Abstract
Two commercially available hypersensitive assays for human immunodeficiency virus type 1 (HIV-1) RNA quantitation, AMPLICOR HIV-1 Monitor Test 1.5 and Quantiplex HIV RNA 3.0, were compared to detect and quantify HIV-1 RNA in the cell-free fraction of cervicovaginal secretions collected by vaginal washing. Three panel specimens were used: pooled cervicovaginal secretions spiked with HIV-1 subtype A or HIV-1 subtype B and cervicovaginal lavages from HIV-positive and HIV-negative women. Compared to the AMPLICOR HIV-1 Monitor Test 1.5 assay, the Quantiplex HIV-1 3.0 assay yielded higher estimates of HIV-1 RNA concentrations in several tested samples spiked with HIV-1 RNA subtype A, as well as subtype B, particularly samples containing low amounts of HIV-1 RNA. The sensitivity and specificity of the AMPLICOR HIV-1 Monitor Test 1.5 assay were 93 and 100%, respectively; the sensitivity and specificity of the Quantiplex HIV RNA 3.0 assay were 97 and 50%, respectively. In conclusion, in quantifying HIV-1 RNA in cervicovaginal secretions, the Quantiplex HIV RNA 3.0 may lack specificity, and the AMPLICOR HIV-1 Monitor Test 1.5 assay, although highly specific, may lack sensitivity.
Little information is available about the accuracy of different nucleic acid or signal amplification techniques in quantifying human immunodeficiency virus type 1 (HIV-1) RNA in the female genital tract (1, 3, 6, 11). The ability to assess the viral burden in genital secretions is essential in understanding the pathophysiology of sexual and mother-to-child transmissions of HIV infection (8) and in predicting the effect of antiretroviral therapy at the level of the genital compartment (6, 12). This will require the availability of standardized assays for the quantification of HIV-1 RNA which should be altogether sensitive, specific, and reproducible. Recently, two commercial hypersensitive assays conceived to detect and quantify low levels of HIV-1 RNA in plasma have become available. These are the AMPLICOR HIV-1 Monitor Test 1.5 (Roche Diagnostics, Branchburg, N.J.), which uses reverse transcription (RT)-PCR and whose threshold of positivity is 50 copies of HIV-1 RNA/ml (13, 14), and the Quantiplex HIV RNA 3.0 assay (Chiron-Bayer, Emeryville, Calif.), which uses the branched-DNA signal amplification technique and whose lower limit of quantification is 50 copies of HIV-1 RNA/ml (4, 5). The aim of the present study was to compare the ability of both assays to quantify HIV-1 RNA in normal female genital secretions spiked with HIV-1 RNA standards and in clinical cervicovaginal lavage samples from HIV-infected women.
MATERIALS AND METHODS
Clinical specimens.
Cervicovaginal secretions were obtained from 30 HIV-1-seropositive women and from 30 HIV-seronegative healthy women not at risk for HIV. Women with vaginal discharge, genital bleeding, or suffering from a sexually transmitted infection were excluded from the study. Participants were asked to avoid sexual intercourse and intravaginal medications for 3 days before enrollment. Samples were processed within 1 h of collection. Genital secretions were collected outside the menstrual period. Following the introduction of a speculum, a standardized 60-s lavage was performed with 3 ml of 1 M phosphate-buffered saline (pH 7.2) as previously described (2). This collection procedure introduces a dilution of the genital secretions of about 1:10 (2). After centrifugation of the cervicovaginal lavage at 1,000 × g for 10 min, the cell supernatant was stored at −80°C. Lavage samples were confirmed to be devoid of a significant amount of contaminating blood by measuring traces of hemoglobin using second derivative spectrophotometry, as described earlier (10, 12).
HIV-1 RNA panels.
Three HIV-1 RNA panels of 1-ml aliquots were prepared with viral stock sources diluted in pooled cell-free cervicovaginal lavages from the HIV-negative control women. The PELICHECK HIV-RNA-97 genotype B kit standards (prepared by the Central Laboratory of the Blood Transfusion Service, Amsterdam, The Netherlands, for the Viral Diagnostic Quality Control [VQC] Programme) was used as viral stock for the first or VQC panel, with the following predicted HIV-1 RNA quantities (copies/ml): 8,333, 2,500, 833, 250, 83, 25, 8, 2, 1, 0.2, 0.1, and 0.03. The second panel was obtained by spiking pooled HIV-negative cervicovaginal lavages with supernatant from a culture of HIV-1 subtype A strain (a gift from Françoise Barré-Sinoussi, Institut Pasteur, Paris, France), quantified by NASBA-QT assay (Organon Teknika Corporation, Durham, N.C.), with the following HIV-1 RNA predicted values (copies/ml): 60,000, 30,000, 12,000, 2,400, 480, 96, 19, and 3. The third panel corresponded to pooled plasma collected from an antiretroviral drug-naive patient infected with HIV-1 subtype B, quantified by NASBA-QT assay, with the following HIV-1 RNA predicted values (copies/ml): 200,000, 100,000, 40,000, 8,000, 1,600, 320, 64, and 12.
Quantitative assays.
Evaluation of the HIV-1 RNA load in cervicovaginal lavage samples was assessed by initial concentration of HIV-1 virions, followed by quantitation of HIV-1 RNA in the resulting pellet, using different initial volumes of vaginal lavage sample for each assay. HIV-1 RNA load in cervicovaginal secretions was finally expressed as RNA copies per milliliter of cervicovaginal lavage sample. Quantitative assays for HIV-1 RNA were used according to the manufacturer's recommendations, with slight modifications. For the AMPLICOR HIV-1 Monitor Test 1.5 assay, the virus concentration was determined by centrifuging 0.5 ml of cervicovaginal lavage samples at 23,600 × g for 60 min at 4°C. The pelleted virus particles were lysed by treatment with 600 μl of working HIV-1 MONITOR lysis buffer containing the quantitation standard, and the released RNA was precipitated with 600 μl of 100% isopropanol. The precipitated RNA was recovered by centrifugation, washed with 1 ml of 70% ethanol, and resuspended in 100 μl of AMPLICOR HIV-1 Monitor specimen diluent. Then, 50 μl of the processed specimen was added to 50 μl of the working AMPLICOR HIV-1 Monitor master mix for the RT-PCR amplification reactions. Further amplification, hybridization, and detection steps were performed according to the manufacturer's instructions. For the Quantiplex HIV RNA 3.0 assay, the virus was concentrated from 1 ml of cervicovaginal lavage samples by centrifugation at 23,500 × g for 60 min at 4°C, and the supernatant was further discarded without disturbing the virus pellet. Pellets were then frozen at −80°C for more than 1 day until they could be tested, as recommended. Further hybridization and detection steps were performed according to the manufacturer's instructions.
Statistical analysis.
Linear regression analysis on log-transformed values was performed to compare observed to expected values obtained with spiked samples and observed values obtained by both assays in clinical samples, by using the software StatView SE+ (Abacus Concepts, Inc., Berkeley, Calif.) and to compare both quantitative assays by using the orthogonal regression Deming method with the EVAL kit software, as described elsewhere (7).
RESULTS
Detection of HIV-1 RNA in spiked samples.
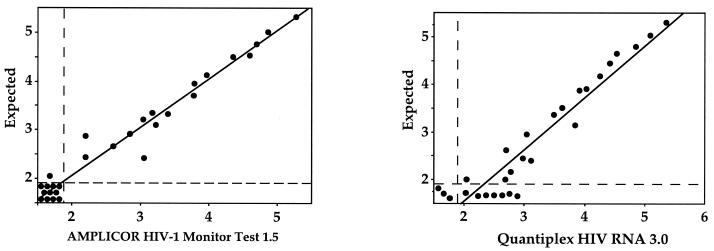
Table 1 and Fig. 1 depict the raw data of HIV-1 RNA estimations tested in duplicate in the three panels of samples of pooled cervicovaginal lavages from HIV-negative women spiked with serial dilutions of HIV-1 subtype A or HIV-1 subtype B standards. Although both assays were able to detect HIV-1 subtype A as well as HIV-1 subtype B standards, differences between the AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex HIV-1 3.0 assays were observed.
TABLE 1.
Quantification of HIV-1 RNA in pooled cervicovaginal secretions lavage samples from HIV-negative women spiked with known amount of HIV-1 subtype A (HIV-1/A) and subtype B (HIV-1/B) by the AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex HIV RNA 3.0 assay
| HIV-1 RNA standard
|
AMPLICOR HIV-1 Monitor 1.5a
|
Quantiplex bDNA 3.0a
|
|||||
|---|---|---|---|---|---|---|---|
| Subtype | Expected (log copies/ml) | Observed (log copies/ml) | Recoveryc (%) | ΔLogd | Observed (log copies/ml) | Recoveryc (%) | ΔLogd |
| HIV-1/B VQCe | 8.3 × 103/3.91 | 6.4 × 103/3.80 | 77 | −0.11 | 9.9 × 103/3.99 | 119 | 0.08 |
| 2.5 × 103/3.40 | 1.4 × 103/3.14 | 56 | −0.24 | 3.2 × 103/3.50 | 91 | −0.11 | |
| 8.3 × 102/2.91 | 1.5 × 102/2.17 | 18 | −0.11 | 1.1 × 103/3.04 | 132 | 0.13 | |
| 2.5 × 102/2.39 | 1.2 × 103/3.07 | 75 | −0.13 | 1.2 × 103/3.07 | 480 | 0.68 | |
| 83/1.91 | 64/1.80 | NAg | −0.10 | 2.6 × 102/2.41 | 313 | 0.61 | |
| 25/1.39 | <50/1.70 | NA | 0.0 | 3.7 × 102/2.56 | 1,480 | 1.17 | |
| 8.3/1.39 | <50/1.70 | NA | 0.0 | 2.8 × 102/2.44 | 3,500 | 1.05 | |
| 2.6/1.39 | <50/1.70 | NA | 0.0 | <50/1.70 | NA | 0.0 | |
| 0.8/−0.09 | <50/1.70 | NA | 0.0 | 5.2 × 102/2.71 | >1,040 | 1.01 | |
| 0.3/−0.52 | <50/1.70 | NA | 0.0 | 8.7 × 102/2.93 | >1,740 | 1.23 | |
| 0.1/−10 | <50/1.70 | NA | 0.0 | 1.1 × 102/2.04 | >120 | 0.34 | |
| 0.03/−1.48 | <50/1.70 | NA | 0.0 | <50/1.70 | NA | 0.0 | |
| 0b | <50/1.70 | NA | 0.0 | 96/1.98 | NA | ||
| HIV-1/Af | 6 × 104/4.77 | 5.1 × 104/4.70 | 85 | −0.07 | 8.9 × 104/4.94 | 103 | 0.17 |
| 3 × 104/4.47 | 2.3 × 104/4.36 | 77 | −0.11 | 5.1 × 104/4.40 | 170 | 0.23 | |
| 1.2 × 104/4.07 | 9.9 × 103/3.99 | 83 | −0.08 | 1.7 × 104/4.25 | 148 | 0.18 | |
| 2.4 × 103/3.38 | 2.0 × 103/3.30 | 85 | −0.08 | 3.9 × 104/3.59 | 162 | 0.21 | |
| 4.8 × 102/2.68 | 4.0 × 102/2.61 | 85 | −0.07 | 4.6 × 102/2.66 | 97 | 0.05 | |
| 96/1.98 | 55/1.74 | 52 | −0.24 | 1.1 × 102/2.04 | 115 | 0.06 | |
| 19/1.27 | <50/1.70 | NA | 0.0 | <50/1.70 | NA | 0.0 | |
| 3/0.47 | <50/1.70 | NA | 0.0 | 2.3 × 102/2.36 | >460 | 0.97 | |
| 0b | <50/1.70 | NA | 0.0 | <50/1.70 | NA | 0.0 | |
| HIV-1/Bf | 2 × 105/5.30 | 1.2 × 105/5.07 | 60 | −0.22 | 2.8 × 105/5.44 | 140 | 0.14 |
| 1 × 105/5.00 | 6.2 × 104/4.79 | 62 | −0.21 | 1.6 × 105/5.20 | 160 | 0.20 | |
| 4 × 104/4.60 | 3.4 × 104/4.53 | 85 | −0.07 | 3.5 × 104/4.54 | 88 | −0.06 | |
| 8 × 103/3.90 | 6.2 × 103/3.79 | 78 | −0.11 | 11 × 103/4.04 | 138 | 0.14 | |
| 1.6 × 103/3.20 | 1.2 × 103/3.07 | 75 | −0.13 | 7.1 × 103/3.85 | 443 | 0.65 | |
| 3.2 × 102/2.50 | 1.6 × 102/2.20 | 51 | −0.30 | 9.2 × 102/2.96 | 287 | 0.46 | |
| 64/1.80 | <50/1.70 | NA | −0.10 | 3.7 × 102/2.56 | 578 | 0.76 | |
| 12/1.39 | <50/1.70 | NA | 0.0 | 1.7 × 102/2.23 | >340 | 0.84 | |
| 0b | <50/1.70 | NA | 0.0 | 1.5 × 102/2.18 | >306 | 0.48 | |
The dynamic range claimed by the manufactures for HIV-1 RNA detection and quantitation in plasma ranged from 50 to 5 × 104 copies/ml for the AMPLICOR HIV-1 Monitor Test 1.5 assay and from 50 to 5 × 105 for the Quantiplex HIV-1 RNA 3.0 assay.
The expected zero value corresponds to the negative control of unspiked, pooled cervicovaginal lavage samples obtained from HIV-seronegative women.
Recovery is defined by the ratio of observed HIV-1 RNA on expected HIV-1 RNA and is expressed as a percentage.
Δ Log is the difference between the log of observed HIV-1 RNA and the log of expected HIV-1 RNA. A log of ≥0.5-log units is considered a significant difference. For statistical analysis, values below 50 copies/ml were assigned a 1.70 value.
PELICHEK HIV-RNA reference panel prepared by Central Laboratory of the Blood transfusion service, Amsterdam, The Netherlands.
Expected values were established by quantifying viral stock of cell culture supernatant by the NASBA assay.
NA, not applicable.
FIG. 1.
Scatter plots of observed versus expected HIV-1 RNA levels, expressed in log copies/milliliter, estimated in parallel by the AMPLICOR HIV-1 Monitor Test 1.5 assay (left) and the Quantiplex HIV RNA 3.0 assay (right) in pooled cervicovaginal lavages obtained from HIV-negative not-at-risk women spiked with known amount of HIV-1 subtype A or subtype B (n = 28). The vertical and horizontal dashed lines indicate the positivity threshold of both assays (1.69 log copies/ml).
Linear regression analysis of observed versus expected HIV RNA load estimations obtained by the AMPLICOR HIV-1 Monitor Test 1.5 assay showed a slope of 0.99 and an r2 value of 0.97 (P < 0.001) (Fig. 1). The mean Δlog values, as defined by the log of observed RNA level minus the log of the expected RNA level, were −0.05 for the VQC panel, −0.08 for the VIH-1/A panel, and −0.10 for the VIH-1/B panel. All Δlog values are below the 0.5-log units, which corresponds to the threshold of significant variation for HIV-1 RNA load determination (9). Of 36 samples, 34 (94%) expected to contain more than 50 HIV-1 RNA copies/ml were detected as positive by the AMPLICOR HIV-1 Monitor Test 1.5, whereas all samples expected to contain fewer than 50 HIV-1 RNA copies/ml were not quantified by this assay. The linear regression analysis of observed versus expected HIV RNA load estimations obtained by the Quantiplex HIV-1 3.0 showed a slope of 1.08 and an r2 value of 0.92 (P < 0.001) (Fig. 1). Furthermore, 22 tested samples of 58 (39%) showed a significant Δlog (>0.5). The mean Δlog values were 0.52 for the VQC panel, 0.23 for the VIH-1/A panel, and 0.39 for the VIH-1/B panel. Of 20 spiked samples, 14 (70%) expected to contain fewer than 50 HIV-1 RNA copies/ml were detected as positive by the Quantiplex HIV-1 3.0 assay. Finally, the HIV RNA load estimations obtained by the AMPLICOR HIV-1 Monitor Test 1.5 assay and those obtained by the Quantiplex HIV-1 3.0 were highly correlated (Deming regression analysis: a slope of 1.055 with a 95% confidence interval [CI] and an intercept of −0.560 with 95% CI).
The upper values of the dynamic ranges of both assays and the lower value of the dynamic ranges of the AMPLICOR HIV-1 Monitor Test 1.5 assay corresponded to those claimed by the manufacturers, when the assays were used to quantify HIV-1 RNA in cervicovaginal samples, the lower value of the dynamic range of the Quantiplex HIV-1 3.0 assay appeared much lower than 50 copies/ml. Compared to the AMPLICOR HIV-1 Monitor Test 1.5 assay, the Quantiplex HIV-1 3.0 assay yielded higher estimates of HIV-1 RNA concentrations in several tested samples spiked with HIV-1 RNA subtype A as well as subtype B, particularly those containing low amounts of HIV-1 RNA.
Assay reproducibility.
To determine intra-assay reproducibility, two pooled cervicovaginal lavages from HIV-seronegative control women spiked with different levels of HIV-1 subtype B RNA representing high and low copy numbers (cervicovaginal secretions samples 1 and 2, respectively) were analyzed simultaneously 10 times by both assays. The results presented in the Table 2 show that the lower-copy-number HIV-1 RNA cervicovaginal secretion sample 2 yielded a slightly larger expected coefficient of variation (CV), compared to the high-copy-number cervicovaginal secretion sample 1 by both assays. However, these values corresponded to a rather narrow within-run standard deviation of <0.33 log, which is <0.5 log. The interassay reproducibilities were estimated for the two spiked cervicovaginal secretions samples 1 and 2, by 10 successive determinations of HIV-1 RNA load by both assays. Although the inter-assay reproducibilities of the Quantiplex HIV-1 3.0 assay were slightly higher than those of the AMPLICOR HIV-1 Monitor Test 1.5 assay, the mean CV values for between-run precision showed a rather low variability and were found to be in the range of 0.09 to 0.16%, which still corresponds to a narrow between-run standard deviation (<0.40 log).
TABLE 2.
Intra-assay and interassay reproducibilities of the AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex HIV RNA 3.0 assays used to quantify HIV-1 RNA in spiked cervicovaginal samplesa
| Cervicovaginal lavage | HIV-1 load (copies/ml) | AMPLICOR HIV-1 MONITOR 1.5
|
Quantiplex HIV-1 RNA 3.0
|
||
|---|---|---|---|---|---|
| Intra-assay (SD) | Interassay (SD) | Intra-assay (SD) | Interassay (SD) | ||
| Sample 1 | 10,000 | 0.10 | 0.10 | 0.09 | 0.11 |
| Sample 2 | 2,500 | 0.12 | 0.15 | 0.13 | 0.16 |
The intra-assay precision represents the standard deviation of the values obtained by 10 simultaneous determinations of HIV-1 RNA load of the same sample of cervicovaginal secretions spiked with known levels of HIV-1 subtype B RNA. The interassay precision represents the standard deviation of the values obtained by 10 successive determinations of HIV-1 RNA load of the same sample of cervicovaginal secretions spiked with known levels of HIV-1 subtype B RNA.
Quantitative analysis of clinical samples.
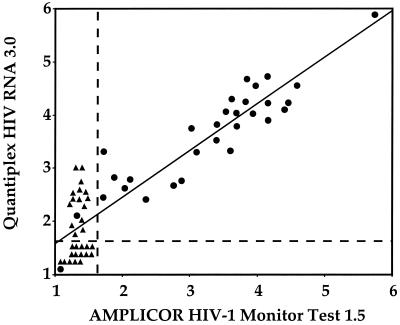
HIV-1 RNA levels were determined in parallel by both ultrasensitive assays in cervicovaginal lavage clinical samples from 30 HIV-1-seropositive women and 30 HIV-seronegative, not-at-risk women (Fig. 2 and Table 3).
FIG. 2.
Correlation of HIV-1 RNA levels, expressed in log copies/milliliter, estimated in parallel by the AMPLICOR HIV-1 Monitor Test 1.5 assay and the Quantiplex HIV RNA 3.0 assay in cervicovaginal lavages obtained from 30 HIV-positive women and 30 HIV-negative not-at-risk controls. ●, Cervicovaginal HIV-1 RNA load from HIV-infected women; ▴, cervicovaginal HIV-1 RNA load from HIV-negative women. The vertical and horizontal dashed lines indicate the positivity thresholds of both assays (1.69 log copies/ml).
TABLE 3.
Quantitative analysis of cervicovaginal lavage samples from 30 HIV-1-seropositive women and 30 HIV-seronegative women by the AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex HIV RNA 3.0 assay
| Cervicovaginal lavage (n) | AMPLICOR HIV-1 MONITOR 1.5a
|
Quantiplex HIV-1 RNA 3.0b
|
||
|---|---|---|---|---|
| No. negative | No. positivec | No. negative | No. positivec | |
| HIV seronegative (30) | 30 | 0 | 15 | 15 |
| HIV seropositive (30) | 2 | 28 | 1 | 29 |
The estimated specificity of the assay for cervicovaginal secretion samples corresponding to the ratio of number of true negative samples out of the total number of negative samples, is 30/30 = 100%.
The estimated specificity of the assay for cervicovaginal secretion samples is 15/30 = 50%.
A cervicovaginal lavage sample was considered positive if HIV-1 RNA level was above the threshold of positivity, i.e., 50 HIV-1 RNA copies/ml for both assays.
The AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex assays were found positive in 28 (93%) and 29 (97%) of the 30 samples from HIV-infected women, respectively (Fig. 2). The levels of HIV-1 RNA in cervicovaginal lavages of HIV-positive women ranged from 52 to 750,000 copies/ml (median, 15,300 copies/ml) by the AMPLICOR HIV-1 Monitor Test 1.5 assay and from 75 to 800,000 copies/ml (median, 18,527 copies/ml) by the Quantiplex HIV RNA 3.0 assay. Overall, the differences between the viral load obtained by the AMPLICOR HIV-1 Monitor Test 1.5 and the Quantiplex HIV RNA 3.0 assays varied for 36 samples by no more than 0.5 log. Conversely, 24 (40%) samples showed a significant, i.e., >0.5-log, discrepancy between the values of HIV-1 RNA level obtained by both assays. The HIV-1 RNA load of these latter samples by the AMPLICOR HIV-1 Monitor Test 1.5 and by the Quantiplex HIV RNA 3.0 assays ranged from <50 to 12,082 copies/ml (median, 25 copies/ml) and from 100 to 50,572 copies/ml (median, 489 copies/ml), respectively, resulting in a mean difference in the HIV-1 RNA values obtained by both assays of 0.75 ± 0.23 log (mean ± standard error of the mean).
All cervicovaginal lavages samples from HIV-negative women were correctly identified by the AMPLICOR HIV-1 Monitor Test 1.5 assay, whereas 15 (50%) of samples were found positive by the Quantiplex HIV RNA 3.0 assay, i.e., they showed an HIV-1 RNA level above the threshold of positivity of the assay (Fig. 2). All of these samples were confirmed as negative in a second determination by an in-house RT-nested PCR for the HIV-1 pol gene, as described earlier (15) (data not shown). The levels of HIV-1 RNA in these 15 false-positive samples by the Quantiplex HIV RNA 3.0 assay ranged from 75 to 1,660 copies/ml (median, 232 copies/ml).
Overall, the 60 cervicovaginal samples analyzed showed a positive correlation (r2 = 0.82) between the values of the HIV-1 RNA loads measured by both assays (P < 0.0001) (Fig. 2). If one considers the threshold of positivity defined by the manufacturers, the sensitivity and specificity of the AMPLICOR HIV-1 Monitor Test 1.5 assay may be estimated to be 93 and 100%, respectively, and the sensitivity and specificity of the Quantiplex HIV RNA 3.0 assay, may be estimated to be 97 and only 50%, respectively.
DISCUSSION
We compared two third-generation hypersensitive assays, the AMPLICOR HIV-1 Monitor Test 1.5 and Quantiplex HIV RNA 3.0, to quantitate HIV-1 RNA levels in cervicovaginal secretions spiked with known amounts of HIV-1 RNA standards and in cervicovaginal lavages clinical samples from HIV-infected and HIV-negative women. Both assays were able to detect similarly HIV-1 subtype A and HIV-1 subtype B RNA mixed in pooled cervicovaginal secretions. The linear ranges of both assays corresponded to those claimed by the manufacturers, except for the lower value of the linear range of the Quantiplex HIV RNA 3.0 assay. Compared to the AMPLICOR HIV-1 Monitor Test 1.5 assay, the Quantiplex HIV-1 3.0 assay yielded higher estimates of HIV-1 RNA concentrations in several tested samples spiked with HIV-1 RNA subtype A or subtype B, particularly those containing low amounts of HIV-1 RNA. The intra-assay and interassay reproducibilities appeared to be nearly similar for both assays. The sensitivities of the AMPLICOR HIV-1 Monitor Test 1.5 assay and of the Quantiplex HIV RNA 3.0 assay were 93 and 97%, respectively, indicating that some samples containing a low amount of HIV-1 RNA could not be detected by the AMPLICOR HIV-1 Monitor Test 1.5 assay. The specificity of the AMPLICOR HIV-1 Monitor Test 1.5 assay and that of the Quantiplex HIV RNA 3.0 assay were 100 and 50%, respectively, indicating clearly that the Quantiplex HIV RNA 3.0 assay gives false-positive results for HIV-negative samples and also overestimates the HIV-1 RNA levels in samples containing copy numbers of HIV-1 RNA under the threshold of detection of the assay. The low specificity of the Quantiplex 3.0 assay used to quantify HIV-1 RNA in cervicovaginal secretions samples suggests that nonspecific hybrization may occur, probably because of the interference with the body fluid matrix. Indeed, we observed that the nonspecific signal was lower when female genital secretions were collected using 10 ml of lavage buffer instead of 3 ml as in the present study (data not shown).
Taken together, our observations demonstrate that the Quantiplex HIV RNA 3.0 assay may lack the specificity to quantify HIV-1 RNA in the acellular fraction of cervicovaginal secretions, the risk of false positivity being particularly marked for the low HIV-1 RNA levels. The AMPLICOR HIV-1 Monitor Test 1.5 assay is highly specific for detecting and quantifying HIV-1 RNA in cervicovaginal secretions, although it may lack sensitivity for some samples with low HIV-1 RNA levels (≤100 copies/ml).
ACKNOWLEDGMENTS
This work was supported by a grant from the Agence Nationale de Recherches sur le SIDA (FF006C).
We thank Florence Thierry and Fréderic Eberlé from Roche Diagnostics Meylan, France, and Françoise Huisse from Bayer-Chiron, Cergy, France, for providing kits for the study; Mathieu Matta and Anne Le Gall for technical assistance and for archival sample retrieval; and the medical staff of the Service d'Immunologie Clinique, Hôpital Broussais, Paris, France, and of the Institut Alfred Fournier, Paris, France, for their support. We also thank François-Xavier Mbopi Kéou from the Central Public Health Laboratory, Colindale, London, United Kingdom, for reviewing the English of the manuscript.
REFERENCES
- 1.Baron P, Bremer J, Wasserman S S, Nowicki M, Driscoll B, Polsky B, Kovacs A, Reichelderfer P S. Detection and quantitation of human immunodeficiency virus type 1 in the female genital tract. J Clin Microbiol. 2000;38:3822–3824. doi: 10.1128/jcm.38.10.3822-3824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belec L, Meillet D, Levy M, Georges A, Tevi-Benissan C, Pillot J. Dilution assessment of cervicovaginal secretions obtained by vaginal washing for immunological assays. Clin Diagn Lab Immunol. 1995;2:57–61. doi: 10.1128/cdli.2.1.57-61.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremer J, Nowicki M, Beckner S, Brambilla D, Cronin M, Herman S, Kovacs A, Reichelderfer P. Comparison of two amplification technologies for detection and quantitation of human immunodeficiency virus type 1 RNA in the female genital tract. J Clin Microbiol. 2000;38:2665–2669. doi: 10.1128/jcm.38.7.2665-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbeik T, Charlebois E, Nassos P, Kahn J, Hecht F M, Yajko D, Ng V, Hadley K. Quantitative and cost comparison of ultrasensitive human immunodeficiency virus type 1 RNA viral load assays: Bayer bDNA Quantiplex versions 3.0 and 2.0 and Roche PCR Amplicor monitor version 1.5. J Clin Microbiol. 2000;38:1113–1120. doi: 10.1128/jcm.38.3.1113-1120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart C E, Lennox L J, Pratt-Palmore M, Wright T C, Schinazi R F, Evans-Strickfaden T, Bush T J, Schnell C, Conley L J, Clancy K A, Ellerbrock T V. Correlation of human immunodeficiency virus type 1 RNA levels in blood and the female genital tract. J Infect Dis. 1999;179:871–882. doi: 10.1086/314656. [DOI] [PubMed] [Google Scholar]
- 7.Linnet K. Performance of Deming regression analysis in case of misspecified analytical error ratio in method comparison studies. Clin Chem. 1998;44:1024–1031. [PubMed] [Google Scholar]
- 8.Mostad S B, Kreiss J K. Shedding of HIV in the genital tract. AIDS. 1996;10:1305–1315. doi: 10.1097/00002030-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Saag M S, Holodniy M, Kuritzkes D R, O'Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 10.Sanderink G J, Van Rijn H J. Quantitative measurement of plasma hemoglobin by second derivative spectrophotometry. Clin Chim Acta. 1985;28:65–73. doi: 10.1016/0009-8981(85)90124-x. [DOI] [PubMed] [Google Scholar]
- 11.Shepard R N, Schock J, Robertson K, Shugars D C, Dyer J, Vernazza P, Hall C, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol. 2000;38:1414–1418. doi: 10.1128/jcm.38.4.1414-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Si-Mohamed A, Kazatchkine M D, Heard I, Goujon C, Prazuck T, Aymard G, Cessot G, Kuo Y H, Bernard M C, Diquet B, Malkin J E, Gutmann L, Belec L. Selection of drug-resistant variants in the female genital tract of human immunodeficiency virus type 1-infected women receiving antiretroviral therapy. J Infect Dis. 2000;182:112–122. doi: 10.1086/315679. [DOI] [PubMed] [Google Scholar]
- 13.Sun R, Ku J, Jayakar H, Kuo J C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Triques K, Coste J, Perret J L, Segarra C, Mpuodi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR tests for quantification of different subtypes of human immunodeficiency virus type 1. J Clin Microbiol. 1999;37:110–116. doi: 10.1128/jcm.37.1.110-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu T, Korber B T, Nahmias A J, Hooper E, Sharp P M, Ho D D. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]