Summary
Traction force microscopy (TFM) is a popular technique for studying cellular stresses; however, the reproducible fabrication of ultrasoft substrates for the reliable detection of weak cellular stresses (below 100 Pa) remains a challenge. Here, we describe a simple in vitro TFM protocol using such ultrasoft protein-coated polyacrylamide gels and wide-field fluorescence microscopy. We complement the protocol with open-source and in-house scripts for data analysis for the easy quantification of traction stresses, which is demonstrated here using peripheral blood mononuclear cells.
Subject areas: Biophysics, Cell Biology, Immunology
Graphical abstract

Highlights
-
•
Reproducible polyacrylamide gel-based TFM protocol to detect weak cellular stresses
-
•
Use of open-source softwares to quantify traction stresses
-
•
Application to measure immune cell stresses
Traction force microscopy (TFM) is a popular technique for studying cellular stresses; however, the reproducible fabrication of ultrasoft substrates for the reliable detection of weak cellular stresses (below 100 Pa) remains a challenge. Here, we describe a simple in vitro TFM protocol using such ultrasoft protein-coated polyacrylamide gels and wide-field fluorescence microscopy. We complement the protocol with open-source and in-house scripts for data analysis for the easy quantification of traction stresses, which is demonstrated here using peripheral blood mononuclear cells.
Before you begin
Despite the numerous exciting developments in the field of Traction Force Microscopy (TFM), the most commonly used system to measure cellular forces remains fairly similar to the one designed by Dembo and Wang (1999): TFM on continuous and linearly elastic substrates embedded with fluorescent beads. The most popular of these substrates are polyacrylamide gels (PAGs) and polydimethylsiloxane silicone (PDMS) elastomers. In the protocol described here, we prefer polyacrylamide gels over their counterparts for two main reasons. First, the stiffness of PAGs can be easily adjusted to mimic that of soft biological tissues and cells (down to the order of 100 Pa), which applies very well to our systems of interest, namely immune cells. Second, PAGs are generally non-fouling, meaning neither cell surface receptors nor adhesive proteins present in the serum can bind directly to the gel, only molecules covalently grafted on the gel surface can act as ligands for the cells.
Here, we implement this optimized protocol for measuring the traction stresses exerted by peripheral blood mononuclear cells (PBMCs), however, we have successfully employed it for different subtypes of primary T cells and Jurkat T cells as well.
Key resources table
Step-by-step method details
Part 1: Production of ultra-soft polyacrylamide gels
Given the toxic and hazardous nature of several of the chemicals mentioned in Part 1, we highly recommend that the following steps are performed under a chemical fume hood.
Preparation of amino-silanized glass-bottom fluorodish(es)
Timing: 1 h
There are two reasons behind specifically choosing these fluorodishes for PAG casting. First, they have an optical quality glass bottom with a thickness of 0.17 mm, making the visualization of the gel on inverted microscopes with high magnification objectives quite easy. Second, they are individually packed and gamma sterilized, which decreases the number of steps and time needed for activating them.
-
1.
Plasma clean the sterile fluorodish for 2 min at high settings.
Note: For our experiments we use residual air plasma. As mentioned above, the fluorodishes are already sterile. This step is an additional precaution to help increase the surface energy of the glass surface to better bond with the reagent added in the next step.
Note: Given that we are using fluorodishes, which are glass-bottom plastic dishes, for casting the PAGs, we cannot use harsh chemicals for activating them. This is why we chose to use a plasma cleaner in this step. In the case of in-availability of a plasma cleaner, we suggest replacing the fluorodishes with glass coverslips and activating them with a piranha solution (sulfuric acid- hydrogen peroxide). We have successfully done this several times.
-
2.
Immediately add 1 mL of 5% APTES diluted in Milli Q water to the activated fluorodish surface. Allow the APTES to react for 5 min. After 5 min, remove the remaining liquid APTES from the fluorodish, flip it upside down on a clean paper towel and allow it to dry for 10 min.
CRITICAL: APTES is highly sensitive to moisture and should be stored by filling the pocket of gas in the bottle with inert gas such as nitrogen or argon, preferentially at 4°C. If it is stored at 4°C, it should be brought to ≈ 25°C before usage. The elevated temperature will enhance the covalent attachment and self-assembly of APTES to the glass during the salinization reaction. The diluted solution should always be prepared fresh. Do not keep any unused solution.
-
3.
Rinse the fluorodish with running Milli Q water for 30 s to remove the remaining APTES from step 2.
Note: It is essential to completely remove any trace of unreacted APTES for it will react with the glutaraldehyde used in step 3 and form an orange-brown precipitate. This precipitate fluoresces under UV light and may therefore interfere with fluorescence imaging of either the beads or the cells (if fluorescently labeled) during the experiment.
-
4.
Add 1 mL of 0.5% glutaraldehyde in Milli Q water to the fluorodish and let it sit for 30 min.
CRITICAL: Using APTES alone to bind the polyacrylamide gel to the glass surface is not reproducible. Thus, following the APTES treatment with glutaraldehyde will help further activate the amino silanized surface and ensure the covalent attachment of the polyacrylamide gel to the glass surface.
-
5.
Aspirate the remaining glutaraldehyde from the fluorodish and rinse it again with running Milli Q water.
Pause point: After this step the fluorodishes can either be (a) dried and used immediately for polyacrylamide gel casting, (b) stored in water at 4°C for up to a week or (c) dried and stored under desiccation for up to a month. We preferentially used options (a) and (b).
Preparation of chloro-silanized coverslip(s);
Timing: 2 h 30 min
-
6.
Sonicate (ELMA ULTRASONIC S 30 H) the 12 mm glass coverslips for 30 min in 2% Hellmanex (diluted in Milli Q water) twice and then for another 30 min in Milli Q water twice. Rinse the coverslips with running Milli Q water for 1 min in between each sonication. Then, dry them using compressed air.
-
7.
Use a plasma cleaner to clean the coverslips for 2 min on the high setting.
Note: For our experiments we use residual air plasma. This step is needed to help increase the surface energy of the coverslips to better bond with the reagent added in the next step.
Alternatives: If a plasma cleaner is not available, we suggest activating the coverslips with a piranha solution (sulfuric acid- hydrogen peroxide). We have successfully done this several times.
-
8.
Immediately cover the activated coverslip surface with undiluted Sigmacote (as provided by the manufacturer) and allow it to react for 5 min. After 5 min, remove the excess Sigmacote solution by capillarity with an absorbing paper (e.g., Kimwipe) and let the coverslips air-dry for 10 min protected from light and dust.
CRITICAL: Sigmacote is a chlorinated silane and thus, similar to APTES, has to be stored under an inert gas atmosphere at 4°C and has to be brought back to ≈ 25°C before usage.
-
9.
Rinse the coverslips with running distilled water
Pause point: After this step, the coverslips can either be dried and used immediately, or stored under desiccation to be used later.
Preparation of soft polyacrylamide gels
Timing: 20 min
There are numerous reports of acrylamide/bis-acrylamide formulations that can be used to produce PAGs with a wide range of stiffness. We have adapted ours from the commonly used and cited protocol published (Tse and Engler, 2010). The Young’s modulus of the PAGs was verified using Atomic Force Microscopy in mapping mode using a JPK Instruments Nanowizard I, equipped with a JPK Instruments Petri Dish heating system set at 37°C. To do so, we used Brucker Instruments MLCT cantilevers that we modified by gluing beads on their tip, by micromanipulation, the size of which is comparable to that of a lymphocyte (diameter ≈ 10 μm, (Sadoun et al., 2021)). We made 48 × 48 μm2 laterally resolved maps of the modulus on several regions (at least 3) on the gels to ensure the lateral homogeneity of this crucial parameter. By performing these measurements regularly, we ensured that our PAG preparation protocol was stable and reproducible.
-
10.
Prepare the polyacrylamide gel premix by mixing acrylamide and bis-acrylamide to their desired concentrations in either Milli Q water or PBS in an eppendorf tube. To obtain ultra-soft polyacrylamide gels (≈ 400 Pa PAG), mix 75 μL Acrylamide (40%) with 30 μL Bis-acrylamide (2%) and 895 μL PBS. To generate gels with different stiffness, refer to Tse and Engler (2010).
Note: It is important to note that using PBS instead of Milli Q water will slightly increase the elastic modulus of the gel. This can be quantified by AFM measurements.
Note: It is preferable to mix the precursors, APS and TEMED, directly before polymerizing the gels since that will limit and control their exposure to oxygen, ensuring a better reproducibility of the final gel stiffness.
-
11.
Add 0.7% total volume of 200 nm fluorescent beads (7 μL) to the premix.
Note: The beads used in this protocol are carboxylate-modified with an orange fluorescence (excitation wavelength of 540/560 nm). Before use, the beads have to be cleaned using a Slide-A-Lyzer Dialysis Device 10 KDa to remove any chemicals present in the bead solution that might interfere with polymerization of the gel. Since the beads used here are of very small size (∼200 nm), dialysis is the safest method to ensure the thorough cleaning of the beads without creating any aggregates.
-
12.
Add 1% total volume of Ammonium persulfate (APS, 10 μL) and 0.1% total volume of Tetramethylethylenediamine (TEMED, 1 μL) to gel solutions.
CRITICAL: APS has to be either freshly prepared or previously aliquoted and stored at −20°C otherwise the polymerization of the gel will be disrupted. Similarly, TEMED that has been stored for a long time or that hasn’t been stored properly will have the same effect.
-
13.
Quickly vortex the polymerizing solution for 20 s and pipette 5 μL of the polymerizing gel into the center of the fluorodish and sandwich it with the chloro-silanized coverslip, with the treated side facing the gel.
Note: If the coverslip was treated properly, the polymerizing gel should wet it perfectly without any need for supplementary manipulation.
Note: The volumes mentioned above should theoretically create a 40 μm thick gel, however the real height of the gel was observed to be ≈ 80 μm when measured using a motorized optical microscope stage system. This discrepancy is due to the fact that gels swell after polymerization, and so the height of the gel cannot be directly calculated from the volume of the cast polymerizing solution. Also, the degree of PAG swelling varies with each acrylamide/bis-acrylamide formulation and cannot be easily predicted, thus, it is important to measure or at least estimate the height of the resulting gel before using it for TFM. The gel must be sufficiently thick such that it can freely deform due to cellular forces without the influence of the underlying glass. As a consequence, the increased gel thickness might impose a limit on the type of microscopy and objectives (depending on their working distance) that can later be used for visualization.
-
14.
Close the fluorodish lid, flip it upside down to allow the beads to move towards the surface, surround it with wet tissue paper, place it in a closed-light protected chamber (e.g., large petri dish covered with aluminum foil), and then allow it to polymerize for 1 h at 4°C.
CRITICAL: Surrounding the fluorodish with wet tissue-papers will provide enough humidity to prevent the gel from drying out and cracking during the polymerization process. Shorter polymerization times may result in insufficient polymerization of all available monomers and may cause the mechanical properties of the hydrogels to vary from the values noted here and/or be heterogeneous over the gel surface. Monitoring the state of the unused solution in the eppendorf, for example by poking it with a needle can help estimate the degree of polymerization of the gel.
Refer to troubleshooting 1: Gels do not polymerize.
-
15.
Fill the fluorodish with PBS (or water) for 20 min before carefully peeling off the coverslip using the tip of a needle or tweezers.
-
16.
Rinse the gel twice by submersion, each time for 5 min in PBS (or water) to remove any unpolymerized acrylamide.
Pause point: At this point, the gels can be stored in PBS (or water) at 4°C for up to a week before usage. Although the gel looks ‘polymerized’ after 1 h, there’s a deeper level of polymerization that takes place within the first 12 h of gel casting. Thus, it is preferable to use the gel only after this time interval has lapsed and the cross-linking has been completed.
Refer to troubleshooting 2: Gels crack.
Gel functionalization
Timing: 2 h
Before the cells can be deposited, the desired proteins that will drive the interactions between the cells and the gel need to be first immobilized onto the gel surface. PAGs are known to be nearly inert, and thus they do not readily adsorb proteins. One way to overcome this is to covalently link the proteins of interest to the PAG surface using a heterobifunctional protein cross-linker called sulfo-SANPAH. Sulfo-SANPAH contains an amine-reactive N-hydroxysuccinimide (NHS) ester on one side and a UV photoactivatable nitrophenyl azide on the other. Once a sulfo-SANPAH-coated gel is exposed to a UV light source with a wavelength between 320 and 365 nm, the nitrophenyl azide group will form a nitrene group that will bind to the polyacrylamide gel, and the sulfosuccinimidyl group will react with the primary amino groups of proteins to form stable amide bonds.
-
17.
Remove PBS or water from pre-silanized fluorodish.
-
18.
Add 250 μL of 0.5 mg/mL sulfo-SANPAH (prepared in 50 mM HEPES, pH 8.5) to the surface for the gel and expose it to 365 nm UV radiation for 2 min at 100% power in a UV-KUB 2 oven. Repeat this step twice with a PBS rinsing step in between.
Note: Sulfo-SANPAH is highly unstable in aqueous solutions and is light sensitive. Thus, it either has to be previously aliquoted and stored at −80°C (where it can be kept for up to 6 months, shielded from light at all times) or stored as powder and prepared immediately before gel functionalization. After sulfo-SANPAH has been exposed to UV, it should change its color from orange red to brown. It is also important to take care when choosing the UV source: we observed that broad-spectrum UV lamps/ovens are likely to bleach the fluorescent beads used here.
Note: When the hydrogels are later used for TFM, non-specific binding of cellular proteins and media proteins to unlinked sulfo-SANPAH sites are not likely to take place because the reaction happens only when the pH is around 8.5, and the pH of the cell medium is usually between 7 and 7.4.
-
19.
Rinse the gel 3 times with PBS to ensure that all unbound sulfo-SANPAH has been removed.
-
20.
Add an appropriate amount of the desired protein onto the surface of the activated gel, and incubate this solution for either 2 h at 25°C or for 12 h at 4°C.
Note: For our experiments, we used a final concentration of 30 μg/mL of the antibody OKT3. A primary concern when using PAGs is the variability of ligand density and the homogeneity of ligand coating, especially given their inert and porous nature. Thus, to optimize the coating protocol to the end user’s specific proteins and conditions, a quantification step is necessary. One way to perform this quantification is by using a fluorescently labeled protein to both relate the measured signal to the amount of protein from reference standards and to inspect the relative lateral homogeneity of the coating (González et al., 2019). Fluorescently labeled proteins can be added in this step instead of their unlabeled counterparts and can then be imaged after incubation using fluorescence microscopy. Nevertheless, it is preferable not to use a fluorescence label for the proteins, especially one with absorption/emission spectra too close to the one of the beads to avoid creating background noise via bleed-through which may compromise the fine detection of bead displacement.
Part 2: Image acquisition
Timing: 15–30 min/sample
-
21.
Once the incubation period is over, rinse the gel gently with a 37°C preheated medium 3 times, leaving the gel immersed in medium after the last rinse (≈ 2 mL of medium), and then transfer the fluorodish to an inverted widefield fluorescence microscope. We used a Zeiss Axiovert 200 microscope equipped with a 40 × NA0.9 air objective, a CoolSnap HQ2 camera (Photometrics), a LED illumination system (Colibri 2, Zeiss) with suitable filter sets (Cazaux et al., 2016), and a petri dish heater module (JPK Instruments) that allows setting the temperature at the desired value, with a stability of a fraction of a degree.
Pause point: Leave the gel to equilibrate at the desired temperature (in our case 37°C) for at least 20 min before starting image acquisition. This reduces the potential drifts due to thermal expansion of the substrate or the microscope stage. The mentioned time interval may need to be adapted to every setup used, in particular for thermal equilibration.
Note: In all TFM experiments, at least two images of the substrate have to be acquired: One image of the bead field while the substrate is subjected to cellular forces (i.e. stressed state) and another image of the bead field in the absence of cellular forces (i.e. relaxed state). In our setup, the fluorodishes are firmly fixed onto the microscope with the petri dish heater module. This enables us to start the image acquisition before the addition of the cells, consequently giving us the relaxed state of the gel at the start of the experiment. This corresponds also to our goal of studying the early stages of immune cell mechanotransduction.
-
22.
Start image acquisition using appropriate software. In our case, we used Zen software (Zeiss) that controls the Colibri 2 diodes system and the camera.
Note: Since we are interested in the dynamics of early immune cell spreading, our image acquisition only lasts between 15 and 30 min, and instead of taking only two images (relaxed and stressed), we acquire time-lapse TFM movies, at a typical interval of 1 frame every 5s, by capturing image doublets of the cells in phase-contrast (20 ms exposure time) and orange fluorescent microspheres (excitation ≈ 488 nm, 200 ms exposure time) over the course of the mentioned time frame. Since the layer of microspheres is only a couple of micrometers beneath the gel surface (due to the flipping of the gel during the polymerization step), the cells will still be easily visualized and tracked while the focus is set on the bead layer.
-
23.
Remove any excess medium (if there is any, depending on the desired final concentration of the cells), and gently pipette the cells onto the gel.
Note: For our experiments, we typically add a final concentration of 0.75 million cells/2 mL in complete RPMI media containing 10% fetal bovine serum (FBS), 1% glutamax, 1% penicillin streptomycin and 2.5% HEPES. Remove 1 mL of medium, add the 0.75 million cells/0.5 mL onto the center of the gel and then add the remaining 1.5 mL of medium around the gel center. Note that the final concentration of cells used needs to be adjusted based on the cell type at hand and the forces they exert; it is crucial that the stress fields exerted by the cells are spatially separate. Also, the cells are always sub-cultured 12 h before the experiment so as to not perturb them the day of the experiment. Keeping culture conditions strict is an important point for ensuring the comparison between different days of experimentation and for subsequent statistical pooling of the data.
Note: A common method to obtain the relaxed reference state of the beads is to carefully detach the cells at the end of the experiment, for example using Triton X-100, SDS, Trypsin, or accutase solutions, wait until substrate deformation is fully reversed, and then capture an image of the beads. In this case, the risk of accidentally moving the sample from its position is greatly augmented, which complicates data analysis, sometimes even rendering it impossible. However, in case of encountering significant drift during step 3, this method could be used instead.
Expected outcomes
The PAGs produced using this protocol exhibit several key specifications. First, they are sufficiently thick (≈ 80 μm) such that the cells feel and respond to the softness of the gels rather than the rigidity of the underlying glass. Second, they have a well characterized and homogeneous Young’s modulus of ≈ 400 Pa, specifically chosen for studying weak forces like those generated by immune cells. This rigidity is not too high that the cells are incapable of significantly deforming the gel, and not so low that the resulting large deformation breaches the linear-response regime, making the linear-elastic theory underlying TFM inapplicable. It is also important to mention that PAGs with this stiffness have a sufficiently small mesh size capable of entrapping sub-resolution (200 nm) fluorescent beads within it. Finally, the gels have a lateral bead density corresponding to about four beads in an area of 2.5 × 2.5 μm2 (16 × 16 px2), carefully chosen in accordance with the lateral distribution and the magnitude of the forces being measured, as well as the quantitative image analysis method employed.
A successful run should have cells interacting with the substrate at the end of the experiment (Figure 1, Method video S1 - after drift correction, see below).
Figure 1.
Transmission image of PBMCs (top) and fluorescent image of beads (bottom)
Inserts: white squares indicating sub-image zones cropped and used for the PIV and FTTC calculations. Scale bar 20 μm.
Quantification and statistical analysis
For the data analysis and the post-processing, we chose to use open source softwares and programming languages. We use the last version of Fiji (Schindelin et al., 2012) which is a meta-package of ImageJ (used as of 13/11/21, v1.53c), with specialized plugins available either on Fiji/ImageJ website, programmer’s website or GitHub repository (see below). For Python, we use the latest release of Anaconda Python distribution (https://www.anaconda.com, used as v3.8.8) which contains all the needed packages for data science applications, together with an IDE (Spyder) and the Jupyter notebook system utilized in this protocol. We perform our analysis on 64 bits Linux PCs (Ubuntu 18.04; Linux Mint 20.2).
Part 3: Data analysis
Timing: 1 h/sample
The computer configuration we used for data analysis was a Dell Precision 3640 Tower, with an Intel i9-10900K (20) @ 5.300 GHz CPU and 64Go RAM under Ubuntu Linux 18.04. We have also successfully performed the analysis on a Dell XPS 13 9370 laptop, with a Intel i7-8550U (8) @ 4.000GHz CPU and 16Go RAM, under Ubuntu Linux 18.04 or Linux Mint 20.2.
Having imaged the sample, one can then extract the cellular traction stresses from the collections of images obtained. This arduous, calculation intensive process encompasses four key steps:
-
1.
Sample Drift Correction
Typically, images taken one after the other may present a slight drift, however, this drift becomes especially evident when acquiring images over a larger time frame and at a relatively high frequency such as in our experiments. Sample drift is of two kinds, either a focus drift (i.e., along the z-axis) caused by small temperature variations between the sample and the objective (in particular in the case of an immersion lens), or a lateral drift (i.e., along the x–y plane), caused by stage instability or dilatation, for example. The z drift is somehow less problematic than the lateral one since it can be either automatically controlled by an autofocus, or z-scan acquisition with further detection of the best focus image (e.g., using the ImageJ macro from https://sites.google.com/site/qingzongtseng/find-focus), or manually adjusted at given time points throughout the acquisition period. The x-y drift on the other hand cannot be easily experimentally predicted or corrected, only minimized by suitable motion and temperature control, and thus can severely corrupt the true displacement data. Consequently, before one can faithfully measure cell-induced bead displacement, the frame-to-frame drift generated due to whole-sample translation, and sometimes rotation, must first be calculated and then subtracted from the total bead displacement to properly align the images.-
a.Install and open Fiji → Import video of the fluorescent beads → Save video as .tif or .czi in a separate folder named “images” → Open Set Scale and properly calibrate the images → Install the TrackMate plugin → Open Plugins → Open Tracking → Run TrackMate → Extract “Track Displacement” → Save the pop-up table titled “Spots in tracks statistics” as “trajectories.csv”Note: TrackMate (Tinevez et al., 2017) is a tool for automated single particle tracking; It assigns each fluorescent bead an identity (based on its coordinates) over the given frames and then reconstructs the trajectories of each bead in the form of a track. We use the full frame 220 × 164 μm2 (1392 × 1040 px2) images (16 bits coded on 12 bits) in this step since they contain a large number of beads that can be detected and tracked, which helps increase the accuracy of the measured drift.
-
b.Create an empty folder named “aligned” and then run the following Jupyter notebook: https://github.com/remyeltorro/SPTAlign. The now aligned frames will appear in the “aligned” folder.Note: The script mentioned above utilizes the trajectories obtained from the previous step to perform a sample drift correction. Since the average shift is often on a subpixel scale, the script applies the shift in Fourier Space: It first direct Fourier transforms the image, applies the shift using Scipy Fourier shift function, and then inverse Fourier transforms the image. The first frame of the movie is chosen as the frame of reference (since the cells are not present yet) and consequently all the following frames are aligned to the coordinates of the fluorescent beads in the first frame.Note: The same trajectories can be used to correct the drift on the cell movie since we do not change the objective and filter-set between the bead-field and cell images.
-
a.
-
2.Cropping Regions of Interest
-
a.Open the drift-corrected bead image sequence in Fiji and search for the areas with bead displacements.Note: Large bead displacements are easily caught by eye (Method video S1), however, low magnitude forces (for example those caused by Jurkat T cells) cause small bead displacements that can be difficult to detect by eye. In these cases, monitoring the position of the cells can be quite beneficial since one could simply project the position of the cells onto the bead images and limit the search for bead displacement to those areas.
-
b.Install the ROI-1click tools (https://imagej.net/plugins/roi-1-click-tools) in Fiji -> Open the ROI-1 click tools → Choose the appropriate selection size (explained below) → Select the regions where the beads are visibly moving by clicking the pointer once at the center of the movement → Leave the ROI manager containing all the selections open (this is crucial for the next step)Note: Choosing the right selection size is highly dependent on the experimental data itself. The chosen region should be centered around the cell, but it should not necessarily be limited to the cell outline; In most cases, substrate deformation by cellular forces is not confined to the area directly underneath the cell, and so some bead movement can be detected in the margins outside the cell outline, due to the continuous elastic nature of the gel. Consequently, constricting the region for force detection to that underneath the cell and neglecting the displacement occurring outside of it will not be a true representation of the real force exerted by the cell. Additionally, if in fact the force does extend outside the cell outline and the bead displacement does not go down to zero at the boundary of the cropped image, some so-called “ringing” artifacts (oscillations) will appear at the boundary and will introduce errors into the final reconstituted force field. Alternatively, if the cropped region is too large, then the stresses will be diluted over an area where a substantial number of beads will be immobile (not experiencing any force, just background noise). In this case, one runs the risk of severely dampening the signal to noise ratio which ultimately would not be a true representation of the real force exerted by the cell (Figure 2).
-
c.Load the Multicrop macro (1-PHP-multicrop.ijm from https://github.com/phpuech/TFM) in Fiji → Click Run → Double click on the “aligned” folder → Click Run. The cropped full-length image sequences of the selections will now appear in the “aligned” folder, each in a separate folder named “segment_n” where n goes from 0 to the total number of selections present.
-
a.
-
3.Calculating Bead Displacement Field and Traction Force Field
-
a.Install the PIV Plugin and the FTTC Plugin from https://sites.google.com/site/qingzongtseng/imagejplugins in Fiji → Open the macro 2-PHP-TFM-loops-modified.ijm from https://github.com/phpuech/TFM in the Macro editor in Fiji → Input the following parameters: Interrogation Windows (IW1, IW2, IW3), Search Windows (SW1, SW2, SW3), Vector spacing between the IWs (VS1, VS2, VS3), Poisson’s ratio (0.5 for PAG), the Young’s modulus of the gel (in Pascal), Pixel size, Width and Height of the cropped image (in pixels), Correlation threshold value and Regularization Factor → Click Run → Double click on the “aligned” folder → Click Run. The PIV and FTTC text files will now appear in a “save” subfolder for each “segment_n” in the “aligned” folder.
-
a.
Figure 2.
Effect of the selected image size (20 × 20 μm2 vs. 40 × 40 μm2) for the PIV and FTTC calculations on the constructed vector map of bead displacement and the gaussian smoothed map of stress norms, using the same PIV parameters and regularization factor
Refer to troubleshooting 3: Poor lateral resolution of the stress maps due to inappropriate choice of PIV and FTTC parameters
Note: The PIV Plugin calculates the bead displacement field from the bead images, and then the FTTC Plugin reconstructs the traction force field from the displacement data. Since the PIV and FTTC Plugins only process two images at a time and our experimental data consists of movies (made up of ≈ 200 frames), this Fiji/ImageJ macro was written to consecutively run the two plugins over the full length image sequences of all the segments in the aligned folder, always taking the first frame in each segment as the reference frame.
Note: If you open the PIV Plugin, three options will appear: Cross-correlation iterative PIV (also known as conventional PIV), Basic iterative PIV (also known as template matching PIV), and Advanced iterative PIV (an extension of the Basic iterative PIV). The macro linked here implements the Advanced iterative PIV: The images are divided into two types of windows, Interrogation Windows (IWs) and larger Search Windows (SWs). In each iteration, the displacement is calculated by a normalized correlation coefficient algorithm, so that an IW from image 1 is compared to the SW in image 2 to find the required shift of image 1 to have the best overlap between images 1 and 2. Up to three iterations can be performed, going from coarse to fine scale, with each iteration taking into account the displacement field measured previously. A correlation threshold, ranging from +1 (exact match) to −1 (zero correlation), is manually defined to filter out low correlation matches resulting from IWs with an insufficient number of beads. Thus, even if the pre-shift given by one of the PIVs is partly inaccurate, the best matched vectors depicted by a significant correlation peak (equal or higher than the threshold) are displayed in the final cross-correlation result, while other vectors with a correlation value lower than the threshold are replaced by the median value of the surrounding neighbors.
-
4.
Post-Processing
To visualize and quantify the bead displacement data and the stress field data in the PIV and FTTC text files:
Open the following Python script: 3-PHP-Reconstruct-Energy-Plots.py from https://github.com/phpuech/TFM in Spyder IDE → Input the source of the segment folders → Input the dt (time interval between frames), VS3, Calibration factor from (to convert px to μm), image size (in px) → Choose which parameters you would like to plot (map of the displacement vectors, histogram of the displacement norm, map of the stress vectors, discreet map of the stress norm, Energy plot, Stress sum plot) → Adjust the output parameters of the plots as needed → Run the script.
Note: The stress norm is defined, at each position of the reconstituted stress field, as the norm of the stress vector obtained by the FTTC macro. The stored energy is calculated as the sum of the scalar products of the displacement vectors and force vector (i.e. the stress times the area on which it applies) as in Butler et al. (2002). The maps show the lateral distribution of a given parameter, and possibly the orientation of the vectors, while an “average” parameter plot focuses more on the time evolution of the global behavior of the cells.
Note: This Python code generates quiver plots (maps of arrows, color coded for their norm values) and histograms for the displacement parameter, as well as heatmaps (color coded for the norm of the vectors at a given position) for the stress parameter. This is recursively done for each “segment_n” present in the “aligned” folder in one go, allowing for a faster processing of a given experiment. It also generates Y vs time plots, where Y is the sum or the mean or the median of the stress norms over the images or the stored energy over the images. The reconstructed maps of the displacement vectors, histograms of the displacement norm and the PIV text files will appear in a ”PIV“ folder while the reconstructed maps of the stress vectors, discreet maps of the stress norm, energy plots, stress sum plots, and the FTTC text files will appear in an “FTTC” folder, both in a subfolder called “save” for each “segment_n”.
Note: We implemented this Python script because we are interested in observing the dynamics of the stress exerted by the cells, however, to generate vector plots for the bead displacement and the stress for a specific time point, a simple alternative would be to use the PIV and FTTC visualization tools available at https://sites.google.com/site/qingzongtseng/imagejplugins.
Note: For the post processing, as for the PIV and FTTC calculations, we choose not to apply a mask corresponding to the projection, at each time point, of the cells. Such a mask is used to exclude the zones where the beads are expected not to move, which, as mentioned before, will impact the stress field calculated by decreasing the signal to noise ratio. Some reports arbitrarily extend the data processing zone to include a margin around the area occupied by the cell, amounting to 10% of the cell area, to account for beads that may move since they are close to the cell margin and hence within the deformation field of the continuous elastic gel. We preferred to minimize the number of assumptions by using a “one image size fits all” approach for a given condition, in order to compare between different cells and cell types.
This is our end point. The maps and plots can be either used as such or the corresponding data can be accessed as text files for further analysis of any kind.
Limitations
First and foremost, since these ultra-soft gels are produced specifically for weak forces, cells that exert high forces are not suitable for study using these gels. Ideally, the rigidity of the PAGs should be optimized for every specific cell type and condition.
Another problem that arises for all PAG-based TFM, due to the hydrophilic, as well as porous nature of polyacrylamide, is that the thickness of the resulting gel is very hard to control. In this protocol, the gels have a relatively stable thickness of ≈ 80 μm. Thick gels preclude low working distance objectives, and as a result many high magnification and high numerical aperture oil objectives cannot be used. Not to mention that PAGs have roughly the same refractive index as water, which consequently also eliminates the possibility of performing surface microscopies (e.g., Reflection Interference Contrast Microscopy, Total Internal Reflection Fluorescence Microscopy).
Additionally, given the high hydrophilicity of PAGs in general, but of soft PAGs, microcontact printing and patterning of proteins onto the gels will be extremely challenging.
Finally, using the set-up described here, a 2D substrate with embedded beads, we are limited to measuring forces exerted in tangential (in-plane) directions (x,y). Forces that may be applied normal to the substrate (out-of-plane) will be very hard, if not impossible, to assess. Some recent reports have proposed to use additional beads to derive these forces (Aramesh et al., 2021).
Troubleshooting
Problem 1
Gels do not polymerize reproducibly
Manufacturing gels of very low elastic moduli is a very sensitive process. Even minor perturbations of the chemical integrity of the reactants can modify, slow or even disrupt the full polymerization reaction. Most commonly, this problem is due to a malfunction of the initiators, APS and TEMED, or to additives present in the fluorescent microsphere solution incorporated in the gel premix.
Potential solution
Solution 1: Prepare fresh APS for each experiment or properly aliquot it and store it at −20°C prior to experimentation. APS is very hygroscopic and begins to degrade almost immediately when dissolved in water.
Solution 2: Renew the TEMED at least every 6 months and make sure it is tightly closed and stored under inert gas (such as Argon and Nitrogen gas) at 4°C during that time interval. TEMED is very hygroscopic and subject to oxidation as well. As it accumulates water and reacts with it over time, it will gradually lose its catalytic activity.
Solution 3: Thoroughly clean the fluorescent beads before they are incorporated in the gel. The fluorescent microsphere solution contains some chemicals that may inhibit the polymerization reaction. We recommend washing by dialysis for 12 h.
Problem 2
Gels crack
The presence of beads in the gels, in combination with the flipping of the fluorodishes, may lead to the formation of visible cracks in the gels. When this happens, the beads will accumulate in the cracks, perturbing the otherwise uniform dispersion of the beads in the surrounding regions and rendering the gel unusable.
Potential solution
Increase the level of humidity during the polymerization process. We recommend surrounding the fluorodishes with wet tissue-papers during that time interval.
Problem 3
Poor lateral resolution of the stress maps due to inappropriate choice of PIV and FTTC parameters.
Potential solution
Choose correct PIV and FTTC parameters as described below.
First, start by choosing the size of the IW in the final iteration (IW3). The smaller the IW, the higher the resolution of the forces. However, if the IW gets too small, the discernable features (fluorescent beads) in the window will become less and less numerous, making the correlation unreliable and prone to error. As a rule of thumb, the smallest IW must contain on average 3–4 beads, or more (Martiel et al., 2015). Second, choose the size of the SW3. In general, the SW should be larger than the IW. The simplest and probably most computationally efficient option is to set the SW3 to be double the size of the IW3. Do not set the SW3 and the IW3 to the same value, as the PIV program will turn automatically into Basic Iterative PIV mode, which is less precise, and hence not recommended (see https://sites.google.com/site/qingzongtseng/tfm). Third, choose the VS, which effectively represents the spacing between the IWs. Again, the simplest and probably most computationally efficient option is to set the VS to be half the size of the IW. Once the final iteration values have been inputted, proceed to the remaining iterations. To run three iterations, one option is to double the values of IW3, SW3 and VS3 to get IW2, SW2 and VS2, and then similarly double the values of IW2, SW2 and VS2 to get IW1, SW1 and VS1. To limit the number of iterations performed, set the IWs and SWs to zero. Remember that the previous PIVs with the larger IWs and SWs only serve as a guide for finding the correlation peak. For example, for the 128 × 128px2 (equivalent to 20 × 20 μm2) bead image shown in Figure 3 and Methods video S2, the following PIVs were used: IW1 = 64 SW1 = 128 VS1 = 32, IW2 = 32 SW2 = 64 VS2 = 16, IW3 = 16 SW3 = 32 VS3 = 8.
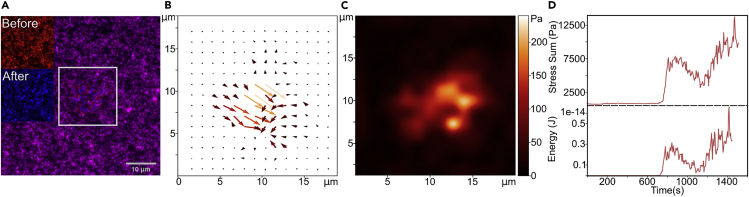
Figure 3.
TFM data processing
(A) Merged images of beads at t = 0 min (red) and t = 20 min (blue). Inserts: reference image in red at t = 0 min, stressed image in blue at t = 20 min, white square indicating sub-image zone cropped and used for the PIV and FTTC calculations.
(B) Vector map of bead displacement obtained for t = 20 min after the PIV calculations over the sub-images.
(C) Gaussian smoothed map of stress norms obtained for t = 20 min after the FTTC calculations over the sub-images.
(D) Curves representing the sum of stresses (top) and the energy (bottom) over the sub-images vs. the time duration of the experiment.
The movie is 1 frame every 5 s.
The movie is 1 frame every 5 s.
Next, set the correlation threshold. This value defines to what extent the program should keep the correlation result. A high correlation threshold value will leave only the vectors resulting from highly matched IWs, while other vectors with correlation value lower than the threshold will be interpolated by the surrounding values (replaced by the median value of the 8 neighbors). Typically, the recommended correlation threshold is 0.6 (Q. Tseng, personal communication and (Martiel et al., 2015)), above which the displacement between the IWs is considered to represent an actual bead displacement.
Finally, set the Regularization Factor for the FTTC. This factor can be viewed as a noise-smoothing parameter (Han et al., 2015).The degree of regularization must be carefully chosen such that it provides a balance between how well it fits the noise-distorted experimental displacement data (coming from erroneous PIV evaluations and/or effective experimental noise) and the overall magnitude of the traction forces. If the data is over-regularized, the stress field will become over-smoothed and the resolution of the recovered stress, both in direction and magnitude, will be lost. Alternatively, if the data is under-regularized, the program will overfit the noise in the displacement and will thus be a false representative of the traction forces. For our data, we tested a large number of possible values coherent with literature (references; starting with 0, as a reference, and then ranging from 10−12 to 10−7). We found that a regularization factor of 9e-10 fits our data best, which corresponds well with the one used in Tseng et al. (2012), upon which our codes are based.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pierre-Henri Puech (pierre-henri.puech@inserm.fr).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
The authors thank the INSERM Cell Culture Facility (PCC). Part of this work was supported by institutional grants from INSERM, CNRS, and Aix-Marseille University to the LAI and CINAM. F.M. was supported by a PhD grant from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 713750, with the financial support of the Regional Council of Provence- Alpes-Côte d’Azur and with of the A∗MIDEX (n° ANR- 11-IDEX-0001-02), funded by the Investissements d'Avenir project funded by the French Government, managed by the French National Research Agency (ANR). We thank Q. Tseng for generously sharing his set of macros, under the form of a shared project in the frame of the CENTURI engineer platform (https://centuri-livingsystems.org/multi-engineering-platform/). The authors want to personally thank him for his dedication and help over the very troubled pandemic times.
Author contributions
F.M. did the experimental work, analyzed the data, and wrote the protocol. K.S. designed the study, performed some pilot experiments, and reviewed the protocol. P.H.P. led the experiments, implemented novel analysis, and reviewed the protocol.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101133.
Contributor Information
Farah Mustapha, Email: farah.mustapha@inserm.fr.
Kheya Sengupta, Email: sengupta@cinam.univ-mrs.fr.
Pierre-Henri Puech, Email: pierre-henri.puech@inserm.fr.
Data and code availability
Images and data sets used here can be obtained from the authors upon request.
Softwares can be obtained from online resources (for Fiji/ImageJ and Plugins, see text). Specific Fiji/ImageJ and Python codes can be found online on https://github.com/phpuech/TFM and https://github.com/remyeltorro/SPTAlign.
References
- Aramesh M., Mergenthal S., Issler M., Plochberger B., Weber F., Qin X.-H., Liska R., Duda G.N., Huppa J.B., Ries J., et al. Functionalized bead assay to measure three-dimensional traction forces during T-cell activation. Nano Lett. 2021;21:507–514. doi: 10.1021/acs.nanolett.0c03964. [DOI] [PubMed] [Google Scholar]
- Butler J.P., Tolić-Nørrelykke I.M., Fabry B., Fredberg J.J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- Cazaux S., Sadoun A., Biarnes-Pelicot M., Martinez M., Obeid S., Bongrand P., Limozin L., Puech P.-H. Synchronizing atomic force microscopy force mode and fluorescence microscopy in real time for immune cell stimulation and activation studies. Ultramicroscopy. 2016;160:168–181. doi: 10.1016/j.ultramic.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Dembo M., Wang Y.-L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González C., Chames P., Kerfelec B., Baty D., Robert P., Limozin L. Nanobody-CD16 catch bond reveals NK cell mechanosensitivity. Biophys. J. 2019;116:1516–1526. doi: 10.1016/j.bpj.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.J., Oak Y., Groisman A., Danuser G. Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods. 2015;12:653–656. doi: 10.1038/nmeth.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiel J.-L., Leal A., Kurzawa L., Balland M., Wang I., Vignaud T., Tseng Q., Théry M. In: Methods in Cell Biology. Paluch Ewa K., editor. Elsevier; 2015. Measurement of cell traction forces with ImageJ; pp. 269–287. [DOI] [PubMed] [Google Scholar]
- Sadoun A., Biarnes-Pelicot M., Ghesquiere-Dierickx L., Wu A., Théodoly O., Limozin L., Hamon Y., Puech P.-H. Controlling T cells spreading, mechanics and activation by micropatterning. Sci. Rep. 2021;11:6783. doi: 10.1038/s41598-021-86133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Tse J.R., Engler A.J. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 2010;47:10–16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- Tseng Q., Duchemin-Pelletier E., Deshiere A., Balland M., Guillou H., Filhol O., Thery M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl. Acad. Sci. U S A. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie is 1 frame every 5 s.
The movie is 1 frame every 5 s.
Data Availability Statement
Images and data sets used here can be obtained from the authors upon request.
Softwares can be obtained from online resources (for Fiji/ImageJ and Plugins, see text). Specific Fiji/ImageJ and Python codes can be found online on https://github.com/phpuech/TFM and https://github.com/remyeltorro/SPTAlign.