Abstract
Background and Objectives
Migraine, stroke, and cervical artery dissection (CeAD) represent a triad of cerebrovascular disorders with pairwise comorbid relationships and vascular involvement. Larger samples and recent advances in methodology invite systematic exploration of their shared genetics.
Methods
Genetic analyses leveraged summary statistics from genome-wide association studies of the largest available samples of each disorder, including subtypes of stroke (ischemic stroke, large artery stroke, small vessel stroke, and cardioembolic stroke) and migraine (with aura and without aura). For each pair of disorders, genetic correlation was assessed both on a genome-wide basis and within independent segments across the genome including known specific loci for each disorder. A cross-trait meta-analysis was used to identify novel candidate loci. Finally, potential causality of migraine susceptibility on stroke and CeAD was assessed by Mendelian randomization.
Results
Among all pairs of disorders, genome-wide genetic correlation was observed only between CeAD and migraine, particularly MO. Local genetic correlations were more extensive between migraine and CeAD than those between migraine and stroke or CeAD and stroke and revealed evidence for novel CeAD associations at rs6693567 (ADAMTSL4/ECM1), rs11187838 (PLCE1), and rs7940646 (MRVI1) while strengthening prior subthreshold evidence at rs9486725 (FHL5) and rs650724 (LRP1). At known migraine loci, novel associations with stroke had concordant risk alleles for small vessel stroke at rs191602009 (CARF) and for cardioembolic stroke at rs55884259 (NKX2-5). Known migraine loci also revealed novel associations but with opposite risk alleles for all stroke, ischemic stroke, and small vessel stroke at rs55928386 (HTRA1), for large artery stroke at rs11172113 (LRP1), and for all stroke and ischemic stroke at rs1535791 and rs4942561 (both LRCH1), respectively. rs182923402 (near PTCH1) was a novel concordant locus for migraine and cardioembolic stroke. Mendelian randomization supported potential causal influences of migraine on CeAD (odds ratio [95% confidence interval] per doubling migraine prevalence = 1.69 [1.24–2.3], p = 0.0009) with concordant risk, but with opposite risk on large artery stroke (0.86 [0.76–0.96], p = 0.0067).
Discussion
The findings emphasize shared genetic risk between migraine and CeAD while identifying loci with likely vascular function in migraine and shared but opposite genetic risk between migraine and stroke subtypes, and a central role of LRP1 in all 3 cerebrovascular disorders.
Migraine, stroke, and extracranial cervical artery dissection (CeAD) represent a triad of brain disorders with vascular involvement and pairwise comorbid relationships that are pertinent to risk assessment and clinical care.1 While the shared clinical features of all 3 disorders point to vasculature as the basis of the comorbidity, precise underlying mechanisms are not established. Understanding the shared and distinct biological mechanisms has the potential to clarify the basis of shared risk while also informing potential prophylactic and treatment strategies.
From this perspective, previous investigations have leveraged the unique properties of human genetics to reveal shared biology among the 3 disorders while limiting the potential influence of reverse causality and confounding that may arise in conventional observational epidemiology. One study found that genome-wide genetic overlap with migraine was most significant for large artery stroke and significant for cardioembolic stroke (CE), contrary to observational associations that had linked migraine to small vessel disease.2,3 Associations were stronger for migraine without aura (MO) than those for either overall migraine or migraine with aura (MA), though the latter is a stronger risk factor of ischemic stroke (IS).4-6 Similarly, genetic associations at specific loci diverged from conventional observational associations. At the 9p21 locus, associations with stroke and MO had concordant direction, but there was no association with MA, and there remained uncertainty about whether the causal variants for MO and stroke at the locus were the same.3 At the FHL5 locus on chromosome 6, the associations with stroke and migraine were in opposite directions, while the same locus has been noted for a concordant association between migraine and CeAD, the latter from a genome-wide association study (GWAS) at subgenome-wide significance.7 The GWAS of CeAD also noted concordant effects with migraine at loci implicating the PHACTR1/EDN1 and LRP1 genes,8 but only the former was replicated in an independent follow-up sample.
Recent GWASs of stroke and migraine incorporating much larger samples than previously available (therefore with much greater power), as well as novel genetic methods and the lack of a systematic comparison among all 3 cerebrovascular disorders, invite a new genetic analysis toward resolving several outstanding questions. First, what is the extent of shared genetics among the 3 disorders? Second, which specific susceptibility loci are shared on a genome-wide basis? Finally, does human genetics support causal relationships underlying the increased risk of stroke and CeAD among individuals susceptible to migraine?
Methods
Overview
Pairwise genetic relationships among migraine, CeAD, and subtypes of stroke were examined using 4 analytic strategies. Genome-wide genetic correlations were calculated to assess for overall genetic sharing. Local genetic correlations were calculated to assess shared genetics within disjoint segments across the genome and at specific candidate loci previously identified for association with at least one of the disorders. A genome-wide cross-trait association analysis was implemented to identify novel variant associations for each trait. Finally, the Mendelian randomization (MR) analysis was performed to assess potential causal influences of migraine on the other cerebrovascular disorders.
Summary Statistics
Analyses used discovery summary statistics from published, consortium-based GWASs of migraine,9 CeAD,7 and stroke.10 The total numbers of samples included in these summary statistics were as follows: any migraine (59,674 cases/316,078 controls), MA (6,332 cases/144,883 controls), MO (8,348 cases/139,622 controls), CeAD (carotoid and vertebral, 1,393 cases/14,416 controls), all stroke (AS) (40,584 cases/406,111 controls), IS (34,217 cases/406,111 controls), large artery stroke (LAS) (4,373 cases/297,290 controls), CE (7,193 cases/355,4468 controls), and small vessel stroke (SVS) (5,386 cases/343,560 controls). All summary statistics were derived from study populations exclusively with European ancestry. The migraine and stroke GWASs were based on 1000 Genomes Project imputed data (hg19) and included approximately 8 million single nucleotide variations ([SNVs], formerly SNPs), while the CeAD GWAS was based on HapMap (hg18) and 1000 Genomes Project (August 2010 release) imputed data and included approximately 6.6 million SNVs. The Women's Genome Health Study (WGHS)11 provided a substantial proportion of cases and controls to the GWAS for MA (1,177 WGHS cases/6,332 total cases) and MO (1,826 WGHS cases/8,348 total cases) while also contributing a smaller proportion of cases to overall migraine (N = 5,122 WGHS cases/59,674 total cases) and stroke (N = 422 cases of all stroke/22,795 controls) GWASs. Other cohorts may have contributed smaller numbers of samples to the GWAS for migraine and stroke. Potential bias due to this overlap was addressed in 2 ways. First, migraine summary statistics from a meta-analysis were as described but omitted the WGHS contribution.9 Second, the genetic correlation and cross-trait association methods intrinsically account for any minimal residual overlap.12-14
Summary statistics for the 23andMe cohort were obtained under an agreement with 23andMe that protects the privacy of the 23andMe participants. The participants of 23andMe provided informed consent and participated in the research under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services. The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Information about access the data from 23andMe can be found at research.23andme.com/collaborate/#dataset-access/. Use of other summary statistics wass consistent with the local IRBs of each of the contributing cohorts or samples. All genomic coordinates refer to genome build hg19.
Genome-wide Genetic Correlation
Two established methods were used to estimate a genome-wide genetic correlation with the GWAS summary statistics: the conventional approach, linkage disequilibrium (LD) score regression (LDSc, version 1.0.0), and a similar but potentially more powerful approach, Genetic Covariation Analyzer (GNOVA) (downloaded in December 2017).12,13 An analysis with LDSc incorporated precomputed LD measures for approximately 1.3 million common SNVs based on the HapMap with minimum minor allele frequency of approximately 10%12 and shared across all of the summary statistics. An analysis with GNOVA included a step to calculate LD relationships among individuals with European ancestry in the 1000 Genomes reference panel including approximately 6.1 million SNVs with minimum minor allele frequency 5%. Genome-wide genetic correlation is the principal estimate in LDSc. By contrast, genetic covariance is the principal estimate in GNOVA, and genetic correlation is derived by scaling with the single-trait heritability estimates. As such, p values refer to the genetic correlation in LDSc and to the genetic covariance in GNOVA. Both LDSc and GNOVA have options to compute genetic correlation or covariance while adjusting for potential sample overlap or other potential causes of inflation. These options were invoked in all analyses. Although LDSc and GNOVA are substantially similar methods and the use of both provides cross-validation, differences in the minor allele frequency thresholds are expected to influence genetic correlation estimates and significance to some extent.
Locally Shared Genetic Effects
Local (as opposed to genome-wide) genetic correlation was estimated by 2 approaches, applied initially to approximately 1,704 prespecified disjoint segments across the genome with minimal intersegment LD within the GWAS summary statistics.15 One approach, ρ Heritability Estimation from Summary Statistics (ρHESS) (version 0.5),16 provided a frequentist estimate of the local covariance of genetic effects, while the other approach, GWAS-pairwise (GWAS-PW) (version 0.21),14 provided a posterior probability of a locally shared genetic association (posterior probability of association in mode 3 [PPA3]) within each segment in an empirical Bayes framework based on GWAS p values. Genome-wide significance across the prespecified segments was 2.9 × 10−5 (=0.05/1,704) in ρHESS and PPA3 >0.9 in GWAS-PW. Candidate genes were assigned for regions showing PPA3 >0.9 based on proximity to potentially shared causal variants. ρHESS was also adapted to examine candidate segments surrounding genome-wide significant loci for stroke or migraine, as previously performed.16 In this study, candidate segments were defined to include the SNVs neighboring each of GWAS index SNV, such that all SNVs outside of the segment had LD r2 < 0.1 to the index SNV. Testing 69 candidate loci for each pairwise comparison, p < 7 × 10−4 (=0.05/69) was considered significant. As needed, pairwise SNV LD was estimated using a European ancestry panel from the 1000 Genomes Project with PLINK or LDlink.17-19
A Cross-Trait Association Analysis With a Multitrait Analysis of GWAS
A cross-trait association analysis was performed using a multitrait analysis of GWAS (MTAG, version 1.0.7), which leverages the pairwise genome-wide trait genetic correlation to boost power in association testing.20 In MTAG, LDSc provided the estimates of pairwise genetic correlation. The significance threshold in the MTAG required pMTAG < 1.67 × 10−8 (=5.00 × 10−8/3 phenotypes) but was also restricted to SNVs that also had nominal significance (p < 0.05) for each phenotype separately in the preexisting univariate GWAS.
MR Instrumental Analysis
Genetic instrumental variable analysis with MR was performed using the 2-sample method, prioritizing the random-effects inverse variance–weighted (IVW) estimator within the package TwoSampleMR in the R computing environment.21,22 Sensitivity analysis included MR-weighted median, MR-Egger, MR-Egger bootstrap, MR-robust adjusted profile score, and MR-PRESSO.23-26 The latter detects and excludes instruments that are consistent with horizontal pleiotropy, a violation of assumptions underlying MR, and then evaluates an overall estimate with the remaining instruments using the IVW method. Pleiotropy was also assessed with Cochran Q for heterogeneity and the MR-Egger intercept as recommended.25 MR was limited to effects of migraine on the other disorders because (1) only the GWAS of any migraine, i.e., not the other disorders, had a sufficient number of genome-wide significant SNVs (N ≥ 10) for use as exposure instruments and (2) because migraine typically precedes stroke or CeAD, potential causal effects of migraine on the other disorders are most consistent with temporal plausibility. Because migraine is a binary exposure, MR effect estimates were scaled by 0.693 for reporting to represent the odds of the outcome for a doubling of the odds of the exposure.27
Standard Protocol Approvals, Registrations, and Patient Consents
The GWAS summary statistics were all derived by a meta-analysis. Participants who contributed to cohort-level summary statistics constituting the meta-analyses provided written informed consent, and each of the cohort protocols was approved by a local institutional review board.
Data Availability
This study used only GWAS summary statistics from published reports as described earlier. The availability of these data or procedures for accessing them is documented in the cited publications. Summary statistics for the GWAS of migraine lacking contribution from the WGHS will be made available by sending an application to the International Headache Genetics Consortium via the corresponding author using the same procedure that governs access to the summary statistics for the published migraine study.9
Results
Genome-wide Genetic Correlation
Genetic correlations involving CeAD and stroke or migraine were generally positive (i.e., concordant) and comparable between LDSc and GNOVA, but significant only with GNOVA for the combinations of migraine (rg [ρ (ρ SE)] = 0.22 [0.048 (0.012)], p = 4.9e-05) or MO (0.29 [0.051 (0.016)], 0.0017) with CeAD, after accounting for multiple testing (Table 1). The estimated genetic correlations were larger but only nominally significant with LDSc (rg = 0.45 and 0.41, respectively).
Table 1.
Genome-wide Genetic Correlations Between Pairs of Brain Disorders
| Pheno 1 | Pheno 2 | LDSc rg (SE), p value | GNOVA rg (cov [cov SE]), p value |
| Any migraine | AS | 0.062 (0.049), 0.20 | 0.042 (0.001 [0.00077]), 0.20 |
| IS | 0.062 (0.047), 0.19 | 0.037 (0.00089 [0.00074]), 0.23 | |
| LAS | −0.36 (0.41), 0.38 | −0.097 (−0.0028 [0.001]), 0.006 | |
| CE | 0.05 (0.069), 0.46 | 0.014 (0.00035 [0.00087]), 0.69 | |
| SVS | 0.049 (0.093), 0.60 | 0.071 (0.0018 [0.00095]), 0.064 | |
| MO | AS | −0.036 (0.089), 0.68 | −0.037 (−0.00073 [0.0011]), 0.50 |
| IS | −0.03 (0.089), 0.74 | −0.065 (−0.0013 [0.0011]), 0.24 | |
| LAS | −0.72 (0.67), 0.28 | −0.15 (−0.0035 [0.0014]), 0.013 | |
| CE | −0.18 (0.11), 0.11 | −0.092 (−0.0019 [0.0012]), 0.13 | |
| SVS | −0.065 (0.16), 0.69 | 0.069 (0.0014 [0.0012]), 0.26 | |
| MA | AS | 0.059 (0.10), 0.57 | 0.098 (0.0019 [0.001]), 0.06 |
| IS | 0.061 (0.10), 0.54 | 0.078 (0.0015 [0.001]), 0.13 | |
| LAS | −0.64 (0.73), 0.39 | −0.018 (−0.00042 [0.0014]), 0.76 | |
| CE | −0.16 (0.13), 0.19 | −0.014 (−0.00029 [0.0012]), 0.81 | |
| SVS | −0.23 (0.20), 0.25 | 0.12 (0.0024 [0.0014]), 0.075 | |
| CeAD | AS | 0.27 (0.15), 0.081 | 0.13 (0.018 [0.0095]), 0.062 |
| IS | 0.22 (0.16), 0.15 | 0.13 (0.018 [0.01]), 0.076 | |
| LAS | 0.35 (0.67), 0.61 | 0.16 (0.025 [0.013]), 0.046 | |
| CE | 0.11 (0.20), 0.58 | 0.075 (0.011 [0.01]), 0.30 | |
| SVS | 0.16 (0.34), 0.64 | 0.093 (0.013 [0.013]), 0.30 | |
| Any migraine | 0.45 (0.17), 0.0077 | 0.22 (0.048 [0.012]), 4.9e-05 | |
| MA | 0.097 (0.23), 0.67 | −0.11 (−0.018 [0.015]), 0.24 | |
| MO | 0.41 (0.21), 0.05 | 0.29 (0.051 [0.016]), 0.0017 |
Abbreviations: AS = all stroke; CE = cardioembolic stroke; CeAD = cervical artery dissection; IS = ischemic stroke; LAS = large artery stroke; MA = migraine with aura; MO = migraine without aura; SVS = small vessel stroke.
Multiple testing significance threshold p = 0.002 (=0.05/23). Both LDSc and GNOVA values, corrected for estimated potential sample overlap and other potential sources of bias.
Local Genetic Correlation
At the experiment-wide significance threshold (PPA3 >0.9), GWAS-PW implicated novel locally concordant associations of migraine and CE on chromosome 9q22.32 (top SNV rs113154802 near PTCH1) (Table 2). The remaining significant segments all include loci previously recognized by GWAS for 1 or more of the disorders, although many are newly implicated for an additional disorder (indicated in bold). These loci were ADAMTSL4/ECM1: CeAD-migraine, CARF: SVS-migraine, NKX2: migraine-CE, HDAC9: migraine-AS/IS, ARMS2: AS/IS/SVS-migraine, LRCH1: migraine-AS/IS, and COL4A1: migraine-AS. At COL4A1, neither migraine nor AS is genome-wide significant in the summary statistics used in this study, which are derived from population of European ancestry alone, but the locus has been recently identified for stroke by a trans-ancestry meta-analysis with index SNV rs9521634.28 However, this variant is not in LD (r2 = 0.02, D′ = 0.52) with the top SNV for the joint analysis of stroke and migraine, rs650724, which is also the top SNV for stroke alone in the current summary statistics derived from European ancestry and in high LD (r2 = 0.86, D′ = 0.94) with the top SNV for migraine alone (rs2000660). Identification of shared, concordant associations involving CeAD in segments encompassing LRP1 and FHL5 is consistent with strong, subgenome-wide significant associations in these regions previously noted.7 Although already known for any migraine, these 2 loci are newly implicated in both MA and MO. None of the segments was significant for combinations of CeAD and stroke. In contrast to the GWAS-PW method, local genetic covariance assessed with ρHESS did not meet experiment-wide significance for any pairwise combination of the cerebrovascular disorders (all p for local rg > 2.9 × 10−5 [=0.05/1704]).
Table 2.
Local Prespecified Segments With Significant Joint GWAS-PW Association Among Cerebrovascular Conditions
| Pheno 1 | Pheno 2 | Segment | Segment top SNV | |||||||||||
| Chr | Start bp | End bp | PPA1a | PPA2a | PPA3a | PPA4a | rsID | bp | P(heno) Z-score | Novel locus for pheno no. | Locus candidate gene(s) | |||
| P1 | P2 | |||||||||||||
| Any migraine | CeAD | 1 | 149788928 | 151538412 | 0.06 | 0.00 | 0.94 | 0.01 | rs6693567 | 150510660 | 5.70 | 3.19 | 2 | Near ADAMTSL4/ECM1 |
| Any migraine | SVS | 2 | 202819643 | 205799152 | 0.00 | 0.00 | 0.99 | 0.00 | rs191602009 | 203795717 | −5.72 | −4.93 | 2 | CARF |
| Any migraine | CE | 5 | 171074292 | 172677991 | 0.00 | 0.00 | 1.00 | 0.00 | rs55884259 | 172642370 | 4.95 | 5.12 | 1 | NKX2-5 |
| Any migraine | CeAD | 6 | 11791351 | 13209144 | 0.00 | 0.00 | 1.00 | 0.00 | rs9349379 | 12903957 | −9.64 | −6.09 | PHACTR1/EDN1 | |
| MO | CeAD | 0.00 | 0.00 | 1.00 | 0.00 | −5.99 | −6.09 | |||||||
| Any migraine | CeAD | 6 | 94441595 | 97093400 | 0.02 | 0.00 | 0.98 | 0.00 | rs9486725 | 97061159 | 10.59 | 4.34 | FHL5 | |
| MA | CeAD | 0.00 | 0.00 | 0.94 | 0.01 | 4.42 | 4.34 | 1 | ||||||
| MO | CeAD | 0.00 | 0.00 | 1.00 | 0.00 | 7.11 | 4.34 | 1 | ||||||
| Any migraine | IS | 7 | 16902510 | 19481290 | 0.00 | 0.02 | 0.96 | 0.01 | rs2107595 | 19049388 | −3.37 | 6.68 | 1 | HDAC9/TWIST1 |
| Any migraine | AS | 0.00 | 0.03 | 0.95 | 0.02 | −3.37 | 6.64 | 1 | ||||||
| Any migraine | CE | 9 | 96671698 | 98921816 | 0.01 | 0.00 | 0.97 | 0.00 | rs182923402 | 98299677 | 4.91 | 4.38 | 1, 2 | Near PTCH1 |
| Any migraine | IS | 10 | 123901203 | 125869042 | 0.02 | 0.00 | 0.95 | 0.02 | rs55928386 | 124220667 | −5.32 | 4.32 | 2 | ARMS2, HTRA1 |
| Any migraine | AS | 0.01 | 0.00 | 0.96 | 0.04 | rs2284665 | 124226630 | 4.73 | −5.42 | 2 | ||||
| Any migraine | SVS | 0.08 | 0.00 | 0.91 | 0.01 | rs72631113 | 124213449 | 5.15 | −4.48 | 2 | ||||
| Any migraine | CeAD | 12 | 55665948 | 57548466 | 0.00 | 0.00 | 1.00 | 0.00 | rs11172113 | 57527283 | −14.72 | −5.45 | LRP1 | |
| MA | CeAD | 0.00 | 0.00 | 1.00 | 0.00 | −4.87 | −5.45 | 1 | ||||||
| MO | CeAD | 0.00 | 0.00 | 1.00 | 0.00 | −8.14 | −5.45 | 1 | ||||||
| Any migraine | IS | 13 | 46496025 | 47430602 | 0.00 | 0.02 | 0.96 | 0.02 | rs4942561 | 47209347 | 3.51 | −5.65 | 1 | LRCH1 |
| Any migraine | AS | 0.00 | 0.02 | 0.96 | 0.02 | rs1535791 | 47165458 | 3.62 | −5.95 | 1 | ||||
| Any migraine | AS | 13 | 109815112 | 111231864 | 0.01 | 0.00 | 0.97 | 0.00 | rs650724 | 110804809 | 4.44 | −4.75 | 1, 2 | COL4A1 |
Abbreviations: AS = all stroke; bp = base pair; CE = cardioembolic stroke; CeAD = cervical artery dissection; GWAS = genome-wide association study; IS = ischemic stroke; MA = migraine with aura; MO = migraine without aura; SNV = single nucleotide variation; SVS = small vessel stroke.
Posterior probability in the segment of association of phenotype 1 only (PPA1), phenotype 2 only (PPA2), shared association of both phenotypes (PPA3), and independent associations of both phenotypes (PPA4).
However, with ρHESS, local genetic covariance was also assessed at candidate regions defined by LD r2 >0.1 around the 69 known genome-wide significant loci for each of the disorders (Methods, Table 3).16 Local genetic covariance was concordant and met significance thresholds for candidate analysis (p < 0.0007 [=0.05/69]) between migraine and CeAD at PHACTR1/EDN1 and LRP1 as previously suggested but not formally demonstrated.7 At nominal significance (p < 0.05), concordant covariance was observed similarly not only at these candidate loci between CeAD and MO but also at FHL5 between CeAD and both any migraine and MO. The nominal associations also suggested shared signals between migraine and various stroke subtypes at known migraine loci mapping to PRDM16 (SVS, opposite directionality), ARMS2/HTRA1 (AS and IS, opposite), LRP1 (LAS, opposite), LRCH1 (AS and IS, opposite), and RNF213 (IS, opposite); and between migraine and CeAD at PLCE1 (concordant) and FGF6 (opposite). None of the nominally significant local correlations implicated MA.
Table 3.
Nominally Significant Local Genetic Covariance (ρg) at 69 Candidate Loci From Previous GWAS
| Locus known phenotype(s) | Pheno 1 | Pheno 2 | Candidate region | Local ρg, p valuea | Candidate gene(s) | |||
| Chr | Start | End | N SNVs | |||||
| Any migraine | Any migraine | SVS | 1 | 3065568 | 3116361 | 97 | −1.1E-04, 0.04584 | PRDM16 |
| Any migraine/CeAD | Any migraine | CeAD | 6 | 12758654 | 13119871 | 486 | 2.3E-03, 0.00021 | PHACTR1/EDN1 |
| Any migraine | MO | CeAD | 2.2E-03, 0.01023 | |||||
| Any migraine | Any migraine | CeAD | 6 | 96682566 | 97082880 | 562 | 2.1E-03, 0.00080 | FHL5 |
| Any migraine | MO | CeAD | 2.5E-03, 0.00337 | |||||
| Any migraine | Any migraine | CeAD | 10 | 95952031 | 97039458 | 1737 | 1.6E-03, 0.04244 | PLCE1 |
| Any migraine | Any migraine | AS | 10 | 123910423 | 124326089 | 891 | −1.3E-04, 0.02415 | ARMS2/HTRA1 |
| Any migraine | Any migraine | IS | −1.2E-04, 0.03811 | |||||
| Any migraine | Any migraine | CeAD | 12 | 4446116 | 4570190 | 232 | −1.0E-04, 0.03432 | FGF6 |
| Any migraine | Any migraine | CeAD | 12 | 57256380 | 57545756 | 360 | 3.1E-03, 0.00003 | LRP1 |
| Any migraine | MO | CeAD | 2.3E-03, 0.00932 | |||||
| Any migraine | Any migraine | LAS | −1.6E-04, 0.02121 | |||||
| Stroke | Any migraine | AS | 13 | 47062093 | 47323718 | 502 | −8.3E-05, 0.03039 | LRCH1 |
| Stroke | Any migraine | IS | −8.1E-05, 0.03382 | |||||
| Any migraine | Any migraine | IS | 17 | 78235300 | 78384523 | 255 | −7.9E-05, 0.04741 | RNF213 |
Abbreviations: AS = all stroke; bp = base pair; CeAD = cervical artery dissection; GWAS = genome-wide association study; IS = ischemic stroke; LAS = large artery stroke; MO = migraine without aura; SNV = single nucleotide variation; SVS = small vessel stroke.
Multiple testing significance threshold is p < 0.0007 (=0.05/69).
Novel Genome-wide Significant SNVs in Cross-Trait Association Analysis
MTAG, which leverages pairwise genome-wide genetic correlations to boost univariate association signals, identified novel genome-wide signals among SNVs that were also nominally significant in single-trait analysis (Methods, Table 4). Combining migraine with CeAD, there were novel associations for CeAD at SNVs mapping to the PLCE1 (chr. 10, rs57866767) and MRVI1 (chr 11, rs7940646) genes. The former was also nominally significant in the candidate local genetic correlation analysis (previous section). MTAG associations also recapitulated the findings from GWAS-PW for CeAD at FHL5 (chr. 6, rs2971603 or rs9486725), LRP1 (chr. 12, rs11172113), and at ADAMTSL4/ECM1 (chr. 1, rs6693567) for SNVs that were previously genome-wide significant for migraine. Two SNVs in moderately high LD (R2 = 0.5, D′ = 1.0), rs2971603 and rs9486725, represent the top associations at FHL5, the former more significant with any migraine and the latter with CeAD. Combining migraine and stroke, MTAG identified 2 novel stroke loci: LRP1 (chr. 12, rs11172113) for LAS at a SNV shared with CeAD (and migraine, opposite effect), and CARF (chr. 2, rs191602009) for SVS also previously genome-wide significant for migraine (concordant effect). No locus was genome-wide significant for CeAD in combination with any of the stroke outcomes.
Table 4.
Pairwise MTAG Genome-wide Significant Associations That Are Also Nominally Significant in Original GWAS
| Pheno 1 | Pheno 2 | SNV rsID | Chr | bp | Segment (bp)a | Coded/ref allele | GWAS association (Z score, p value) | MTAG association (beta [SE], pMTAG) | Novel locus for pheno no. | Candidate gene | ||
| Pheno 1 | Pheno 2 | Pheno 1 | Pheno 2 | |||||||||
| Any migraine | CeAD | rs6693567 | 1 | 150510660 | 150065704–150714741 | C/T | 5.7, 1.2E-08 | 3.2, 1.4E-03 | 0.016 (0.003), 3.1E-09 | 0.163 (0.028), 3.4E-09 | 2 | ADAMTSL4/ECM1 |
| Any migraine | SVS | rs191602009 | 2 | 203795717 | 203439395–204264839 | G/A | −5.7, 1.1E-08 | −4.9, 8.2E-07 | −0.022 (0.004), 6.9E-09 | −0.025 (0.004), 8.1E-09 | 2 | ALS2CR8 (CARF) |
| Any migraine | CeAD | rs9349379 | 6 | 12903957 | 12568218–13148388 | G/A | −9.6, 5.8E-22 | −6.1, 1.2E-09 | −0.024 (0.002), 6.2E-24 | −0.251 (0.024), 1.8E-25 | PHACTR1/EDN1 | |
| MO | CeAD | rs9349379 | 6 | 12903957 | 12681855–13145093 | G/A | −6.0, 2.1E-09 | −6.1, 1.2E-09 | −0.026 (0.004), 1.2E-12 | −0.257 (0.031), 8.0E-17 | ||
| Any migraine | CeAD | rs2971603 | 6 | 97035418 | 96319657–97267047 | T/C | 10.8, 2.8E-27 | 3.4, 6.4E-04 | 0.031 (0.003), 5.8E-28 | 0.267 (0.028), 8.8E-22 | 2 | FHL5 |
| Any migraine | CeAD | rs9486725 | 6 | 97061159 | 96319657–97267047 | T/C | 10.6, 3.5E-26 | 4.3, 1.4E-05 | 0.027 (0.002), 2.4E-27 | 0.251 (0.025), 1.4E-23 | 2 | |
| MO | CeAD | rs9486725 | 6 | 97061159 | 96643134–97267047 | T/C | 7.1, 1.3E-12 | 4.3, 1.4E-05 | 0.030 (0.004), 4.5E-15 | 0.245 (0.032), 2.7E-14 | 1, 2 | |
| Any migraine | CeAD | rs57866767 | 10 | 96023077 | 95798179–96274157 | C/T | −7.6, 2.3E-14 | −2.1, 3.6E-02 | −0.018 (0.002), 1.3E-14 | −0.157 (0.024), 4.9E-11 | 2 | PLCE1 |
| Any migraine | CeAD | rs11187838 | 10 | 96038686 | 95798179–96274157 | A/G | −7.6, 3.0E-14 | −2.4, 1.8E-02 | −0.018 (0.002), 1.4E-14 | −0.161 (0.024), 1.8E-11 | 2 | |
| Any migraine | CeAD | rs7940646 | 11 | 10669228 | 10454911–10899696 | T/C | −7.5, 5.0E-14 | −2.6, 1.0E-02 | −0.019 (0.002), 1.9E-14 | −0.174 (0.026), 1.1E-11 | 2 | MRVI1 |
| Any migraine | CeAD | rs11172113 | 12 | 57527283 | 57057912–57745756 | C/T | −14.7, 5.6E-49 | −5.4, 5.1E-08 | −0.035 (0.002), 8.5E-51 | −0.328 (0.024), 9.2E-42 | 2 | LRP1 |
| Any migraine | LAS | rs11172113 | 12 | 57527283 | 57056380–57745756 | C/T | −14.0, 1.9E-44 | 2.9, 3.8E-03 | −0.034 (0.002), 3.6E-44 | 0.023 (0.003), 4.4E-13 | 2 | |
| MO | CeAD | rs11172113 | 12 | 57527283 | 57302981–57734912 | C/T | −8.1, 4.3E-16 | −5.4, 5.1E-08 | −0.033 (0.004), 1.2E-19 | −0.282 (0.031), 1.0E-19 | 1, 2 | |
Abbreviations: bp = base pair; CeAD = cervical artery dissection; GWAS = genome-wide association study; LAS = large artery stroke; MO = migraine without aura; MTAG = multi-trait analysis of GWAS; SNV = single nucleotide variation; SVS = small vessel stroke.
Span of genome-wide significant MTAG associations for either phenotype and maximum distance 200 kb.
Mendelian Randomization
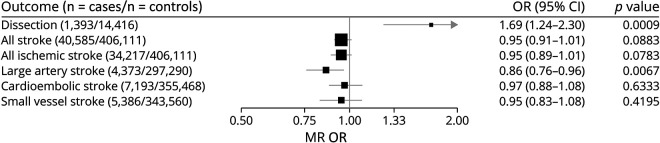
In MR analysis (i.e., genetic instrumental analysis), liability to migraine was supported as causal for increased CeAD risk (odds ratio [OR] [95% confidence interval (CI)] = 1.69 [1.24–2.3], p = 0.0009) but protective for LAS (0.86 [0.76–0.96], p = 0.007) (Figure 1). There were no significant effects on either AS or other stroke subtypes, including all IS. There was significant heterogeneity detected for the migraine-CeAD effect (Cochran Q = 109, df = 39, p = 1.48 × 10−8), which was diminished but not eliminated by exclusion of 2 clearly pleiotropic SNVs, rs11172113 (LRP1) and rs9349379 (PHACTR1/EDN1) (Q = 65, df = 37, p = 0.003), leading to an attenuated but still nominally significant effect (OR [95% CI] = 1.33 [1.02–1.73], p = 0.04) (eTables 1 and 2, links.lww.com/NXG/A511). The MR-Egger intercept test for directional pleiotropy was null (p = 0.85), as suggested also by the largely consistent estimates of the effect in the sensitivity analyses (eTable 3, links.lww.com/NXG/A511). By contrast, there was no significant heterogeneity in the effect of migraine on LAS (Cochran Q = 46, df = 39, p = 0.21), and sensitivity analyses for pleiotropy yielded consistent and significant estimated protective effects of migraine, with the exception of MR-Egger (1.01 [0.73–1.39], p = 0.97). However, the effect obtained from the MR-Egger bootstrap test (0.85 [0.66–1.08], p = 0.091) was directionally consistent with the primary analysis, suggesting a potential undue influence of outliers on the estimate from MR-Egger when including all instruments without bootstrapping.
Figure 1. Instrumental Effects by MR of Migraine on the Other Cerebrovascular Disorders.
MR was performed using the random-effects inverse variance–weighted method. ORs and 95% CIs are scaled to reflect the effect of migraine liability on CeAD and stroke per doubling of migraine prevalence (see Methods). CeAD = cervical artery dissection; CI = confidence interval; MR = Mendelian randomization; OR = odds ratio.
Discussion
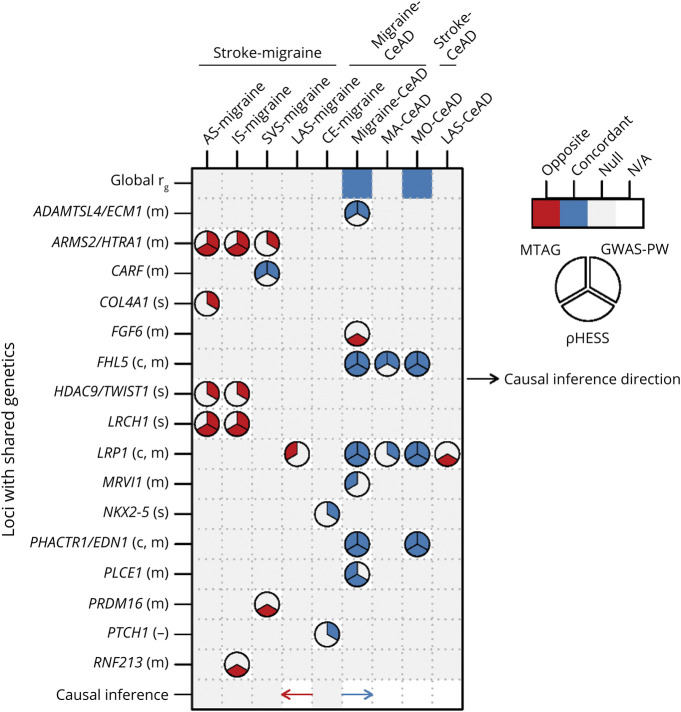
The preceding analysis was undertaken to investigate the etiologic basis of comorbidity among each pair of 3 brain disorders with known vascular involvement through the unique properties of genetics. Both genome-wide and at specific loci, the findings emphasized extensive sharing of biology between migraine and CeAD. Genetic sharing was less for migraine and stroke but still implicated a few loci, while still less sharing was detected for stroke and CeAD. Figure 2 summarizes the significant pairwise associations from all analyses.
Figure 2. Summary of Pairwise Genetic Comparisons for the 3 Disorders.
Only pairs with significant associations are shown. For Mendelian randomization, the direction of each arrow indicates whether the direction of inferred causality is from the first disorder of each pair to the second (rightward arrow) or the opposite (leftward arrow) with concordant (blue) or opposite (red) functional relationship. Symbols following gene name labels for each locus refer to the disorder(s) for which association has been previously reported, if any, as follows: m = migraine, c = CeAD, s = stroke, and - = no known association. CeAD = cervical artery dissection; GWAS-PW = genome-wide association study-pairwise; MTAG = multitrait analysis of GWAS; ρHESS = ρ Heritability Estimation from Summary Statistics.
While the challenge of recruiting large samples of CeAD cases for genome-wide genetic analysis had limited power for previous genome-wide analysis, local genetic correlation with migraine boosted genetic signals to highlight novel candidate loci for CeAD and reinforced existing candidates, all with concordant effects on migraine. Novel genomic regions on at chr.1q21.3 (ADAMTSL4/ECM1 candidate genes), chr10q23.33 (PLCE1), and chr11p15.4 (MRVI1) were all previously associated with migraine9 and are now implicated also with CeAD. All reached genome-wide significance in the MTAG, and the chromosome 1 and 10 loci were further supported by GWAS-PW and ρHESS analyses, respectively. The extracellular matrix protein 1 (ECM1) gene at the chr1q21.3 locus has been suggested for involvement in vascular development.29 The association at the MRVI1 gene, encoding murine retrovirus integration site 1 homolog, a tumor suppressor, arose previously as a candidate influencing both migraine and IS.3 The lead SNV, rs7940646, and its LD proxies associated with migraine in the region are also associated with blood pressure, arterial stiffness, airway inflammatory diseases, platelet aggregation, brain region volume, and measures for white matter integrity.30-34 PLCE1 encodes the phospholipase C, epsilon 1 protein, at which GWAS has implicated other vascular conditions including CVD and blood pressure.33,35 The remaining CeAD loci shared with migraine at the PHACTR1/EDN1, LRP1, and FHL5 genes were all noted in the original CeAD GWAS (FHL5 at suggestive significance), but only PHACTR1/EDN1 was formally genome-wide significant in replication, again likely due to limited sample.7 All have been previously annotated as playing a role in vascular development or function.29 At the PHACTR1/EDN1 locus, recent functional work has not fully resolved which of 2 candidate genes may underlie the functional effects.8,36 Though there is no strong genetic overlap between stroke and CeAD, the genetic overlap involving LRP1 implicates both with migraine. Owing to the vascular etiology of CeAD and stroke, the preceding shared loci may be particularly relevant to vascular etiologies of migraine.
Despite the modest genome-wide correlations, local comparisons with GWAS-PW revealed potential new candidate loci with concordant effects for various stroke subtypes and any migraine. CARF (rs191602009), a known migraine locus encoding the calcium-response transcription factor, likely mediates calcium signaling in neurons, including regulation of the brain-derived neurotrophic factor,37 and was implicated in SVS; NKX2-5 (rs55884259), a known CE stroke locus encoding a homeobox protein for which variations cause congenital heart defects, was implicated in migraine; and PTCH1 (rs182923402), encoding patched 1, is a member of a protein family of receptors that are ligands for sonic hedgehog signaling peptides in development and was implicated in both migraine and CE. Other pairwise shared loci revealed novel associations for 1 disorder but with opposite risk alleles compared with the known association at the second disorder: (1) HDAC9/TWIST1 (rs2107595), a known LAS locus for which the candidate gene is not yet definitively identified,38,39 is implicated in migraine; (2) ARMS2/HTRA1 (3 SNVs [all D′ = 1, low R2 with each other and with rs10490924]), a known a migraine locus for which HTRA1 encoding a serine peptidase may be the best candidate gene, is implicated in AS, IS, and SVS, reinforcing previous subthreshold associations.10 Mendelian associations with small vessel disease also support this association because monogenic variations in HTRA1 lead to a rare autosomal dominant form of SVD, CARASIL40,41; (3) LRCH1 (2 SNVs, LD R2 = 1), a migraine locus, is now implicated in AS and IS; and (4) COL4A1 (rs650724), encoding collagen type 4 alpha 1, is now implicated in migraine having been previously identified for stroke by GWAS and Mendelian genetics of SVD.10,42 Signals at ARMS2/HTRA1 and LRCH1 were also supported at nominal significance by the ρHESS candidate local genetic correlations, while the signal at rs191602009 (CARF) was supported in the MTAG.
The less extensive genetic sharing of migraine (particularly MA) and IS across the genome is contrary to their strong comorbidity in epidemiologic studies.4 Genome-wide genetic correlations were not only modest but also emphasized an opposite relationship rather than concordance, particularly between migraine and LAS. Similarly, causality in an opposite relationship of migraine liability to LAS was supported by the instrumental analysis. This finding is consistent with prior MR analyses, which identified opposite instrumental relationships of migraine liability with coronary artery disease, a disorder that shares pathophysiology with LAS.43 Similarly, the findings are reminiscent of previous analysis of shared genetics between migraine, especially MO, and stroke that also found overlap, especially for large artery and CE types.3 Although the genetic correlation of migraine with SVS, which has been suspected in the mechanism of migraine comorbidity,2 was concordant with findings from conventional epidemiology and with migraine being an important feature of monogenic forms of small vessel disease, the estimate was only marginally significant. This observation may be qualified, however, by the low power of GWAS for IS subtypes, as well as likely imperfect ascertainment of SVS in many studies. The genetic relationship between migraine and small vessel disease deserves further investigation using more specific MRI markers of SVD, such as white matter hyperintensity burden.44
An MTAG-based genome-wide significant association at rs11172113 for LAS that was supported by local correlation at nominal significance in ρHESS with migraine implicates LRP1, which is the only locus influencing risk of all 3 cerebrovascular disorders, although opposite in its effect on stroke compared with that on migraine or CeAD. This same locus has recently also been implicated in aortic and coronary dissection and abdominal aortic aneurysm with the same directionality for CeAD and migraine, placing LRP1 at the center of shared biology and deserving further study.45-47 LRP1, a member of the LDL receptor family, has been implicated by GWAS also in pulmonary function and CHD, the latter likely related in pathophysiology to the association with LAS.33,48,49 LRP1 protein is involved in endocytosis of a wide variety of ligands, including lipoproteins, and understanding mechanism(s) of its contribution to the shared susceptibilities will require further research.50
The strengths of this study are the very large sample sizes and therefore power represented by the GWAS summary statistics for migraine and stroke. The study is limited in its restriction to populations of European ancestry, although multiancestry meta-analysis for stroke subtypes supports the top loci, implying that relevant biological functions are shared among European and other ancestries. However, it remains possible that genetic relationships in non-European ancestries among the 3 disorders would highlight additional relationships, including those that may contribute to health disparities. The study is also limited by the modest sample size underlying the summary statistics for CeAD, a consequence of the challenge in accumulating genome-wide genetic data for extremely low prevalence events. The incidence of CeAD is only on the order of approximately 2.6 per 100,000 per year.51 Similarly, despite the relatively large total sample for the stroke GWAS, heterogeneity in stroke mechanism and intrinsic difficulties in assigning stroke subclassifications may have limited the ability to detect genetic overlap with either migraine or CeAD. An additional consequence of the limitations in the CeAD and stroke GWASs was an insufficient number of qualifying instruments to perform MR for assessing potential causal effects of liability to these disorders on migraine.
Taken together, the results thus provide novel support for the contribution of vascular functions to migraine and enhance understanding of the comorbidity among migraine, CeAD, and stroke. Future functional studies prioritizing specific loci identified through this genetic analysis may reveal deeper insights into corresponding vascular mechanisms leading to susceptibility to the 3 brain disorders.
Acknowledgment
The authors thank the IHGC researchers and member cohort participants, the staff, and participants of all CADISP centers and the research participants and employees of 23andMe for their important contributions that made this work possible. See supplementary material for listings of IHGC and CADISP researchers (links.lww.com/NXG/A511).
Glossary
- ρHESS
Heritability Estimation from Summary Statistics
- AS
all stroke
- CE
cardioembolic stroke
- CeAD
cervical artery dissection
- CI
confidence interval
- GNOVA
Genetic Covariation Analyzer
- GWAS
genome-wide association study
- GWAS-PW
GWAS-pairwise
- IS
ischemic stroke
- IVW
inverse variance–weighted
- LAS
large artery stroke
- LD
linkage disequilibrium
- LDSc
linkage disequilibrium score regression
- MA
migraine with aura
- MR
Mendelian randomization
- MO
migraine without aura
- MTAG
multitrait analysis of GWAS
- OR
odds ratio
- PPA3
posterior probability of association in mode 3
- SNV
single nucleotide variation
- SVS
small vessel stroke
- WGHS
Women's Genome Health Study
Appendix. Authors
| Name | Location | Contribution |
| Iyas Daghlas, MD | Harvard Medical School, Boston, MA; Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA | Drafting/revision of the article for content, including medical writing for content; study concept or design; and analysis or interpretation of data |
| Muralidharan Sargurupremraj, PhD | University of Bordeaux, Inserm, Bordeaux Population Health Research Center, Team VINTAGE, UMR 1219, Bordeaux, France; Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases, University of Texas Health, San Antonio | Drafting/revision of the article for content, including medical writing for content; major role in the acquisition of data; study concept or design; and analysis or interpretation of data |
| Rebecca Danning, MS | Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA | Drafting/revision of the article for content, including medical writing for content, and analysis or interpretation of data |
| Padhraig Gormley, PhD | Massachusetts General Hospital, Boston | Drafting/revision of the article for content, including medical writing for content, and analysis or interpretation of data |
| Rainer Malik, PhD | Institute for Stroke and Dementia Research, Klinikum der Universität München, Ludwig-Maximilians-University, Germany | Drafting/revision of the article for content, including medical writing for content, and major role in the acquisition of data |
| Philippe Amouyel, MD, PhD | LabEx DISTALZ-U1167, RID-AGE-Risk Factors and Molecular Determinants of Aging-Related Diseases, University of Lille, France; Inserm U1167, Lille, France; Centre Hospitalier Universitaire Lille, France; Institut Pasteur de Lille, France | Drafting/revision of the article for content, including medical writing for content, and major role in the acquisition of data |
| Tiina Metso, PhD | Department of Neurology, Helsinki University Central Hospital, Finland | Drafting/revision of the article for content, including medical writing for content, and major role in the acquisition of data |
| Alessandro Pezzini, MD | Department of Clinical and Experimental Sciences, Neurology Clinic, Brescia University Hospital, Italy | Drafting/revision of the article for content, including medical writing for content, and major role in the acquisition of data |
| Tobias Kurth, MD, ScD | Institute of Public Health, Charité—Universitätsmedizin Berlin, Germany | Drafting/revision of the article for content, including medical writing for content; study concept or design; and analysis or interpretation of data |
| Stéphanie Debette, MD, PhD | University of Bordeaux, Inserm, Bordeaux Population Health Research Center, Team VINTAGE, UMR 1219, Bordeaux, France; Department of Neurology, CHU de Bordeaux, France | Drafting/revision of the article for content, including medical writing for content; major role in the acquisition of data; and study concept or design |
| Daniel I. Chasman, PhD | Harvard Medical School, Boston, MA; Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA | Drafting/revision of the article for content, including medical writing for content; major role in the acquisition of data; and study concept or design; analysis or interpretation of data |
Contributor Information
Iyas Daghlas, Email: iyas.daghlas@ucsf.edu.
Muralidharan Sargurupremraj, Email: sargurupremr@uthscsa.edu.
Rebecca Danning, Email: beckydanning@gmail.com.
Padhraig Gormley, Email: padhraig.j.gormley@gsk.com.
Rainer Malik, Email: rainer.malik@med.uni-muenchen.de.
Philippe Amouyel, Email: philippe.amouyel@pasteur-lille.fr.
Tiina Metso, Email: tiina.m.metso@helsinki.fi.
Alessandro Pezzini, Email: alessandro.pezzini@unibs.it.
Tobias Kurth, Email: tobias.kurth@charite.de.
Stéphanie Debette, Email: stephanie.debette@u-bordeaux.fr.
Study Funding
This work was supported by grants from the NINDS of the NIH (R21NS092963 and R21NS104398 to D.I.C.). The CADISP study has been supported by Inserm, Lille 2 University, Institut Pasteur de Lille, and Lille University Hospital and received funding from the ERDF (FEDER funds) and Région Nord-Pas de Calais in the frame of Contrat de Projets Etat-Region 2007–2013 Région Nord-Pas-de-Calais—Grant No. 09120030, Centre National de Genotypage, Emil Aaltonen Foundation, Paavo Ilmari Ahvenainen Foundation, Helsinki University Central Hospital Research Fund, Helsinki University Medical Foundation, Päivikki and Sakari Sohlberg Foundation, Aarne Koskelo Foundation, Maire Taponen Foundation, Aarne and Aili Turunen Foundation, Lilly Foundation, Alfred Kordelin Foundation, Finnish Medical Foundation, Orion Farmos Research Foundation, Maud Kuistila Foundation, the Finnish Brain Foundation, Biomedicum Helsinki Foundation, Projet Hospitalier de Recherche Clinique Régional, Fondation de France, Génopôle de Lille, Adrinord, Basel Stroke-Funds, Käthe-Zingg-Schwichtenberg-Fonds of the Swiss Academy of Medical Sciences, and Swiss Heart Foundation. Murali Sargurupremraj has been supported by a grant from the European Commission under H2020 Grant no. 640643.
Disclosure
P. Gormley is a current employee and/or stockholder of Glaxo SmithKline, but the work was conducted while employed by Massachusetts General Hospital, Boston, MA. T. Kurth reports to have received personal compensation for outside the submitted work from Eli Lilly & Company, Teva, Total S.E., and the BMJ. The other authors report no disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Malik R, Winsvold B, Auffenberg E, Dichgans M, Freilinger T. The migraine-stroke connection: a genetic perspective. Cephalalgia. 2016;36(7):658-668. doi: 10.1177/0333102415621055. [DOI] [PubMed] [Google Scholar]
- 2.Arkink EB, Terwindt GM, de Craen AJ, et al. Infratentorial microbleeds: another sign of microangiopathy in migraine. Stroke. 2015;46(7):1987-1989. doi: 10.1161/STROKEAHA.115.009604. [DOI] [PubMed] [Google Scholar]
- 3.Malik R, Freilinger T, Winsvold BS, et al. Shared genetic basis for migraine and ischemic stroke: a genome-wide analysis of common variants. Neurology. 2015;84(21):2132-2145. doi: 10.1212/WNL.0000000000001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurth T, Rist PM, Ridker PM, Kotler G, Bubes V, Buring JE. Association of migraine with aura and other risk factors with incident cardiovascular disease in women. JAMA. 2020;323(22):2281-2289. doi: 10.1001/jama.2020.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Androulakis XM, Kodumuri N, Giamberardino LD, et al. Ischemic stroke subtypes and migraine with visual aura in the ARIC study. Neurology. 2016;87(24):2527-2532. doi: 10.1212/WNL.0000000000003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurth T, Slomke MA, Kase CS, et al. Migraine, headache, and the risk of stroke in women: a prospective study. Neurology. 2005;64(6):1020-1026. doi: 10.1212/01.WNL.0000154528.21485.3A. [DOI] [PubMed] [Google Scholar]
- 7.Debette S, Kamatani Y, Metso TM, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47(1):78-83. doi: 10.1038/ng.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RM, Hadaya J, Trehan A, et al. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170(3):522-533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gormley P, Anttila V, Winsvold BS, et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet. 2016;48(8):856-866. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chasman DI, Schurks M, Anttila V, et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet. 2011;43(7):695-698. doi: 10.1038/ng.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q, Li B, Ou D, et al. A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am J Hum Genet. 2017;101(6):939-964. doi: 10.1016/j.ajhg.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48(7):709-717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berisa T, Pickrell JK. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32(2):283-285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Mancuso N, Spendlove S, Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am J Hum Genet. 2017;101(5):737-751. doi: 10.1016/j.ajhg.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555-3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turley P, Walters RK, Maghzian O, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229-237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R: A Language and Environment for Statistical Computing [Computer Program]. R Foundation for Statistical Computing; 2019. [Google Scholar]
- 22.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Q, Chen Y, Wang J, Small DS. Powerful three-sample genome-wide design and robust statistical inference in summary-data Mendelian randomization. Int J Epidemiol. 2019;48(5):1478-1492. doi: 10.1093/ije/dyz142. [DOI] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018;33(10):947-952. doi: 10.1007/s10654-018-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik R, Rannikmae K, Traylor M, et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann Neurol. 2018;84(6):934-939. doi: 10.1002/ana.25369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyholt DR, Borsook D, Griffiths LR. Migrainomics—identifying brain and genetic markers of migraine. Nat Rev Neurol. 2017;13(12):725-741. doi: 10.1038/nrneurol.2017.151. [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Zhang J, Ibrahim JG, et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol Psychiatry. 2021;26(12):3943-3955. doi: 10.1038/s41380-019-0569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung K, Ramirez J, Warren HR, et al. Genome-wide association study identifies loci for arterial stiffness index in 127,121 UK Biobank participants. Sci Rep. 2019;9(1):9143. doi: 10.1038/s41598-019-45703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira MAR, Mathur R, Vonk JM, et al. Genetic architectures of childhood- and adult-onset asthma are partly distinct. Am J Hum Genet. 2019;104(4):665-684. doi: 10.1016/j.ajhg.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65-75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42(7):608-613. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103-109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Musunuru K. Confirmation of causal rs9349379- PHACTR1 expression quantitative trait locus in human-induced pluripotent stem cell endothelial cells. Circ Genom Precis Med. 2018;11(10):e002327. doi: 10.1161/CIRCGEN.118.002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West AE. Biological functions and transcriptional targets of CaRF in neurons. Cell Calcium. 2011;49(5):290-295. doi: 10.1016/j.ceca.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asare Y, Campbell-James TA, Bokov Y, et al. Histone deacetylase 9 activates IKK to regulate atherosclerotic plaque vulnerability. Circ Res. 2020;127(6):811-823. doi: 10.1161/CIRCRESAHA.120.316743. [DOI] [PubMed] [Google Scholar]
- 39.Nurnberg ST, Guerraty MA, Wirka RC, et al. Genomic profiling of human vascular cells identifies TWIST1 as a causal gene for common vascular diseases. PLoS Genet. 2020;16(1):e1008538. doi: 10.1371/journal.pgen.1008538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara K, Shiga A, Fukutake T, et al. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N Engl J Med. 2009;360(17):1729-1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- 41.Verdura E, Herve D, Scharrer E, et al. Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain. 2015;138(8):2347-2358. doi: 10.1093/brain/awv155. [DOI] [PubMed] [Google Scholar]
- 42.Gould DB, Phalan FC, van Mil SE, et al. Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med. 2006;354(14):1489-1496. doi: 10.1056/NEJMoa053727. [DOI] [PubMed] [Google Scholar]
- 43.Daghlas I, Guo Y, Chasman DI. Effect of genetic liability to migraine on coronary artery disease and atrial fibrillation: a Mendelian randomization study. Eur J Neurol. 2020;27(3):550-556. doi: 10.1111/ene.14111. [DOI] [PubMed] [Google Scholar]
- 44.Sargurupremraj M, Suzuki H, Jian X, et al. Cerebral small vessel disease genomics and its implications across the lifespan. Nat Commun. 2020;11(1):6285. doi: 10.1038/s41467-020-19111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turley TN, O'Byrne MM, Kosel ML, et al. Identification of susceptibility loci for spontaneous coronary artery dissection. JAMA Cardiol. 2020;5(8):929-938. doi: 10.1001/jamacardio.2020.0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo DC, Grove ML, Prakash SK, et al. Genetic variants in LRP1 and ULK4 are associated with acute aortic dissections. Am J Hum Genet. 2016;99(3):762-769. doi: 10.1016/j.ajhg.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bown MJ, Jones GT, Harrison SC, et al. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89(5):619-627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43(11):1082-1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122(3):433-443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108(6):779-784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee VH, Brown RD Jr, Mandrekar JN, Mokri B. Incidence and outcome of cervical artery dissection: a population-based study. Neurology. 2006;67(10):1809-1812. doi: 10.1212/01.wnl.0000244486.30455.71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study used only GWAS summary statistics from published reports as described earlier. The availability of these data or procedures for accessing them is documented in the cited publications. Summary statistics for the GWAS of migraine lacking contribution from the WGHS will be made available by sending an application to the International Headache Genetics Consortium via the corresponding author using the same procedure that governs access to the summary statistics for the published migraine study.9