Abstract
Staphylococcal scalded-skin syndrome is usually diagnosed clinically by its characteristic exfoliating rash. Isolation of Staphylococcus aureus from the patient further supports the diagnosis. Several detection systems have been developed to determine whether the isolated strain produces exfoliative toxin, but none are routinely available in hospital laboratories. In a novel approach, we used computer models to predict the structure of the exfoliative toxins based on other serine proteases and to identify surface epitopes for the production of antibodies that specifically bound the exfoliative toxin A (ETA) serotype. Several rapid immunologically based diagnostic tests for ETA were developed with these antibodies and compared with existing systems. Our results showed that Western blot analysis using these antibodies was in complete correlation with PCR, which has been validated against the “gold standard” mouse model. On the other hand, the double-antibody enzyme-linked immunosorbent assay (ELISA) and Ouchterlony immunodiffusion assay gave unacceptably high false-positive results due to interference by staphylococcal protein A. This problem was successfully overcome by the development of a F(ab′)2 fragment ELISA, which was rapid and reproducible and was as sensitive and specific as PCR and Western blot analysis. The F(ab′)2 fragment ELISA is superior to existing diagnostic systems because it is quantitative, which may be related to the severity of the condition, and can detect amounts of exfoliative toxin in the picogram range directly from serum. This is the first detection system with the potential to confirm the diagnosis of staphylococcal scalded-skin syndrome from a routine blood test within 3 h of presentation.
Staphylococcal scalded-skin syndrome (SSSS) is the clinical term used for a collection of blistering skin diseases caused by the exfoliative toxins A (ETA) and B (ETB) of Staphylococcus aureus (22). The disease primarily affects infants and young children (20, 21), but occasional cases have been reported in adults (6). SSSS is characterized by erythema and fever, followed by the formation of large fragile superficial blisters, which rupture to leave extensive areas of denuded skin (22). Disease severity ranges from a few localized blisters to generalized exfoliation affecting the entire body surface (23).
Currently, diagnosis of SSSS relies mainly on the clinical picture, supported by a favorable response to antistaphylococcal antibiotics (20–23). However, early symptoms and signs are often nonspecific, which can lead to long delays in diagnosis that could lead to a fatal outcome, particularly among immunocompromised patients and those with renal failure (6, 22). Even among both healthy children who are also at risk and their parents, the unsightly appearance of the disease can cause much distress and anxiety (20–23). Early and appropriate antibiotic treatment has been shown to prevent progression of exfoliation (25), which in turn may reduce subsequent complications that can be fatal in neonates and young children, such as dehydration, poor temperature control, and secondary infection (8, 17).
Isolation of S. aureus from blood cultures or skin lesions of the patient often supports the diagnosis of SSSS (11, 18, 20, 22, 23), and production of exfoliative toxin by these strains of S. aureus can be determined using a range of currently available diagnostic tests, including PCR, radioimmunoassay, Ouchterlony immunodiffusion assay, and reverse passive latex agglutination assay (1, 10, 19, 30). However, these tests are not routinely available in hospital laboratories. Furthermore, they all (even the “gold standard” neonatal-mouse model) require isolation of the causative S. aureus strain from the patient (1, 19, 20, 22, 26, 30), which occurs only in a small proportion of SSSS patients (6, 14). Even when the infective agent is isolated, the time required to obtain and process blood and superficial cultures as well as to isolate and identify the organism (usually 24 to 72 h) makes any further tests to detect the presence of the exfoliative toxins useful only for retrospective confirmation of the diagnosis.
In this study, computer models of the toxins predicted from models of other known serine proteases were used as the basis for producing serotype-specific antibodies. These antibodies were then used to develop several immunologically based detection systems for ETA, which were then compared for their sensitivity and specificity with Ouchterlony immunodiffusion assays and PCR. We also report development of the first system that can detect exfoliative toxin directly from serum.
MATERIALS AND METHODS
Staphylococcal strains.
An ETA-producing strain of S. aureus SSS 681 isolated from a baby with generalized SSSS and shown in the neonatal mouse model to produce exfoliation was used as a positive control (8). More than 300 strains of S. aureus isolated from patients suspected to have SSSS were subjected to PCR as described below. Forty ETA-positive strains and 40 ETA-negative strains (10 of which produced ETB and 5 of which produced toxic shock syndrome toxin 1 [TSST-1]) were then used to compare the various detection systems.
S. aureus strains were maintained on tryptic soy agar plates and grown using TSKG culture medium according to the method of Sheehan et al. (32). The TSKG medium consisted of 17 g of tryptone (Difco), 3 g of soy peptone (Oxoid), 2.5 g of K2HPO4, 12.5 ml of 40% glucose, and 0.5 ml of a 1-μg/ml solution of Novobiocin (Sigma)—all made up to 1 liter with distilled water (32). Six single colonies of S. aureus were transferred from each tryptic soy agar medium plate into separate 25-ml universal bottles containing 10 ml of TSKG culture medium and were left overnight on an orbital shaker at 37°C. Culture bottles were then centrifuged, and the cells were washed twice in Tris-EDTA buffer.
Bacterial cell culture.
One hundred milliliters of TSKG culture medium was added to lengths of 30- by 5-cm dialysis tubing, which were then placed in separate 1-liter conical flasks and sterilized by autoclaving at 110°C, at 10 lb/in2 for 20 min. Then, 0.5 ml of washed S. aureus SSS 681 cells was added to each flask, and flasks were left overnight on an orbital shaker (150 rpm) at 37°C for 20 to 24 h.
Harvesting the culture supernatant.
The bathing fluid outside the dialysis tubes in the 10 flasks was collected and centrifuged at 10,000 × g at 4°C for 30 min, in a Beckman XL-90 ultracentrifuge. The supernatant was collected, passed through a 0.22-μm-pore-size filter to remove any bacteria, and kept on ice.
ETA purification.
Solid ammonium sulfate was added to the supernatant to give a 45% saturation according to the method of Dawson et al. (9) and stirred for 1 h at 4°C before being centrifuged at 18,000 × g for 45 min. The pellet was discarded, and the supernatant was brought to a 90% ammonium sulfate saturation, stirred for 1 h at 4°C, and centrifuged as described above to obtain the pellet. The pellet was dissolved in 20 mM Tris-HCl buffer (pH 7.2) and extensively dialyzed against the same buffer. The dialyzed protein was loaded onto a DEAE-Sepharose column washed with 20 mM Tris-HCl, and the unbound protein was collected and concentrated using an Amicon ultrafiltration unit with a PM10 filter. Gel filtration chromatography was then carried out using a Pharmacia fast-performance liquid chromatography system on a Superose 12 column equilibrated with 20 mM Tris-HCl (pH 8.0). Proteins were eluted using the same buffer at a rate of 1.0 ml/min, and 1-ml fractions were collected. The ETA fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and pooled and concentrated using an Amicon Centriprep 10 concentrator. Finally, the ETA was purified by ion-exchange chromatography using a Mono-Q column on a Pharmacia fast-performance liquid chromatography system. The column was equilibrated with Tris-HCl (pH 8.0), and the ETA was eluted at 0.15 M NaCl using a 0 to 0.3 M NaCl gradient. Fractions of 1 ml were collected and the ETA was identified using SDS-PAGE and concentrated using an Amicon Centriprep 10 concentrator. The identity of the toxin was confirmed by (i) SDS-PAGE, which revealed a >98% pure 28-kDa protein consistent with ETA (4, 35); (ii) Western blot analysis, which correctly identified the purified protein as ETA; (iii) N-terminal sequencing of the purified protein, using the amino-acid-sequencing facility at the Medical Research Council (Mill Hill, London, United Kingdom), which correctly identified the first 10 N-terminal amino acids; and (iv) a positive Nikolsky sign in the newborn-mouse model, as described previously (8).
SDS-PAGE.
An SDS–10 to 15% PAGE was run on the PhastSystem (Pharmacia; PhastSystem development technique file 210), with low-molecular-weight markers (Bio-Rad).
Anti-ETA antibody production.
ETA-specific antibodies were generated using computer modeling to identify surface antigenic epitopes on ETA and using the known amino acid sequence of the toxin and information derived from the known structure of other serine proteases (2). These models were used to identify the external peptide sequences of each toxin. ETA epitopes were chosen by taking into account the differences between ETA and ETB, antigenic indices, and protrusion indices. Two peptide sequences were then synthetically made and used as potential ETA-specific epitopes. J1 (amino acids 77 to 87, HIAKFANGDPSKVSFR) is is exposed on the surface of the ETA molecule and consists of two α-helices lying between the third and fourth β-strands in the first domain (4, 34, 35). J2 (amino acids 210 to 223, HSSKVSHLDREHQI) belongs to a loop known as loop 2 on ETA and is located vetween the fifth and sixth β-strands of the second domain. The corresponding ETB sequences for J1 and J2 are HVAREAAKNPSNIIFT and HSGKGG, respectively. The peptides were synthesized and conjugated, using lysine bridges, by EPITOPE Custom Peptides Co. Ltd. (University of Newcastle, Newcastle, United Kingdom). Antibodies against the peptides were raised in sheep by Polyclonal Antibodies Ltd. (Cardiff, Wales). The antibody used to detect anti-J1/J2 in the enzyme-linked immunosorbent assay (ELISA) and the Western blots was an anti-sheep antibody conjugated with horseradish peroxidase (HRP) supplied by Jackson Immuno Research Laboratories Ltd. Anti-ETA immunoglobulin G (IgG) antibody was purified using the Pierce Immunopure IgG purification kit. Polyclonal antibodies to ETB were kindly provided by the Public Health Laboratory, London, United Kingdom.
Western blot.
Protein samples were resolved on discontinuous SDS-PAGE using the Mini Protean system (Bio-Rad) by the Laemmli method (24) and electrophoresed for 45 min at 200 V. The proteins on the SDS-PAGE gel were electrophoretically transferred (overnight at 30 mA) onto a nitrocellulose membrane using a Semi-Dry Transblot apparatus (Hoefer Scientific Instruments). The membrane was washed in phosphate-buffered saline (PBS), and unbound sites were blocked by incubating with 3% bovine serum albumin (BSA) in PBS for 15 min. Anti-ETA IgG in 1% BSA in PBS with 0.1% Tween 20 (PBS-Tween) was added to the membrane and incubated for 45 min. The membrane was washed twice in PBS-Tween for 10 min per time. Anti-sheep IgG conjugated to HRP was then added and incubated for 45 min, washed as described above, and developed using 3,3-diaminobenzidine tetrahydrochloride plus urea/hydrogen peroxide (Sigma). Detection sensitivity was 100 ng.
PCR.
PCR was performed according to the method of Sakurai et al. (30). Two ETA primers, 5′-CTATTTACTGTAGGAGCTAG-3′, corresponding to nucleotides 46 to 65, and 5′-ATTTATTTGATGCTCTCTAT-3′, corresponding to nucleotides 767 to 786 of the eta sequences counted from the initiation codon, and two ETB primers, 5′-ATACACACATTACGGATAAT-3′, corresponding to nucleotides 167 to 186, and 5′-CAAAGTGTCTCCAAAAGTAT-3′, corresponding to nucleotides 776 to 795 of the etb sequences counted from the initiation codon, were synthesized (Genoysis; Sigma). The eta and etb fragments were amplified from 1 μg of nucleic acid or one colony of S. aureus as the template in a 50-μl reaction mixture with 50 mM KCl–10 mM Tris-HCl (pH 8.3)–1.5 mM MgCl2–0.01% gelatin–200 μM concentrations each of the deoxynucleoside triphosphates–0.25 μM concentrations each of the eta- and etb-specific oligonucleotide primers–1.25 U of Taq polymerase (Promega). Two drops of mineral oil (Sigma) was added to overlay each sample, followed by denaturation at 95°C for 4 min and 20 cycles of 94°C for 1.5 min, 50°C for 1.5 min, and 73°C for 1.5 min with autoextension in a thermal DNA cycler. Ten microliters of the PCR product was transferred to a 1.5% agarose minigel containing 0.5 μg of ethidium bromide per ml. Electrophoresis was performed at 100 V for 40 min in TAE buffer (pH 8.0).
Ouchterlony immunodiffusion assay.
Culture supernatants were tested against anti-ETA IgG and pure ETA using the immunodiffusion assay described by Arbuthnott and Billcliffe (1). Wells were cut in plates coated with 1% agarose (Bio-Rad) in PBS. Precipitin lines were visualized after 48 h of incubation at room temperature.
ELISA.
To determine whether anti-ETA antibody also bound native ETA, an antibody ELISA was performed according to the method of Harlow and Lane (15) using plastic microtiter 96-well plates (PolySorb; Nunc) as the solid phase. ETA at a 1-mg/ml concentration was serially diluted in 50 mM sodium bicarbonate buffer (pH 9.5). PolySorb microtiter plates were coated with 100 μl of the diluted antigen and incubated at room temperature for 2 h. Each well was washed twice in PBS, and the remaining wall area was blocked with 3% BSA in PBS for 2 h at room temperature. A dilution of the anti-ETA IgG (100 ng/100 μl) in 1% BSA in PBS-Tween was added and incubated for 2 h. Each well was washed three times with PBS-Tween. Anti-sheep IgG conjugated to HRP was added and incubated for 1 h, washed three times, and developed using o-phenylenediamine dihydrochloride (Sigma) and H2O2 in 0.15 M phosphate citrate buffer (pH 5), and the color was read spectrophotometrically at a wavelength of 450 nm.
Double-antibody capture ELISA.
Anti-ETA antibody conjugation with HRP was accomplished using the Pierce Immunopure Plus activated peroxidase kit. MaxiSorb microtiter plates were coated with anti-ETA IgG in PBS (100 ng/100 μl) for 1 h at 37°C. The plates were washed three times with PBS-Tween, and the remaining wall area was blocked with 3% BSA in PBS-Tween and incubated for 1 h at 37°C before washing twice in PBS-Tween. One hundred microliters of diluted ETA protein (made up in PBS-Tween) or culture medium or a serum sample was added and incubated at 37°C for 2 h. The wells were washed three times in PBS-Tween, and anti-ETA IgG conjugated to HRP was added and incubated for a further 2 h at 37°C before washing and developing with o-phenylenediamine dihydrochloride, as described previously. The detection limit was 100 pg.
F(ab′)2 fragment ELISA.
F(ab′)2 fragments were prepared using the Pierce Immunopure F(ab′)2 preparation kit. MaxiSorb microtiter plates were coated with F(ab′)2 fragments in PBS for 1 h at 37°C, and the subsequent procedure was as described above. The detection limit was 500 pg.
Testing for ETA in serum.
Both the double-antibody and the F(ab′)2 fragment ELISA were used to detect ETA in serum by spiking pure ETA of known concentrations into 30% sheep, rabbit, or human serum. The subsequent procedure was as described above with equivalent detection limits for the ETA.
RESULTS
Production and characterization of ETA-specific antibody.
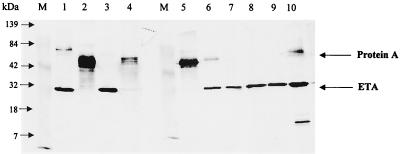
Using surface epitope mapping, we were able to prepare synthetic peptides that produced high-titer antibodies that were sensitive and specific for both native and denatured ETA. Western blot analysis of the supernatant of an overnight culture of the positive control strain SSS681 showed a single dark band with a molecular mass of about 28 kDa (Fig. 1), which is consistent with the predicted ETA molecular weight (4, 35). The minor band at 15 kDa is a degradation product of ETA caused by repeated freeze-thawing of samples, and this band highlights the importance of using fresh samples. No such bands were detected with either ETB-producing or TSST-producing strains. This method gives a clear and visual indication of the presence of ETA when compared to the positive control. When examining strains by this method, it can be seen that some of the strains that had previously been identified as being ETA-positive by Ouchterlony immunodiffusion in fact did not produce ETA (Fig. 1, lanes 2, 4, and 5).
FIG. 1.
Detection of ETA using anti-ETA IgG on Western blots from culture supernatants. Eight strains of S. aureus found to produce ETA by the Ouchterlony immunodiffusion assay were grown in TSKG medium, cells were removed by centrifugation, and the supernatant was resolved on an SDS-12.5% PAGE, transferred onto nitrocellulose, and probed with anti-ETA IgG as described in Materials and Methods. Lane M is prestained molecular-weight markers. Lanes 1 through 8 are S. aureus strains suspected of being ETA producers by Ouchterloney immunodiffusion assay. Lane 9 is 0.1 μg of pure ETA, and lane 10 is our reference strain SSS681. Nonspecific binding of IgG is due to protein A.
In Fig. 1, lanes 1, 2, 4, 5, 6, and 10 show a nonspecific broad cross-reacting band of molecular masses between 45 and 50 kDa, which was present even when only the developing IgG-HRP conjugate was used without the anti-ETA antibody. This band is most probably staphylococcal protein A, which binds nonspecifically to immunoglobulins and is responsible for the false-positive results with the Ouchterlony immunodiffusion method (12, 13). Furthermore, the production of protein A was not related to the presence of the exfoliative toxins (Fig. 1), and the level of protein A varied unpredictably from none (Fig. 1, lanes 3, 7, 8, and 9) to excessive amounts (Fig. 1, lanes 2, 4, 5, 6, and 10).
Development of ELISA.
Although Western blot analysis, which separates out proteins, could clearly distinguish between true ETA-producing strains and those producing protein A, it was considered impractical for routine clinical use. Two different ELISA-based systems were therefore developed that could be used in both clinical and research laboratories. They would also have the advantage of quantitating ETA levels produced by different S. aureus strains, which may be related to the varying severity of SSSS. The double-antibody direct-sandwich ELISA detected ETA from culture supernatants at levels as low as 100 pg/ml, while the ELISA using F(ab′)2 fragments as the first capture antibody detected the toxin at 500 pg/ml. In addition, the suitability of ELISA for detecting ETA in serum was assessed by spiking pure ETA into 30% sheep, rabbit, or human serum. Both ELISA-based systems were able to quantitate levels of ETA with detection limits that were similar to the culture supernatants.
Evaluation of the detection systems.
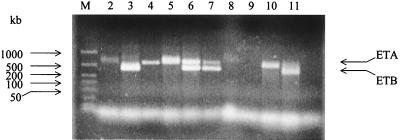
Forty known ETA-producing and 40 ETA-negative staphylococcal isolates, as determined by PCR, were used to compare various different detection assays for ETA, including (i) Western blot analysis, (ii) Ouchterlony immunodiffusion assay, (iii) double-antibody ELISA, and (iv) F(ab′)2 fragment ELISA. The results are summarized in Table 1. PCR had the advantage of allowing a clear distinction between ETA and ETB producers as well as double producers (Fig. 2), and this method has been validated in the newborn-mouse model (30). There was a 100% correlation between PCR and the Western blot II assay. The immunodiffusion assay had relatively poor sensitivity and unacceptably high false-positive rates. When the different assays were compared strain by strain, the Western blots showed that 28 of 40 (70%) of the ETA-positive and 24 of 40 (60%) of the ETA-negative strains produced various quantities of protein A. Furthermore, only those strains with significant protein A gave false-positive results with the immunodiffusion assay (Fig. 1).
TABLE 1.
Sensitivity and specificity of various tests for identification of ETA-positive strainsa
| Detection system | Sensitivity for ETA (%) (n = 40) | Specificity for ETA (%) (n = 40) |
|---|---|---|
| Western blot | 100 | 100 |
| Ouchterlony assay | 75 | 55 |
| Double-antibody ELISA | 90 | 65 |
| F(ab′)2 fragment ELISA | 100 | 95 |
Forty ETA-positive and 40 ETA-negative strains of S. aureus (10 of which produced ETB, and 5 of which produced TSST-1), as determined by PCR, were subjected to Western blot analysis, Ouchterlony immunodiffusion, double-antibody sandwich ELISA, and F(ab′)2 fragment ELISA. The sensitivity of the test was defined as the proportion of ETA-positive strains correctly identified by the test, while specificity was defined as the proportion of ETA-negative strains correctly identified by the test.
FIG. 2.
PCR of S. aureus strains showing ETA and ETB genes. Single-cell colonies of S. aureus strains were subjected to PCR analysis, and the products were analyzed on agerose gel electrophoresis, as described in the Materials and Methods section. Lane M contains kilobase standards. Lanes 2, 4, 5, and 10 are samples from ETA producers. Lanes 3 and 11 are samples from ETB producers. Lanes 6 and 7 are samples that are both ETA and ETB producers. Lanes 8 and 9 are samples from TSST-1 producers (negative controls).
The double-antibody ELISA also failed due to interference by protein A, despite the use of several blocking procedures previously described in the literature (see Discussion) (3, 5, 31). On the other hand, the F(ab′)2 fragment ELISA correlated well with the Western blots and PCR and was further able to quantitate ETA concentrations for all of the ETA-positive strains. There was, however, one false-positive result, which was subsequently shown by Western blotting to be due to a strain of S. aureus that produced excessive amounts of protein, which suggests that some protein A still bound F(ab′)2 fragments (12, 13, 31).
DISCUSSION
Although SSSS was first described over a century ago, our understanding of it began only after Melish and Glasgow (26) demonstrated that a staphylococcal exotoxin was the cause of SSSS, using a neonatal-mouse model that remains the gold standard for the disease. Since then, a range of different diagnostic tests has been developed for the diagnosis of SSSS (1, 10, 19, 20, 22, 26, 30). However, all of these tests require isolation of the causative staphylococcal strain, which takes time, and the tests are not routinely available in hospital laboratories (7, 28). Our main aim was to develop a detection system that would effectively and rapidly diagnose SSSS in suspected cases within a hospital setting. This detection system would also have other important uses, including rapid identification of carriers of toxin-producing staphylococcal strains in order to prevent outbreaks and further research on the toxins and their epidemiology (16). Our previous attempts at producing polyclonal ETA antibodies using purified toxin showed that there was significant cross-reactivity with ETB. In order to develop serotype-specific antibodies, we used computer models of ETA and ETB (2) based on the three-dimensional structures of known glutamate-specific serine proteases to identify epitopes on ETA that were least similar to ETB. This approach of surface epitope mapping to produce highly specific antibodies has successfully been used in the past, but invariably the three-dimensional structure of the protein was known. In this case, the three-dimensional crystallographic structures of the toxins were not known at the time, and their structures have only recently been published (4, 34, 35). We took a novel approach by using molecular modeling to produce a structure (2), and this turned out to correlate well with the crystallographic structure of ETA. The use of surface epitope mapping and the subsequent use of peptides to generate antibodies led to the successful production of antibody that bound specifically to ETA but not ETB or TSST-1. Furthermore, it was able to detect denatured toxin in Western blots and native toxin with the F(ab′)2 fragment ELISA. The results also show that Western blotting must be considered as the laboratory gold standard for ETA detection because it provides unequivocal evidence for the presence or absence of the toxin, which in turn must be related to the diagnosis.
Having characterized the anti-ETA antibody and demonstrated its ability to specifically detect ETA in minute quantities from culture supernatants, an attempt was made to develop an assay that was quantitative and could routinely be used to diagnose SSSS in hospital laboratories. The Ouchterlony immunodiffusion test was included in this study because, until recently, it was the selected detection method for diagnosing SSSS in the United Kingdom (23), while the PCR has been validated against the gold standard newborn mouse model. We elected to develop an ELISA system because it is quick, easy to use, cheap, and reliable; it gives both qualitative and quantitative results; and it is a well-established procedure in hospital laboratories (27, 29, 31).
A direct comparison of the double-antibody ELISA, immunodiffusion, and Western blots soon revealed that the majority of strains that gave false-positive results produced protein A on the Western blot, which always correlated well with the PCR. Protein A is a 40- to 60-kDa polymorphic polypeptide produced by up to 90% of all strains of S. aureus (12, 13, 31). It has the ability to bind the second and third constant regions of the Fc domain of immunoglobulins from many different species, which explains why a Western blot with the detecting IgG-HRP conjugate and no anti-ETA antibody also gave a positive result. The presence of protein A is a major problem in developing detecting systems for any staphylococcal exotoxin, including staphylococcal TSST-1 and the enterotoxins (27, 29, 31). In developing a noncompetitive ELISA for TSST-1, for example, Rosten et al. (29) discovered that 42 of 60 isolates (70%) that were negative for TSST-1 would have given a false-positive result if tested by Ouchterlony immunodiffusion because of the presence of protein A in the supernatant.
False-positive results due to protein A also occur with whole-IgG double-antibody ELISA (31), and the double-antibody ELISA that we developed also suffered from the same disadvantage. Readings showed no correlation with the presence or amount of ETA, most values being over the measurable range and related more to the amount of protein A than ETA in the supernatant.
Several methods have been proposed, with variable success, to circumvent the interfering effect of protein A, including (i) pretreatment of the culture supernatant with 10% rabbit serum to bind protein A nonspecifically; (ii) using affinity chromatography with a Sepharose-rabbit IgG column; (iii) the use of sheep IgG instead of rabbit IgG because it has a lower affinity for protein A; and (iv) the use of rabbit anti-protein A (3, 5, 31). On the other hand, F(ab′)2 fragment ELISAs have already been successfully used to detect other staphylococcal toxins and have been shown to increase sensitivity when compared to a whole-IgG ELISA (31). Compared to the double-antibody ELISA, the sensitivity and specificity of F(ab′)2 fragment ELISA were increased from 90 to 100% and from 65 to 95%, respectively. However, there was still one false-positive with the F(ab′)2 fragment ELISA, due to a strain that produced excessive protein A. Berdal et al. have also shown that the presence of excessive amounts (>0.5 mg/ml) of protein A could significantly affect detection of staphylococcal enterotoxins using noncompetitive ELISA (3). It is possible that some protein A polymorphisms may be able to bind the F(ab′)2 fragments.
We have compared several immunologically based detection systems for ETA and assessed their suitability for routine use in the hospital setting. Of these, both the Ouchterlony and the double-antibody ELISA are clearly unsuitable because of their poor specificity. The PCR, Western blot analysis, and the F(ab′)2 fragment ELISA are all promising. However, the latter has two major advantages over all other existing diagnostic systems in that it can be adapted to detect ETA directly from biological fluids and is quantitative. Our results demonstrate that the double-antibody ELISA detects ETA in serum at levels as low as 100 pg/ml, while the F(ab′)2 fragment ELISA has a sensitivity of around 500 pg/ml, similar to F(ab′)2 fragment ELISAs developed for other staphylococcal toxins (31). Although the double-antibody ELISA is fivefold more sensitive than the F(ab′)2 fragment ELISA, the latter has the added advantage of being able to detect ETA in vitro from staphylococcal supernatants as well as serum. Unfortunately, there have been no studies to determine the level of exfoliative toxins in human serum. We are currently investigating the use of the F(ab′)2 fragment ELISA for rapid diagnosis of SSSS in the clinical setting, and we aim to measure the level of exfoliative toxins in serum at the same time.
Conclusions.
We have used computer modeling to predict the structure of the exfoliative toxins and to develop serotype-specific antibodies, which were then used to develop several detection systems for ETA. We have demonstrated that the F(ab′)2 fragment ELISA is as sensitive and specific as current diagnostic tests and has the further advantages of being quantitative, which may be related to the severity of the condition, and being able to detect ETA in the picogram range, directly from serum. Such a detection system would not only provide rapid diagnosis from simple blood tests but would also rapidly identify carriers in the event of an outbreak (8) and improve our understanding of the epidemiology and biological effects of the toxins. In particular, studies on the role of the toxins in diseases in which staphylococci and superantigens are suspected—including psoriasis, atopic dermatitis, Kawasaki disease, and sudden infant death syndrome—are now warranted (20–23).
ACKNOWLEDGMENTS
We thank the special trustees of St. Thomas' Hospital, the Wellcome Trust student scholarship, and the Nuffied student scholarship for financial support.
We are also grateful to Ty Pitt, Public Health Laboratory Service Colindale, for providing some of the staphylococcal strains with respective case histories and antibodies to ETB, and to A. Atkin, Medical Research Council, Mill Hill, for N-terminal sequencing of the pure ETA protein.
REFERENCES
- 1.Arbuthnott J P, Billcliffe B. Qualitative and quantitative methods for detecting staphylococcal epidermolytic toxin. J Med Microbiol. 1976;9:191–201. doi: 10.1099/00222615-9-2-191. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa J A R G, Saldanha J W, Garratt R C. Novel features of serine protease active sites and specificity pockets: sequence analysis and modelling studies of glutamate-specific endopeptidases and epidermolytic toxins. Protein Eng. 1996;9:591–601. doi: 10.1093/protein/9.7.591. [DOI] [PubMed] [Google Scholar]
- 3.Berdal B P, Olsvik O, Omland T. A sandwich ELISA method for detection of Staphylococcus aureus enterotoxins. Acta Pathol Microbiol Scand Sect B. 1981;89:411–415. [PubMed] [Google Scholar]
- 4.Caverelli J, Prevost G, Bourguet W, Moulinier L, Chevrier B, Delagoutte B, Bilwes A, Mourey L, Rifai S, Piemont Y, Moras D. The structure of Staphylococcus aureus epidermolytic toxin A, an atypic serine protease, at 1.7A resolution. Structure. 1997;5:813–824. doi: 10.1016/s0969-2126(97)00235-9. [DOI] [PubMed] [Google Scholar]
- 5.Chu M C, Kreiswirth B N, Pattee P A, Novick R P, Melish M E, James J F. Association of toxic shock syndrome toxin 1 determinant with a heterologous insertion at multiple loci in the Staphylococcus aureus chromosome. Infect Immun. 1988;56:2702–2708. doi: 10.1128/iai.56.10.2702-2708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cribier B, Piemont Y, Grosshans E. Staphylococcal scalded skin syndrome in adults: a clinical review illustrated with a case. J Am Acad Dermatol. 1984;30:319–324. doi: 10.1016/s0190-9622(94)70032-x. [DOI] [PubMed] [Google Scholar]
- 7.Dancer S J, Noble W C. Nasal, axillary and perineal carriage of Staphylococus aureus among pregnant women: identification of strains producing epidermolytic toxin. J Clin Pathol. 1991;44:681–684. doi: 10.1136/jcp.44.8.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dancer S J, Simmons N A, Poston S M, Noble W C. Outbreak of staphylococcal scalded skin syndrome among neonates. J Infect. 1988;16:87–103. doi: 10.1016/s0163-4453(88)96249-4. [DOI] [PubMed] [Google Scholar]
- 9.Dawson R M C, Elliott D C, Elliot W H, Jones K M. Data for biochemical research. Oxford, United Kingdom: Oxford Science Publications; 1986. pp. 537–539. [Google Scholar]
- 10.De Azavedo A, Arbuthnott J P. Assays for epidermolytic toxin of Staphylococcus aureus. Methods Enzymol. 1988;165:333–337. doi: 10.1016/s0076-6879(88)65049-x. [DOI] [PubMed] [Google Scholar]
- 11.Falk D K, King L E. Criteria for the diagnosis of staphylococcal scalded skin syndrome. CUTIS. 1983;21:421–424. [PubMed] [Google Scholar]
- 12.Forsgren A. Significance of protein A production by staphylococci. Infect Immun. 1970;2:672–673. doi: 10.1128/iai.2.5.672-673.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsgren A, Nordstrom K, Philipson L, Szoquist J. Protein A mutants of Staphylococcus aureus. J Bacteriol. 1971;107:245–250. doi: 10.1128/jb.107.1.245-250.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg N S, Ahmed T, Robinson B, Ascensao H, Horwitz H. Staphylococcal scalded skin syndrome mimicking acute graft-versus-host disease in a bone marrow transplant recipient. Arch Dermatol. 1989;15:385–389. [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 16.Hoefnagels-Schuermans A, Peetermans W E, Jorissen M, Van Lierde S, van Den Oord J, De Vos R, Van Eldere J. Staphylococcus aureus adherence to nasal epithelial cells in a physiological in vitro model. In Vitro Cell Dev Biol Anim. 1999;35:472–480. doi: 10.1007/s11626-999-0054-0. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann R R, Lohner M, Bohm N, Schaefer H E, Letitis J. Staphylococcal scalded skin syndrome and consecutive septicaemia in a preterm infant. Pathol Res Pract. 1994;190:77–81. doi: 10.1016/S0344-0338(11)80499-1. [DOI] [PubMed] [Google Scholar]
- 18.Itani O, Crump R, Mimouni F, Tunnessen W W. Ritter's disease (neonatal staphylococcal scalded skin syndrome) Am J Dis Child. 1992;146:424–426. [PubMed] [Google Scholar]
- 19.Kawabata A, Ichiyama S, Tinuma Y, Hasegawa Y, Ohta M, Shimokata K. Exfoliative toxin detection using reversed passive latex agglutination: clinical and epidemiologic applications. J Clin Microbiol. 1997;35:1984–1987. doi: 10.1128/jcm.35.8.1984-1987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladhani S, Joannou C L. Difficulties in the diagnosis and management of staphylococcal scalded skin syndrome. Pediatr Infect Dis J. 2000;142:1251–1255. doi: 10.1097/00006454-200009000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Ladhani S, Newson T. A familial outbreak of staphylococcal scalded skin syndrome. Pediatr Infect Dis J. 2000;19:578–579. doi: 10.1097/00006454-200006000-00021. [DOI] [PubMed] [Google Scholar]
- 22.Ladhani S, Joannou C L, Lochrie D P, Evans R W, Poston S M. Clinical, microbial and biochemical aspects of the exfoliative toxins causing staphylococcal scalded skin syndrome. Clin Microbiol Rev. 1999;12:224–242. doi: 10.1128/cmr.12.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladhani S, Evans R W. Staphylococcal scalded skin syndrome. Arch Dis Child. 1998;78:85–88. doi: 10.1136/adc.78.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Melish M E, Glasgow L A, Turner M D, Lillibridge C B. The staphylococcal epidermolytic toxin: its isolation, characterisation and site of action. Ann N Y Acad Sci. 1974;236:317–342. doi: 10.1111/j.1749-6632.1974.tb41501.x. [DOI] [PubMed] [Google Scholar]
- 26.Melish M E, Glasgow L A. The staphylococcal scalded skin syndrome: development of an experimental model. N Engl J Med. 1970;282:1114–1119. doi: 10.1056/NEJM197005142822002. [DOI] [PubMed] [Google Scholar]
- 27.Miwa K, Fukuyama M, Kunitomo T, Igarashi H. Rapid assay for detection of toxic-shock syndrome toxin-1 from human sera. J Clin Microbiol. 1994;32:539–542. doi: 10.1128/jcm.32.2.539-542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble W C. The micrococci. London, United Kingdom: Lloyd-Luke; 1981. pp. 152–181. [Google Scholar]
- 29.Rosten P M, Bartlett K H, Chow A W. Detection and quantitation of toxic shock syndrome toxin-1 in vitro and in vivo by noncompetitive enzyme-linked immunosorbent assay. J Clin Microbiol. 1987;25:327–332. doi: 10.1128/jcm.25.2.327-332.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai S, Suzuki H, Machida K. Rapid identification by polymerase chain reaction of staphylococcal exfoliative toxin serotype A and B genes. Microbiol Immunol. 1995;39:379–386. doi: 10.1111/j.1348-0421.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 31.See R H, Adilman S, Bartlett K H, Chow A W. Colony immunoblot assay for the detection of staphylococcal toxic shock syndrome toxin 1 (TSST-1) with anti-TSST-1 F(ab′)2 fragments. J Clin Microbiol. 1989;27:2050–2053. doi: 10.1128/jcm.27.9.2050-2053.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheehan B J, Foster T J, Dorman C J, Park S, Stewart G S. Osmotic and growth-phase dependent regulation of the eta gene of Staphylococcus aureus: a role for DNA supercoiling. Mol Gen Genet. 1992;232:49–57. doi: 10.1007/BF00299136. [DOI] [PubMed] [Google Scholar]
- 33.Shuter J, Hatcher V B, Lowy F D. Staphylococcus aureus binding to human mucin. Infect Immun. 1996;64:310–318. doi: 10.1128/iai.64.1.310-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vath G M, Earhart C A, Monie D D, Iandolo J J, Schlievert P M, Ohlendorf D H. The crystal structure of exfoliative toxin B: a superantigen with enzymatic activity. Biochemistry. 1999;38:10239–10246. doi: 10.1021/bi990721e. [DOI] [PubMed] [Google Scholar]
- 35.Vath G M, Earhart C A, Rago J V, Kim M H, Bohach G A, Schlievert P M, Ohlendorf D H. The structure of the superantigen exfoliative toxin A suggests novel regulation as a serine protease. Biochemistry. 1997;36:1559–1566. doi: 10.1021/bi962614f. [DOI] [PubMed] [Google Scholar]