Abstract
Although the COVID-19 pandemic has been the defining global health crisis of our time, public health officials have been sounding the alarm of another ominous threat for years: an impending antimicrobial resistance crisis.
In dermatology, antibiotics are often used for prolonged courses in the treatment of skin and soft tissue infections and common inflammatory skin conditions, increasing the risk of microbiome alteration and antibiotic-related adverse effects, all while exerting consequential selective pressures on both pathogenic and bystander bacteria. In this review, we hope to raise awareness of the crisis of antimicrobial resistance and review resistance concerns related to dermatology-relevant bacterial pathogens.
Key words: antibiotic resistance, antimicrobial resistance, bacteria, clindamycin-resistant Streptococcus pyogenes, dermatology, drug-resistant Mycobacterium tuberculosis, methicillin-resistant Staphylococcus aureus, multidrug-resistant Pseudomonas aeruginosa, resistant Cutibacterium acnes
Abbreviations used: AMR, antimicrobial resistance; CA-MRSA, community-acquired methicillin-resistant Staphylococcus aureus; GAS, group A Streptococcus; HA-MRSA, hospital-acquired methicillin-resistant Staphylococcus aureus; LA-MRSA, livestock-associated methicillin-resistant Staphylococcus aureus; MDR, multidrug-resistant; MRSA, methicillin-resistant Staphylococcus aureus; SSTI, skin and soft tissue infections; TB, Mycobacterium tuberculosis; PBP, penicillin-binding proteins
Learning objectives
After completing this learning objective, the reader will be able to better discuss the causes and clinical implications of antimicrobial resistance among bacterial pathogens of dermatologic significance.
Disclosures
None.
Editors
The editors involved with this CME activity and all content validation/peer reviewers of the journal-based CME activity have reported no relevant financial relationships with commercial interest(s).
Authors
The authors involved with this journal-based CME activity have reported no relevant financial relationships with commercial interest(s).
Planners
The planners involved with this journal-based CME activity have reported no relevant financial relationships with commercial interest(s). The editorial and education staff involved with this journal-based CME activity have reported no relevant financial relationships with commercial interest(s).
Introduction
One of the greatest global public health threats of the 21st century is growing antimicrobial resistance (AMR). Concerns for a looming AMR crisis may have been temporarily overshadowed by the COVID-19 pandemic, but many experts worry that pandemic-induced increases in antibiotic exposures, hospitalizations, and diverted resources have only accelerated its arrival.1, 2, 3, 4
Since penicillin’s discovery in 1928, antibiotic therapy has been a foundational pillar of modern medicine. Unfortunately, the rate at which bacterial pathogens are becoming resistant to currently available antibiotics has consistently outpaced the rate of new antibiotic development.5 The Centers for Disease Control and Prevention estimate that AMR infections already cause 1 death every 15 minutes in the United States.6 Globally, AMR has become a top public health concern7 , 8 and is now responsible for more deaths than HIV/AIDS or malaria; In 2019 alone, drug resistant bacterial infections were associated with an estimated 4.95 million deaths worldwide, including 1.27 million deaths that were directly attributable to antibiotic resistance.8 Resistance among Staphylococcus aureus, Streptococcus. pyogenes, and Pseudomonas aeruginosa are among those that the Centers for Disease Control and World Health Organization consider prioritized threats.6 , 7
The development of an AMR crisis, fueled largely by the overuse and misuse of antibiotics, should be a rallying call for increased antibiotic stewardship.9, 10, 11 The Centers for Disease Control and Prevention estimate that 50% of outpatient antibiotics are inappropriately prescribed based on agent selection, dosing, or duration and at least 30% of outpatient antibiotics are given unnecessarily.12 , 13 Since over 80% of all human antibiotic use occurs in the outpatient setting,14 outpatient prescribing patterns are a critical target for stewardship efforts. The overuse of broad-spectrum antibiotics is particularly concerning in terms of their potential to induce wider microbiome disturbances and resistance among bacteria. Broad-spectrum antibiotics such as mupirocin, doxycycline, minocycline, and trimethoprim-sulfamethoxazole are valuable therapeutic agents in dermatology, but the consequences of their widespread use for narrow-spectrum indications must be considered. In this article, we review key dermatology-relevant issues related to bacterial AMR.
Discussion
Methicillin-resistant Staphylococcus aureus
Key points
-
•
Methicillin resistance in S aureus (MRSA) occurs due to alterations in penicillin-binding proteins (PBPs) and can be transferred on a mobile genetic element.
-
•
Hospital-acquired, community-acquired, and livestock-associated strains of MRSA are distinct strains, but their clinical and epidemiologic distinctions are blurring.
S aureus colonizes 20% to 40% of the population and causes infections of almost every body site, with skin involvement being the most common (Table I ).15, 16, 17 S aureus exemplifies the remarkable ability of bacteria to adapt to selective antibiotic pressures.18 Within just 2 years of penicillin’s introduction, penicillinase - producing - resistant S aureus clones emerged, and by the 1970s, 95% of S aureus isolates were penicillin resistant.16, 17, 18, 19 Efforts to synthesize agents impervious to penicillinase led to the discovery of methicillin in 1959, to which S aureus responded with resistance within a year.20
Table I.
Methicillin-resistant Staphylococcus aureus antibiotic options and resistance mechanisms17, 18, 19, 20, 21, 22, 23
| Agent | Mechanism of action | Resistance mechanism |
|---|---|---|
| TMP-SMX | Sequential inhibition of nucleic acid synthesis: SMX inhibits synthesis of dihydrofolic acid by dihydropteroate synthetase (DHPS) and TMP inhibits synthesis of tetrahydrofolate by dihydrofolate reductase (DHFR) | Altered drug target and decreased drug binding via point mutations in DHFR and DHPS |
| Doxycycline Minocycline |
Binding to 16S rRNA within 30S ribosomal subunit → inhibition of protein synthesis | 1) Active drug efflux via tetracycline efflux pumps encoded by plasmid-borne genes Tet (K) and Tet (L) 2) Ribosomal target protection via transposon-encoded genes tet (M) and tet (O) that catalyze tetracycline dissociation from the ribosome |
| Clindamycin | Binding to 23S rRNA of 50S ribosomal subunit → inhibition of protein synthesis | Modification of drug-binding site via erm gene, an enzyme that methylates the 23S rRNA → variable cross-resistance to macrolides, lincosamides, and streptogramin B (MLSb phenotype) |
| Vancomycin | Binding to D-ala-D-ala residues → inhibition of peptidoglycan cell-wall synthesis | VISA: inhibition of drug binding to cell-wall target via mutations in genes regulating cell-wall synthesis VRSA: Reduced drug binding to target via acquisition of vanA, a plasmid element from Enterococcus spp that encodes a modified D-ala-D-lac precursor |
| Teicoplanin | Binding to D-ala-D-ala residues → inhibition of peptidoglycan cell-wall synthesis | Decreased drug affinity to target via modified peptidoglycan precursors |
| Fluoroquinolones | Inhibition of enzymes involved in DNA replication, DNA gyrase (topoisomerase II) and topoisomerase IV | 1) Alteration of target site via mutations in genes encoding topoisomerase II (gyrA and gyrB) and topoisomerase IV (grlA and grlB) 2) Active drug efflux via overexpression of efflux pumps NorA and NorB |
| Linezolid | Binding to of the 23S rRNA of 50S ribosomal subunit → protein synthesis inhibition | 1) Decreased drug binding via point mutations in the ribosomal binding site 2) Modification of the ribosomal binding site via enzymatic methylation mediated by cfr gene |
| Daptomycin | Binding and inserting into the bacterial cytoplasmic membrane → damage to the cell membrane and to the membrane potential | 1) Modification in the peptidoglycan layer via mpfF, an enzyme that incorporates a lysine residue → inhibits binding of daptomycin 2) Changes in phospholipid metabolism via genes encoding cardiolipin synthetases (pgsA, cls) → decreased drug binding |
| Aminoglycosides | Binding to 16S rRNA of the 30S ribosomal subunit → inhibition of protein synthesis | Enzymatic modification of the drug → decreased drug binding to its target |
| Fifth generation cephalosporins (Ceftaroline, Ceftobiprole) | Beta-lactams with high affinity for PBP2a/PBP2c proteins | Resistance to ceftaroline has been reported but the mechanisms are unclear |
| Quinupristin/Dalfopristin | Sequential binding to 50S ribosomal subunit → inhibition of protein synthesis | 1) Modification of ribosomal subunit via acquisition of erm genes encoding methylases → decreased drug binding 2) Enzymatic degradation of the drug via acquisition of acetyltransferases |
MLSb, Macrolide-lincosamide-streptogramin B; MRSA, methicillin-resistant Staphylococcus aureus; PBP, penicillin-binding protein; TMP-SMX, trimethoprim-sulfamethoxazole; VISA, vancomycin-intermediate Staphylococcus aureus; VRSA, vancomycin-resistant Staphylococcus aureus.
Methicillin resistance arises via the alteration of PBPs. Methicillin and other ß-lactam antibiotics bind to PBPs to block key cross-linking steps in bacterial cell-wall synthesis. MRSA isolates express low-affinity PBPs, PBP2a and PBP2c, that are encoded by the mecA and mecC genes, respectively, and carried on a mobile genetic element known as the staphylococcal chromosomal cassette.18 , 20 , 22 Not only can the staphylococcal chromosomal cassette be readily transferred among staphylococcal species, but it can also carry other resistance genes, partially accounting for the multidrug-resistant (MDR) phenotype of MRSA.20
In the 1960s and 1970s, MRSA infections occurred primarily as isolated hospital outbreaks among patients with comorbidities. By the 1980s, hospital-acquired MRSA (HA-MRSA) clones had spread worldwide. Current hospital isolates demonstrate resistance rates of over 50% in the United States and exceed 70% in select East Asian countries.17 , 23 HA-MRSA is now a leading cause of nosocomial bacteremia, pneumonia, and vascular-access- and catheter-associated infections.17 HA-MRSA strains are typically resistant to fluoroquinolones, macrolides, trimethoprim, and tetracyclines, necessitating the increased use of agents such as vancomycin, daptomycin, and linezolid.16 , 17 , 20 , 23 Unfortunately, resistance to even these newer agents has already been documented (Table I).
In the 1990s, reports of community outbreaks of skin and soft tissue infections (SSTIs) due to MRSA among otherwise healthy indivuduals with no hospital exposure began appearing. These community- acquired (CA-MRSA) strains were subsequently found to be genetically and phenotypically distinct strains from HA-MRSA and to express virulence toxins such as Panton-Valentine leucocidin that enabled them to infect otherwise healthy hosts.20 , 23 Whereas HA-MRSA is most often associated with nosocomial infections like bacteremia and endocarditis, over 90% of CA-MRSA infections are purulent SSTIs.24 Unlike HA-MRSA, CA-MRSA is also typically sensitive to several non-ß-lactam drugs, including clindamycin, trimethoprim-sulfamethoxazole, and tetracyclines.20 , 23 Despite these differences, distinguishing CA-MRSA and HA-MRSA on epidemiologic grounds has become increasingly difficult. Since the early 2000s, CA-MRSA strains have been implicated in a significant proportion of hospital-acquired infections, including bacteremia, postsurgical infections, and necrotizing infections in newborns.24 , 25 Growing antibiotic exposure has also led to the development of MDR CA-MRSA, thereby blurring the susceptibility differences between CA-MRSA and HA-MRSA isolates.16 , 23 , 26
Another new MRSA lineage, livestock-associated MRSA (LA-MRSA), was first reported in pigs in 2005 and has since been documented among other domesticated animals, veterinarians, farmers, and patients from rural settings.16 , 17 Due to the routine antibiotics used in livestock farming, LA-MRSA is widespread among livestock and readily capable of spreading to humans.27 Livestock workers are at a significantly higher risk for LA-MRSA colonization and subsequent infection than the general population.28 Currently, LA-MRSA accounts for 15% of community-acquired SSTIs and 1% to 2% of nosocomial infections; multidrug resistance is also observed in LA-MRSA.16 As with other S aureus strains, carriers are at risk of subsequent infection and serve as important reservoirs for person-to-person transmission.17 The emergence of LA-MRSA highlights concerns about the development of AMR with widespread antibiotic use in animal husbandry and the zoonotic transmission of infection and resistance.
The successful dissemination of MRSA poses significant challenges for the management of SSTIs (Fig 1 ).29 Although CA-MRSA are often sensitive to tetracyclines, trimethoprim-sulfamethoxazole, and clindamycin, resistance to these agents has been reported, and antibiotic selection should be guided by susceptibility testing.30 Clindamycin’s oral formulation and activity against both S aureus and S pyogenes make it especially useful for the empiric outpatient treatment of SSTIs. Clindamycin resistance occurs with the acquisition of erythromycin-ribosomal-methylase, erm, a gene that encodes for the methylation of the bacterial 23S ribosomal subunit, the shared action site of both macrolides and clindamycin. The resulting cross-resistant phenotype, known as macrolide-lincosamide-streptogramin B (MLSB), is seen in MRSA, methicillin-susceptible S aureus, and S pyogenes.20 Expression of the erm gene can be either constitutive or induced by macrolide exposure. As would be expected, isolates with constitutive erm expression demonstrate both clindamycin and erythromycin resistance by standard in vitro bacterial resistance testing. Bacteria with macrolide-inducible erm expression test as erythromycin resistant but clindamycin sensitive. Clinically, however, treatment failure with clindamycin occurs as treatment pressure in vivo rapidly selects for mutants that constitutively express erm and are clindamycin resistant.31 Since clindamycin resistance also occurs via non-erm mediated mechanisms, a specialized susceptibility test, known as the double-disk diffusion, can help identify inducible erm-mediated resistance.20
Fig 1.
Outpatient management of skin and soft tissue infections from the Centers for Disease Control and Prevention.
I&D, Incision and drainage; FDA, Food and Drug Administration; MRSA, methicillin-resistant Staphylococcus aureus; SSTI, skin and soft tissue infection.
Clindamycin-resistant Streptococcus pyogenes
Key points
-
•
Unlike S aureus, group A Streptococcus (GAS) remains exquisitely sensitive to penicillin-class antibiotics.
-
•
In invasive GAS infections, cotreatment with clindamycin is recommended, but rates of resistance to clindamycin are rising.
Streptococcus pyogenes (GAS) is a gram-positive coccus that causes a variety of infections and immune-mediated sequelae due to antigenic mimicry, interactions with human immunity, and the expression of virulence factors, often with prominent skin findings (Table II ).15 , 32, 33, 34, 35 Asymptomatic carriage has been reported in 4% to 5% of adults and 2% to 20% of children.36
Table II.
Spectrum of streptococcal skin infections, treatment recommendations, and notable associated complications33, 34, 35, 36
| Disease | Virulence factor | Recommended management (Strength of recommendation, quality of evidence) | Possible Sequelae |
|---|---|---|---|
| Pharyngitis | M-types 1, 3, 5, 6, 12, 14, 17, 19, 24 | Rapid antigen detection test or throat culture for confirmation of GAS (strong, high) Oral penicillin V OR amoxicillin OR intramuscular penicillin G (strong, high) If Penicillin-allergic: oral cephalexin OR cefadroxil (strong, high) Other options for penicillin-allergy: oral azithromycin OR erythromycin OR clindamycin (strong, moderate) |
Guttate psoriasis Acute rheumatic fever Erythema nodosum Polyarteritis nodosa Small-vessel vasculitis Type I scleroderma Acute febrile neutrophilic dermatosis Peritonsillar abscess Cervical lymphadenitis Invasive GAS infection |
| Acute rheumatic fever† | M-types 1, 3, 5, 6, 11, 12, 14, 17, 18, 19, 24, 27, 29, 30, 32, 41 |
Treatment, regardless of throat culture results IM benzathine penicillin OR Oral penicillin OR Oral amoxicillin OR Oral macrolide in penicillin-allergy |
Arthritis (60-80%) Carditis∗ (30-45%) Chorea (10%) Erythema marginatum (2%) Subcutaneous nodules |
| Scarlet fever | M-types 1, 4, 12, 49, 55, 57, 60 (nephritogenic) SPE-A, SPE-B, SPE-C |
Oral penicillin or amoxicillin If penicillin-allergic: 1st generation cephalosporin OR clindamycin OR macrolide |
Acute rheumatic fever† Post streptococcal glomerulonephritis when associated with nephritogenic strains |
| Ecthyma | - | C&S to identify if GAS or S. aureus is the causative pathogen (strong, moderate) Treatment without C&S is reasonable and should include anti-staphylococcal coverage (strong, moderate) Treatment with oral penicillin if GAS identified on culture (strong, high) |
- |
| Impetigo | M-types 33, 41, 42, 52, 53, 70 | C&S recommended for pathogen identification (strong, moderate) Treatment without C&S is reasonable and should include anti-staphylococcal coverage (strong, moderate) Localized: Topical mupirocin OR retapamulin (strong, high) Widespread: If GAS identified on culture treat with oral penicillin (strong, high) |
If associated with nephritogenic M proteins 1, 4, 12, 49, 55, 57, 60 there is an increased risk of post streptococcal glomerulonephritis |
| Erysipelas | - | Blood cultures, aspirates, biopsy, or swab not routinely recommended (strong, moderate) If mild with no systemic symptoms outpatient regimen (strong, moderate) with Penicillin OR Cephalosporin OR Dicloxacillin OR Clindamycin |
- |
| Streptococcal toxic shock syndrome (STSS) | M-types 1 and 3 Streptolysin O SPE-A > SPE-B, SPE-C Antigen-independent activation of T-cells by SPEs, SSA, SmeZ → massive production of IL-1, IL-6, TNF-α |
Early surgical intervention First line for documented GAS: Penicillin PLUS Clindamycin Linezolid may be an alternative to clindamycin to inhibit toxin production IVIG may be used as an adjunct |
Widespread tissue damage Disseminated intravascular thrombosis End-organ dysfunction |
| Necrotizing fasciitis | Uncontrolled T-cell response to superantigens → proinflammatory cytokine storm | Prompt surgical consultation (strong, low) Empirical therapy with broad coverage as infection can be poly- or mono-bacterial (strong, low) For documented GAS treat with penicillin PLUS clindamycin (strong, low) |
STSS Multi-organ failure |
ARF, Acute rheumatic fever; C&S, culture and sensitivity; GAS, group A streptococcus; IL-1, interleukin-1; IL-6, interleukin-6; IVIG, Intravenous immunoglobulins; SmeZ, streptococcal mitogenic exotoxin Z; SPE, streptococcal pyrogenic exotoxin; SSA, streptococcal superantigen; STSS, streptococcal toxic shock syndrome; TNF-α, tumor necrosis factor α.
ARF can still be prevented if antibiotics for GAS pharyngitis are initiated within 9 days of symptom onset.
Rheumatic heart disease is a complication of untreated acute rheumatic fever and is the leading cause of pediatric heart disease worldwide.
GAS most commonly causes pharyngitis and impetigo.35 Invasive diseases, including bacteremia, cellulitis, necrotizing fasciitis, pneumonia, and puerperal infections, carry mortality rates of 7% to 30% and can be complicated by progression to streptococcal toxic shock syndrome in one-third of the cases, with an associated mortality of up to 81%.34 , 35
Unlike S aureus, GAS is universally sensitive to penicillin, which remains the treatment of choice due to its efficacy, safety, narrow spectrum, and low cost.32 , 34 , 35 , 37 In invasive GAS infections, however, penicillin monotherapy is associated with higher mortality, and the addition of clindamycin is recommended.34 The high inoculum of invasive infection means that a bacterial stationary growth phase in which PBPs are relatively underexpressed is reached quickly making penicillin, but not clindamycin, less effective in invasive GAS infection.32 , 34 , 35 Clindamycin’s ability to inhibit bacterial protein synthesis also helps reduce the GAS toxin production known to complicate these infections.32 , 34 , 37
Since the 1980s, there has been an increase in the incidence and severity of invasive GAS infections.34 , 35 More than 20% of invasive GAS infections are due to clindamycin- and erythromycin-resistant strains in the United States. Increasing rates of resistance have also been reported in Europe and Asia, with as many as 94% of isolates in China demonstrating clindamycin resistance.38, 39, 40 The rise in clindamycin resistance can be partially attributed to the soaring rates of macrolide resistance over the past 40 years that have paralleled the preferential use of macrolides to treat localized GAS infections.35 , 41, 42, 43 Since concomitant macrolide and clindamycin resistance arises through the acquisition of erm, the overuse of macrolides for localized GAS infections also promotes clindamycin resistance and jeopardizes the ability of clindamycin to serve as an important therapeutic option for serious, life-threatening GAS infections. Comparatively, linezolid may be an effective alternative to clindamycin, as it provides broad-spectrum, gram-positive coverage and has toxin inhibition properties.44 , 45
Resistant Cutibacterium acnes
Key points
-
•
Broad-spectrum antibiotic therapy in acne promotes resistance in C acnes and other skin commensals.
-
•
C acnes can cause biofilm-associated infections of prosthetic joints, heart valves, and other indwelling implants.
C acnes is a commensal within the pilosebaceous unit thought to contribute to several steps in acne pathogenesis, including follicular occlusion.46, 47, 48, 49 Due to their antibacterial and anti-inflammatory properties, antibiotics are widely used in acne therapy.50 , 51 Prolonged treatment courses, the bacteriostatic (and not bactericidal) effects of many agents, and the availability of over-the-counter antibiotics in some countries have promoted global C acnes resistance.46 , 48 , 49
C. acnes resistance was first reported in the 1970s, with the introduction and widespread use of topical erythromycin and clindamycin.46 , 52 , 53 By the late 1990s, global resistance rates had risen to 62%.53 Approximately 50% of patients with acne develop resistance after oral or topical therapy, and 1 in 4 C acnes strains are currently resistant to tetracyclines, macrolides, or clindamycin.46 , 52 , 53 C acnes has historically remained sensitive to doxycycline and minocycline, but recent studies demonstrating increasing rates of resistance suggest that this may be changing.54, 55, 56, 57, 58
C acnes resistance mechanisms are tied to the mode of action of specific antimicrobials. Both macrolides and clindamycin inhibit bacterial protein synthesis by binding to the 23S ribosomal RNA (rRNA) subunit. Point mutations and base substitutions in the 23S rRNA binding site confer variable degrees of resistance to clindamycin, macrolides, or both.46 , 49 , 52 , 59 C acnes can also acquire erm from other C acnes strains or Corynebacterium species, leading to the macrolide-lincosamide-streptogramin B (MLSB) phenotype with high-level macrolide-clindamycin cross-resistance.49 , 59 Tetracycline resistance occurs with point mutations in the 16S rRNA subunit, affecting antibiotic binding, or via efflux pumps.46 , 49 , 52 , 54 , 60
The development of antibiotic resistance in C acnes is consequential for several reasons. In terms of therapeutic outcomes in acne, resistance has been associated with more recalcitrant lesions and treatment failure.54, 55, 56, 57, 58 , 61 Additionally, resistance complicates the already-challenging treatment of C acnes biofilm-associated infections of prosthetic joints, heart valves, and other indwelling implants.46 , 62 When broad-spectrum antibiotics, such as doxycycline and clindamycin, are used for acne, their effects can extend beyond C acne s to cause inadvertent damage to “bystander” bacteria of the microbiome. For example, minocycline treatment in acne has been associated with significant dysbiosis of the skin and gut.63
Broad-spectrum antibiotic use for acne exerts selection pressure on a wide range of bacteria to promote the survival of strains that have acquired resistance mechanisms. The result is increased resistance among C acnes as well as other pathogenic bacteria such as S aureus, GAS, and coagulase-negative Staphylococcus strains, which compromises the effectiveness of these antibiotics for other treatment indications. For example, clindamycin-resistant coagulase-negative Staphylococcus has been isolated from more than 80% of patients with clindamycin-resistant C acnes.48 , 50 Furthermore, 35% of patients with acne on antibiotics were found to carry oropharyngeal GAS, 85% of which was tetracycline resistant.64 The use of clindamycin and doxycycline in acne therapy can also select for resistant MRSA strains, limiting the use of these agents in MRSA infections.50 , 65 Recent evidence suggests that even low-dose, or subantimicrobial doses of, antibiotics like doxycycline or minocycline may still potentiate resistance through a variety of mechanisms.66, 67, 68
Novel narrow-spectrum antibiotics for acne may help limit resistance. Sarecycline, a tetracyline antibiotic developed for the treatment of acne, maintains high activity against gram-positive bacteria, including C acnes, and demonstrates a low propensity for inducing resistance. Its greatly diminished activity against gram-negative bacteria compared to doxycycline and minocycline allows it to achieve a similar therapeutic efficacy in acne but with a lower risk of gut microbiome and resistome alterations.54 , 60
Multidrug-resistant Pseudomonas aeruginosa
Key points
-
•
P aeruginosa’s extensive, intrinsic resistance mechanisms make antipseudomonal drug development challenging.
-
•
Multidrug-resistant pseudomonal infections are an urgent threat due to the life-threatening nature of invasive infections and very limited treatment options.
P aeruginosa is a ubiquitous, gram-negative, aerobic bacillus implicated in several localized cutaneous and invasive hospital-acquired infections. Life-threatening infections disproportionately affect the immunocompromised and those with extensive skin breakdown.69
While localized skin infections have good prognoses, invasive infections have high mortality rates and require prompt and careful antimicrobial selection.69 , 70 The treatment of a Pseudomonas SSTI typically relies on an antipseudomonal ß-lactam or a fluoroquinolone in combination with surgical intervention.71 Due to concerns for resistance, the empiric treatment of life-threatening infections involves 2 antipseudomonal agents from different classes while awaiting culture and sensitivities.69 , 71
Owing to intrinsic resistance mechanisms, the available armamentarium against wild-type P aeruginosa includes only a few antibiotic classes (Table III ).69, 70, 71, 72 P aeruginosa has developed resistance mechanisms to all available antipseudomonal antibiotic classes, largely due to the increased use of these agents coupled with the efficient transmission of resistance traits.71 Since quinolones upregulate and serve as substrates for all 4 known pseudomonas efflux systems, continuous quinolone exposure is an important risk factor for the generation of cross-resistance.
Table III.
| Antibacterial class | Specific agents | Mechanism of action | Acquired resistance mechanisms |
|---|---|---|---|
| Monobactams | Aztreonam | Binding to PBP3 → inhibition of peptidoglycan synthesis | 1) Class A serine ESBLs (PER, VEB, GES, BEL, SHV, CTX-M, TEM) hydrolyze the beta-lactam 2) Class D β-lactamases (OXA-type enzymes: OXA-2, OXA-10) hydrolyze the β-lactam |
| Aminoglycosides | Tobramycin Gentamicin Amikacin Netilmicin |
Binding to 16S rRNA of the 30S ribosomal subunit → inhibition of protein synthesis | 1) Aminoglycoside modifying enzymes (AME) inactivate the drug via attachment of acetyl, phosphate or adenyl groups. Most AMEs tend to spare Amikacin 2) MexXY-OprM efflux pump reduces drug concentrations 3) Genes RMtA or RMtB encoded in mobile genetic elements methylate 16S rRNA leading to an altered drug target |
| Carbapenem | Imipenem | Binding to PBPs → inhibition of cell-wall synthesis | 1) Class D metallo-β-lactamases (IMP, VIM, SPM, GIM) hydrolyze the beta-lactam∗ 2) MexEF-OprN efflux pump reduces drug concentrations 3) Reduced expression of membrane porin OprD leads to decreased drug uptake |
| Meropenem | 1) Class D metallo-beta-lactamases (IMP, VIM, SPM, GIM) hydrolyze the beta-lactam∗ 2) MexEF-OprN efflux pump reduces drug concentrations |
||
| Cephalosporins | Ceftazidime (3rd generation) Cefepime (4th generation) |
Binding to PBPs → inhibition of cell-wall synthesis | 1) Class A serine ESBL (PER, VEB, GES, BEL, SHV, CTX-M, TEM) can hydrolyze the beta-lactam 2) Class D β-lactamases (OXA-type enzymes: OXA-2, OXA-10) can hydrolyze the beta-lactam |
| Fluoroquinolones | Ciprofloxacin Levofloxacin |
Inhibition of enzymes involved in DNA replication DNA gyrase (topoisomerase II) and topoisomerase IV | 1) Efflux pumps MexAB-OprM, MexXY-OprM, MexCD-OprJ and MexEF-OprN reduce drug concentrations† 2) Point mutations in the quinolone resistance determining region (QRDR) of genes gyrA and gyrB, encoding subunits of the DNA gyrase, and the genes parC and parE, encoding subunits of topoisomerase IV lead to an altered drug target |
| Penicillins | Carboxipenicillin Ureidopenicillin |
Binding to penicillin-binding proteins → inhibition of peptidoglycan cross-linking | 1) Amber Class A serine β-lactamases of types TEM, PSE or CARB hydrolyze the beta-lactam 2) Class A serine ESBL (PER, VEB, GES, BEL, SHV, CTX-M, TEM) hydrolyze the β-lactam 3) Class D β-lactamases (OXA-type enzymes:(OXA-1, OXA-2, OXA-10)) hydrolyze the β-lactam |
| Polymyxins | Colistin and polymyxin B | Binding to lipopolysaccharide and disruption of the cell membrane | 1) Enzymatic alteration of the lipopolysaccharide alters drug binding |
| Fosfomycin | Fosfomycin | Inhibition of MurA enzyme → inhibition of the first step in peptidoglycan synthesis | 1) FosA modifying enzyme can lead chemical modification and inactivation of drug 2) Mutations in gltP permease, a transporter enzyme, lead to reduced drug uptake |
The global rise in MDR P aeruginosa, defined as resistance to 1 drug agent in at least 3 classes, has reached rates of 15% to 50%, leaving limited therapeutic options for invasive disease.70 , 71 , 73 In cases of MDR or pan-resistant strains, last-resort agents include polymyxins (colistin and polymyxin B), which have very narrow therapeutic windows due to nephrotoxicity.73
Drug-resistant Mycobacterium tuberculosis
Key points
-
•
Tuberculosis is difficult to treat, even under ideal conditions.
-
•
Several factors have contributed to the global threat of increasingly drug-resistant Mycobacterium tuberculosis (TB).
TB is an acid-fast, alcohol-fast, aerobic bacillus that spreads via airborne transmission and causes more deaths globally than any other infectious agent.74 Though cutaneous tuberculosis accounts for only 1% of extrapulmonary cases, the past few decades have seen a resurgence of both pulmonary and cutaneous TB and a rise in drug-resistant strains.74, 75, 76, 77
The clinical factors contributing to resistance include an insufficient duration of therapy, nonadherence, interrupted active phase of treatment, lack of access to therapy, and a rise in HIV coinfection. The treatment of drug-susceptible TB requires a 6-month, multidrug treatment with first-line agents and, under ideal conditions, has a success rate of only 85%.78 The treatment of MDR-TB necessitates more expensive and more toxic second-line agents given for an extended duration, sometimes up to 20 months, to achieve a success rate of 57% (Table IV ).79
Table IV.
| Resistance | Definition |
|---|---|
| Drug-resistant tuberculosis | Resistance to 1 of 4 first-line agents, more commonly isoniazid |
| Multidrug-resistant tuberculosis (MDR-TB) | Resistance to 2 of 4 first-line agents, at least rifampicin and isoniazid |
| Extensively drug-resistant tuberculosis (XDR-TB) | Resistance to first-line agents (MDR-TB) PLUS at least 1 fluoroquinolone and a second-line injectable agent (amikacin, kanamycin, or capreomycin) |
| First-line agents | Rifampin Isoniazid Ethambutol Pyrazinamide |
| Second-line agents | Streptomycin/amikacin/kanamycin, Levofloxacin/moxifloxacin/ofloxacin Para-amino-salicylic acid Capreomycin/viomycin Cycloserine Terizidone Ethionamide/Prothionamide |
| Third line agents | Linezolid Clofazimine Amoxicillin-Clavulanate Imipenem Thiacitazone Clarithromycin Bedaquiline Delamanid |
Due to the devastating effects of the COVID-19 pandemic on health care resources in lower-income countries, there has been a decline in TB detection and notification and a reallocation of resources away from the TB sector. The World Health Organization estimates that the impact of the pandemic will lead to an additional 6.2 million global TB cases between 2020 and 2025.74
Emerging approaches to limit antimicrobial resistance
As demonstrated by the COVID-19 pandemic response, antimicrobial strategies must contend with the relentless ability of microbes to resist and adapt. Conventional antimicrobial drug development has struggled to keep pace with the development of AMR. Simple yet effective resistance reduction strategies include the avoidance of antibiotic monotherapy in acne treatment and the limitation of the duration of oral antibiotic use. Innovative therapies currently being investigated, such as nanoparticles and phage therapy, have actions unlike those of conventional antimicrobials and may be particularly promising strategies to address the AMR crisis.82
Phage therapy
Phage therapy utilizes bacterial viruses to (1) selectively attack a bacterial population, (2) deliver drug molecules to a bacterial target, and (3) insert enzymes that inactivate or reverse resistance genes. Given their ubiquitous nature and high specificity for bacterial receptors, bacteriophages can be engineered to target a specific pathogenic bacterial population without damage to collateral flora.81 , 83
Phage therapy is already being applied in the investigational treatment of ulcers, burns, and chronic wounds (Table V ).84, 85, 86, 87, 88, 89 In addition to their specificity, their efficacy against S aureus and Pseudomonas biofilms make bacteriophages an especially promising modality for the management of chronic wounds, ulcers, and acne.83 C acnes bacteriophages have shown in vitro efficacy in lysing C acnes colonies without affecting collateral microbes, a significant advantage compared to currently used antibacterials.90 , 91
Table V.
Clinical trials of dermatologic interest utilizing phage therapy as registered on clinicaltrials.gov
| Brief study description | Status | Dermatologic condition | Primary outcome measure | Study identifier |
|---|---|---|---|---|
| Phase I/II trial comparing the efficacy of standard treatment with a topical anti-staphylococcal bacteriophage cocktail versus placebo for diabetic foot ulcers infected with MRSA or MSSA (127) | Recruitment planned June 2020 Aims: 60 adult participants with type 1 or type 2 diabetes and a wound below the ankle (evolving for >2 weeks) mono-infected with MRSA |
Diabetic foot ulcers | Relative reduction in wound surface area (%) at 12 weeks. | NCT02664740 |
| Phase I, randomized, open-label study investigating the safety/tolerability of phage-cocktail spray as an adjunct to standard therapy (xeroform dressing and Kenacomb cream for localized signs of infection) for the prevention and treatment of burns susceptible to infection/or infected by S aureus, Paeruginosa, or K pneumoniae | Not yet recruiting | Wound infection | Incidence of treatment-related adverse events and incidence of treatment discontinuation due to adverse events | NCT04323475 |
| Phase I/II, randomized, open-label trial comparing tolerance and efficacy of local bacteriophage treatment (using Pherecydes Pharma anti-Escherichia coli and anti-Pseudomonas aeruginosa bacteriophage cocktails) to standard therapy (silver sulfadiazine) of burn wounds infected with E coli or P aeruginosa | Closed Findings: The primary endpoint was reached in a median of 144 h (95% CI 48–not reached) in the PP1131 group versus a median of 47 h (23–122) in the standard of care group |
Burn wound infection | Median time to sustained reduction of bacterial burden | NCT02116010 |
| Phase I, randomized, controlled double-blind study evaluating the safety of WPP-201 in full thickness venous leg ulcers > 30 days (WPP-201 in a pH neutral phage preparation with 8 bacteriophages lytic for P aeruginosa, S aureus, and E coli) | Completed recruitment | Venous leg ulcers | Safety of product use | NCT00663091 |
| Phase II randomized trial comparing the efficacy of T4N5 liposomal lotion (topical bacteriophage T4 endonuclease V) versus placebo in preventing recurrence of NMSC in patients with previous history of NMSC who have undergone kidney transplant | Completed recruitment Last update in 2015 |
Nonmelanoma skin cancer (NMSC) | Incidence of NMSC per patient on the sun-exposed skin of renal transplant recipients with a history of NMSC treated with T4N5 vs placebo | NCT00089180 |
| Phase I randomized, double-blind trial assessing adverse events to 2 ascending concentrations of topical administration of AB-SA01 (topical 3 phage cocktail against S. aureus) | Completed. Application was well tolerated |
- | Incidence of adverse events, including change in clinical lab tests from baseline and skin reaction, from first dose through the end of the study, including | NCT02757755 |
E. coli, Escherichia coli; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; NMSC, Nonmelanoma skin cancer; S aureus, Staphylococcus aureus; P aeruginosa, Pseudomonas aeruginosa; K pneumoniae.
Nanoparticles
Nanoparticles are metals or metal oxides that combat microbes via a number of simultaneous mechanisms, including the disruption of the cell membrane, generation of reactive oxygen species, and damage to intracellular contents, making them low-risk for resistance and particularly attractive against MDR pathogens. Nanoparticles hold great potential as protective coatings, such as on wound dressings or implants, to prevent biofilm-associated infections.92, 93, 94
Despite their theoretical and early investigational promise, most of these innovative strategies are still in early stages of development with further investigations needed to establish the safety, in vivo efficacy, interactions with human immunity, and pharmacokinetics before they can be used clinically to address AMR.83 , 92, 93, 94
Conclusion
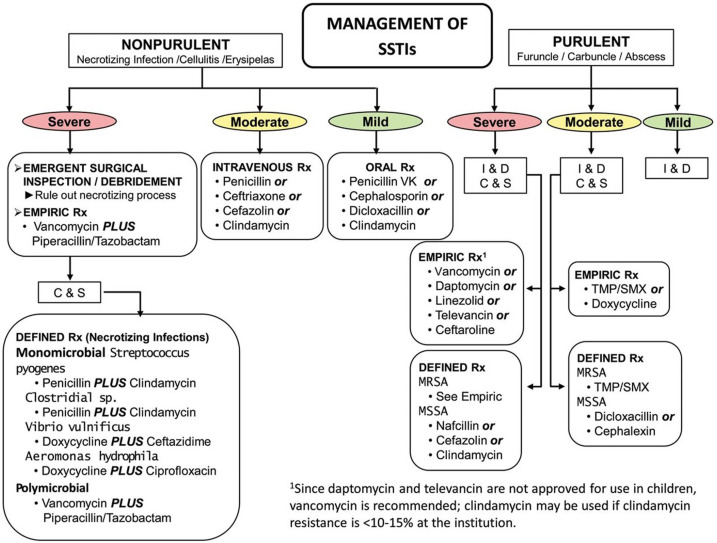
Dermatology treatment guidelines issued by expert panels over the past 2 decades consistently emphasize the need for judicious antibiotic use. Overall antibiotic prescribing by dermatologists decreased by 36.6% between 2008 and 2016. During the same time frame, however, oral antibiotic use associated with surgical visits increased by 69.6%, suggesting that there may still be areas for improvement.64 In the treatment of SSTIs, dermatologists should adhere to evidence-based infectious diseases guidelines for appropriate antibiotic selection (Fig 2 ) and be cognizant of local resistance patterns for key pathogens. Continued support for research into the off-target effects of antibiotics on the cutaneous microbiome and innovative strategies for the treatment of skin disease are needed to generate evidence-based interventions that prevent AMR.
Fig 2.
Outpatient management of skin and soft tissue infections from the Infectious Diseases Society of America.29
C&S, Culture and sensitivity; I&D, incision and drainage; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; TMP/SMX, trimethoprim-sulfamethoxazole; SSTI, skin and soft tissue infection; Rx, treatment.
Conflicts of interest
None disclosed.
Footnotes
Funding sources: None.
IRB approval status: Not applicable.
Date of release: June 2022.
Expiration date: June 2025.
References
- 1.Cantón R., Gijón D., Ruiz-Garbajosa P. Antimicrobial resistance in ICUs: an update in the light of the COVID-19 pandemic. Curr Opin Crit Care. 2020;26(5):433–441. doi: 10.1097/MCC.0000000000000755. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S., Bornman C., Zafer M.M. Antimicrobial resistance threats in the emerging COVID-19 pandemic: where do we stand? J Infect Public Health. 2021;14(5):555–560. doi: 10.1016/j.jiph.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Founou R.C., Blocker A.J., Noubom M., et al. The COVID-19 pandemic: a threat to antimicrobial resistance containment. Future Sci OA. 2021;7(8):FSO736. doi: 10.2144/fsoa-2021-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C.C., Chen S.Y., Ko W.C., Hsueh P.R. Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents. 2021;57(4):106324. doi: 10.1016/j.ijantimicag.2021.106324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alanis A.J. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res. 2005;36(6):697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention . U.S. Department of Health and Human Services; 2019. Antibiotic Resistance Threats in the United States: 2019. [Google Scholar]
- 7.New report calls for urgent action to avert antimicrobial resistance crisis: International organizations unite on critical recommendations to combat drug-resistant infections and prevent staggering number of deaths each year. World Health Organization. https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis
- 8.Murray C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022 doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goossens H. Antibiotic consumption and link to resistance. Clin Microbiol Infect. 2009;15(Suppl 3):12–15. doi: 10.1111/j.1469-0691.2009.02725.x. [DOI] [PubMed] [Google Scholar]
- 10.Goossens H., Ferech M., Vander Stichele R., Elseviers M., ESAC Project Group Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365(9459):579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 11.Martens E., Demain A.L. The antibiotic resistance crisis, with a focus on the United States. J Antibiot (Tokyo) 2017;70(5):520–526. doi: 10.1038/ja.2017.30. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez GV, Fleming-Dutra KE, Roberts RM, Hicks LA. Core elements of outpatient antibiotic stewardship. MMWR Recomm Rep. 2016;65(No. RR-6):1-12. [DOI] [PubMed]
- 13.Fleming-Dutra K.E., Hersh A.L., Shapiro D.J., et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA. 2016;315(17):1864–1873. doi: 10.1001/jama.2016.4151. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention . 2017. Outpatient antibiotic prescriptions—United States. [Google Scholar]
- 15.Stevens D.L., Bisno A.L., Chambers H.F., et al. Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the infectious diseases society of America. Clin Infect Dis. 2014;59(2):147–159. doi: 10.1093/cid/ciu296. [DOI] [PubMed] [Google Scholar]
- 16.Gajdács M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 2019;8(2):52. doi: 10.3390/antibiotics8020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee A.S., de Lencastre H., Garau J., et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 18.Chambers H.F., Deleo F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobanovska M., Pilla G. Penicillin's discovery and antibiotic resistance: lessons for the future? Yale J Biol Med. 2017;90(1):135–145. [PMC free article] [PubMed] [Google Scholar]
- 20.Pantosti A., Sanchini A., Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2(3):323–334. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 21.Novick R.P., Bouanchaud D. The problems of drug-resistant pathogenic bacteria. Extrachromosomal nature of drug resistance in Staphylococcus aureus. Ann N Y Acad Sci. 1971;182:279–294. doi: 10.1111/j.1749-6632.1971.tb30664.x. [DOI] [PubMed] [Google Scholar]
- 22.Ito T., Katayama Y., Asada K., et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45(5):1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers H.F. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7(2):178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kale P., Dhawan B. The changing face of community-acquired methicillin-resistant Staphylococcus aureus. Indian J Med Microbiol. 2016;34(3):275–285. doi: 10.4103/0255-0857.188313. [DOI] [PubMed] [Google Scholar]
- 25.Seybold U., Kourbatova E.V., Johnson J.G., et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42(5):647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 26.Preeja P.P., Kumar S.H., Shetty V. Prevalence and characterization of methicillin-resistant Staphylococcus aureus from community- and hospital-associated infections: a tertiary care center study. Antibiotics (Basel) 2021;10(2):197. doi: 10.3390/antibiotics10020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aires-de-Sousa M. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. 2017;23(6):373–380. doi: 10.1016/j.cmi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Chen C., Wu F. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) colonisation and infection among livestock workers and veterinarians: a systematic review and meta-analysis. Occup Environ Med. 2021;78(7):530. doi: 10.1136/oemed-2020-106418. [DOI] [PubMed] [Google Scholar]
- 29.Stevens D.L., Bisno A.L., Chambers H.F., et al. Executive summary: practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 update by the Infectious Diseases Society of America. Clin Infect Dis. 2014;59(2):147–159. doi: 10.1093/cid/ciu296. [DOI] [PubMed] [Google Scholar]
- 30.Outpatient† management of skin and soft tissue infections in the era of community-associated MRSA‡. Centers for Disease Control and Prevention. https://www.cdc.gov/mrsa/pdf/flowchart_pstr.pdf
- 31.Siberry G.K., Tekle T., Carroll K., Dick J. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin Infect Dis. 2003;37(9):1257–1260. doi: 10.1086/377501. [DOI] [PubMed] [Google Scholar]
- 32.Manders S.M. Toxin-mediated streptococcal and staphylococcal disease. J Am Acad Dermatol. 1998;39(3):383–398. doi: 10.1016/s0190-9622(98)70314-7. quiz 399-400. [DOI] [PubMed] [Google Scholar]
- 33.Shulman S.T., Bisno A.L., Clegg H.W., et al. Clinical practice guideline for the diagnosis and management of group A streptococcal pharyngitis: 2012 update by the Infectious Diseases Society of America. Clin Infect Dis. 2012;55(10):1279–1282. doi: 10.1093/cid/cis847. [DOI] [PubMed] [Google Scholar]
- 34.Waddington C.S., Snelling T.L., Carapetis J.R. Management of invasive group A streptococcal infections. J Infect. 2014;69 Suppl 1:S63–S69. doi: 10.1016/j.jinf.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Walker M.J., Barnett T.C., McArthur J.D., et al. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev. 2014;27(2):264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunnarsson R.K., Holm S.E., Soderstrom M. The prevalence of beta-haemolytic streptococci in throat specimens from healthy children and adults. Implications for the clinical value of throat cultures. Scand J Prim Health Care. 1997;15(3):149–155. doi: 10.3109/02813439709018506. [DOI] [PubMed] [Google Scholar]
- 37.Johansson L., Thulin P., Low D.E., Norrby-Teglund A. Getting under the skin: the immunopathogenesis of Streptococcus pyogenes deep tissue infections. Clin Infect Dis. 2010;51(1):58–65. doi: 10.1086/653116. [DOI] [PubMed] [Google Scholar]
- 38.Gooskens J., Neeling A.J., Willems R.J., Wout J.W., Kuijper E.J. Streptococcal toxic shock syndrome by an iMLS resistant M type 77 Streptococcus pyogenes in the Netherlands. Scand J Infect Dis. 2005;37(2):85–89. doi: 10.1080/00365540510027192. [DOI] [PubMed] [Google Scholar]
- 39.Pesola A.K., Sihvonen R., Lindholm L., Patari-Sampo A. Clindamycin resistant emm33 Streptococcus pyogenes emerged among invasive infections in Helsinki metropolitan area, Finland, 2012 to 2013. Euro Surveill. 2015;20(18):21117. doi: 10.2807/1560-7917.es2015.20.18.21117. [DOI] [PubMed] [Google Scholar]
- 40.Lu B., Fang Y., Fan Y., et al. High prevalence of macrolide-resistance and molecular characterization of Streptococcus pyogenes isolates circulating in China from 2009 to 2016. Front Microbiol. 2017;8:1052. doi: 10.3389/fmicb.2017.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fyfe C., Grossman T.H., Kerstein K., Sutcliffe J. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb Perspect Med. 2016;6(10):a025395. doi: 10.1101/cshperspect.a025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34(4):482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 43.DeMuri G.P., Sterkel A.K., Kubica P.A., Duster M.N., Reed K.D., Wald E.R. Macrolide and clindamycin resistance in group A Streptococci isolated from children with pharyngitis. Pediatr Infect Dis J. 2017;36(3):342–344. doi: 10.1097/INF.0000000000001442. [DOI] [PubMed] [Google Scholar]
- 44.Bryant A.E., Bayer C.R., Aldape M.J., McIndoo E., Stevens D.L. Emerging erythromycin and clindamycin resistance in group A streptococci: efficacy of linezolid and tedizolid in experimental necrotizing infection. J Glob Antimicrob Resist. 2020;22:601–607. doi: 10.1016/j.jgar.2020.04.032. [DOI] [PubMed] [Google Scholar]
- 45.LaPlante K.L., Leonard S.N., Andes D.R., Craig W.A., Rybak M.J. Activities of clindamycin, daptomycin, doxycycline, linezolid, trimethoprim-sulfamethoxazole, and vancomycin against community-associated methicillin-resistant Staphylococcus aureus with inducible clindamycin resistance in murine thigh infection and in vitro pharmacodynamic models. Antimicrob Agents Chemother. 2008;52(6):2156–2162. doi: 10.1128/AAC.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dessinioti C., Katsambas A. Propionibacterium acnes and antimicrobial resistance in acne. Clin Dermatol. 2017;35(2):163–167. doi: 10.1016/j.clindermatol.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Dreno B., Pecastaings S., Corvec S., Veraldi S., Khammari A., Roques C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32(Suppl 2):5–14. doi: 10.1111/jdv.15043. [DOI] [PubMed] [Google Scholar]
- 48.Farrah G., Tan E. The use of oral antibiotics in treating acne vulgaris: a new approach. Dermatol Ther. 2016;29(5):377–384. doi: 10.1111/dth.12370. [DOI] [PubMed] [Google Scholar]
- 49.Xu H., Li H. Acne, the skin microbiome, and antibiotic treatment. Am J Clin Dermatol. 2019;20(3):335–344. doi: 10.1007/s40257-018-00417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh T.R., Efthimiou J., Dreno B. Systematic review of antibiotic resistance in acne: an increasing topical and oral threat. Lancet Infect Dis. 2016;16(3):e23–e33. doi: 10.1016/S1473-3099(15)00527-7. [DOI] [PubMed] [Google Scholar]
- 51.Zaenglein A.L., Pathy A.L., Schlosser B.J., et al. Guidelines of care for the management of acne vulgaris. J Am Acad Dermatol. 2016;74(5):945–973.e33. doi: 10.1016/j.jaad.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 52.Eady E.A., Gloor M., Leyden J.J. Propionibacterium acnes resistance: a worldwide problem. Dermatology. 2003;206(1):54–56. doi: 10.1159/000067822. [DOI] [PubMed] [Google Scholar]
- 53.Leccia M.T., Auffret N., Poli F., Claudel J.P., Corvec S., Dreno B. Topical acne treatments in Europe and the issue of antimicrobial resistance. J Eur Acad Dermatol Venereol. 2015;29(8):1485–1492. doi: 10.1111/jdv.12989. [DOI] [PubMed] [Google Scholar]
- 54.Graber E.M. Treating acne with the tetracycline class of antibiotics: a review. Dermatol Rev. 2021;2(6):321–330. doi: 10.1002/der2.49. [DOI] [Google Scholar]
- 55.Leyden J.J., McGinley K.J., Cavalieri S., Webster G.F., Mills O.H., Kligman A.M. Propionibacterium acnes resistance to antibiotics in acne patients. J Am Acad Dermatol. 1983;8:41–45. doi: 10.1016/s0190-9622(83)70005-8. [DOI] [PubMed] [Google Scholar]
- 56.Dreno B., Thiboutot D., Gollnick H., et al. Antibiotic stewardship in dermatology: limiting antibiotic use in acne. Eur J Dermatol. 2014;24:330–334. doi: 10.1684/ejd.2014.2309. [DOI] [PubMed] [Google Scholar]
- 57.Sheffer-Levi S., Rimon A., Lerer V., et al. Antibiotic susceptibility of Cutibacterium acnes strains isolated from Israeli acne patients. Acta Derm Venereol. 2020;100(17) doi: 10.2340/00015555-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alkhawaja E., Hammadi S., Abdelmalek M., Abdelmalek M., Mahasneh N., Alkhawaja B., Abdelmalek S.M. Antibiotic resistant Cutibacterium acnes among acne patients in Jordan: a cross sectional study. BMC Dermatol. 2020;20(1):17. doi: 10.1186/s12895-020-00108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aoki S., Nakase K., Hayashi N., Noguchi N. Transconjugation of erm(X) conferring high-level resistance of clindamycin for Cutibacterium acnes. J Med Microbiol. 2019;68(1):26–30. doi: 10.1099/jmm.0.000875. [DOI] [PubMed] [Google Scholar]
- 60.Batool Z., Lomakin I.B., Polikanov Y.S., Bunick C.G. Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome. Proc Natl Acad Sci U S A. 2020;117(34):20530–20537. doi: 10.1073/pnas.2008671117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eady E.A., Cove J.H., Holland K.T., Cunliffe W.J. Erythromycin resistant propionibacteria in antibiotic treated acne patients: association with therapeutic failure. Br J Dermatol. 1989;121(1):51–57. doi: 10.1111/j.1365-2133.1989.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 62.Achermann Y., Goldstein E.J., Coenye T., Shirtliff M.E. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev. 2014;27(3):419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson K.G., Rainer B.M., Antonescu C., et al. Minocycline and its impact on microbial dysbiosis in the skin and gastrointestinal tract of acne patients. Ann Dermatol. 2020;32(1):21–30. doi: 10.5021/ad.2020.32.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbieri J.S., Bhate K., Hartnett K.P., Fleming-Dutra K.E., Margolis D.J. Trends in oral antibiotic prescription in dermatology, 2008 to 2016. JAMA Dermatol. 2019;155(3):290–297. doi: 10.1001/jamadermatol.2018.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller Y.W., Eady E.A., Lacey R.W., Cove J.H., Joanes D.N., Cunliffe W.J. Sequential antibiotic therapy for acne promotes the carriage of resistant staphylococci on the skin of contacts. J Antimicrob Chemother. 1996;38(5):829–837. doi: 10.1093/jac/38.5.829. [DOI] [PubMed] [Google Scholar]
- 66.Wistrand-Yuen E., Knopp M., Hjort K., Koskiniemi S., Berg O.G., Andersson D.I. Evolution of high-level resistance during low-level antibiotic exposure. Nat Commun. 2018;9(1):1599. doi: 10.1038/s41467-018-04059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gullberg E., Cao S., Berg O.G., et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7(7) doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westhoff S., van Leeuwe T.M., Qachach O., Zhang Z., van Wezel G.P., Rozen D.E. The evolution of no-cost resistance at sub-MIC concentrations of streptomycin in Streptomyces coelicolor. ISME J. 2017;11(5):1168–1178. doi: 10.1038/ismej.2016.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu D.C., Chan W.W., Metelitsa A.I., Fiorillo L., Lin A.N. Pseudomonas skin infection: clinical features, epidemiology, and management. Am J Clin Dermatol. 2011;12(3):157–169. doi: 10.2165/11539770-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 70.Ruiz-Garbajosa P., Canton R. Epidemiology of antibiotic resistance in Pseudomonas aeruginosa. Implications for empiric and definitive therapy. Rev Esp Quimioter. 2017;30 Suppl 1:8–12. [PubMed] [Google Scholar]
- 71.Bassetti M., Vena A., Croxatto A., Righi E., Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7:212527. doi: 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silver L.L. Fosfomycin: mechanism and resistance. Cold Spring Harb Perspect Med. 2017;7(2):a025262. doi: 10.1101/cshperspect.a025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horcajada J.P., Montero M., Oliver A., et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4) doi: 10.1128/CMR.00031-19. e00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Global Tuberculosis Report 2020. World Health Organization. Licence: CC BY-NC-SA3.OIGO.
- 75.Barbagallo J., Tager P., Ingleton R., Hirsch R.J., Weinberg J.M. Cutaneous tuberculosis: diagnosis and treatment. Am J Clin Dermatol. 2002;3(5):319–328. doi: 10.2165/00128071-200203050-00004. [DOI] [PubMed] [Google Scholar]
- 76.Hill M.K., Sanders C.V. Cutaneous tuberculosis. Microbiol Spectr. 2017;5(1) doi: 10.1128/microbiolspec.TNMI7-0010-2016. [DOI] [PubMed] [Google Scholar]
- 77.MacGregor R.R. Cutaneous tuberculosis. Clin Dermatol. 1995;13(3):245–255. doi: 10.1016/0738-081x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 78.Nahid P., Dorman S.E., Alipanah N., et al. Executive summary: official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical practice guidelines: treatment of drug-susceptible tuberculosis. Clin Infect Dis. 2016;63(7):853–867. doi: 10.1093/cid/ciw566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lange C., Chesov D., Heyckendorf J., Leung C.C., Udwadia Z., Dheda K. Drug-resistant tuberculosis: an update on disease burden, diagnosis and treatment. Respirology. 2018;23(7):656–673. doi: 10.1111/resp.13304. [DOI] [PubMed] [Google Scholar]
- 80.Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis. 2001;32(11):1608–1614. doi: 10.1086/320532. [DOI] [PubMed] [Google Scholar]
- 81.Gordillo Altamirano F.L., Barr J.J. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 2019;32(2) doi: 10.1128/CMR.00066-18. e00066-e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cattoir V., Felden B. Future antibacterial strategies: from basic concepts to clinical challenges. J Infect Dis. 2019;220(3):350–360. doi: 10.1093/infdis/jiz134. [DOI] [PubMed] [Google Scholar]
- 83.Nikolich M.P., Filippov A.A. Bacteriophage therapy: developments and directions. Antibiotics (Basel) 2020;9(3):135. doi: 10.3390/antibiotics9030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Standard treatment associated with phage therapy versus placebo for diabetic foot ulcers infected by S. aureus (PhagoPied). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02664740
- 85.Phage therapy for the prevention and treatment of wound infections in burned patients. Clinical Trials.gov. https://clinicaltrials.gov/ct2/show/NCT04323475
- 86.Evaluation of phage therapy for the treatment of Escherichia coli and Pseudomonas aeruginosa wound infections in burned patients (PHAGOBURN). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02116010
- 87.A prospective, randomized, double-blind controlled study of WPP-201 for the safety and efficacy of treatment of venous leg ulcers (WPP-201). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT00663091
- 88.T4N5 liposomal lotion in preventing the recurrence of nonmelanoma skin cancer in patients who have undergone a kidney transplant. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT00089180
- 89.Ascending dose study of the safety of AB-SA01 when topically applied to intact skin of healthy adults. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02757755
- 90.Castillo D.E., Nanda S., Keri J.E. Propionibacterium (Cutibacterium) acnes bacteriophage therapy in acne: current evidence and future perspectives. Dermatol Ther (Heidelb) 2019;9(1):19–31. doi: 10.1007/s13555-018-0275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jończyk-Matysiak E., Weber-Dąbrowska B., Żaczek M., et al. Prospects of phage application in the treatment of acne caused by Propionibacterium acnes. Front Microbiol. 2017;8:164. doi: 10.3389/fmicb.2017.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L., Hu C., Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baptista P.V., McCusker M.P., Carvalho A., et al. Nano-strategies to fight multidrug resistant bacteria-“a battle of the titans. Front Microbiol. 2018;9:1441. doi: 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allahverdiyev A.M., Kon K.V., Abamor E.S., Bagirova M., Rafailovich M. Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev Anti Infect Ther. 2011;9(11):1035–1052. doi: 10.1586/eri.11.121. [DOI] [PubMed] [Google Scholar]