Abstract
Omics methodologies are widely used in toxicological research to understand modes and mechanisms of toxicity. Increasingly, these methodologies are being applied to questions of regulatory interest such as molecular point-of-departure derivation and chemical grouping/read-across. Despite its value, widespread regulatory acceptance of omics data has not yet occurred. Barriers to the routine application of omics data in regulatory decision making have been: 1) lack of transparency for data processing methods used to convert raw data into an interpretable list of observations; and 2) lack of standardization in reporting to ensure that omics data, associated metadata and the methodologies used to generate results are available for review by stakeholders, including regulators. Thus, in 2017, the Organisation for Economic Co-operation and Development (OECD) Extended Advisory Group on Molecular Screening and Toxicogenomics (EAGMST) launched a project to develop guidance for the reporting of omics data aimed at fostering further regulatory use. Here, we report on the ongoing development of the first formal reporting framework describing the processing and analysis of both transcriptomic and metabolomic data for regulatory toxicology. We introduce the modular structure, content, harmonization and strategy for trialling this reporting framework prior to its publication by the OECD.
Keywords: transcriptomics, metabolomics, toxicology, OECD, transcriptomics reporting framework, TRF, metabolomics reporting framework, MRF, regulatory, QA/QC
1. Introduction
Omics methodologies are widely used in toxicological research to understand mechanisms of toxicity. Strategies for regulatory applications of omics data have continued to evolve in parallel, particularly with regard to informing read-across and identifying potential point(s)-of-departure for use in chemical risk assessment (Krewski et al. 2020), for example as submitted to the European Chemicals Agency under the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) legislation (Sperber et al. 2019). Although a suite of omics technologies has been developed over the last two decades, each to measure a specific class of biological molecules, transcriptomics and metabolomics have emerged as the principal approaches in toxicology. Today, for example, high-throughput transcriptomic platforms are available to rapidly and cost-effectively produce toxicogenomic data (Alpern et al. 2019; Mav et al. 2018; Verbist et al. 2019; Yeakley et al. 2017) on an unprecedented scale (Harrill et al. 2019; Igarashi et al. 2015; Subramanian et al. 2017). These data can be analysed to identify molecular initiating events and key events in adverse outcome pathways (AOPs) (Brockmeier et al. 2017; Corton et al. 2020; Heusinkveld et al. 2018; Ramaiahgari et al. 2019). In addition, diverse studies have consistently shown that points-of-departure derived from changes in gene expression are predictive of points-of-departure from conventional ‘endpoint’ studies, supporting their use in risk assessment (Gwinn et al. 2020; Johnson et al. 2020; Thomas et al. 2013). Similarly, high-throughput metabolomic platforms can generate data at scale (Dunn et al. 2011; Southam et al. 2016), enabling the discovery of metabolic biomarkers that are predictive of adverse outcomes in human and environmental toxicology (Hines et al. 2010; Palmer et al. 2020; Taylor et al. 2018; Zurlinden et al. 2020). In read-across, chemical grouping is based on an a priori hypothesis that substances in the group cause toxicity via a common mechanism. Transcriptomic and metabolomic measurements can be used to provide a biologically-based rationale for forming a chemical group, thereby facilitating robust read-across (De Abrew et al. 2019; Low et al. 2013; Sperber et al. 2019; van Ravenzwaay et al. 2016). For transcriptomics and metabolomics, case studies developed among the research and regulatory communities are beginning to build consensus on their use in different decision-making contexts (Cote et al. 2016; Kavlock et al. 2018; Krewski et al. 2020).
Despite this progress, regulatory acceptance of omics data is not widespread. The European Centre for Ecotoxicology and Toxicology of Chemicals (ECETOC) held a workshop in October 2016 to identify barriers to regulatory acceptance and directly discuss how to overcome these obstacles to facilitate regulatory acceptance and usage of omics data. The workshop discussions confirmed that following best practices when performing omics studies for regulatory purposes will be important for increasing regulatory acceptance and use. However, participants felt that overly prescriptive guidance/protocols for collection and analysis of omics data might not be helpful, as omics technologies and analysis workflows are constantly evolving, and fit-for-purpose approaches are often required. Instead, a reporting framework was suggested as a way forward, to describe how omics data are generated, processed, analysed and stored. Meeting participants believed that such a reporting framework would contribute to establishing a baseline for best practice, thereby facilitating the regulatory applicability of omics data (Buesen et al. 2017; Gant et al. 2017; Sauer et al. 2017). Aside from this benefit, there is also an immediate need for a reporting framework to facilitate the ongoing submission of omics data to regulatory agencies. Such a framework would ensure consistent, transparent and complete reporting of omics data, as is routinely applied to more traditional toxicity assays.
To address these needs, in 2017, the Organisation for Economic Co-operation and Development (OECD) Extended Advisory Group on Molecular Screening and Toxicogenomics (EAGMST) launched a project to develop guidance for reporting omics data types. It started with the development of a Transcriptomics Reporting Framework (TRF) and was followed in 2018 by the Metabolomics Reporting Framework (MRF). The project focuses specifically on reporting omics studies in toxicology and is not intended to recommend best practices. Its primary purpose is to address the aforementioned barriers to the adoption of omics data in regulatory toxicology and to foster and encourage international acceptance and use of omics data, for example in the context of the OECD’s programme on chemical safety (https://www.oecd.org/chemicalsafety/). Both the TRF and MRF were undertaken by international teams of experts from government agencies, regulatory bodies, industry and academia, who were tasked with developing reporting templates and supporting guidance for completing those templates, and then to trial them extensively. Since the inception of the project, the content of the TRF and MRF has been harmonized where possible, and they now exist as a single, integrated, modular framework. In the sections below we introduce the scope (section 2.1), modular structure of the integrated TRF/MRF framework (section 2.2), technology-specific details of the TRF (section 2.3) and MRF (section 2.4), and finally the approach used to trial this framework (section 2.5).
2. OECD omics reporting framework
There are a variety of previously established frameworks for reporting of data from transcriptomic and metabolomic studies (Brazma et al. 2001; Conesa et al. 2016; Haug et al. 2020; Parkinson et al. 2005). These frameworks focus primarily upon annotation of data (raw and normalized), samples, meta-data, sample-to-data relationships and technology-specific feature annotation, not the specific analysis steps required to obtain an interpretable result. The steps recommended to produce an interpretable result are the focus of initiatives such as the Omics Data Analysis Framework for Regulatory Application (R-ODAF) (Verheijen et al. 2020) and the National Toxicology Program’s (NTP) Approach to Genomic Dose-Response Modeling (NTP 2018). These initiatives prescribe steps for the analysis of omics data toward results of potential regulatory interest. However, the focus of R-ODAF and the NTP’s activities was not on reporting.
For metabolomics, the ECETOC MEtabolomics standaRds Initiative in Toxicology (MERIT) project brought together a team of international experts from industry, government agencies, regulators and academia in 2017 to begin to address this reporting need. Best practice guidelines and minimal reporting standards for the acquisition, processing and statistical analysis of untargeted metabolomics and targeted metabolite data in the context of regulatory toxicology were developed (Viant et al. 2019). While the MERIT project successfully drew attention to the metabolomics platform, it did not attempt to develop a reporting framework under the auspices of an international organisation.
Currently, no formal reporting structure exists that captures all of the steps and resources needed to perform analyses and foster reproducibility, transparency and eventual uptake of single or multi-omics data in the regulatory toxicology arena.
2.1. Scope
The scope of the OECD omics reporting framework was defined by its intended application in regulatory toxicology, specifically to report laboratory-based toxicological experiments. Furthermore, the omics reporting framework must integrate alongside the existing reporting of non-omics toxicity data, hence relevant databases and reporting templates (e.g., OECD harmonised reporting templates, OHTs) were carefully considered. Specifically, reporting fields relevant to the toxicology experiment were developed with the aim of aligning with existing OHTs such as OHT 201 (intermediate effects) (https://www.oecd.org/ehs/templates/harmonised-templates-intermediate-effects.htm) and OHT 211 (non-guideline in vitro studies) (https://ntp.niehs.nih.gov/iccvam/suppdocs/feddocs/oecd/oecd-gd211-2014-508.pdf). To the greatest extent possible, the reporting elements relative to both transcriptomics and metabolomics have been harmonized (e.g., for in vivo and in vitro experimental design and common data analysis applications) to increase familiarity and reduce the complexity of the framework for the data submitters and end users. Technology-specific reporting modules have been developed for mature, stable and proven omics technologies, but can be updated with newer omics technologies as they become available. The scope includes both untargeted omics (i.e., broad profiling of gene expression or the metabolic responses of a given sample) and targeted analysis (i.e., measurement of a small number of pre-defined genes or metabolic biomarkers). Ultimately, the framework is intended to describe the essential information that should be reported when an omics technology is applied in the context of regulatory decision-making. This information comprises the omics data and metadata, methodologies used for producing, processing and analysing the data, and the results of these analyses being used in the regulatory assessment. The reporting should allow a regulator to check compliance standards and omics study quality, from its design through to the collection, processing and statistical analysis of the data. There are also benefits to the submitter, including a clear expectation of what reporting is expected by the regulators as well as clarity on how the omics data will be assessed. One goal of the framework is to maximise the likelihood that the findings of regulatory relevance can be reproduced. Adherence to such a reporting framework is also intended to promote transparency, data sharing and the meta-analysis of omics datasets for all stakeholders.
2.2. Modular structure of reporting framework
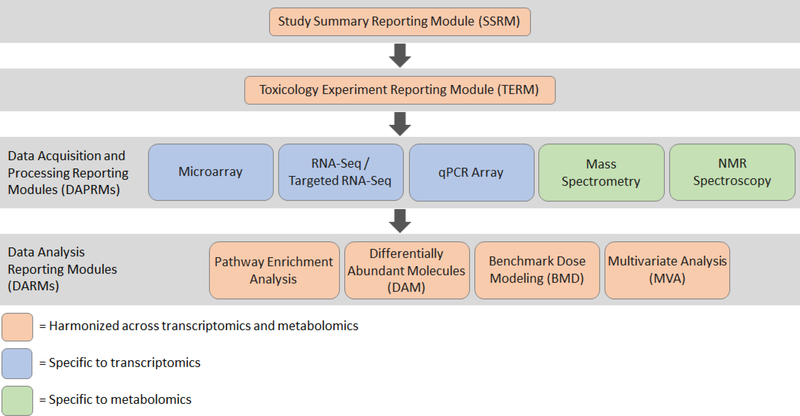
The framework is based on a modular structure that facilitates the updating of existing technologies and data analysis strategies, and that allows the development of additional modules for new technologies, data analysis approaches or omics disciplines such as proteomics or epigenomics. The modular structure is presented in Figure 1. Where possible, reporting elements that are relevant to both transcriptomics and metabolomics studies were harmonized and employ the same reporting modules.
Figure 1. Modular structure of the omics reporting framework for transcriptomics and metabolomics.
Four principal types of modules are included: Study Summary Reporting Module (SSRM) describing a subset of reporting elements to provide a high level overview of the whole study; Toxicology Experiment Reporting Module (TERM) describing the in vivo or in vitro toxicology study; Data Acquisition and Processing Reporting Modules (DAPRM) describing the omics assays, data acquisition and processing; and Data Analysis Reporting Modules (DARM) describing the statistical analysis of the omics data. Orange modules are harmonized across transcriptomics and metabolomics, blue modules are specific to transcriptomics, and green modules are specific to metabolomics.
Four types of modules are included:
Study Summary Reporting Module (SSRM) - describes a subset of reporting elements in order to provide a high-level overview of a regulatory toxicology and omics experiment; used for transcriptomics and metabolomics.
Toxicology Experiment Reporting Module (TERM) - captures and reports the key descriptors of an in vivo or in vitro toxicology study from which samples are derived for omics analysis; used for transcriptomics and metabolomics.
Data Acquisition and Processing Reporting Modules (DAPRMs) - capture and report descriptions of the omics assays, data acquisition and associated data processing prior to statistical analysis; these modules are unique to each omics data type.
Data Analysis Reporting Modules (DARMs) - capture and report descriptions of the statistical analysis that is often undertaken in an omics study, e.g., for the purposes of discovering differentially abundant transcripts or metabolites; these are used for transcriptomics and/or metabolomics.
The framework consists of a narrative guidance document that describes the modules in detail, with a complementary reporting template for each module (currently in the form of a multi-workbook Microsoft Excel file) (https://www.oecd.org/chemicalsafety/testing/omics.htm). Furthermore, each reporting element within the modules is listed as either ‘required’ or ‘optional’, the former defining the essential elements for the minimum reporting standards of an omics study. Practically, to report an omics study, scientists should select the relevant reporting modules, minimally comprising one each of the SSRM, TERM, DAPRM and DARM. The narrative guidance should be used in parallel with the reporting templates to guide their completion.
2.3. Transcriptomics Reporting Framework (TRF)
Transcriptomics technologies included in this work to date are those generally considered to be most broadly used in toxicogenomic studies. At present, DAPRM modules have been developed for DNA microarrays, RNA-sequencing (RNA-Seq; including both global and targeted RNA-Seq technologies), and quantitative real-time PCR arrays. These modules typically begin with a brief description of the technology used, followed by RNA extraction and quality control evaluation of sample integrity (if applicable). The experimental design including sample pooling and batch processing is reported as well as the specific types of controls, how they are used and descriptions of their limits of acceptability. Sample preparation (e.g., complementary RNA labeling or RNA-Seq library preparation) is described along with links to or submission of standard operating procedures where available. Following this, data acquisition, processing (e.g., normalization), quality evaluation, and gene quantification protocols are reported, followed by post-normalization filtering (e.g., identification and removal of outliers). Finally, reporting fields for linking to publicly available code and accession numbers to public omics data repositories are included.
The processed gene expression data are intended to be paired with a DARM to produce data sets for use in regulatory evaluation. This could include a variety of analyses: derivation of differentially expressed genes, pathway or gene set enrichment analysis, clustering approaches for exploring correlation in transcriptomic profiles (e.g., hierarchical clustering or principal component analysis), transcriptomic biomarker analyses, or benchmark dose (BMD) analysis. In recognition of the fact that many of these analyses are applicable to both transcriptomics and metabolomics workflows, the DARMs were designed to accommodate both. For example, because the identification of differentially expressed genes (i.e., expression changes relative to matched controls) is analogous to the identification of differentially abundant metabolites, a single DARM was developed termed the Differentially Abundant Molecules (DAM) module.
2.4. Metabolomics Reporting Framework (MRF)
The MRF has been designed to enable the reporting of untargeted metabolomics and targeted metabolite analysis, both of which are important in regulatory toxicology. In addition, the framework includes the ability to report hybrid approaches that include both targeted and untargeted analyses of biological samples. Based on international surveys (Weber et al. 2015; Weber et al. 2017), the four most widely applied analytical methods in metabolomics have been described: liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), direct infusion mass spectrometry (DI-MS) and nuclear magnetic resonance (NMR) spectroscopy. The essential information to be reported includes the experimental design, quality assurance and quality control, sampling of biological specimens, extraction of metabolites, data acquisition and processing of untargeted, targeted or hybrid assays, metabolite annotation and/or identification of the metabolites, and a range of statistical analyses. To date, the following modules have been developed for metabolomics studies: SSRM, TERM, MS and NMR Spectroscopy DAPRMs, and DAM, BMD and Multivariate Analysis (MVA) DARMs.
2.5. Approach to trialling the reporting framework
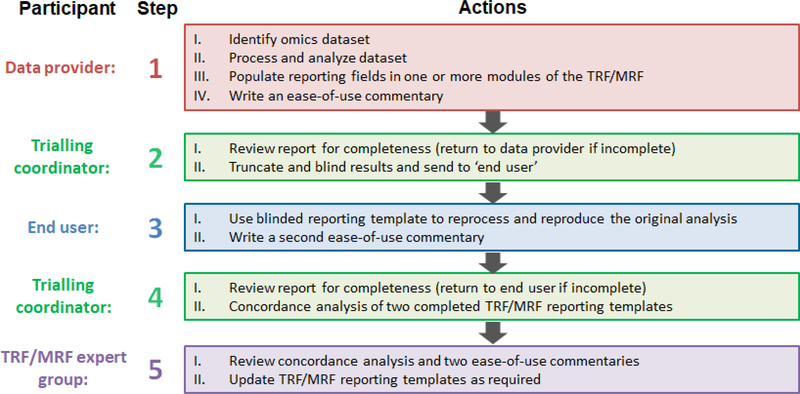
To ensure that the reporting framework is fit-for-purpose, each completed reporting module is being reviewed for clarity, completeness, utility and ease of use, prior to publication by the OECD. To do this, the framework is being evaluated via multiple trials, each run in five phases (Figure 2). Phase 1 is designed to be an initial evaluation of the reporting framework by a ‘data provider’, in which the relevant reporting fields in one or more modules are populated with methods, data, metadata and results from an omics study, and an ease-of-use commentary is written. In Phase 2, the ‘trial coordinators’ check the reporting framework for completeness, and then truncate and blind the results. The completed documentation and truncated, blinded results are then sent to an ‘end user’, and in Phase 3 they attempt to reproduce the data processing and/or statistical analyses (dependent on the module(s) being trialled) according to the information and data originally populated by the ‘data provider’. The ‘end user’ also writes an ease-of-use commentary. Next, in Phase 4, the ‘trial coordinators’ check this second reporting framework for completeness, and then consolidate all of the reports to enable a concordance analysis of the two sets of results. Finally, the TRF/MRF expert groups review the concordance analysis and ease-of-use commentaries and amend the reporting templates and guidance as needed to ensure their clarity and utility. Each concordance analysis (from each trial) will be used to determine whether a further trial is required, based on the similarity of the two sets of results. We regard the demonstration of reproducibility of the data processing and analysis (e.g. achieving the same result when data is processed by two (or more) independent entities), based only on the information provided within the reporting template, as an important step toward confirming the completeness of the framework and hence the acceptance by the OECD. Six trials are currently underway, four focused on transcriptomics and two focused on metabolomics, with publications to follow.
Figure 2. Approach to trialling the reporting framework.
The modules within the framework are currently being reviewed for clarity, completeness, utility and ease of use through six trials. Each trial comprises five phases, involving a data provider (initial analysis –red box), end user (re-analysis – blue box), a trialling coordinator (green boxes) and the TRF or MRF expert groups (purple box). For a module to be approved, it must enable an end user to reproduce the analysis of an omics dataset, relative to the initial analysis by the data provider.
3. Conclusions
We report the ongoing development of the first formal reporting framework to describe the processing and analysis of both transcriptomics and metabolomics data in regulatory toxicology. The completed framework is now undergoing trials to evaluate utility and performance. The findings from these trials will be used to refine the reporting modules to ensure that they achieve the goals of clarity and utility, demonstrated by the reproducibility of results from omics studies. These reporting modules will then be reviewed by the OECD before publication as formal OECD Guidance Documents. Future activities include the development of reporting modules that describe how results from omics studies should be summarized and reported in the context of specific regulatory applications such as chemical grouping / read-across and point-of-departure derivation.
Highlights.
Omics reporting framework established for regulatory toxicology
Modular structure accommodates various technologies and analyses
Harmonization of reporting fields for transcriptomics and metabolomics
Tool for documenting analysis steps used to generate interpretable results from omics data
Funding
TWG was supported by the National Institutes of Health Research under the Environmental Exposures and Health Protection Research Unit co-led from Imperial College London and Public Health England (https://eeh.hpru.nihr.ac.uk/). ML was supported by the Dutch Ministry of Health, Welfare and Support. This work was supported in part by a European Chemicals Agency contract to Michabo Health Science (ECHA/2018/135) and by the European Chemical Industry Council (Cefic; awarded to BASF, Imperial College London, Syngenta, University of Birmingham and Vrije Universiteit Amsterdam).
Abbreviations
- AOP
Adverse Outcome Pathway
- BMD
Benchmark Dose
- DAM
Differentially Abundant Molecules
- DAPRMs
Data Acquisition and Processing Reporting Modules
- DARMs
Data Analysis Reporting Modules
- DI-MS
Direct Infusion Mass Spectrometry
- EAGMST
Extended Advisory Group on Molecular Screening and Toxicogenomics
- ECETOC
European Centre for Ecotoxicology and Toxicology of Chemicals
- GC-MS
Gas Chromatography-Mass Spectrometry
- LC-MS
Liquid Chromatography-Mass Spectrometry
- MERIT
MEtabolomics standaRds Initiative in Toxicology
- MRF
Metabolomics Reporting Framework
- MS
Mass Spectrometry
- MVA
Multivariate Analysis
- NMR
Nuclear Magnetic Resonance spectroscopy
- NTP
National Toxicology Program
- OECD
Organisation for Economic Co-operation and Development
- OHT
OECD Harmonised Template
- QA/QC
Quality Assurance / Quality Control
- qPCR
Quantitative Polymerase Chain Reaction
- RNA-Seq
RNA sequencing
- R-ODAF
Omics Data Analysis Framework for Regulatory Application
- SSRM
Study Summary Reporting Module
- TERM
Toxicology Experiment Reporting Module
- TRF
Transcriptomics Reporting Framework
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Disclaimer
The opinions expressed and arguments employed herein are those of the authors and do not necessarily reflect the official views of the OECD or of the governments of its member countries. This manuscript has been reviewed in accordance with official U.S. EPA policy and approved for publication. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The findings and conclusions in this article do not necessarily represent the views or policies of the U.S. EPA. In addition, this article should not be construed to represent FDA’s views or policies.
References
- Alpern D, Gardeux V, Russeil J, Mangeat B, Meireles-Filho ACA, Breysse R, Hacker D, Deplancke B. 2019. Brb-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk rna barcoding and sequencing. Genome Biol. 20(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC et al. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 29(4):365–371. [DOI] [PubMed] [Google Scholar]
- Brockmeier EK, Hodges G, Hutchinson TH, Butler E, Hecker M, Tollefsen KE, Garcia-Reyero N, Kille P, Becker D, Chipman K et al. 2017. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol Sci. 158(2):252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesen R, Chorley BN, da Silva Lima B, Daston G, Deferme L, Ebbels T, Gant TW, Goetz A, Greally J, Gribaldo L et al. 2017. Applying ‘omics technologies in chemicals risk assessment: Report of an ECETOC workshop. Regul Toxicol Pharmacol. 91 Suppl 1:S3–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JC, Hill T, Sutherland JJ, Stevens JL, Rooney J. 2020. A set of six gene expression biomarkers identify rat liver tumorigens in short-term assays. Toxicol Sci. 177(1):11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, Bois FY, Burgoon LD, Chiu WA, Crawford-Brown D et al. 2016. The next generation of risk assessment multi-year study-highlights of findings, applications to risk assessment, and future directions. Environ Health Perspect. 124(11):1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Abrew KN, Shan YK, Wang X, Krailler JM, Kainkaryam RM, Lester CC, Settivari RS, LeBaron MJ, Naciff JM, Daston GP. 2019. Use of connectivity mapping to support read across: A deeper dive using data from 186 chemicals, 19 cell lines and 2 case studies. Toxicology. 423:84–94. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN et al. 2011. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 6(7):1060–1083. [DOI] [PubMed] [Google Scholar]
- Gant TW, Sauer UG, Zhang SD, Chorley BN, Hackermuller J, Perdichizzi S, Tollefsen KE, van Ravenzwaay B, Yauk C, Tong W et al. 2017. A generic transcriptomics reporting framework (trf) for ‘omics data processing and analysis. Regul Toxicol Pharmacol. 91 Suppl 1:S36–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn WM, Auerbach SS, Parham F, Stout MD, Waidyanatha S, Mutlu E, Collins B, Paules RS, Merrick BA, Ferguson S et al. 2020. Evaluation of 5-day in vivo rat liver and kidney with high-throughput transcriptomics for estimating benchmark doses of apical outcomes. Toxicol Sci. 176(2):343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J, Shah I, Setzer RW, Haggard D, Auerbach S, Judson R, Thomas RS. 2019. Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr Opin Toxicol. 15:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, O’Donovan C. 2020. Metabolights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48(D1):D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusinkveld HJ, Wackers PFK, Schoonen WG, van der Ven L, Pennings JLA, Luijten M. 2018. Application of the comparison approach to open tg-gates: A useful toxicogenomics tool for detecting modes of action in chemical risk assessment. Food Chem Toxicol. 121:115–123. [DOI] [PubMed] [Google Scholar]
- Hines A, Staff FJ, Widdows J, Compton RM, Falciani F, Viant MR. 2010. Discovery of metabolic signatures for predicting whole organism toxicology. Toxicol Sci. 115(2):369–378. [DOI] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H. 2015. Open tg-gates: A large-scale toxicogenomics database. Nucleic Acids Res. 43(Database issue):D921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KJ, Auerbach SS, Costa E. 2020. A rat liver transcriptomic point of departure predicts a prospective liver or non-liver apical point of departure. Toxicol Sci. 176(1):86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock RJ, Bahadori T, Barton-Maclaren TS, Gwinn MR, Rasenberg M, Thomas RS. 2018. Accelerating the pace of chemical risk assessment. Chem Res Toxicol. 31(5):287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D, Andersen ME, Tyshenko MG, Krishnan K, Hartung T, Boekelheide K, Wambaugh JF, Jones D, Whelan M, Thomas R et al. 2020. Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch Toxicol. 94(1):1–58. [DOI] [PubMed] [Google Scholar]
- Low Y, Sedykh A, Fourches D, Golbraikh A, Whelan M, Rusyn I, Tropsha A. 2013. Integrative chemical-biological read-across approach for chemical hazard classification. Chem Res Toxicol. 26(8):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mav D, Shah RR, Howard BE, Auerbach SS, Bushel PR, Collins JB, Gerhold DL, Judson RS, Karmaus AL, Maull EA et al. 2018. A hybrid gene selection approach to create the s1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS One. 13(2):e0191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. 2018. NTP research report on national toxicology program approach to genomic dose-response modeling: Research report 5. Durham (NC). [PubMed]
- Palmer JA, Smith AM, Gryshkova V, Donley ELR, Valentin JP, Burrier RE. 2020. A targeted metabolomics-based assay using human induced pluripotent stem cell-derived cardiomyocytes identifies structural and functional cardiotoxicity potential. Toxicol Sci. 174(2):218–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M et al. 2005. Arrayexpress--a public repository for microarray gene expression data at the ebi. Nucleic Acids Res. 33(Database issue):D553–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, Auerbach SS, Saddler TO, Rice JR, Dunlap PE, Sipes NS, DeVito MJ, Shah RR, Bushel PR, Merrick BA et al. 2019. The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicol Sci. 169(2):553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer UG, Deferme L, Gribaldo L, Hackermuller J, Tralau T, van Ravenzwaay B, Yauk C, Poole A, Tong W, Gant TW. 2017. The challenge of the application of ‘omics technologies in chemicals risk assessment: Background and outlook. Regul Toxicol Pharmacol. 91 Suppl 1:S14–S26. [DOI] [PubMed] [Google Scholar]
- Southam AD, Weber RJ, Engel J, Jones MR, Viant MR. 2016. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat Protoc. 12(2):310–328. [DOI] [PubMed] [Google Scholar]
- Sperber S, Wahl M, Berger F, Kamp H, Lemke O, Starck V, Walk T, Spitzer M, Ravenzwaay BV. 2019. Metabolomics as read-across tool: An example with 3-aminopropanol and 2-aminoethanol. Regul Toxicol Pharmacol. 108:104442. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 171(6):1437–1452 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Gavin A, Viant MR. 2018. Metabolomics discovers early-response metabolic biomarkers that can predict chronic reproductive fitness in individual daphnia magna. Metabolites. 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Wesselkamper SC, Wang NC, Zhao QJ, Petersen DD, Lambert JC, Cote I, Yang L, Healy E, Black MB et al. 2013. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci. 134(1):180–194. [DOI] [PubMed] [Google Scholar]
- van Ravenzwaay B, Sperber S, Lemke O, Fabian E, Faulhammer F, Kamp H, Mellert W, Strauss V, Strigun A, Peter E et al. 2016. Metabolomics as read-across tool: A case study with phenoxy herbicides. Regul Toxicol Pharmacol. 81:288–304. [DOI] [PubMed] [Google Scholar]
- Verbist B, Adriaensen E, Keersmaekers V, Putri D, Crabbe M, Derks M, Bagdziunas R, Laenen G, De Wolf H. 2019. Analyzing magnetic bead qQuantiGene® Plex 2.0 gene expression data in high throughput mode using QGprofiler. BMC Bioinformatics. 20(1):378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen M, Tong W, Shi L, Gant TW, Seligman B, Caiment F. 2020. Towards the development of an omics data analysis framework. Regul Toxicol Pharmacol. 112:104621. [DOI] [PubMed] [Google Scholar]
- Viant MR, Ebbels TMD, Beger RD, Ekman DR, Epps DJT, Kamp H, Leonards PEG, Loizou GD, MacRae JI, van Ravenzwaay B et al. 2019. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat Commun. 10(1):3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RJ, Winder CL, Larcombe LD, Dunn WB, Viant MR. 2015. Training needs in metabolomics. Metabolomics. 11(4):784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RJM, Lawson TN, Salek RM, Ebbels TMD, Glen RC, Goodacre R, Griffin JL, Haug K, Koulman A, Moreno P et al. 2017. Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through galaxy. Metabolomics. 13(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Shepard PJ, Goyena DE, VanSteenhouse HC, McComb JD, Seligmann BE. 2017. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One. 12(5):e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurlinden TJ, Saili KS, Rush N, Kothiya P, Judson RS, Houck KA, Hunter ES, Baker NC, Palmer JA, Thomas RS et al. 2020. Profiling the toxcast library with a pluripotent human (h9) stem cell line-based biomarker assay for developmental toxicity. Toxicol Sci. 174(2):189–209. [DOI] [PMC free article] [PubMed] [Google Scholar]