulates amyloid-beta-mediated neurodegeneration. Front. Cell Dev. Biol. 2020;8:117. doi: 10.3389/fcell.2020.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccall K., Peterson J.S. Detection of apoptosis in Drosophila. Methods Mol. Biol. 2004;282:191–205. doi: 10.1385/1-59259-812-9:191. [DOI] [PubMed] [Google Scholar]
- Mehta A.S., Deshpande P., Chimata A.V., Tsonis P.A., Singh A. Newt regeneration genes regulate Wingless signaling to restore patterning in Drosophila eye. iScience. 2021;24:103166. doi: 10.1016/j.isci.2021.103166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran M.T., Tare M., Kango-Singh M., Singh A. Homeotic Gene teashirt (tsh) has a neuroprotective function in amyloid-beta 42 mediated neurodegeneration. PLoS ONE. 2013;8:e80829. doi: 10.1371/journal.pone.0080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Chimata A.V., Singh A. Motif 1 binding protein suppresses wingless to promote eye fate in Drosophila. Sci. Rep. 2020;10:17221. doi: 10.1038/s41598-020-73891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Gogia N., Glenn N., Singh A., Jones G., Powers N., Srivastava A., Kango-Singh M. A soy protein Lunasin can ameliorate amyloid-beta 42 mediated neurodegeneration in Drosophila eye. Sci. Rep. 2018;8:13545. doi: 10.1038/s41598-018-31787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Chan J., Chern J.J., Choi K.W. Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics. 2005;171:169–183. doi: 10.1534/genetics.105.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Choi K.W. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351–6360. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Singh A., Gopinathan K.P. Confocal microscopy: a powerful technique for biological research. Curr. Sci. 1998;74:841–851. [Google Scholar]
- Singh A., Kango-Singh M., Sun Y.H. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Singh A., Shi X., Choi K.W. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development. 2006;133:4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed] [Google Scholar]
- Steffensmeier A.M., Tare M., Puli O.R., Modi R., Nainaparampil J., Kango-Singh M., Singh A. Novel neuroprotective function of apical-basal polarity gene crumbs in amyloid beta 42 (abeta42) mediated neurodegeneration. PLoS ONE. 2013;8:e78717. doi: 10.1371/journal.pone.0078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M., Modi R.M., Nainaparampil J.J., Puli O.R., Bedi S., Fernandez-Funez P., Kango-Singh M., Singh A. Activation of JNK signaling mediates amyloid-ss-dependent cell death. PLoS ONE. 2011;6:e24361. doi: 10.1371/journal.pone.0024361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare M., Sarkar A., Bedi S., Kango-Singh M., Singh A. Cullin-4 regulates Wingless and JNK signaling-mediated cell death in the Drosophila eye. Cell Death Dis. 2016;7:e2566. doi: 10.1038/cddis.2016.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.L., Hawkins C.J., Yoo S.J., Muller H.A., Hay B.A. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- White K., Grether M.E., Abrams J.M., Young L., Farrell K., Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Woodfield S.E., Graves H.K., Hernandez J.A., Bergmann A. De-regulation of JNK and JAK/STAT signaling in ESCRT-II mutant tissues cooperatively contributes to neoplastic tumorigenesis. PLoS ONE. 2013;8:e56021. doi: 10.1371/journal.pone.0056021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates C.J., Sarkar A., Deshpande P., Kango-Singh M., Singh A. A two-clone approach to study signaling interactions among neuronal cells in a pre-clinical Alzheimer's disease model. iScience. 2020;23:101823. doi: 10.1016/j.isci.2020.101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Palliyil S., Ran C., Kumar J.P. Drosophila Pax6 promotes development of the entire eye-antennal disc, thereby ensuring proper adult head formation. Proc. Natl. Acad. Sci. U S A. 2017;114:5846–5853. doi: 10.1073/pnas.1610614114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated or analyzed during this study are included in the protocol. This paper does not report any original code.