Abstract
The interplay between the activities of lytic transglycosylases (LTs) and penicillin-binding proteins (PBPs) is critical for the health of bacterial cell wall. Bulgecin A (a natural-product inhibitor of LTs) potentiates the activity of β-lactam antibiotics (inhibitors of PBPs), underscoring this intimate mechanistic interdependence. Bulgecin A in the presence of an appropriate β-lactam causes bulge deformation due to the formation of aberrant peptidoglycan at the division site of the bacterium. As Pseudomonas aeruginosa, a nefarious human pathogen, has 11 LT paralogs, the answer as to which LT activity correlates with β-lactam potentiation is important and is currently unknown. Growth of P. aeruginosa PAO1 strains harboring individual transposon-insertion mutants at each of the 11 genes for LTs, in the presence of the β-lactam antibiotic ceftazidime or meropenem, implicated the gene products of slt, mltD, and mltG (of the eleven), in bulge formation and potentiation. Hence, the respective enzymes would be the targets of inhibition by bulgecin A, which was indeed documented. We further demonstrated by imaging in real time and by SEM that cell lysis occurs by the structural failure of this bulge. Upon removal of the β-lactam antibiotic prior to lysis, P. aeruginosa experiences delayed recovery from the elongation and bulge phenotype in the presence of bulgecin A. These observations argue for a collaborative role for the target LTs in the repair of the aberrant cell wall, the absence of activities of which in the presence of bulgecin A results in potentiation of the β-lactam antibiotic.
The bacterial cell wall is a polymer formed by the crosslinking of the peptide stems of glycan strands.1 The glycan strands consist of the repeating N-acetyl glucosamine (GlcNAc) β-1,4-tethered to N-acetyl muramic acid (MurNAc) disaccharide (Figure 1A). The peptide stem is found on the MurNAc saccharide. The enzymes inactivated by the β-lactam antibiotics are the penicillin-binding proteins (PBPs), the catalysts of the transpeptidation reaction that crosslinks these stems. Ceftazidime (CAZ) is a cephalosporin β-lactam that in Gram-negative bacteria inactivates primarily a specific PBP required for the synthesis of the septum.2 In Pseudomonas aeruginosa a particular PBP inhibited by CAZ is PBP3, an essential enzyme of this pathogen.3 PBP3 is an enzyme of the divisome, the multi-protein assembly that spatially and temporally controls the synthesis of the septal peptidoglycan cell wall. Meropenem (MEM) is a carbapenem β-lactam that inactivates both the PBP2 and PBP3 of P. aeruginosa. PBP2 is an enzyme of the elongasome (the multi-protein assembly involved in the creation of the side-wall peptidoglycan) and of the nascent divisome. Although the sequence of structural transformations catalyzed by the elongasome and the divisome is unknown, an early observation was that net peptidoglycan synthesis required concomitant peptidoglycan disassembly and reassembly.4 A recent observation is that the bactericidal event induced by the loss of PBP function results from the failed synchronization of these two seemingly opposing processes.5 These observations have drawn sharp attention to the role(s) played by a second key enzyme family, that of the lytic transglycosylases (LTs) (Figure 1A), which catalyze peptidoglycan disassembly.6,7
Figure 1.
(A) The reaction of the LTs is shown (R’ = d-Ala-d-Ala for the full-length peptide). LTs catalyze the non-hydrolytic cleavage of the β-1,4-glycosidic bond. (B) The bulgecins (A, B, and C) mimic the oxocarbenium transition species of the LT reaction.18
A confounding aspect to the understanding of the role of the LTs is the plurality of LTs. There are eight LTs in Escherichia coli, up to eight LTs in Neisseria gonorrhoeae, and 11 LTs in P. aeruginosa.7,8 The entire LT family of 11 enzymes is conserved in virtually all sequenced P. aeruginosa genomes.9 Single-gene knockouts of each LT of P. aeruginosa give viable phenotypes.10 Evolution of paralogous LT genes can result in the formation of families of proteins within a genome that share highly conserved—both at the protein sequence and three-dimensional structure level—catalytic domains. Notwithstanding the acquisition of additional domains in various LTs—presumably for the purposes of regulation, sequestration, and/or substrate breadth—the members of the LT family upon in vitro assay appear to have overlapping catalytic activities.11,12 If viability is lost as a result of a single-gene knockout, the gene is deemed as “essential.” A single gene is deemed “non-essential” if viability is not lost. However, this reasoning fails when the gene function is so critical that evolution builds redundancy within the genome, so as to preserve the function. The LT superfamily exemplifies this phenomenon.7,13 As overall LT function is critical, LTs are worthy targets for interference in manifestation of antibacterial effect.
The bulgecins are natural-product inhibitors of LTs (Figure 1B).14,15 The bulgecins cause septal bulges in Gram-negative bacteria in the presence of β-lactam antibiotics, resulting in enhanced bactericidal activity.16 Bulgecin A was shown first to inhibit a soluble LT (Slt70) of E. coli.17 At the time of this observation Slt70 was the only known LT. With the current recognition that each Gram-negative bacterium has a superfamily of LTs, we undertook the assessment of the LT superfamily of the nefarious human pathogen P. aeruginosa for inhibition by bulgecin A. The key question was identification of the subset of the LTs inhibited by bulgecin A to achieve bactericidal potentiation of the β-lactams. We addressed this question using the two β-lactam antibiotics, CAZ and MEM, having different PBP selectivities.
RESULTS AND DISCUSSION
Potentiation of β-lactams by Swarm-motility Assays
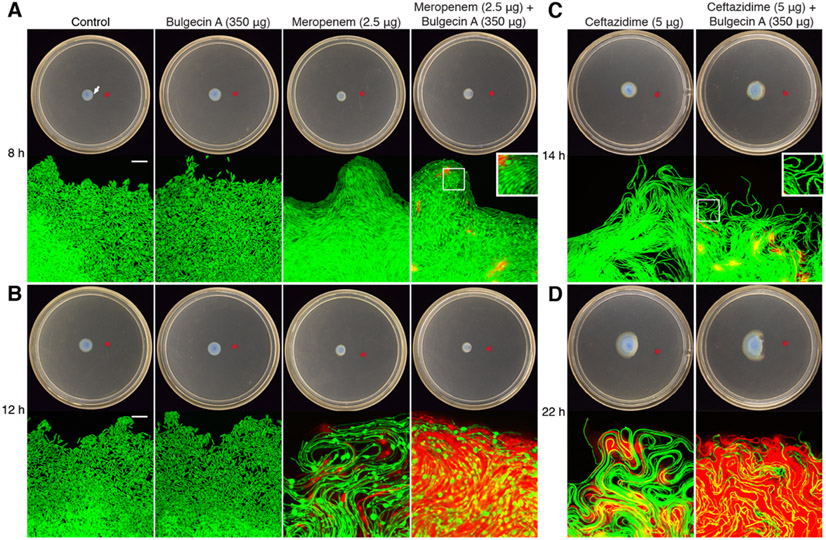
We previously reported that bulgecin A potentiates CAZ against P. aeruginosa as visualized by confocal microscopy of swarm-motility assays.16,19 In this assay the β-lactam antibiotic (alone, or with bulgecin A) is spotted distant from a P. aeruginosa colony. As the bacteria swarm outward, the swarm-colony edge that faces the antibiotic spot encounters an increasing antibiotic gradient. These bacteria undergo structural deformation (in response to the antibiotics) and eventual cell lysis. Cell lysis is visualized by the red fluorescence arising from encounter of the bacterial DNA, spilled into the agar upon cell lysis, with propidium iodide embedded in the agar. In this assay CAZ alone induces an elongated cell phenotype as a result of the failure of septum formation due to the loss of PBP3 function by CAZ inactivation. In the presence of both CAZ and bulgecin A, a different shape phenotype was observed, that of a periodic structural bulge within the elongated cell phenotype. This bulged phenotype correlated with a more rapid progression to cell lysis.
We performed this same experiment with MEM (Figure 2A). A bulgecin A–MEM mixture was plated 20 mm away from a P. aeruginosa colony. The swarm edge (white arrow, top left panel) of the colony was imaged at 8 h and at 12 h. We saw again an elongated cell phenotype with MEM alone (MEM also inhibits PBP3). Bulge formation in the elongated cell phenotype was not observed in the cells exposed to MEM alone, but was observed with the bulgecin A–MEM combination at 8 h (Figure 2A). At 12 h both conditions showed bulge structures (Figure 2B). Notwithstanding that the MEM-induced bulge structures formed in both conditions, we again saw earlier and more extensive cell lysis of bacteria encountering the MEM–bulgecin A combination compared to MEM alone. The previously reported results for the CAZ–bulgecin A combination at 14 h (Figure 2C) and 22 h (Figure 2D) are shown for comparison.
Figure 2.
Bulgecin A potentiation of the bactericidal activity of MEM. GFP-labeled P. aeruginosa was inoculated at the center of the swarm plate. At the red dot was placed a solution containing 2.5 μg MEM or 2.5 μg MEM–350 μg bulgecin A. The bacteria were imaged at the facing edge (100× magnification) at (A) 8 h and (B) 12 h post-inoculation. An arrow marks the imaging site in the top left plate as representative. The left-most panels show a 10-μm scale bar. Red fluorescence indicates the presence of DNA due to cell lysis. Representative data at 14 h (C) and 22 h (D) for both CAZ alone and for a CAZ–bulgecin A mixture shown for comparison.
These observations showed that loss of LT function by the presence of bulgecin A enhanced the bactericidal action of both β-lactams. We acquired from the Manoil Library the transposon (Tn)-insertion mutant for each of the 11 LTs of P. aeruginosa.20,21 The background wild-type P. aeruginosa PAO1 strain for this library, and the 11 Tn-insertion strains in this same background, each were transformed for visualization by the mini-Tn7 chromosomal, constitutive, GFP-expressing insertion (Table S1).22
The first experiment was the individual challenge of each strain by CAZ and by MEM separately in the swarming motility plate assay. The data for CAZ are given in Figure S1. Of the 11 mutants, early (6 h) septal bulge formation was detected foremost in the slt::Tn strain (Figure S1A). Bulge formation was also evident in the membrane-bound lytic transglycosylase D (MltD) Tn-insertion strain mltD::Tn. Slt is a soluble LT of the periplasm, while MltD is a lipoprotein of the periplasm. The nine remaining mutants (for the lipoprotein LTs MltA, MltB, MltF, MltF2, MltG, RlpA; and the soluble LTs SltB1, SltB2, and SltB3) showed no discernable bulge formation. At 9 h minor bulge formation was detected for the mltA::Tn, mltF::Tn, mltG::Tn, sltB1::Tn, and sltB3::Tn strains. However, the degree of bulge formation in these five strains was significantly less than that seen for slt::Tn and mltD::Tn (Figure S2). The results of the Tn-insertion mutants for the genes sltB1 and rlpA (for LTs SltB1 and RlpA, respectively) are shown in Figure S1A as representative data. After 9 h, cell lysis was seen in both the slt::Tn and mltD::Tn strains (Figure S1B) as assessed by the formation of red fluorescence. Cell lysis was not detected in the wild-type control at this time point, nor in the nine other LT Tn mutants (Figure S2; Figure S1 shows the mltG::Tn, sltB1::Tn, and rlpA::Tn strains as representative data). These observations implicate functional redundancy between Slt and MltD, with the appearance of septal bulges in both of their Tn-insertion strains as a foreshadow of progression to bactericidal lysis (vide infra) in the presence of the challenge by β-lactam antibiotic CAZ.
This identical experiment was repeated with MEM as the β-lactam. Two notable experimental changes were made with MEM compared to CAZ experiments. The first is a spatial change as MEM was spotted 20 mm (compared to 15 mm for CAZ) placement of β-lactam from the site of inoculation of the bacterium. A longer distance between the colony and MEM was required as a result of the significantly faster MEM-induced cell lysis compared to CAZ. Accordingly, the second change was temporal—the MEM experiments required different observation times. The results from the wild-type strains in the presence of MEM (Figure 2A) show that MEM induces large bulge-like structures in cells. In the slt::Tn and mltG::Tn strains (Figure S3; but not the remaining nine Tn-insertion mutants, Figure S4) bulge formation was seen at 8 h. At 10 h significant bulge formation was detected in the slt::Tn, mltG::Tn, and mltD::Tn strains. Cell lysis was most prevalent in the slt::Tn strain. Although these bulges have not formed after 10 h in the other Tn-insertion mutant strains, they form if the experiment is given time to continue. Progression to MEM-induced bulge formations is most rapid in the slt::Tn, mltD::Tn, and mltG::Tn Tn-mutant strains. This result identified Slt, MltD, and MltG as the targets of bulgecin A.
Bulgecin A Binding Affinity for LT Targets
We purified these enzymes to homogeneity (P. aeruginosa MltD and MltG were prepared as the soluble enzymes MltDsol and MltGsol by genetic deletion of their respective lipoprotein signal sequences). Their binding ability for bulgecin A was assessed by mass spectrometry analyses (Figure 3). RlpAsol and SltB1, two LTs whose respective Tn-insertion strains lacked bulge formation for either CAZ or MEM, were used for comparison. As the Pseudomonas MltGsol gave a weak MS signal, we used E. coli MltGsol (Ec MltGsol, ECK1083: 35,864 Da) in place of the MltGsol of P. aeruginosa (38% sequence identity, 97% query coverage). Bulgecin A binds to Slt and MltDsol at single-digit micromolar concentrations (Slt and MltDsol, Kd values of 8.5 ± 1.1 μM and 1.4 ± 0.3 μM, respectively). Bulgecin A also binds well to Ec MltGsol (Kd of 24 ± 5.1 μM). The Kd values for SltB1 (Kd of 160 ± 21 μM) and RlpA (1200 ± 280 μM) were higher. The lack of bulge formation upon exposure to CAZ in P. aeruginosa lacking the activities of MltG, SltB1, and RlpA (the respective Tn-insertion mutants) indicates that the loss of function of these three enzymes individually is not linked directly to the loss of function of PBP3 (the primary PBP target of CAZ). In contrast, the loss of function of Slt, MltD, and MltG is linked to the loss of function of PBP2, the primary PBP target of MEM.
Figure 3.

Evaluation of the dissociation constants of bulgecin A for (A) Slt, (B) MltDsol, (C) E. coli MltGsol, (D) SltB1, and (E) RlpAsol by non-denaturing mass spectrometry. The fraction bound (%) corresponds to the fraction of protein bound to bulgecin A relative to the total protein signal (bound and unbound) calculated from the deconvoluted extracted-ion chromatogram (Figure S5-S9).
Potentiation of β-lactams by Growth-curve Assays
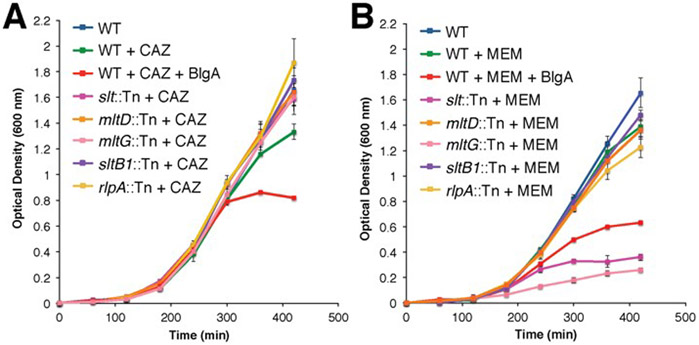
We explored the effect of CAZ and MEM challenges to the individual Tn-insertion strains by growth-curve assays in liquid medium. We showed previously that bulgecin A potentiates CAZ (MIC = 0.5 μg mL−1) at a sub-MIC concentration (0.28 μg mL−1) of CAZ. Wild-type P. aeruginosa in the presence of bulgecin A and CAZ, and also in the presence of bulgecin A and MEM, showed normal growth (not delayed growth) until mid-log phase, at which time a lysis event occurred (Figure 4, red trace). None of the 11 single-Tn-insertion strains showed potentiation in the presence of sub-MIC CAZ (data for the slt::Tn, mltD::Tn, mltG::Tn, sltB1::Tn, and rlpA::Tn strains are shown in Figure 4). The only difference (compared to wild type) among the 11 strains in the presence of sub-MIC of CAZ was a reproducible delay of growth (but without bulge formation) for the mltF::Tn strain (Figure S10). The reason for this delay is unknown. The important conclusion is that the loss of activity of any one LT in P. aeruginosa—be it by Tn insertion into the gene (or by logical extension, inhibition of a single given LT) does not potentiate CAZ at the selected sub-MIC concentration of 0.28 μg mL−1. The MIC for MEM of the parent strain is 0.5 μg mL−1. In the presence of a sub-MIC concentration of MEM (0.14 μg mL−1) potentiation is observed for either the slt::Tn or mltG::Tn strain. As was seen with CAZ, the delay-of-growth (but without bulge formation) phenomenon is observed with MEM for the mltF::Tn strain (Figure S10).
Figure 4.
Bacteria growth-curve assays performed for wild-type PAO1 without (blue line) and with (other lines) a β-lactam antibiotic. (A) CAZ at 0.28 μg mL−1 and (B) MEM at 0.14 μg mL−1; green line). Growth-curve data are given for the slt::Tn (dark pink line), mltD::Tn (orange line), mltG::Tn (light pink line), sltB1::Tn (purple line), and rlpA::Tn (yellow line) strains with CAZ or MEM at the same respective concentrations. The data with the remaining six Tn-insertion strains are given in Figure S10. At these concentrations of the two β-lactams, wild-type PAO1 exhibited normal growth. Wild-type strain PAO1 with β-lactam and bulgecin A (50 μg mL−1) exhibited early (mid-log phase) cell lysis (red line) for both. In the case of sub-MIC MEM, notable potentiation is seen with two LT Tn-insertion strains (slt::Tn and mltG::Tn).
The observation for sub-MIC CAZ seemingly contrasts with the observation of both bulge formation and cell lysis in the Tn-insertion strains for mltD (similar to slt::Tn) as shown in Figure S1. We attribute the lysis shown in Figure S1 to exposure to an increasing concentration gradient of drug as the colony swarms toward the antibiotic spot. In contrast, when the strains are grown in liquid medium at a fixed sub-MIC concentration of CAZ, cell growth is unhindered (Figure 4). The antibiotic efficacy of CAZ against P. aeruginosa is diminished by β-lactam induction of expression of the native AmpC β-lactamase.23, 24 Therefore, the CAZ concentration will diminish over time. In contrast, with sub-MIC MEM (a poorer substrate of AmpC than CAZ) we see potentiation with two single Tn mutants (slt::Tn and mltG::Tn). This result is consistent with a more stable titer of MEM in the MEM-exposed growth-curve assays. The results of both the swarm assays and the growth-curve assays, in the presence of either CAZ or MEM challenge, implicate bulge formation as a pre-requisite for early lysis. We conclude that the potentiation by bulgecin at the sub-MIC of CAZ used (0.28 μg mL−1) results from concurrent loss of a subset (that of Slt and MltD) LT activities. Potentiation at the sub-MIC of MEM (0.14 μg mL−1) is seen upon Tn-insertion inactivation of either the slt or mltG genes.
Inhibition of Cell Wall Repair
Lytic transglycosylases of Gram-negative bacteria have a central role in sensing the cell-wall damage resulting from encounter with β-lactam antibiotics. In addition, LTs may have a key role in the repair of this damage, such as by removal of the accumulation of nascent non-crosslinked glycan strands.5 Failure of this repair of the peptidoglycan contributes to the eventual cell lysis. Indeed, the products (referred to as anhydromuropeptides) of LT catalysis are enriched in non-crosslinked pentapeptide stems.12 We obtained support for a function of the LTs in the repair of damaged peptidoglycan. A bacterial swarm assay used the parent P. aeruginosa PAO1 strain inoculated at a 20 mm distance from a co-spot of CAZ (5 μg) and bulgecin A (350 μg). Confocal-microscopic imaging at 14 h verified that the bacteria at the swarm edge facing the antibiotic spot were elongated and showed septal bulging. These damaged cells were collected by aspiration using 5 μL of FAB-glucose broth. These cells were diluted 3-fold so as to decrease the concentration of CAZ. The diluted cells were imaged on a glass slide to verify a homogenous culture of septal-bulge-containing cells (data not shown). A portion (1 μL) of these cells was placed onto a swarm plate without either CAZ or bulgecin A. A second 1 μL portion was mixed with a solution of 350 μg of bulgecin A, and was added to a second plate. The two plates were imaged at their swarm edges at 2 h and 5 h time after inoculation (Figure S11). Bacteria premixed with bulgecin A exhibit delayed recovery compared to bacteria on the control plate (Figure S11A). Moreover, the bacteria plated on the bulgecin A plate displayed significantly more bulges and less recovery from the damage caused by CAZ leading to more cell death (Figure S11B). Bulgecin A delays recovery of the bacteria during the period following antibiotic exposure. These observations affirm, at the whole-cell level, an important role of LTs in cell-wall repair.
Bulge Deformation is the Site of Cell Lysis
We propose two mechanisms that may contribute to the ability of bulgecin A to potentiate CAZ. LTs initiate the signaling pathway that results in induction of the expression of the AmpC β-lactamase resistance enzyme.5,25,26 Accordingly, bulgecin A-treated cells may arrest (or delay) this potent resistance mechanism. Second, we have experimental evidence that the bulge structure seen in cells exposed to both CAZ and bulgecin A is a point of structural weakness. A movie of the slt Tn-insertion mutant exposed to CAZ on a swarm plate captures this vulnerability (Movie1). Figure 5 shows four snapshots from this movie. The white arrow in Figure 5A (at 5 o’clock) points to a bulge that is in the process of expansion. This bulge grows (Figure 5B) and bursts subsequently (Figure 5C). The red fluorescence that appears (Figure 5C, 33 s into the movie) indicates spillage of the DNA from the bacterium upon the burst. The culminating image implicates loss of the structural integrity of the bacterium, seen now only in the form of the red fluorescence plume (at 74 s, Figure 5D). Examination of other bacteria in the field of these images suggests the possibility of compromised viability prior to the burst of their bulges. In Figure 5B the green fluorescence of the cells immediately flanking (above) the bulge at 5 o’clock appears to fade, presumably as a result of cessation of GFP biosynthesis (Figure 5B). These images attest that the coordinated actions of a PBP inhibitor (the β-lactam) and of an LT inhibitor (bulgecin A) compromise, in the form of osmotically fragile bulges, the integrity of the peptidoglycan of the bacterium.
Figure 5.
Rupture of the bacterium initiates at the bulge structure (A). The white arrow points to the septal bulge that identifies damaged cells. (B) As the cell begins to die, the green fluorescent signal dissipates from the cell (less intense green silhouetting of the affected bacteria). (C) The red color indicates cell lysis in the image of DNA exiting the cell at the point of the bulge (D). The cytosolic GFP dissipates rapidly. A 10-μm white scale bar is given in panel A.
The structural character of the bulge defect was examined by scanning electron microscopy (SEM). In the absence of either β-lactams or bulgecin A, the images of the PAO1 strain of P. aeruginosa strain show rods with the typical (2 µm length, 1 μm width) dimensions and with the customary crenellated exterior surface (Figure 6A). The presence of bulgecin A alone gives an image that is indistinguishable (Figure 6B). The presence of CAZ alone gives lengthened rods as a result of blocked septation (Figure 6C). The presence of both CAZ and bulgecin A gives an initial mid-cell bulge (Figure 6D) that progresses to substantial size (Figure 6E) prior to structural failure (Figure 6F). The presence of both MEM and bulgecin A effects an initial mid-cell bulge (Figure 6G) that is (other than the very different cell lengths) qualitatively similar to the image of Figure 6D. The MEM/bulgecin A combination progresses to a cell whose appearance (Figure 6H) suggests greater structural defect than is seen with CAZ/bulgecin A (Figure 6E) prior to cell lysis (Figure 6I). Prior to lysis both images of the β-lactam/bulgecin A-treated cells (Figure 6E and 6H) show an exterior surface similar to the control images (Figure 6A and 6B). This similarity suggests that bulge growth coincides with the underlying presence of the entirety of the outer-membrane-creating biosynthetic apparatus. As the visualization of cell lysis indicates that the interior of the bulge contains DNA (Figure 5), bulge formation is interpreted as a fundamentally derailed pattern of bacterial growth, as a consequence of the synergistic perturbation of peptidoglycan biosynthesis resulting from the simultaneous loss of critical PBP and LT functions.
Figure 6.
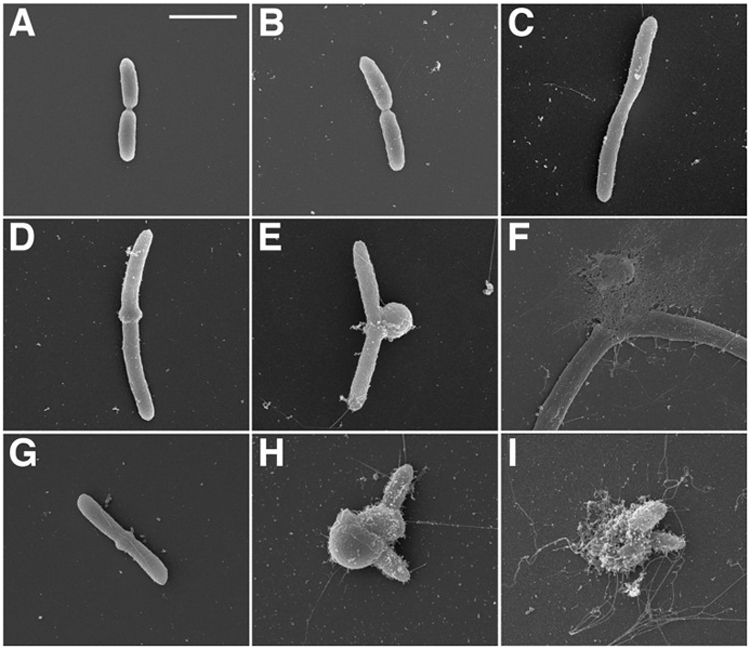
SEM images are shown for P. aeruginosa (A) wild-type, (B) wild-type + bulgecin A, (C) wild-type + CAZ, (D-F) wild-type + CAZ + bulgecin A, (G-I) wild-type + MEM + bulgecin A. The concentration of MEM and CAZ used in the experiment was 8x MIC (4 μg ml−1). The concentration of bulgecin A was 50 μg ml−1. A 2-μm white scale bar is given in panel A.
MIC Determinations
P. aeruginosa is an opportunistic pathogen.27-30 As all of the above experiments used the non-pathogenic P. aeruginosa PAO1 strain, the relevance of bulgecin potentiation of β-lactam antibiotic efficacy was assessed by MIC determinations for ten clinical P. aeruginosa strains (Table S2). The preponderance of the MIC data of this table shows a 2-fold to 4-fold reduction in the β-lactam MIC value in the presence of 100 μg mL−1 bulgecin A. The potentiation of CAZ, and of MEM, by bulgecin A is not unique to P. aeruginosa PAO1. These experiments address three fundamental questions. The first question is whether this relationship has clinical implication. While the determination of MIC values against pathogenic strains is not a complete answer, the data of Table S2 indicate that the fundamental, molecular-level basis for this potentiation exists among a range of P. aeruginosa strains. Complementary study on LT inhibition in other Gram-negative bacteria indicates that an intimate molecular-level relationship between the PBPs and the LTs is retained in other Gram-negative bacteria.31-33 The second question is the nature of the bulge. The SEM images (Figure 6) support an interpretation of the bulge as a misshape arising from fundamentally flawed cell growth. Given the pronounced curvature of the bulge, and the emerging recognition of the outer membrane as a load-bearing component of the bacterial cell envelope,34 it is hardly surprising that cell lysis coincides with structural failure of the bulge (Figure 5). Given the extensive covalent tethering of the peptidoglycan to the proteins and lipids of the Gram-negative outer membrane,35 structural derailing of ordered peptidoglycan biosynthesis could concurrently derail the many events of outer-membrane biosynthesis.36
The third question is the molecular origin of the LT inhibitor-PBP inhibitor potentiation. This study identifies in particular three LTs (Slt, MltD, and MltG) of the 11 found in P. aeruginosa as complicit in β-lactam-induced bulge formation. Both the MS analyses (Figure 3) and the selectivity seen with respect to the structural consequence of β-lactams with the LT-Tn mutants (Figure S1 and S3), pair the Slt and MltD LTs as complementary targets and quite possibly LTs with redundancy of physiological function. Moreover, the restoration of LT activity (by withholding bulgecin A) coincident with the progressive regain of the normal cell shape of P. aeruginosa (Figure S11) further affirms a contribution of LT activity to cell-shape creation.37 The absence of a discernible morphological phenotype with respect to most of the other LTs could follow from the absence of a direct PBP-LT paired function, and (with respect to bulgecin A as an LT inhibitor) the fact that bulgecin A is a selective and not a universal inhibitor of the LT family.38
The points of similarity and difference between the two β-lactams used in this study, ceftazidime and meropenem, attest to the complexity of the paired LT-PBP functions. A key similarity for both pair is bulge formation at the apparent mid-cell location (where the biosynthesis of the septum initiates). This location for bulge formation arguably is not surprising with respect to ceftazidime as the PBP inhibitor. Ceftazidime inhibits preferentially PBP1a and PBP3.39 PBP3 is a component of the divisome machinery. Sub-inhibitory concentrations of selective PBP3-inactivating β-lactams give dramatically elongated/filamentous cells (Figure 2) as a result of unimpeded elongation and failed septation. Sub-MIC concentrations of meropenem preferentially inhibit both PBP2 and PBP3,40 resulting in an initial mid-cell bulge in the rod bacterium (Figure 6) that ultimately transitions to a spherical cell, likely as a result of the loss of function of at least the two PBPs. Although PBP2 is most often described as a key PBP of the elongasome of the Gram-negative bacteria,41,42 PBP2 also co-localizes with PBP3 at the nascent septum.41,43 This concurrence at the apparent site of bulge synthesis may indicate that the loss of LT function (by bulgecin A inhibition) compromises the transition between elongasome and divisome peptidoglycan biosynthesis. The seam of peptidoglycan corresponding to this transition is significantly more structurally fragile than either the peptidoglycan of the sidewall, or the peptidoglycan of the poles.44 Meropenem additionally inactivates l,d-transpeptidase-dependent peptidoglycan synthesis.45,46 Its multiple targets may account for the ability of meropenem–bulgecin A to more rapidly progress to lysis compared to the case of ceftazidime–bulgecin A.
An unexpected observation with meropenem (but not ceftazidime) was the appearance of a bulge phenotype with the mltG::Tn strain (Figure S3). Although this observation is consistent with our evaluation of bulgecin A as an inhibitor of (E. coli) MltG (Figure 3), the observation does not reconcile with the inchoate understanding of the function of the MltG LT. MltG is the most recently identified member of the Gram-negative LT family, and is suggested to cut to size the glycan strands of nascent septal peptidoglycan.31 The assignment of a molecular basis for bulge formation upon MIC exposure of the mltG::Tn strain to meropenem is not possible at this time.
The key accomplishment of this study is its sharp identification of three of the 11 LTs of P. aeruginosa—Slt, MltD, and MltG—for intensive experimental study using β-lactam–bulgecin A pairs. The β-lactam family is structurally diverse with a corresponding diversity of PBP inactivation patterns.47,48 These studies must also reconcile the fact that loss of function of particular LTs can coincide also with increased β-lactam resistance.10,32,49,50 The experimental approach of this study provides a pathway to identify the β-lactam structures having a PBP profile that aligns optimally with concurrent loss of LT function while minimizing AmpC β-lactamase induction;25,51,52 that minimizes AmpR- and LT-dependent cross-talk with other resistance networks;53 that favors the LT loss-of-function appearance of peptidoglycan defects facilitating antibiotic permeation;10 that directs the structural manipulation of the LT inhibitor toward optimal potency and selectivity;32,54 and that will allow a molecular-level understanding as to basis for elongasome and/or divisome misfunction upon simultaneous PBP and LT incapacitation.
METHODS
Complete descriptions of the strains used in the study, the confocal imaging, non-denaturing mass spectrometry chromatograms, growth curve assay negative data for the remaining Tn-insertion mutant strains, cell-wall recovery study, and MIC determinations is provided in the SI Materials and Methods.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants GM61629 (SM) and AI113219 (JDS) from the National Institute of Health (NIH). DAD was supported by NIH Training Grant T32GM075762 and by the ECK Institute for Global Health at the University of Notre Dame. ST was supported by a postdoctoral fellowship from the Uehara Memorial Foundation. We thank S. Chapman and T. Orlova for their assistance with the SEM experiments.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- (1).Typas A, Banzhaf M, Gross CA, and Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol 10, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bush K (2012) Improving known classes of antibiotics: an optimistic approach for the future. Curr. Opin. Pharmacol 12, 527–534. [DOI] [PubMed] [Google Scholar]
- (3).Chen W, Zhang YM, and Davies C (2017) Penicillin-binding protein (PBP) 3 is essential for growth of P. aeruginosa. Antimicrob. Agents Chemother 61, e01651–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Höltje JV (1996) Molecular interplay of murein synthases and murein hydrolases in Escherichia coli. Microb. Drug Resist 2, 99–103. [DOI] [PubMed] [Google Scholar]
- (5).Cho H, Uehara T, and Bernhardt TG (2014) β-Lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Herlihey FA, and Clarke AJ (2017) Controlling autolysis during flagella insertion in Gram-negative bacteria. Adv. Exp. Med. Biol 925, 41–56. [DOI] [PubMed] [Google Scholar]
- (7).Dik DA, Marous DR, Fisher JF, and Mobashery S (2017) Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol 52, 503–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chan YA, Hackett KT, and Dillard JP (2012) The lytic transglycosylases of Neisseria gonorrhoeae. Microb. Drug Resist 18, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, and Brinkman FS (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RN, and Burrows LL (2015) Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. MicrobiologyOpen 4, 879–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, and Mobashery S (2013) Reactions of all Escherichia coli lytic transglycosylases with bacterial cell wall. J. Am. Chem. Soc 135, 3311–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lee M, Hesek D, Dik DA, Fishovitz J, Lastochkin E, Boggess B, Fisher JF, and Mobashery S (2017) From genome to proteome to elucidation of reactions for all eleven-known lytic transglycosylases from Pseudomonas aeruginosa. Angew. Chem. Int. Ed 56, 2735–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Scheurwater E, Reid CW, and Clarke AJ (2008) Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol 40, 586–591. [DOI] [PubMed] [Google Scholar]
- (14).Imada A, Kintaka K, Nakao M, and Shinagawa S (1982) Bulgecin, a bacterial metabolite which in concert with β-lactam antibiotics causes bulge formation. J. Antibiot 35, 1400–1403. [DOI] [PubMed] [Google Scholar]
- (15).Williams AH, Wheeler R, Thiriau C, Haouz A, Taha MK, and Boneca IG (2017) Bulgecin A: the key to a broad-spectrum inhibitor that targets lytic transglycosylases. Antibiotics 6, 8. [Google Scholar]
- (16).Tomoshige S, Dik DA, Akabane-Nakata M, Madukoma CS, Fisher JF, Shrout JD, and Mobashery S (2018) Total syntheses of bulgecins A, B, and C and their bactericidal potentiation of the β-lactam antibiotics. ACS Infect. Dis 4, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Templin MF, Edwards DH, and Holtje JV (1992) A murein hydrolase is the specific target of bulgecin in Escherichia coli. J. Biol. Chem 267, 20039–20043. [PubMed] [Google Scholar]
- (18).Byun B, Mahasenan KV, Dik DA, Marous DR, Speri E, Kumarasiri M, Fisher JF, Hermoso JA, and Mobashery S (2018) Mechanism of the Escherichia coli MltE lytic transglycosylase, the cell-wall-penetrating enzyme for Type VI secretion system assembly. Sci. Rep 8, 4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, and Shrout JD (2014) Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A 111, 18013–18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, and Manoil C (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A 100, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Held K, Ramage E, Jacobs M, Gallagher L, and Manoil C (2012) Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol 194, 6387–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Koch B, Jensen LE, and Nybroe O (2001) A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45, 187–195. [DOI] [PubMed] [Google Scholar]
- (23).Livermore DM (1995) β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev 8, 557–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zamorano L, Reeve TM, Juan C, Moya B, Cabot G, Vocadlo DJ, Mark BL, and Oliver A (2011) AmpG inactivation restores susceptibility of pan-β-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother 55, 1990–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dik DA, Fisher JF, and Mobashery S (2018) Cell-wall recycling of the Gram-negative bacteria and the nexus to antibiotic resistance. Chem. Rev 118, 5952–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jacobs C, Huang LJ, Bartowsky E, Normark S, and Park JT (1994) Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13, 4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Burrows LL (2018) The therapeutic pipeline for Pseudomonas aeruginosa infections. ACS Infect. Dis 4, 1041–1047. [DOI] [PubMed] [Google Scholar]
- (28).López-Causapé C, Cabot G, Del Barrio-Tofiño E, and Oliver A (2018) The versatile mutational resistome of Pseudomonas aeruginosa. Front. Microbiol 9, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Nguyen L, Garcia J, Gruenberg K, and MacDougall C (2018) Multidrug-resistant Pseudomonas infections: hard to treat, but hope on the horizon. Curr. Infect. Dis. Rep 20, 23. [DOI] [PubMed] [Google Scholar]
- (30).Schick A, and Kassen R (2018) Rapid diversification of Pseudomonas aeruginosa in cystic fibrosis lung-like conditions. Proc. Natl. Acad. Sci. U. S. A 115, 10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yunck R, Cho H, and Bernhardt TG (2016) Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol 99, 700–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, and Burrows LL (2013) Changes to Its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 57, 3078–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Skalweit MJ, and Li M (2016) Bulgecin A as a β-lactam enhancer for carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii clinical isolates containing various resistance mechanisms. Drug Des. Devel. Ther 10, 3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, and Huang KC (2018) The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Asmar AT, and Collet JF (2018) Lpp, the Braun lipoprotein, turns 50 - Major achievements and remaining issues. FEMS Microbiol. Lett 365, fny199. [DOI] [PubMed] [Google Scholar]
- (36).Konovalova A, Kahne DE, and Silhavy TJ (2017) Outer membrane biogenesis. Annu. Rev. Microbiol 71, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lommatzsch J, Templin MF, Kraft AR, Vollmer W, and Holtje JV (1997) Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J. Bacteriol 179, 5465–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Romeis T, Vollmer W, and Höltje JV (1993) Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol. Lett 111, 141–146. [DOI] [PubMed] [Google Scholar]
- (39).Davies TA, Page MGP, Shang W, Andrew T, Kania M, and Bush K (2007) Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother 51, 2621–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang Y, Bhachech N, and Bush K (1995) Biochemical comparison of imipenem, meropenem and biapenem: permeability, binding to penicillin-binding proteins, and stability to hydrolysis by β-lactamases. J. Antimicrob. Chemother 35, 75–84. [DOI] [PubMed] [Google Scholar]
- (41).van der Ploeg R, Verheul J, Vischer NOE, Alexeeva S, Hoogendoorn E, Postma M, Banzhaf M, Vollmer W, and den Blaauwen T (2013) Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol. Microbiol 87, 1074–1087. [DOI] [PubMed] [Google Scholar]
- (42).Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, Walker S, Kahne DE, and Bernhardt TG (2018) A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).den Blaauwen T, Aarsman ME, Vischer NO, and Nanninga N (2003) Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol 47, 539–547. [DOI] [PubMed] [Google Scholar]
- (44).Yao X, Jericho M, Pink D, and Beveridge T (1999) Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol 181, 6865–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Hugonnet JE, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerlé C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, and Arthur M (2016) Factors essential for l,d-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife 5, e19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Silva AM, Otten C, Biboy J, Breukink E, VanNieuwenhze M, Vollmer W, and den Blaauwen T (2018) The fluorescent d-amino acid NADA as a tool to study the conditional activity of transpeptidases in Escherichia coli. Front. Microbiol 9, 2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kocaoglu O, and Carlson EE (2015) Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob. Agents Chemother 59, 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sutaria DS, Moya B, Green KB, Kim TH, Tao X, Jiao Y, Louie A, Drusano GL, and Bulitta JB (2018) First penicillin-binding protein occupancy patterns of β-lactams and β-lactamase inhibitors in Klebsiella pneumoniae. Antimicrob. Agents Chemother 62, e00282–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zeng X, Gillespie B, and Lin J (2015) Important role of a putative lytic transglycosylase Cj0843c in β-lactam resistance in Campylobacter jejuni. Front. Microbiol 6, 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Yin J, Sun Y, Sun Y, Yu Z, Qiu J, and Gao H (2018) Deletion of lytic transglycosylases increases β-lactam resistance in Shewanella oneidensis. Front. Microbiol 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Dhar S, Kumari H, Balasubramanian D, and Mathee K (2018) Cell-wall recycling and synthesis in Escherichia coli and Pseudomonas aeruginosa–their role in the development of resistance. J. Med. Microbiol 67, 1–21. [DOI] [PubMed] [Google Scholar]
- (52).Juan C, Torrens G, Barceló IM, and Oliver A (2018) Interplay between peptidoglycan biology and virulence in Gram-negative pathogens. Microbiol. Mol. Biol. Rev 82, e00033–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Balasubramanian D, Schneper L, Kumari H, and Mathee K (2013) A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kraft AR, Prabhu J, Ursinus A, and Höltje JV (1999) Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J. Bacteriol 181, 7192–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.