Summary
This protocol details the induction of inflammation and acute myelin degeneration in larval zebrafish with a duration of <10 days. We describe the use of this model to screen the effects of candidate compounds on inflammation, followed by RNA isolation, and qPCR-based quantification of gene expression. We then outline the steps for bioinformatic analysis of the mechanisms associated with the compounds. This protocol can be used in combination with drugs and genetic targeting to identify pathways that contribute to neurodegeneration.
For complete details on the use and execution of this profile, please refer to Wheeler et al. (2019).
Subject areas: Immunology, Model Organisms, Molecular Biology, Neuroscience
Graphical abstract

Highlights
-
•
Protocol for rapid screening of candidate compounds on inflammation in zebrafish
-
•
Protocol enables the identification of transcriptional changes
-
•
Bioinformatic analysis of the pathways associated with drug candidates
This protocol details the induction of inflammation and acute myelin degeneration in larval zebrafish with a duration of <10 days. We describe the use of this model to screen the effects of candidate compounds on inflammation, followed by RNA isolation, and qPCR-based quantification of gene expression. We then outline the steps for bioinformatic analysis of the mechanisms associated with the compounds. This protocol can be used in combination with drugs and genetic targeting to identify pathways that contribute to neurodegeneration.
Before you begin
This protocol describes a zebrafish model of systemic inflammation and whole-animal qPCR. However, we have also used this protocol for isolation of specific cell types, such as astrocyte-related radial glia (Bernardos and Raymond, 2006; Linnerbauer et al., 2020; Sanmarco et al., 2021; Wheeler and Quintana, 2019), by increasing the number of zebrafish used for the experiment.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Cuprizone (Bis(cyclohexanone)oxaldihydrazone) | Sigma-Aldrich | #C9012 |
| LPS-EK | InvivoGen | #tlrl-eklps |
| TRIzol LS | Thermo Fisher Scientific | #10296010 |
| PBS | Thermo Fisher Scientific | #14190250 |
| 0.5M EDTA | Amresco | #E177-100ML |
| Bovine serum albumin (BSA) | MilliporeSigma | #A3294 |
| Critical commercial assays | ||
| Direct-zol RNA kit | Zymo Research | #R2050 |
| High-Capacity cDNA Reverse Transcription Kit | Life Technologies | #4368813 |
| Taqman Fast Universal PCR Master Mix |
Life Technologies | #4367846 |
| Zebrafish: actb1 probe | Life Technologies | #Dr03432610_m1 |
| Zebrafish: ccl20 probe | Life Technologies | #Dr03431608_m1 |
| Zebrafish: il1b probe | Life Technologies | #Dr03114369_m1 |
| Zebrafish: il10 probe | Life Technologies | #Dr03103209_m1 |
| Zebrafish: il17a/f1 probe | Life Technologies | #Dr03096843_g1 |
| Zebrafish: mpz probe | Life Technologies | #Dr03131914_m1 |
| Zebrafish: nos2a probe | Life Technologies | #Dr03124753_m1 |
| Zebrafish: actb1 probe | Life Technologies | #Dr03432610_m1 |
| Experimental models: Organisms/strains | ||
| Zebrafish: Danio rerio/AB | Wheeler et al., (2019) | ZFIN: ZDB-GENO-960809-7; RRID:ZIRC_ZL1438 |
| Zebrafish: Danio rerio/ Tg(gfap::egfp) | Bernardos and Raymond (2006) | ZFIN: ZDB-TGCONSTRCT-070117-154 |
Materials and equipment
E3 water
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 5 mM | 14.61 mg |
| KCl | 170 μM | 0.634 mg |
| CaCl2 | 433 μM | 2.403 mg |
| MgSO4 | 675 μM | 4.062 mg |
| HEPES | 80 μM | 0.953 mg |
| ddH2O | n/a | To 50 mL |
| Total | n/a | 50 mL |
Store at 20°C for >1 month.
Cuprizone stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Cuprizone | 40 μg/mL | 0.02 g |
| E3 water | n/a | 500 mL |
| Total | n/a | 500 mL |
Note: Prepare cuprizone solution fresh immediately before use. Dissolve cuprizone in zebrafish system water by warming the solution to 42°C.
CRITICAL: Cuprizone is a hazardous chemical which can cause irritation, brain and liver damage. Handle in a ventilated fume hood. Use gloves, a lab coat, and goggles when handling.
LPS stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| LPS-EK | 1 mg/mL | 5 mg |
| Endotoxin free water | n/a | 5 mL |
| Total | n/a | 5 mL |
Note: Prepare LPS stock solution in endotoxin free water. Aliquots can be stored at 4°C for 1 month or −20°C for 6 months. Avoid repeated freeze-thaw cycles.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA 500 mM | 500 μM | 500 μL |
| BSA | 0.5% | 2.5 g |
| PBS | n/a | to 500 mL |
| Total | n/a | 500 mL |
Store at 4°C for >1 month.
Cuprizone + LPS treatment solution
| Reagent | Final concentration | Amount |
|---|---|---|
| LPS-EK stock solution | 150 μg/mL | 900 μL |
| Cuprizone stock solution | 12.5 μg/mL | 1875 μL |
| Zebrafish system water | n/a | 3225 μL |
| Total | n/a | 6000 μL |
Store at 20°C for 1 day.
| Target gene | qPCR probe ID |
|---|---|
| ccl20 | Dr03431608_m1 |
| il1b | Dr03114369_m1 |
| il10 | Dr03103209_m1 |
| il17a/f1 | Dr03096843_g1 |
| mpz | Dr03131914_m1 |
| nos2a | Dr03124753_m1 |
| actb1 | Dr03432610_m1 |
Step-by-step method details
Zebrafish breeding
Timing: 2 h
In this step, we describe the parameters used to collect zebrafish for these experiments
-
1.
Prepare the E3 water (see “materials and equipment” for detailed recipe).
-
2.Breed the zebrafish to acquire the embryos and larvae for the experiment. (If the number of embryos is inadequate, see troubleshooting 1).
-
a.To maximize embryo production, females and males can be kept in separate tanks before breeding.
-
b.Prior to the day when you wish to obtain embryos, transfer adult male and female zebrafish into the same tank, at a 1:2 ratio, respectively. Use breeding tanks or use marbles to cover the bottom of the tank.
-
a.
Pause point: Allow the fish to stay in the breeding tank for next ∼12 h
-
3.After the beginning of the next light cycle, collect embryos into a fresh petri dish by siphoning and place the adult fish back in their home tanks.
-
a.Remove unfertilized and dead embryos from the petri dish and place 50 live embryos per dish in E3 water at 28°C for 7 days to develop.
 Pause point: Allow larvae to develop at 28°C for 7 days.
Pause point: Allow larvae to develop at 28°C for 7 days. -
b.During the growth period, remove dead embryos daily and change E3 water if needed.
-
a.
Induction of inflammation and demyelination
Timing: 1 h
In this step, we describe the use of cuprizone and LPS to induce inflammation and demyelination
-
4.
Prepare cuprizone stock solution (see “materials and equipment” for detailed recipe).
-
5.
Thaw or prepare LPS stock solution (see “materials and equipment” for detailed recipe).
-
6.
Prepare Cuprizone + LPS treatment solution (see “materials and equipment” for detailed recipe) and control (zebrafish system water) conditions in a 12-well plate and transfer the 7 dpf fish into the plate with 3 fish/well and 3 wells/condition.
-
7.
To screen the effects of additional compounds on inflammation (such as environmental pollutants as described in (Wheeler et al., 2019)), additional groups can be included which include compound only, and Cuprizone + LPS + compound.
-
8.
Place the 12 well plate containing larvae into a 28°C incubator for 48 h.
Pause point: Keep larvae in the Cuprizone + LPS solution at 28°C for 48 h.
Sample collection
Timing: 1 h
In this step, we describe how to isolate zebrafish samples.
Note: Animals have to be euthanized according to the permission and ethical rules of local authorities.
-
9.
Euthanize larvae by placing the 12-well plate on ice.
-
10.
Collect at least n=3 larvae per group for the desired end point measurement.
RNA isolation from zebrafish
Timing: 1–2 h
In this step, we describe how to isolate RNA from zebrafish larvae
-
11.
Isolate RNA from at least n=3 larvae per group with TRIzol LS per the manufacturerś protocol.
CRITICAL: TRIzol is a hazardous chemical, process samples in a ventilated hood and use gloves, a lab coat, and goggles when handling.
-
12.
Purify RNA with Direct-zol RNA kit according to the manufacturer’s protocol.
Measuring inflammation and demyelination markers by qPCR
Timing: 3–4 h
In this step, we describe how to quantify gene expression in zebrafish
-
13.
Synthesize cDNA with High-Capacity cDNA Reverse Transcription Kit according to manufacturerś protocol.
| Reverse transcription thermocycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Pre-primer extension | 25°C | 10 min | 1 |
| DNA polymerization | 37°C | 120 min | 1 |
| Enzyme deactivation | 85°C | 5 min | 1 |
| Hold | 4°C | forever | |
-
14.
Measure gene expression by qPCR with TaqMan Fast Universal PCR Master Mix according to manufacturerś protocol.
| Quantitative PCR thermocycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 20 s | 1 |
| Denaturation | 95°C | 1 s | 40–60 cycles |
| Annealing/Extension | Variable | 20 s | |
-
15.
Analyze qPCR data using the DDDDCt method by comparing the expression of target genes of interest (ccl20, il1b, il10, il17a/f1, mpz, nos2a) relative to actb1. (troubleshooting 2).
Bioinformatic identification of environmental compound mechanisms
Timing: Variable
In this step, we describe how to bioinformatically analyze the pathways regulated by each compound
-
16.
To identify mechanisms associated with compound hits from the EPA ToxCast inventory, go to the Dashboard, here. These data are derived from the EPA ToxCast compound inventory studies (Dix et al., 2007; Sipes et al., 2013).
-
17.
Search for a compound of interest.
-
18.
Identify the molecules which interact with the compound of interest using the “Bioactivity” ↪ “ToxCast: Summary”.
-
19.
To construct interaction networks, go to https://www.networkanalyst.ca/ and input the gene set of interest derived from the compound’s interactions.
-
20.
Select the species of interest as well as the database of interest to identify interactions between the interacting molecules (Figure 1E).
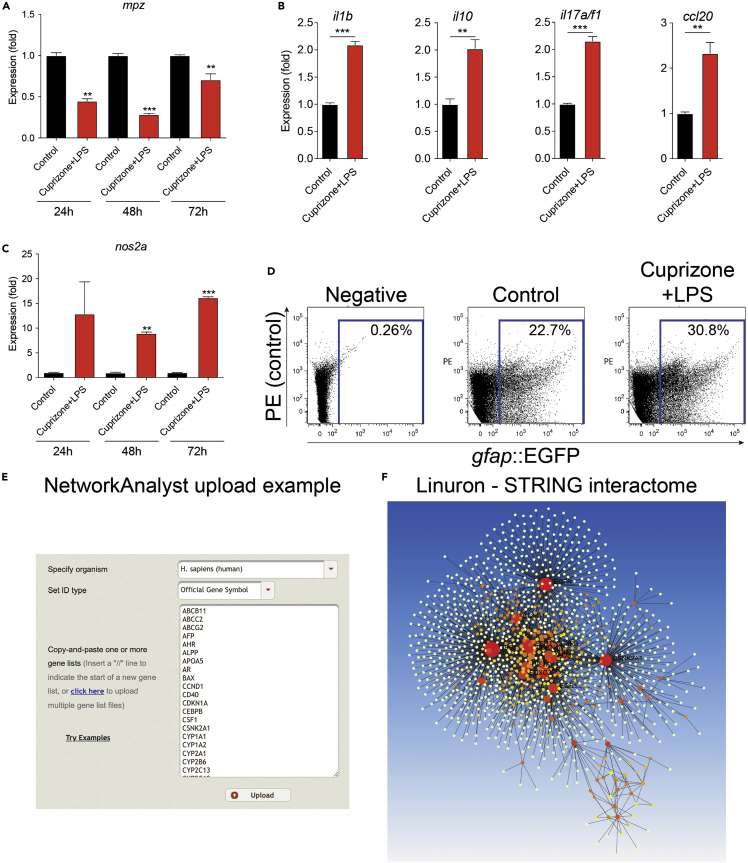
Figure 1.
Expected outcomes of screening environmental factors in zebrafish
(A) Expression of myelin protein zero (mpz) in different exposure time points. LPS + cuprizone exposure results in significant demyelination in all measured time points.
(B) LPS + cuprizone induces expression of inflammatory markers. Expression of il1b, il10, il17a/f1, and ccl20 is significantly increased after 48 h cuprizone exposure.
(C) Expression of inducible nitric oxide synthase (nos2a) in different exposure time points.
(D) LPS + cuprizone causes astrocyte activation. Dot plot analysis of gfap::EGFP expression after 48 h LPS + cuprizone exposure demonstrates increased gfap::EGFP expression. Expression of nos2a in gfap::EGFP+ cells is increased after 48 h LPS + cuprizone exposure.
(E and F) Example upload and (F) visualization in NetworkAnalyst of geneset identified by compound screening. ∗∗p<0.001; ∗∗∗ p<0.0001. Data shown as mean±SEM.
Expected outcomes
After induction of inflammation and demyelination as described above, there should be a clear downregulation of myelination associated genes (Figure 1A) and upregulation of inflammation related genes (Figures 1B and 1C) as measured by qPCR. Tested compounds should show effects on canonical markers of cellular activation specific to each experimental system (Figures 1C and 1D). When analyzing CNS inflammation for example, we focused on cytokines and chemokines il1b, il10, il17a/f1, ccl20, nos2a, as well as GFAP when studying astrocytes (Figures 1B–1D). Putative gene expression networks can be constructed using a combined analysis of the EPA ToxCast Dashboard and any bioinformatic tools of interest, for example, Network Analyst (Figures 1E and 1F) (Xia et al., 2015).
Note: For details on cell isolation and sorting, including transgenic lines (Figure 1D) please see the original paper (Wheeler et al., 2019).
Quantification and statistical analysis
Timing: Variable
All qPCR data should be analyzed by comparing treatment condition to controls. For bioinformatic network analysis, only genes with significant effects in the EPA ToxCast database should be included.
Limitations
Other brands and types of LPS reagents have been demonstrated not to result in a similar inflammation and demyelination as seen in Figures 1A–1D. To avoid this, use only reagents listed in the Key Resources table.
Troubleshooting
Problem 1
Zebrafish are not producing the desired number of embryos for experiments (step 2).
Potential solution
This problem might arise from zebrafish husbandry. To resolve the issue, pay attention to proper and reproducible husbandry and well-being of the adult fish. Another option is to pair more fish by setting up additional spawning tanks. Ensure that adult females are not spawned more frequently than once weekly, with 2–3 week breaks between spawnings being ideal.
Problem 2
Upregulation of the inflammation and demyelination markers by qPCR are not observed at the end point (step 15).
Potential solution
Ensure that cuprizone is completely dissolved (step 3). If needed, a heated shaker can be utilized.
Problem 3
cDNA concentration is low (step 13).
Potential solution
Ensure that RNA purification is performed in an RNase free environment. Combine more zebrafish to increase RNA yield.
Problem 4
No interactions between compound target genes detected bioinformatically (step 20).
Potential solution
Select a different network analysis and incorporate more genes into your network.
Problem 5
A large number of interactions are detected in the bioinformatic networks (step 20).
Potential solution
Decrease the number of genes used as input or increase the threshold criteria defining interactions.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Francisco Quintana, fquintana@rics.bwh.harvard.edu.
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by the Sigrid Jusélius Foundation as well as grants NS102807, ES02530, ES029136, AI126880, and AI149699 from the NIH; RG4111A1 from the National Multiple Sclerosis Society (to F.J.Q.); and PA-1604-08459 from the International Progressive MS Alliance (to F.J.Q.). M.J. was supported by a postdoctoral fellowship from Sigrid Juselius and postdoctoral grants from Saastamoinen Foundation, Paulo Foundation, The Finnish MS-Foundation, Orion Farmos Research Foundation, and Maud Kuistila Memory Foundation. M.A.W. was supported by the NIH (1K99NS114111, F32NS101790), a training grant from the NIH and Dana-Farber Cancer Institute (T32CA207201), a travelling neuroscience fellowship from the Program in Interdisciplinary Neuroscience at the Brigham and Women’s Hospital, and the Women’s Brain Initiative at the Brigham and Women’s Hospital. We thank all members of the Quintana laboratory for helpful advice and discussions. In addition, we thank E. Buys, T. Pedulla, S. ÓLoughlin, and A. Cintolo for assistance with zebrafish husbandry.
Author contributions
M.J., M.A.W., and F.J.Q. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate/analyze datasets/code.
References
- Bernardos R.L., Raymond P.A. GFAP transgenic zebrafish. Gene Expr. Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Dix D.J., Houck K.A., Martin M.T., Richard A.M., Setzer R.W., Kavlock R.J. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- Linnerbauer M., Wheeler M.A., Quintana F.J. Astrocyte crosstalk in CNS inflammation. Neuron. 2020 doi: 10.1016/j.neuron.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmarco L.M., Polonio C.M., Wheeler M.A., Quintana F.J. Functional immune cell-astrocyte interactions. J. Exp. Med. 2021;218:e20202715. doi: 10.1084/jem.20202715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes N.S., Martin M.T., Kothiya P., Reif D.M., Judson R.S., Richard A.M., Houck K.A., Dix D.J., Kavlock R.J., Knudsen T.B. Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol. 2013;26:878–895. doi: 10.1021/tx400021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.A., Jaronen M., Covacu R., Zandee S.E.J., Scalisi G., Rothhammer V., Tjon E.C., Chao C.C., Kenison J.E., Blain M., et al. Environmental control of astrocyte pathogenic activities in CNS inflammation. Cell. 2019;176:581–596 e518. doi: 10.1016/j.cell.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.A., Quintana F.J. Regulation of astrocyte functions in multiple sclerosis. Cold Spring Harbor Perspect. Med. 2019;9:a029009. doi: 10.1101/cshperspect.a029009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Gill E.E., Hancock R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.