Abstract
Hypoxia has been linked with increased resistance to treatment in various solid tumors, including head and neck squamous cell carcinoma (HNSCC). The aim of the present study was to identify genes involved in hypoxia-mediated responses to radiotherapy in HNSCC. A total of three HNSCC cell lines with an epithelial phenotype were selected for this study and cultured under normoxic (21% O2) or hypoxic (1% O2) conditions. The sensitivity of the HNSCC cells to radiotherapy was assessed by a crystal violet assay. Western blotting (for protein expression), cDNA microarrays and reverse transcription-quantitative PCR (for gene expression) were also applied. Small interfering RNA silencing was used to knock down target genes. The results revealed that hypoxia negatively affected the response of HNSCC cells to radiotherapy. Of note, increased levels of N-cadherin, vimentin and fibronectin, as well as stem cell-associated transcription factors, were observed under hypoxia. The microarray analysis revealed a number of hypoxia-regulated genes that were involved in multiple biological functions. However, downregulation of hypoxia-regulated genes did not affect sensitivity to radiotherapy of the investigated cell lines. Taken together, the present findings indicated several important pathways and genes that were involved in hypoxia and radiotherapy resistance. It is hypothesized that panels of reported hypoxia-regulated genes may be useful for the prediction of radiotherapy responses in patients with HNSCC.
Keywords: hypoxia, epithelial-mesenchymal transition, head and neck cancer, radiotherapy, microarray
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common cancer worldwide (1,2). Treatment depends largely on the stage of the tumor and is based on a multimodality strategy primarily involving surgery and radiotherapy. Patients with advanced disease are offered a cisplatin-based chemotherapy or combination of radiotherapy with cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR) (3). Despite the progress in therapy, both surgery and radiotherapy, similar to chemotherapy, patient survival has not substantially improved in the last few decades. The 5-year overall survival in patients with advanced HNSCC remains <50% (4).
Hypoxia in HNSCC has been linked with poor response to treatment, increased metastasis and tumor aggressivity (5,6). Adaptive responses of cells to hypoxia include stimulation of angiogenesis and alteration of cellular metabolism (7). The reduced oxygen tension in cells leads to activation of the hypoxia-inducible factor (HIF)-1 transcription factor, which is constitutively produced and degraded by the ubiquitin-proteasome system under normoxic conditions but becomes stabilized and transcriptionally active under hypoxic conditions (8). HIF-1 regulates the transcription of hundreds of genes that code for proteins involved in various aspects of cancer biology (9), such as angiogenesis (10), metabolism (11), genetic instability (12), invasion and metastasis (13,14), resistance to chemotherapy and radiation (9,10,15). Ionizing radiation is used extensively and is an integral part of cancer treatment. The mechanism underlying radiation resistance in human cancer is not fully understood. However, research into tumor radioresistance in HNSCC has suggested distinct mechanisms that have a negative impact on tumor irradiation, including hypoxia (16). Tumor recurrence after radiotherapy is a major obstacle to recovery in HNSCC (17).
During metastasis, tumor cells lose the cell-cell adhesion capacity and acquire the capability of cell motility for invasion via the epithelial-mesenchymal transition (EMT). In a majority of tumors, including HNSCC, EMT is tightly linked to hypoxia, leading to an increased radioresistance (18,19).
Due to the importance of hypoxia in tumors, there is a requirement for reliable biomarkers for this condition to serve as a diagnostic marker of hypoxia and a potential therapeutic target. To date, a number of tumor tissue markers have been described as potential biomarkers in HNSCC (20). Among them, the expression of hypoxia markers, including HIF-1a, carbonic anhydrase 9 (CA9) and glucose transporter (GLUT)1, in HNSCC is associated with poor prognosis for the patients (21,22).
Several studies using gene expression microarrays have demonstrated that the expression levels of various genes can be used to characterize different tumor types and to identify diagnostic and therapeutic biomarkers (23,24). In the present study, cDNA microarrays were used to assess the impact of hypoxia (1% O2) on the global gene expression in HNSCC and to identify hypoxia-induced differentially expressed genes possessing potential therapeutic impact.
Materials and methods
Cell lines and culture conditions
In this study, the UT-SCC-14 cell line from the University of Turku (provided by Professor Reidar Grénman at the Department of Otorhinolaryngology, Head and Neck Surgery, Turku University Central Hospital), and two cell lines from University of Linköping, LK0858 and LK0863 (25), were used. All cell lines were derived from tissue specimens from patients diagnosed with HNSCC. The UT-SCC-14 and LK0858 cell lines originate from the tongue, whereas LK0863 originates from the larynx. The present study was approved by the Research Ethics Committee of the Linköping University (approval no. 03–537). Written consent was obtained from the patients involved in the study.
All cell lines were cultured in keratinocyte serum-free medium supplemented with antibiotics (50 U/ml penicillin, 50 µg/ml streptomycin) and 1% FBS (all from Gibco; Thermo Fisher Scientific, Inc.).
The cells were given fresh culture media twice per week and were subcultured to confluence after detaching the cells with 0.25% trypsin + 0.02% EDTA at a weekly split ratio of ~1:2. Cultures from passages 10 to 25 were used in all experiments. The cells were screened periodically for mycoplasma contamination using a Mycoplasma Detection kit (MycoAlert™; Lonza Group, Ltd.).
Introduction of hypoxia
Hypoxic conditions were achieved in a New Brunswick Galaxy® CO2 incubator 14S at 37°C with atmospheric conditions of 5% CO2 and 1% O2. The cells were cultured in hypoxic conditions for 24 h prior to planned experiments.
Cell proliferation assay
Tumor cells were seeded in 12-well plates at densities of 300–800 cells/cm2, depending on the plating efficiency of each cell line. Selected cells were irradiated (2, 4 or 6 Gy) with 4 MeV photons generated by a linear accelerator (Clinac 4/100; Varian Medical Systems), delivering a dose-rate of 2.0 Gy/min. The cytostatic/cytotoxic effect was determined after another 9 days. After fixation in 4% paraformaldehyde (20 min; room temperature), the cells were stained with crystal violet (0.04% in 1% ethanol; room temperature) for 20 min at room temperature, and were then washed and air-dried. After solubilization in 1% SDS, the optical density at 550 nm was measured using a Victor plate reader (EG&G Wallac).
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR analysis was performed on a 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Total RNA was extracted from the cells using a RNeasy Mini kit (Qiagen GmbH), the cDNA was synthesized using a High-Capacity RNA-to-cDNA kit according to the manufacturer's instructions (Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression of hypoxia-responsive genes was analyzed with a panel of TaqMan® Gene Expression assays [cadherin 1 (CDH1; Hs01023895_m1); cadherin 2 (CDH2; Hs00983056_m1); vimentin (VIM; Hs00958111_m1); fibronectin 1 (FN1; Hs01549976_m1); FOXC2 (Hs00270951_s1); TWIST1 (Hs04989912_s1); CD44 (Hs01075864_m1); SOX2 (Hs04234836_s1); NANOG (Hs02387400_g1); GLUT3 (Hs00359840_m1); CA9 (Hs00154208_m1); caspase 14 (CASP14; Hs00201637_m1); serpin family E 1 (SERPINE1; Hs00167155_m1); lysyl oxidase (LOX; Hs00942480_m1); amphiregulin (AREG; Hs00950669_m1); epiregulin (EREG; Hs00914313_m1); all labelled with FAM and purchased from Thermo Fisher Scientific, Inc.] and amplified using a TaqMan real-time PCR protocol according to manufacturer's instructions (Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: Initial denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min.
Amplification of both GAPDH [TaqMan Gene Expression assay (Hs02758991_g1); Thermo Fisher Scientific, Inc.] and ACTB [TaqMan Gene Expression assay (Hs99999903_m1); Thermo Fisher Scientific, Inc.] was used as an internal standard. The comparative Cq method was applied to determine the fold-difference in expression levels relative to a control sample (26).
Western blot analysis
Whole cell extracts were prepared from hypoxic and normoxic HNSCC cells. The cells were lysed with RIPA buffer (cat. no. 89900) supplemented with protease and phosphatase inhibitor (cat. no. A32959) (both from Thermo Fisher Scientific, Inc.). Protein concentration was determined using the DC protein assay (cat. no. 5000111; BioRad Laboratories, Inc.). For each sample, 20 µg protein was separated using 10% SDS-PAGE (CBS Scientific), blotted onto a nitrocellulose membrane using an iBlot 2 transfer device (Thermo Fisher Scientific, Inc.), followed by blocking for 60 min in TBS-Tween-20 (0.1%; TBS-Tween) containing 5% skimmed milk and 1% BSA (Sigma-Aldrich; Merck KGaA). The membranes were incubated with anti-HIF-1α (cat. no. 610959; 1:500; BD Biosciences), anti-CDH2 (cat. no. ab18203; 1:1,000; Abcam), anti-CA9 (cat. no. NB100-417; 1:1,000; Novus Biologicals, Ltd.) or anti-SERPINE1 (cat. no. ab125687; 1:1,000; Abcam) antibodies in TBS-0.1% Tween20 containing 0.1% skimmed milk at overnight at 4°C. The membranes were then washed with TBS-Tween (0.1%) and incubated for 1 h at room temperature with a HRP-conjugated anti-mouse (cat. no. AP127P) or anti-rabbit (AP156P) antibodies (both 1:3,000 and from MilliporeSigma). The bands were visualized with Western Blotting Luminol Reagent (Bio-Rad Laboratories, Inc.) and captured digitally using the Chemidoc XRS system (Bio-Rad Laboratories, Inc.). Equal loading was verified by reprobing the membranes for 60 min at room temperature with an HRP-conjugated anti-β-actin antibody (cat. no. A5441; 1:10,000 Sigma-Aldrich; Merck KGaA).
RNA interference
Cells were seeded at a density of 12,000 cells/cm2 and transfected 24 h later with FlexiTube small interfering (si)RNA (cat. no. 1027416; Qiagen GmbH) against AREG (cat. nos. SI00299845 and SI03049683), EREG (cat. no. SI04199244), HIF-1α (cat. nos. SI02664053 and SI04262041), SERPINE1 (cat. nos. SI00012628 and SI03039715), CDH2 (cat. nos. SI02757335 and SI04434619) and CA9 (cat. nos. SI00023541 and SI00023534), or a non-targeting siRNA with no homology to any known human gene (AllStars Negative Control siRNA; cat. no. 1027280) with HiPerFect transfection reagent (Qiagen GmbH). The list of siRNA target sequences is provided in Table SI. A total of two siRNA clones to gene of interest in 1:1 ratio were used for each transfection, except for EREG siRNA. The UT-SCC-14 cell line was transfected with the HIF-1α, CA9, SERPINE1, AREG and EREG siRNA. The final siRNA concentration in the culture medium was 10 nmol/l. At 24 h after transfection, the cells were sparsely seeded into 12-well plates (Falcon; Corning Life Sciences). After another 24 h, half of the cultures were moved to 1% O2 and the rest of the cultures were cultured under standard conditions (20% O2); after an additional 24 h, the cells were irradiated (2, 4 or 6 Gy). The treated cells were grown for another 9 days, after which they were fixed and stained with crystal violet as aforementioned. Knockdown was verified via RT-qPCR. A reduction in the mRNA level by ≥70% was achieved in all experiments.
Microarray analysis
Three HNSCC cell lines (UT-SCC-14, LK0859 and LK0863) were selected for the microarray analysis. Cells were cultured to 70–80% confluence and exposed to either normoxic (21% O2) or hypoxic (1% O2) conditions for 24 h prior to total RNA extraction. The independently derived cell lines were considered as biological replicates for each experimental condition. Each experimental condition included a single hybridization of the same sample in our microarray design. Total RNA was extracted from the HNSCC cell lines cultured in normoxic and hypoxic conditions using the RNeasy Mini kit and quantified using a Nanodrop ND-100 Spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNA (150 ng) was used to process the Affymetrix GeneChip® Human Transcriptome 2.0 arrays (Affymetrix; Thermo Fisher Scientific, Inc.) using a GeneChip WT Plus Reagent kit (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. Hybridized arrays were scanned with an Affymetrix GeneChip 3000 fluorescent scanner (Affymetrix; Thermo Fisher Scientific, Inc.).
Robust multiarray analysis was used for normalization of the microarray data. The raw expression data was log2 transformed, and the probes with a variance <0.1 were filtered out before statistical analyses. Differential expression analyses were performed using the Limma package (version 3.38.3) in R (version 3.5) using a pairwise comparison between normoxia (n=3) vs. hypoxia (n=3) conditions. A P-value adjustment was performed using the Benjamini-Hochberg procedure (27,28). The P<0.05 and minimum log2 fold change of 2 from the analyzed data set (normoxia vs. hypoxia) was used to identify hypoxia-associated genes. Pathway analysis was performed using Ingenuity Pathways Analysis (IPA) software (build version, 486617M; content version, 46901286; Qiagen Inc.; http://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). Only significantly (adjusted P<0.05) upregulated and downregulated genes were considered for IPA-supported analysis.
Statistical analysis
All values obtained were represented as the mean ± SD of at least three independent experiments. Statistical analysis was performed with GraphPad Prism software v. 7.0 (GraphPad Software, Inc.). An unpaired Student's t-test or one-way ANOVA, followed by a Bonferroni post hoc test was used to analyze the data. P<0.05 was considered to indicate a statistically significant difference.
Results
Hypoxia-induced changes in treatment response
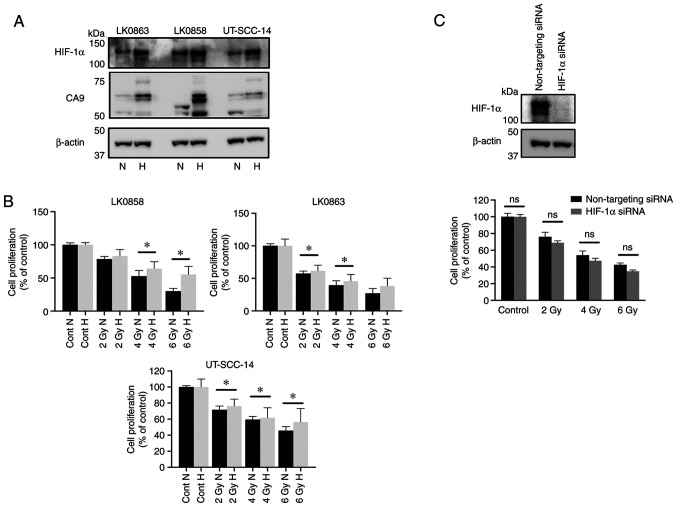
Our previous study demonstrated that hypoxia was associated with the enhanced survival of HNSCC cells in response to different treatments (29). To further evaluate the impact of hypoxia on radiation treatment, three HNSCC cell lines (UT-SCC-14, LK0858 and LK0863) were cultured in the presence of 21% O2 (normoxia) or 1% O2 (hypoxia), followed by exposure of the cells to various doses of ionizing γ-irradiation (2, 4 and 6 Gy). To ensure the presence of hypoxic conditions during the experimental settings, the expression of two hypoxia-associated proteins, namely, HIF-1α and CA9, were assessed using western blotting (Fig. 1A). The cells cultured in the hypoxic conditions exhibited an increased survival capacity in response to irradiation (Fig. 1B). Furthermore, siRNA-mediated downregulation of HIF-1α was not associated with sensitization of hypoxic HNSCC cells towards radiation treatment (Fig. 1C).
Figure 1.
Hypoxia-induced response to radiotherapy in three head and neck squamous cell carcinoma cell lines. The LK0858, LK0863 and UT-SCC-14 cell lines were cultured in normoxic (21% O2) and hypoxic (1% O2) conditions. (A) Western blot analysis of HIF-1α and CA9 expression in cell lines cultured in normoxic and hypoxic conditions. β-actin was used as the loading control. (B) LK0858, LK0863 and UT-SCC-14 cells were irradiated (2, 4 and 6 Gy) at 48 h after seeding and subsequently returned to normoxic or hypoxic conditions. The hypoxic cells were exposed to hypoxia for 24 h prior to irradiation. After 9 days, the cytotoxic/cytostatic effect on cell proliferation was determined by a crystal violet assay. (C) UT-SCC-14 cells transiently transfected with either non-targeting siRNA or HIF-1α-targeting siRNA were exposed to hypoxia for 24 h prior to transfection and placed back under hypoxic conditions after transfection. The efficiency of HIF-1α downregulation was assessed via western blotting at 48 h post-transfection. In parallel experimental settings, the UT-SCC-14 cells were irradiated at 2, 4 and 6 Gy 24 h post-transfection with the respective siRNAs, followed by re-exposure to hypoxic conditions (1% O2). After 9 days, the cytotoxic/cytostatic effect on cell proliferation was determined by a crystal violet assay. Cell proliferation is presented as the percentage of the untreated controls, and data are presented as the mean ± SD from three experiments performed in triplicate. *P<0.05. The data was analyzed using an unpaired Student's t-test. HIF, hypoxia-inducible factor; CA9, carbonic anhydrase 9; siRNA, small interfering RNA; N, normoxia; H, hypoxia; ns, not significant.
Hypoxia-induced changes in EMT- and cancer stem cell (CSC)-associated genes
The role of hypoxia in the acquisition of EMT and cancer stemness as a leading cause of metastasis has been suggested (19). To investigate this, the mRNA expression levels of various EMT-associated genes and CSC-associated genes in hypoxic HNSCC cell lines compared with cells cultured in normoxic conditions were analyzed via RT-qPCR.
As presented in Table I, hypoxia induced significantly increased levels of EMT-associated CDH2 (UT-SCC-14 and LK0863), vimentin (UT-SCC-14, LK0858 and LK0863) and fibronectin 1 (FN1; LK0858 and LK0863). In addition, the CDH1 mRNA expression level was significantly decreased in UT-SCC-14 in response to hypoxia, whereas no significant changes were observed in LK0858 or LK0863 cells. Moreover, hypoxia induced increases in CSC-associated gene expression in all analyzed cell lines, with significantly elevated mRNA levels of SOX2 and NANOG transcription factors.
Table I.
Relative mRNA expression of epithelial-mesenchymal transition and cancer stem cell markers in hypoxic head and neck squamous cell carcinoma cells.
| Cell line, mean ± SD expression level | |||
|---|---|---|---|
|
|
|||
| Gene | UT-SCC-14 | LK0858 | LK0863 |
| CDH1 | 0.3±0.08a | 1.5±0.19 | 0.9±0.29 |
| CDH2 | 8.5±2.08a | 0.9±0.12 | 3.8±1.06a |
| VIM | 6±2.11a | 2.6±0.44a | 4.6±0.77a |
| FN1 | 1.5±0.25 | 2.4±0.47a | 2.6±0.28a |
| FOXC2 | 1.8±0.53 | 0.7±0.10 | 1±0.39 |
| TWIST1 | 2.5±1.11 | 2.3±0.19 | 1.4±0.23 |
| CD44 | 1.4±0.28 | 0.7±0.08 | 0.7±0.12 |
| SOX2 | 6.8±0.98a | 0.8±0.05 | 2.7±0.35a |
| NANOG | 7.2±0.77a | 4.7±0.75a | 3.2±0.14a |
P<0.05 vs. normoxia. Data are presented as the mean ± SD relative to cells cultured in normoxia. CDH, cadherin; VIM, vimentin; FN1, fibronectin 1.
Of note, silencing of CDH2 with siRNA did not affect the proliferation of hypoxic HNSCC cells in response to ionizing γ-irradiation (Fig. S1).
Microarray analysis of hypoxia-regulated gene expression in HNSCC cell lines
To identify hypoxia-regulated genes, HNSCC cells cultured in normoxic and hypoxic conditions were analyzed with an mRNA microarray. The expression profiles generated from the HNSCC cells (UT-SCC-14, LK0859 and LK0863) cultured in hypoxic conditions were compared with the HNSCC cells cultured in normoxic conditions. Following normalization and variance filtering, 20,778 probes were investigated for further statistical analyses. A total of 71 genes were found to be upregulated and 147 were found to be downregulated by hypoxia compared with normoxia (adjusted P<0.05; ±log2 fold change >2). The profile of hypoxia-regulated gene expression in the three HNSCC cell lines is shown in Fig. 2.
Figure 2.
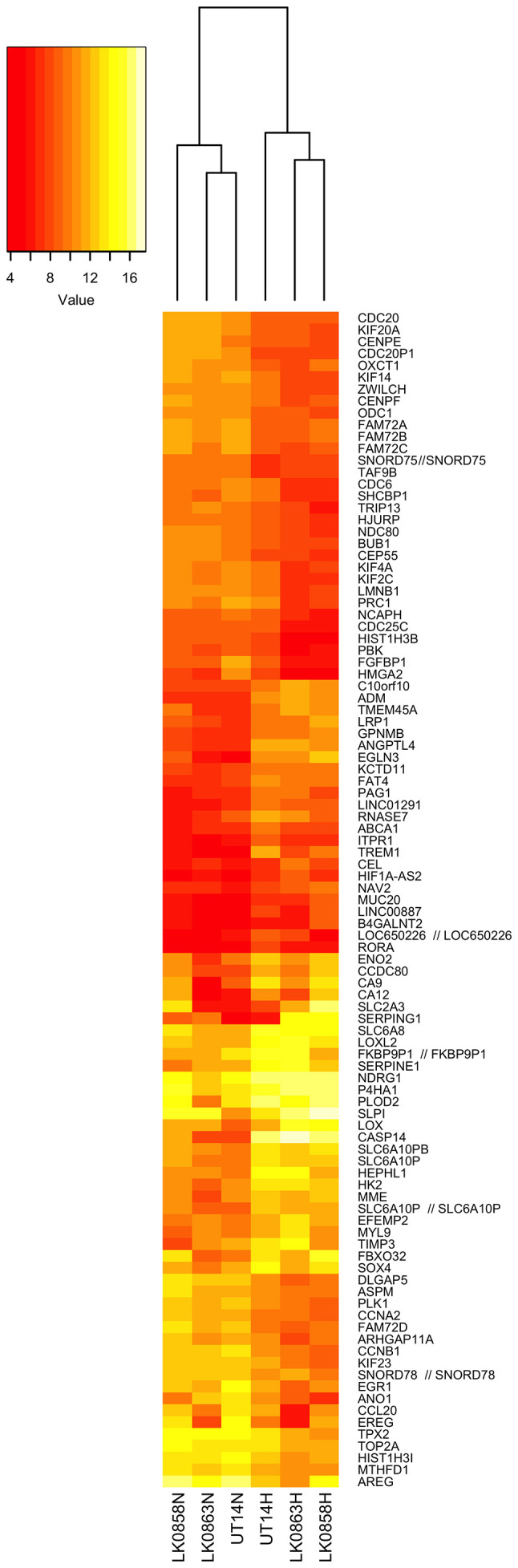
Heatmap representing the expression of hypoxia-regulated genes compared with normoxia in head and neck squamous cell carcinoma cell lines. Differentially expressed genes are indicated on the right, and the legend key is on the left. Each row represents one probe set, and each column represents one experimental sample. N, normoxia; H, hypoxia.
The genes highly upregulated by hypoxia included CASP14, EGLN3, TREM1, CA9, ANGPTL4, SERPING1, ADM, LOX, SLC2A3 (GLUT3) and SERPINE1. The genes that were the most hypoxia-repressed included AREG, FAM72C/FAM72D, EREG, CCNB1, ANO1, FGFBP1, HIST1H3B, PLK1, CDC20 and KIF14 (Table II).
Table II.
Top significantly dysregulated associated genesa.
| A, Upregulated genes | ||
|---|---|---|
| Gene symbol | Full gene name | Log2 fold change |
| CASP14 | Caspase 14 | 7.372 |
| EGLN3 | Egl-9 family hypoxia-inducible factor 3 | 5.029 |
| TREM1 | Triggering receptor expressed on myeloid cells 1 | 4.323 |
| CA9 | Carbonic anhydrase 9 | 4.267 |
| ANGPTL4 | Angiopoietin like 4 | 4.252 |
| SERPING1 | Serpin family G member 1 | 3.975 |
| ADM | Adrenomedullin | 3.856 |
| LOX | Lysyl oxidase | 3.362 |
| SLC2A3 (GLUT3) | Solute carrier family 2 member 3 | 3.321 |
| SERPINE1 | Serpin family E member 1 | 3.054 |
|
| ||
| B, Downregulated genes | ||
|
| ||
| Gene symbol | Full gene name | Log2 fold change |
|
| ||
| AREG | Amphiregulin | −3.255 |
| FAM72C/FAM72D | Family with sequence similarity 72 member C/D | −3.252 |
| EREG | Epiregulin | −3.044 |
| CCNB1 | Cyclin B1 | −2.975 |
| ANO1 | Anoctamin 1 | −2.974 |
| FGFBP1 | Fibroblast growth factor binding protein 1 | −2.906 |
| HIST1H3B | Histone cluster 1, H3b | −2.740 |
| PLK1 | Polo like kinase 1 | −2.671 |
| CDC20 | Cell division cycle 20 | −2.626 |
| KIF14 | Kinesin family member 14 | −2.602 |
adjusted P<0.05 and Log2Fold change >2.
A number of hypoxia-responsive genes from microarray analysis were selected for validation with RT-qPCR. The cumulative mRNA data analysis (LK0858, LK0863 and UT-SCC-14 cells) confirmed upregulation of SERPINE1, CA9, CASP14, LOX and SLC2A3 (GLUT3), as well as downregulation of AREG and EREG (Fig. 3). The statistically significant deregulation of hypoxia-induced genes was found with CA9, CASP14 and EREG.
Figure 3.
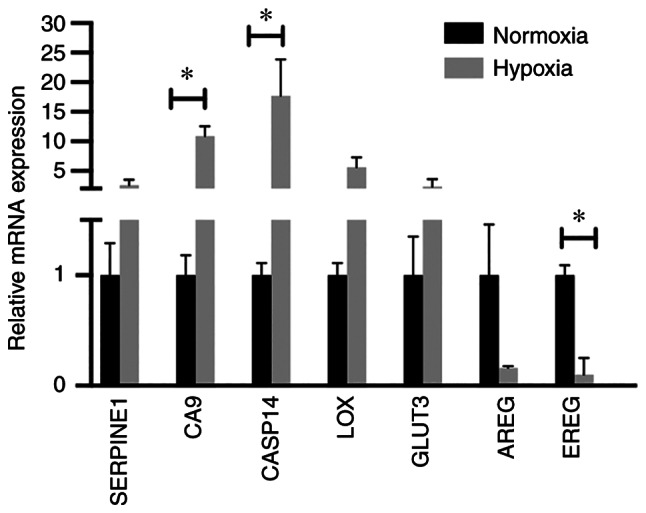
Validation of microarray analysis. Reverse transcription-quantitative PCR analysis was used to analyze the mRNA expression levels of hypoxia-responsive genes in the LK0858, LK0863 and UT-SCC-14 cells using microarray. The relative amounts of SERPINE1, CA9, CASP14, LOX, GLUT3, AREG and EREG mRNA was calculated using the 2−ΔΔCq method and amplification of both GAPDH and β-actin were used as an internal standard. The cumulative, relative mRNA levels from the LK0858, LK0863 and UT-SCC-14 cells are presented as the mean ± SD relative to cells cultured in normoxia. *P<0.05. The data was analyzed using an unpaired Student's t-test. SERPINE1, serpin family E 1; CA9, carbonic anhydrase 9; CASP14, caspase 14; LOX, lysyl oxidase; GLUT3, glucose transporter 3; AREG, amphiregulin; EREG, epiregulin.
With the use of IPA, the top networks assembled from the hypoxia-regulated genes were cellular growth and proliferation, cell cycle, cellular assembly and organization, cancer, cell death and survival, and cellular function and maintenance (Table III). The top regulator effect network with a consistency score of 22.15 consisted of 14 upstream regulators (CSF2, E2F2, E3F3, EP400, INSR, JAK2, LIN9, MED1, NTRK2, TBX2, RARA, RBL2 and RB) that targeted a number of differentially expressed genes under hypoxic conditions in the HNSCC cell lines. The connected downstream functions included cell proliferation of carcinoma cell lines, metabolism of DNA and segregation of chromosomes, among others (Fig. S2).
Table III.
Top networks generated by Ingenuity IPA software.
| Score | Focus genes | Associated network functions |
|---|---|---|
| 49 | ABCE1a, BLMHa, BZW2a, CDCA7a, CENPEa, CENPFa, CST6b, CTPS1a, DCTPP1a, DCXRa, DIMT1a, IDH3Ba, MRPL24a, MTFR2a, MYCa, NUF2a, PAICSa, PLBD2b, PSMA3a, PSMB2a, PSMB3a, RBMS3b, SLC25A19a, SRMa, STX3b, ZWINTa | Cellular growth and proliferation, cell cycle |
| 41 | AIFM1a, C1QBPa, CASP14b, COQ2a, COQ3a, DNASE2b, GARTa, GPNMBb, GSSa, MBOAT1a, MCCC2a, NDUFA9a, NDUFAF1a NDUFV1a, NUAK1b, OXCT1a, PPATa, SIGMAR1a, STOML2a, TUFMa, UCA1b | Cellular assembly and organization, nucleic acid metabolism |
| 37 | AAASa, BAG2a, BTG1b, BUB1a, CCT2a, CCT3a, CCT5a, CCT7a, CDC20a, CLCA2b, DKC1a, ELAC2a, INCENPa, NDC1a, NUP88a, NUP93a, PCGF5b, PFKFB3b, PRMT3a, RAB8Bb, SEH1La, TSC22D2b, VLDLRb, WDR77a | Cellular assembly and organization, molecular transport, RNA trafficking |
| 19 | ALDH1A3a, CA9b, CHRNA5a, DHX33a, ETS2b, GSRa, HAT1a, HIST1H2BJa, HIST1H3Ba, HMGA2a, PNPT1a, PPP1R13Lb, SAAL1a, SLPIb, TIMP3b, WSB1b, ZNF160b | Cancer, cell death and survival, cellular function and maintenance |
| 19 | ABCC4a, ASPMa, FBXO32b, FHa, FNIP1b, GCLCa, KRT18a, NQO1a, PDHBa, SERPINE1b, SLC25A10a, SLC6A8b, TALDO1a, TP53INP1b | Cancer, cell death and survival, organismal injury and abnormalities |
Downregulated in hypoxia;
upregulated in hypoxia.
Effect of microarray-revealed target genes' downregulation on the radiotherapy response
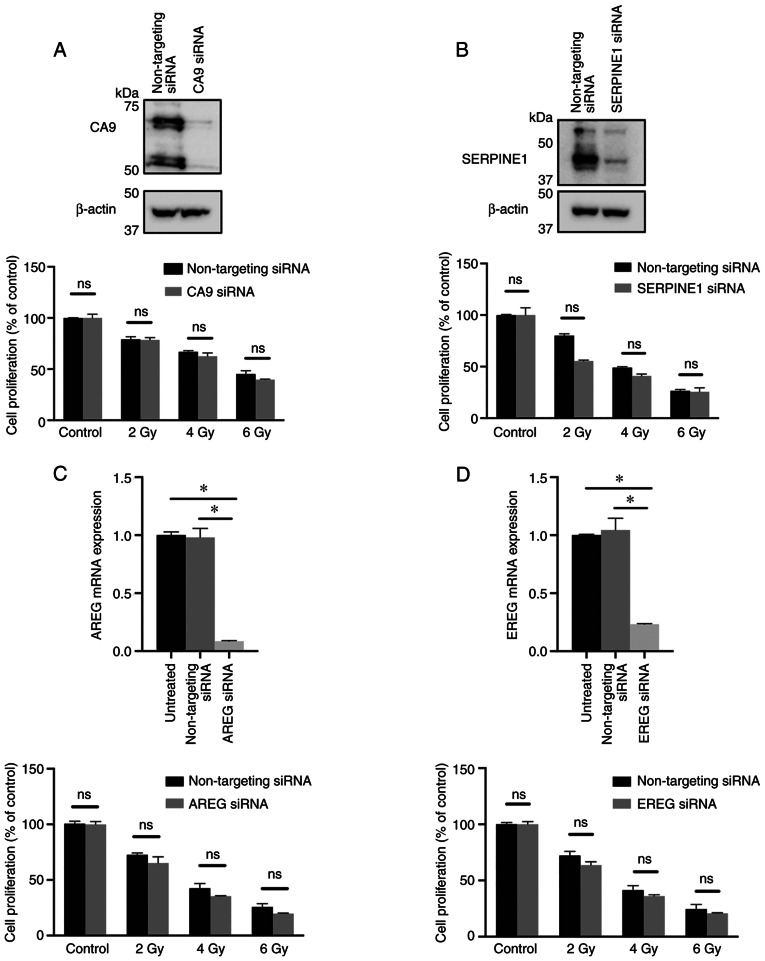
As radiation resistance is associated with hypoxia, whether the highly upregulated genes in hypoxia, namely, CA9 and SERPINE1, affected radioresistance in HNSCC cells was explored. The results showed that silencing CA9 or SERPINE1 with siRNA did not sensitize hypoxic HNSCC cells to ionizing γ-irradiation (Fig. 4A and B).
Figure 4.
Effect of CA9, SERPINE1, AREG and EREG downregulation on the radiation response in head and neck squamous cell carcinoma cells. UT-SCC-14 cells were transiently transfected with either non-targeting siRNA or siRNA interfering with expression of (A) CA9, (B) SERPINE1, (C) AREG and (D) EREG. Cells transfected with either non-targeting siRNA or siRNA targeting CA9 or SERPNE1 were exposed to hypoxia for 24 h prior transfection and placed back under hypoxic (1% O2) conditions after transfection. The protein expression was evaluated 48 h post-transfection using western blotting. β-actin was used as the loading control. In parallel experimental settings, cells transfected with CA9 or SERPINE1 siRNA were irradiated at 2, 4 or 6 Gy 24 h post-transfection, followed by re-exposure to hypoxic conditions, whereas cells transfected with AREG or EREG siRNA were exposed to normoxic conditions (21% O2) after irradiation. The efficiency of AREG and EREG downregulation with specific siRNA was assessed via reverse transcription-quantitative PCR. After 9 days, the cytotoxic/cytostatic effect on cell proliferation was determined by a crystal violet assay. Cell proliferation is presented as the percentage of the untreated controls, and the data are presented as the mean ± SD from three experiments performed in triplicate. All results are shown as the mean ± SD. *P<0.05 vs. non-targeting siRNA. The data was analyzed either using one-way ANOVA followed by Bonferroni's multiple comparison test or an unpaired Student's t-test. SERPINE1, serpin family E 1; CA9, carbonic anhydrase 9; AREG, amphiregulin; EREG, epiregulin; siRNA, small interfering RNA; ns, not significant.
To further explore an association between hypoxia-mediated downregulation of genes and radiotherapy response, the study subsequently focused on the two genes notably repressed by hypoxia: AREG and EREG. UT-SCC-14 cells were transfected with AREG or EREG siRNA prior to culture under normoxic conditions, as the expression of these genes was downregulated under hypoxic conditions. Significant changes in the proliferation rate of HNSCC cells in response to ionizing γ-irradiation upon AREG or EREG targeting was not observed when compared with cells transfected with non-targeting siRNA (Fig. 4C and D). This suggested that the hypoxia-mediated downregulation of AREG and EREG expression did not have a direct effect on the response of HNSCC cells to radiation therapy.
Discussion
Several studies have indicated that hypoxia is associated with cancer progression and the development of resistance to chemotherapy and radiotherapy (30–32). Oxygenation status of head and neck tumors appears to be important, as hypoxic tumors exhibit a poorer response to surgery and radiotherapy (33,34). Identification of biomarkers for hypoxia possessing predictive parameters is essential and targeting tumor hypoxia could therefore improve the response to radiotherapy in HNSCC. HIF-1α is a well-recognized member of the HIF family that mediates a cellular response to hypoxia, and numerous reports have identified HIF-1α as a leading factor promoting radioresistance in tumor cells (35,36). The present study observed a link between hypoxia and the response of HNSCC cells to radiation treatment; however, in the present experimental settings, the increased survival capacity of hypoxic HNSCC cells compared with cells cultured in normoxic conditions after radiation did not appear to be HIF-1α-dependent. Thus, targeting hypoxia via preirradiation inhibition of HIF-1α in order to improve the response of HNSCC to radiotherapy may not be sufficient. Furthermore, findings by Harada et al (37) suggested that the timing of HIF-1α inhibition determined whether a HIF-1 inhibitor can suppress or enhance the effect of radiation treatment, with the authors highlighting the therapeutic significance of inhibiting HIF-1α action after radiation treatment. Hypoxia is known to modulate radiation response and may have an effect on the expression of hypoxia-responsive genes, particularly HIF-1α. There are reports showing a slight increase in HIF-1α after irradiation (38,39). In the current experimental design, the impact of downregulation of selected hypoxia-associated genes on radiotherapy response was explored via siRNA-mediately silencing prior to exposure to hypoxia and irradiation. The siRNA-mediated downregulation was validated via RT-qPCR and western blotting prior to irradiation. However, irradiation-induced increases in HIF-1α expression as a response to oxidative stress cannot be excluded.
It is widely hypothesized in cancer research that hypoxia triggers the EMT, which has been associated with metastasis in cancer, including HNSCC (19,40). There are a number of established EMT-associated markers, and their role in tumorigenesis has been well described. Our previous study reported that the EMT marker FN1 can serve as a biomarker for intrinsic radiosensitivity in HNSCC (41). As hypoxia and HIF-1α signaling drives the EMT, the role of CDH2 in the hypoxia-mediated response to radiation treatment was investigated. CDH2 is one of the EMT-associated markers that promotes tumor cell survival, migration and invasion, and high levels are often associated with a poor prognosis (40,42). However, sensitization of hypoxic HNSCC cells towards radiation was not observed upon the silencing of CDH2 with siRNA. This suggested that CDH2, one of numerous important players in such a dynamic process as EMT, cannot be considered as a determinant of the radiation response upon hypoxia treatment.
Next, the effect of hypoxia on global gene expression in the HNSCC cell lines was investigated via microarray analysis. Implementation of a hypoxia-associated gene expression signature in predicting the radiotherapy response is desirable in HNSCC treatment (43–45). Similar to other studies regarding tumor hypoxia, the commonly accepted marker for hypoxic cells, CA9, was also found to be highly upregulated in the present experimental setting (46–48). Among the 71 upregulated genes observed in hypoxic HNSCC cell lines, CASP14, EGLN3, TREM1, CA9, ANGPTL4, SERPING1, ADM, LOX, SLC2A3 (GLUT3) and SERPINE1 were most prominent. CASP14 has been most extensively described in breast cancer, where its high expression is a marker of breast cancer aggressiveness in association with proliferation and cancer stemness (49). Induction of ADM by hypoxia in turn is associated with increased invasiveness of pancreatic cancer cells and may influence angiogenesis (50), but its role in hypoxic HNSCC cells has not been described. Moreover, hypoxia signaling also controls the establishment of the premetastatic niche, for which LOX activity is indispensable (51,52).
Increased expression of hypoxia-regulated GLUT3 has been observed in different types of tumors, including head and neck cancer. GLUT1 and GLUT3 protein expression is associated with a poor prognosis in oral squamous cell carcinoma, most likely due to the increased glycolytic metabolism of more aggressive cancer cells (53). CA9 is highly expressed in response to hypoxia and is very often associated with increased invasiveness of tumors (54). However, the prognostic and predictive role of CA9 in radiotherapy of HNSCC is quite inconclusive (55–57). Moreover, dysregulation of SERPINE1 has been linked to the activation of hypoxia-related factors and radiation resistance in head and neck cancer (58–60). As CA9 and SERPINE1 were upregulated in all analyzed HNSCC cell lines when cultured under hypoxic conditions, it was hypothesized that they had a direct impact on the radiosensitivity of HNSCC cells. However, subsequent experiments did not support this hypothesis, as the targeting of CA9 and SERPINE1 with siRNA did not lead to resensitization of hypoxic HNSCC cells to irradiation when compared with cells transfected with non-targeting siRNA. Taking into consideration the number of upregulated genes under hypoxic conditions and the complexity of the hypoxia-mediated radioresistance, it is proposed that a panel of hypoxia-responsive genes should be considered as a predictive factor of radiotherapy in HNSCC.
Among the genes repressed by hypoxia, AREG and EREG have been reported to serve a role in cancer progression; AREG and EREG act as ligands of EGFR, and their tumorigenic action and implication in treatment resistance have been already reported (61,62). Our previous study reported that the expression of AREG and EREG influences the treatment (cetuximab and cisplatin) sensitivity of HNSCC cells and may be useful as predictive markers (25). For functional analyses, AREG and EREG downregulation with siRNA under normoxic conditions was performed in order to assess their role in mediating radioresistance in HNSCC cells. The results showed that hypoxia-mediated downregulation of AREG and EREG expression was not a determinant of radiotherapy resistance in HNSCC.
As AREG and EREG were downregulated in hypoxia-exposed cells, their interactions with other signaling molecules may be different than those in normoxic conditions. It would be informative to test in the future if the forced expression of these genes in hypoxia-exposed and irradiated cells affects their survival. However, these genes cannot be excluded from the panel of genes that affect the radiotherapy response under hypoxia.
In conclusion, a number of hypoxia-regulated genes were identified in HNSCC that may promote tumorigenesis and radiotherapy resistance. In addition, the identified genes were involved in multiple biological functions, such as the cell cycle, DNA replication, cellular development, cellular growth and proliferation. The hypoxia-associated gene pattern observed in the present study, in combination with previous reports, may be useful for outcome prediction in HNSCC.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Reidar Grénman (Department of Otorhinolaryngology, University of Turku, Finland) for providing the UT-SCC-14 cell line and Dr Fredrik Levander (National Bioinformatics Infrastructure, SciLifeLab, Lund University, Sweden) for their assistance with the pre-processing of the microarray data.
Funding Statement
This study was supported by the Swedish Cancer Society (grant no. 2017/301), the County Council of Östergötland, the Research Funds of Linköping University Hospital and the Cancer Foundation of Östergötland.
Availability of data and materials
The datasets generated and/or analyzed during the current study (https://www.ncbi.nlm.gov/geo/query/acc.cgi?acc=GSE182734) are available in the Gene Expression Omnibus repository.
Authors' contributions
EW and KR designed the study. EW, NM and KR performed the experiments. EW, NM, AA and KR analyzed the data. EW, NM and KR wrote the manuscript. All authors have read and approved the final manuscript. EW, NM and KR confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of the Linköping University (approval no. 03-537). Written consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Saman DM. A review of the epidemiology of oral and pharyngeal carcinoma: Update. Head Neck Oncol. 2012;4:1. doi: 10.1186/1758-3284-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koukourakis G, Kouloulias V, Koukourakis M, Kouvaris J, Zacharias G, Gouliamos A. The efficacy of combined treatment with cetuximab (erbitux) and radiation therapy in patients with head and neck cancer. J BUON. 2009;14:19–25. [PubMed] [Google Scholar]
- 4.Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: A period analysis. Oncologist. 2010;15:994–1001. doi: 10.1634/theoncologist.2009-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsahafi E, Begg K, Amelio I, Raulf N, Lucarelli P, Sauter T, Tavassoli M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019;10:540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredell MG, Ernst J, El-Kochairi I, Dahlem Y, Ikenberg K, Schumann DM. Current relevance of hypoxia in head and neck cancer. Oncotarget. 2016;7:50781–50804. doi: 10.18632/oncotarget.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv X, Li J, Zhang C, Hu T, Li S, He S, Yan H, Tan Y, Lei M, Wen M, Zuo J. The role of hypoxia-inducible factors in tumor angiogenesis and cell metabolism. Genes Dis. 2016;4:19–24. doi: 10.1016/j.gendis.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 9.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos C, Turley H, Talks K, Gatter KC, Harris AL. Hypoxia-inducible factor (HIF1A and HIF2A), angiogenesis, and chemoradiotherapy outcome of squamous cell head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2002;53:1192–1202. doi: 10.1016/S0360-3016(02)02848-1. [DOI] [PubMed] [Google Scholar]
- 11.Al Tameemi W, Dale TP, Al-Jumaily RMK, Forsyth NR. Hypoxia-modified cancer cell metabolism. Front Cell Dev Biol. 2019;7:4. doi: 10.3389/fcell.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luoto KR, Kumareswaran R, Bristow RG. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013;4:5. doi: 10.1186/2041-9414-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rankin EB, Nam JM, Giaccia AJ. Hypoxia: Signaling the metastatic cascade. Trends Cancer. 2016;2:295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erler JT, Bennewith KL, Nicolau M, Dornhöfer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 15.Burroughs SK, Kaluz S, Wang D, Wang K, Van Meir EG, Wang B. Hypoxia inducible factor pathway inhibitors as anticancer therapeutics. Future Med Chem. 2013;5:553–572. doi: 10.4155/fmc.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perri F, Pacelli R, Della Vittoria Scarpati G, Cella L, Giuliano M, Caponigro F, Pepe S. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck. 2015;37:763–770. doi: 10.1002/hed.23837. [DOI] [PubMed] [Google Scholar]
- 17.Higgins GS, O'Cathail SM, Muschel RJ, McKenna WG. Drug radiotherapy combinations: Review of previous failures and reasons for future optimism. Cancer Treat Rev. 2015;41:105–113. doi: 10.1016/j.ctrv.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Dudas J, Ladanyi A, Ingruber J, Steinbichler TB, Riechelmann H. Epithelial to mesenchymal transition: A mechanism that fuels cancer radio/chemoresistance. Cells. 2020;9:428. doi: 10.3390/cells9020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo CD, Kang N, Choi SY, Kim BN, Park CK, Kim JW, Kim YK, Kim SJ. The role of hypoxia on the acquisition of epithelial-mesenchymal transition and cancer stemness: A possible link to epigenetic regulation. Korean J Intern Med. 2017;32:589–599. doi: 10.3904/kjim.2016.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh JC, Wang HM, Wu MH, Chang KP, Chang PH, Liao CT, Liau CT. Review of emerging biomarkers in head and neck squamous cell carcinoma in the era of immunotherapy and targeted therapy. Head Neck. 2019;41((Suppl 1)):S19–S45. doi: 10.1002/hed.25932. [DOI] [PubMed] [Google Scholar]
- 21.Swartz JE, Pothen AJ, Stegeman I, Willems SM, Grolman W. Clinical implications of hypoxia biomarker expression in head and neck squamous cell carcinoma: A systematic review. Cancer Med. 2015;4:1101–1116. doi: 10.1002/cam4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos K, Pastorek J, Wykoff CC, Gatter KC, Harris AL. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin Cancer Res. 2001;7:3399–3403. [PubMed] [Google Scholar]
- 23.Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene. 2003;22:6497–6507. doi: 10.1038/sj.onc.1206865. [DOI] [PubMed] [Google Scholar]
- 24.Kurahashi I, Fujita Y, Arao T, Kurata T, Koh Y, Sakai K, Matsumoto K, Tanioka M, Takeda K, Takiguchi Y, et al. A microarray-based gene expression analysis to identify diagnostic biomarkers for unknown primary cancer. PLoS One. 2013;8:e63249. doi: 10.1371/journal.pone.0063249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jedlinski A, Ansell A, Johansson AC, Roberg K. EGFR status and EGFR ligand expression influence the treatment response of head and neck cancer cell lines. J Oral Pathol Med. 2013;42:26–36. doi: 10.1111/j.1600-0714.2012.01177.x. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiechec E, Hansson KT, Alexandersson L, Jonsson JI, Roberg K. Hypoxia mediates differential response to Anti-EGFR therapy in HNSCC cells. Int J Mol Sci. 2017;18:943. doi: 10.3390/ijms18050943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorensen BS, Horsman MR. Tumor hypoxia: Impact on radiation therapy and molecular pathways. Front Oncol. 2020;10:562. doi: 10.3389/fonc.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebestyen A, Kopper L, Danko T, Timar J. Hypoxia signaling in cancer: From basics to clinical practice. Pathol Oncol Res. 2021;27:1609802. doi: 10.3389/pore.2021.1609802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isa AY, Ward TH, West CM, Slevin NJ, Homer JJ. Hypoxia in head and neck cancer. Br J Radiol. 2006;79:791–798. doi: 10.1259/bjr/17904358. [DOI] [PubMed] [Google Scholar]
- 34.Swartz JE, Pothen AJ, van Kempen PM, Stegeman I, Formsma FK, Cann EM, Willems SM, Grolman W. Poor prognosis in human papillomavirus-positive oropharyngeal squamous cell carcinomas that overexpress hypoxia inducible factor-1α. Head Neck. 2016;38:1338–1346. doi: 10.1002/hed.24445. [DOI] [PubMed] [Google Scholar]
- 35.Harada H. Hypoxia-inducible factor 1-mediated characteristic features of cancer cells for tumor radioresistance. J Radiat Res. 2016;57((Suppl 1)):i99–i105. doi: 10.1093/jrr/rrw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moeller BJ, Dewhirst MW. HIF-1 and tumour radiosensitivity. Br J Cancer. 2006;95:1–5. doi: 10.1038/sj.bjc.6603201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada H, Itasaka S, Zhu Y, Zeng L, Xie X, Morinibu A, Shinomiya K, Hiraoka M. Treatment regimen determines whether an HIF-1 inhibitor enhances or inhibits the effect of radiation therapy. Br J Cancer. 2009;100:747–757. doi: 10.1038/sj.bjc.6604939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Q, He Y, Ji J, Yao Y, Shen W, Luo J, Zhu W, Cao H, Geng Y, Xu J, et al. Hypoxia-inducible factor 1α (HIF-1α) and reactive oxygen species (ROS) mediates radiation-induced invasiveness through the SDF-1α/CXCR4 pathway in non-small cell lung carcinoma cells. Oncotarget. 2015;6:10893–10907. doi: 10.18632/oncotarget.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/S1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 40.Thierauf J, Veit JA, Hess J. Epithelial-to-Mesenchymal transition in the pathogenesis and therapy of head and neck cancer. Cancers (Basel) 2017;9:76. doi: 10.3390/cancers9070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jerhammar F, Ceder R, Garvin S, Grenman R, Grafstrom RC, Roberg K. Fibronectin 1 is a potential biomarker for radioresistance in head and neck squamous cell carcinoma. Cancer Biol Ther. 2010;10:1244–1251. doi: 10.4161/cbt.10.12.13432. [DOI] [PubMed] [Google Scholar]
- 42.Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. 2004;48:463–476. doi: 10.1387/ijdb.041793ld. [DOI] [PubMed] [Google Scholar]
- 43.Toustrup K, Sorensen BS, Alsner J, Overgaard J. Hypoxia gene expression signatures as prognostic and predictive markers in head and neck radiotherapy. Semin Radiat Oncol. 2012;22:119–127. doi: 10.1016/j.semradonc.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Yang L, West CM. Hypoxia gene expression signatures as predictive biomarkers for personalising radiotherapy. Br J Radiol. 2019;92:20180036. doi: 10.1259/bjr.20180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toustrup K, Sørensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, Overgaard J. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011;71:5923–5931. doi: 10.1158/0008-5472.CAN-11-1182. [DOI] [PubMed] [Google Scholar]
- 46.Sung FL, Hui EP, Tao Q, Li H, Tsui NB, Lo YM, Ma BB, To KF, Harris AL, Chan AT. Genome-wide expression analysis using microarray identified complex signaling pathways modulated by hypoxia in nasopharyngeal carcinoma. Cancer Lett. 2007;253:74–88. doi: 10.1016/j.canlet.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Beasley NJ, Wykoff CC, Watson PH, Leek R, Turley H, Gatter K, Pastorek J, Cox GJ, Ratcliffe P, Harris AL. Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res. 2001;61:5262–5267. [PubMed] [Google Scholar]
- 48.Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- 49.Handa T, Katayama A, Yokobori T, Yamane A, Horiguchi J, Kawabata-Iwakawa R, Rokudai S, Bao P, Gombodorj N, Altan B, et al. Caspase14 expression is associated with triple negative phenotypes and cancer stem cell marker expression in breast cancer patients. J Surg Oncol. 2017;116:706–715. doi: 10.1002/jso.24705. [DOI] [PubMed] [Google Scholar]
- 50.Keleg S, Kayed H, Jiang X, Penzel R, Giese T, Büchler MW, Friess H, Kleeff J. Adrenomedullin is induced by hypoxia and enhances pancreatic cancer cell invasion. Int J Cancer. 2007;121:21–32. doi: 10.1002/ijc.22596. [DOI] [PubMed] [Google Scholar]
- 51.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, Le QT, Giaccia AJ. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165–178. [PMC free article] [PubMed] [Google Scholar]
- 53.Ayala FR, Rocha RM, Carvalho KC, Carvalho AL, da Cunha IW, Lourenço SV, Soares FA. GLUT1 and GLUT3 as potential prognostic markers for oral squamous cell carcinoma. Molecules. 2010;15:2374–2387. doi: 10.3390/molecules15042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benej M, Pastorekova S, Pastorek J. Carbonic anhydrase IX: Regulation and role in cancer. Subcell Biochem. 2014;75:199–219. doi: 10.1007/978-94-007-7359-2_11. [DOI] [PubMed] [Google Scholar]
- 55.Kwon OJ, Park JJ, Ko GH, Seo JH, Jeong BK, Kang KM, Woo SH, Kim JP, Hwa JS, Carey TE. HIF-1α and CA-IX as predictors of locoregional control for determining the optimal treatment modality for early-stage laryngeal carcinoma. Head Neck. 2015;37:505–510. doi: 10.1002/hed.23620. [DOI] [PubMed] [Google Scholar]
- 56.Hwa JS, Kwon OJ, Park JJ, Woo SH, Kim JP, Ko GH, Seo JH, Kim RB. The prognostic value of immunohistochemical markers for oral tongue squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2015;272:2953–2959. doi: 10.1007/s00405-014-3254-5. [DOI] [PubMed] [Google Scholar]
- 57.Eriksen JG, Overgaard J Danish Head and Neck Cancer Study Group (DAHANCA), corp-author Lack of prognostic and predictive value of CA IX in radiotherapy of squamous cell carcinoma of the head and neck with known modifiable hypoxia: An evaluation of the DAHANCA 5 study. Radiother Oncol. 2007;83:383–388. doi: 10.1016/j.radonc.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Lee YC, Yu CC, Lan C, Lee CH, Lee HT, Kuo YL, Wang PH, Chang WW. Plasminogen activator inhibitor-1 as regulator of tumor-initiating cell properties in head and neck cancers. Head Neck. 2016;38((Suppl 1)):E895–E904. doi: 10.1002/hed.24124. [DOI] [PubMed] [Google Scholar]
- 59.Pavon MA, Arroyo-Solera I, Cespedes MV, Casanova I, Leon X, Mangues R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget. 2016;7:57351–57366. doi: 10.18632/oncotarget.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayer C, Schilling D, Hoetzel J, Egermann HP, Zips D, Yaromina A, Geurts-Moespot A, Sprague LD, Sweep F, Baumann M, et al. PAI-1 levels predict response to fractionated irradiation in 10 human squamous cell carcinoma lines of the head and neck. Radiother Oncol. 2008;86:361–368. doi: 10.1016/j.radonc.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Busser B, Sancey L, Brambilla E, Coll JL, Hurbin A. The multiple roles of amphiregulin in human cancer. Biochim Biophys Acta. 2011;1816:119–131. doi: 10.1016/j.bbcan.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 62.Cluckey A, Perino AC, Yunus FN, Leef GC, Askari M, Heidenreich PA, Narayan SM, Wang PJ, Turakhia MP. Efficacy of ablation lesion sets in addition to pulmonary vein isolation for paroxysmal atrial fibrillation: Findings from the SMASH-AF meta-analysis study cohort. J Am Heart Assoc. 2019;8:e009976. doi: 10.1161/JAHA.118.009976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study (https://www.ncbi.nlm.gov/geo/query/acc.cgi?acc=GSE182734) are available in the Gene Expression Omnibus repository.