Abstract
Viral structural proteins are emerging as effective targets for new antivirals. In a viral lifecycle, the capsid must assemble, disassemble, and respond to receptors, all at the right time and place. These reactions work within a narrow range of conditions, making them susceptible to small molecule interference. In at least three specific viruses, this approach has had met with preliminary success. In rhinovirus and poliovirus, compounds like pleconaril bind capsid and block RNA release. Bevirimat binds to Gag protein in HIV, inhibiting maturation. In Hepatitis B virus, core protein allosteric modulators (CpAMs) promote spontaneous assembly of capsid protein leading to empty and aberrant particles. Despite the biological diversity between viruses and the chemical diversity between antiviral molecules, we observe common features in these antivirals’ mechanisms of action. These approaches work by stabilizing protein-protein interactions.
Introduction
One advantage of targeting viral structural proteins is that they are unique to the pathogen and generally have no human homologs. Another advantage of structural proteins is their propensity to form oligomers, leading to a dominant negative effect: a drug does not necessarily need to act on every viral protein subunit to disrupt the oligomer’s function (••1). The challenge of targeting viral structural proteins is that they are not enzymes, where the substrate provides a straightforward starting point for transition state analogs. The regions of functionality for structural proteins are often macromolecular interfaces, and small molecules which bind to these interfaces must be identified through naïve screens. A further challenge in targeting a molecular interface is allostery, where the site of drug binding and the site of its effect can be spatially distinct. However, by understanding the requirements for a structural protein to function properly, we concomitantly identify the targets for antiviral effectors.
Physical Chemistry of Capsid Assembly: A Qualitative Summary
The four critical lessons to be learned from modelling capsid assembly are (••2–6):
Subunits are multivalent. They may interact one subunit at a time to build capsid shells, or hierarchically first making small oligomers. Multivalency gives them avidity.
The pairwise association energy between any two subunits during the assembly reaction is weak, but summing the energy of a large number of contacts results in a particle that can be very stable (7).
Assembly reactions follow a steep downhill energy gradient. More subunits mean more contacts. Like a car rolling downhill, such reactions are very difficult to stop.
Assembly and disassembly are not always mirror images. Capsid closure and postassembly conformational maturation can both contribute to capsids that are much more stable than might be expected (8).
In the simplest model, an assembly reaction consists of subunits adding one at a time to a starting nucleus. To model this reaction, one need only consider the geometry of subunits, pairwise per-contact association energy, and on-rate (••2–6). Overly strong association energy can lead to kinetic traps where the reaction runs out of subunits before completing the maximum possible number of capsids or, worse, any defects that are accidentally incorporated get cemented in place by subsequent additions (9, ••10). Weak association energy allows the reversibility needed to repair defects and escape dead-end kinetic traps.
Nucleation factors are also important to consider (11, 12). Nucleation regulates the number of assembling particles, thus decreasing the potential for kinetic traps. Nucleation may also direct the path of assembly, affecting the size and shape of the resulting particle. The viral genome is often involved in nucleation, and the role(s) of specific sites on the genome may direct assembly and genome organization (6, 13, ••14)
Though capsids assemble based on weak interactions, the end products may be remarkably stable (6, 8). Some of this stability arises because the first subunit to leave a capsid must break many contacts simultaneously, creating a kinetic and thermodynamic barrier to dissociation. Capsid stability may also arise from post-assembly conformational and chemical changes (e.g. proteolytic maturation, see below) (15).
Enteroviruses and Capsid Stabilization
The enterovirus genus of the picornavirus family includes rhinovirus, poliovirus, and Coxsackie virus. They are small unenveloped RNA viruses with pseudo-T=3, or P=3, symmetry (16). The immature capsid is comprised of trimers of Vp0, Vp1, and Vp3 subunits, initially part of a polyprotein. These trimers assemble into stable pentamers that can reversibly assemble to empty capsids (17) or package RNA. Assembled capsids autoproteolytically mature by cleavage of Vp0 to Vp2 and the ~70-residue Vp4 peptide. In response to receptor binding or to heat, mature picornaviruses undergo a conformational change to release viral RNA and Vp4, a reaction termed “uncoating” though the capsid may remain largely intact. Unless stimulated to uncoat, mature enteroviruses can be remarkably stable.
The “WIN” series of compounds (after Sterling-Winthrop) contributed to the development of pleconaril and pirodavir and were found to inhibit the uncoating reaction of rhinovirus (18, 19). They show the same effects in many, but not all enteroviruses. Structurally first observed in 1986 (•20), these stabilizing compounds fill a long, tubular pocket that is entirely within Vp1, distal to sites of pentamer-pentamer interaction (Figure 1). To examine mechanism, the temperature dependence of poliovirus uncoating was determined in the presence and absence of WIN compounds (••21). Via an Arrhenius analysis, it was determined that the entropy of the complex decreased, increasing the barrier to uncoating. Consistent with this result, molecular dynamics showed that WIN compounds could decrease the volume filled by rhinovirus capsid proteins and alter concerted movements of the capsid proteins (22). Similarly, it was observed that WIN compounds suppress transient exposure of Vp4 outside the capsid, an indicator of virus “breathing” (23). Though it was an effective antiviral, pleconaril did not ultimately gain FDA approval because of off-target effects. Furthermore, not all enteroviruses possess a pocket accessible for this class of drugs (24).
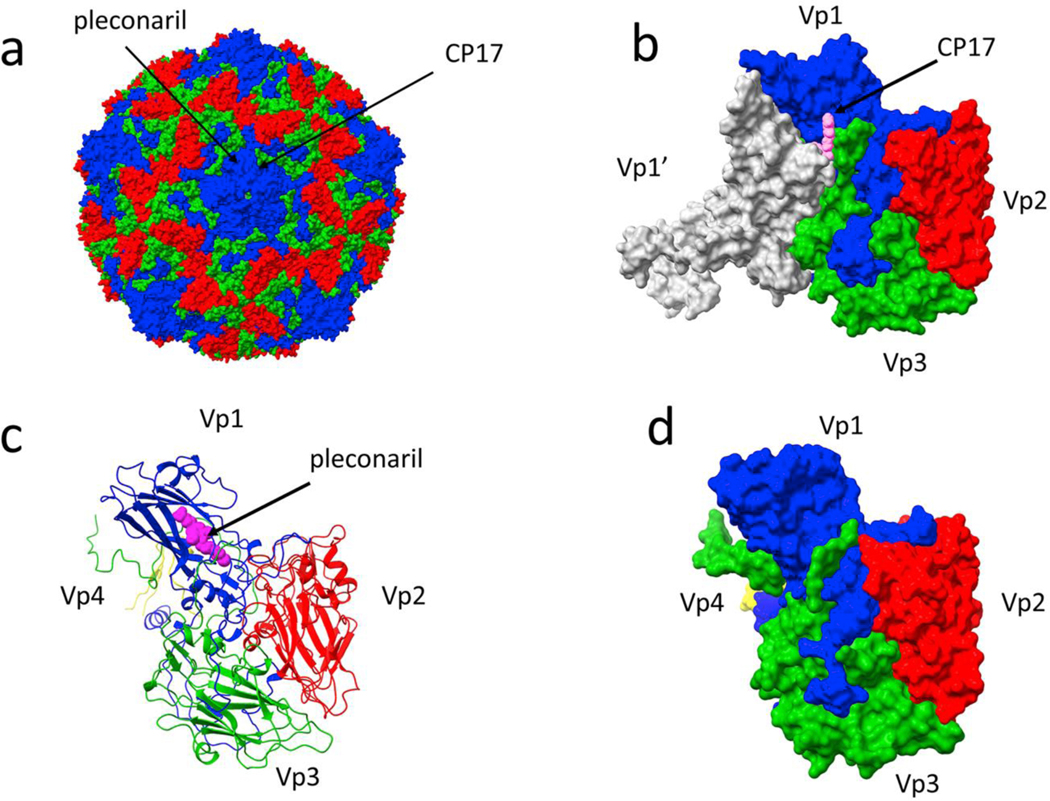
Figure 1. Antiviral binding sites on a picornavirus capsid.
(a) A coxsackie virus capsid (1cov), with the viral structural proteins Vp1–3 colored blue, red, and green, respectively; Vp4 is inside the capsid and not visible in this image. Two distinct sites for capsid-directed antiviral compounds are denoted with arrows. (b) The binding site for compound 17 (CP17, pink), from Abdelnabi et al. 2019 (6gzv), is at an interface of a Vp3 and the two adjacent Vp1 molecules (one shown in white). The view is from the outside of the capsid. (c) In contrast, the binding location for pleconaril (magenta), binding the same site as a WIN compound, only contacts a single copy of Vp1 and is not located at an intermolecular interface (1ncr). (d) A surface representation of the same complex as (c) emphasizes that pleconaril is completely enclosed by Vp1.
More recently a new class of uncoating inhibitor was reported for Coxsackie virus (25). These molecules, exemplified by “compound 17” (CP17), bind at the junction of three subunits, two VP1s and a VP3. This pocket is distal to both the WIN pocket and the interaction between pentameric subunits that forms during capsid assembly (Figure 1). Yet, like the WIN compounds, CP17 inhibits uncoating. Because the interface between proteins is a geometric necessity, the structure of the pocket is well-conserved. Thus, such molecules are likely to show activity across many enteroviruses. In summary, stabilization of enterovirus capsids interferes with uncoating and RNA release. A likely mechanism is that they damp capsid dynamics, increasing the energetic barrier to uncoating.
CpAMs: Targeting Hepatitis B Virus Capsid Assembly
Despite an effective vaccine, Hepatitis B virus (HBV) remains a global health problem, particularly in Asia and Africa where chronic infection is underdiagnosed (26). HBV is a small enveloped virion with an icosahedral core containing a circular viral genome of partially double stranded DNA. Because the genome is not completely double stranded, it has been named relaxed circular DNA (rcDNA). After cell entry, the rcDNA genome is deposited in the nucleus where it is repaired by host enzymes to yield a covalently closed circular DNA chromosome. Viral RNA transcripts are then exported to the cytoplasm, which include a pre-genomic RNA (pgRNA). Core protein dimers (Cp) assemble around a pgRNA-reverse transcriptase complex, usually forming a T=4 icosahedral capsid. In addition to the genome-filled particles, a large fraction of newly assembled capsids in infected cells are empty. Reverse transcription of pgRNA to rcDNA occurs inside the capsid and the mature particles are notably fragile, partly due to the fact that DNA is a stiff polymer compared to the 25nm inner diameter of an HBV capsid (27, 28). Arguably, HBV Cp plays roles in almost every step of the viral lifecycle (29), and Cp has become an important target of HBV-specific antivirals. Variants of capsid-specific molecules have a proven capability to suppress virus replication in cell culture, and more than ten are currently undergoing clinical trials (30). We prefer to call these molecules core protein allosteric modulators (CpAMs); other names include assembly agonists, capsid assembly modulators (CAMs), capsid assembly affectors (CAEs), and core protein inhibitors (CIs).
Initially, phenylpropenamides (PPAs) were found to reduce HBV replication in cell culture (31, 32) and led to accumulation of empty particles by somehow inhibiting RNA packaging (33). Soon after, heteroaryldihydropyrimidines (HAPs) were described in the literature, where the authors established that their mechanism of action involved Cp and led to reduced viral DNA and reduced intracellular Cp levels (34, 35). Much later, sulfamoylbenzamides (SBAs) emerged, which also reduced cytoplasmic pgRNA capsids, but did so without significantly decreasing total capsid protein (36).
Efforts to identify the target of HAP activity quickly converged on the process of capsid assembly (37). Mechanistically, rather than inhibit assembly, CpAMs stimulate assembly, increasing both the rate and extent of assembly reactions with purified core protein (38). Super-stoichiometric concentrations of HAP (more HAP molecules than Cp sites) caused Cp to assemble into larger, flatter structures with distinctly hexameric features (Figure 2) (38, 39). The shape of these assemblies appears to be tunable, forming the larger flatter structures when assembling slowly, and more spherical structures when assembling quickly (•40). A cryo-EM study of HAP assembly products was able to selectively reconstruct tubular assemblies, and while the HAP was not resolved, the protein subunits exhibited a striking pattern of regular repeating hexamers.(41)
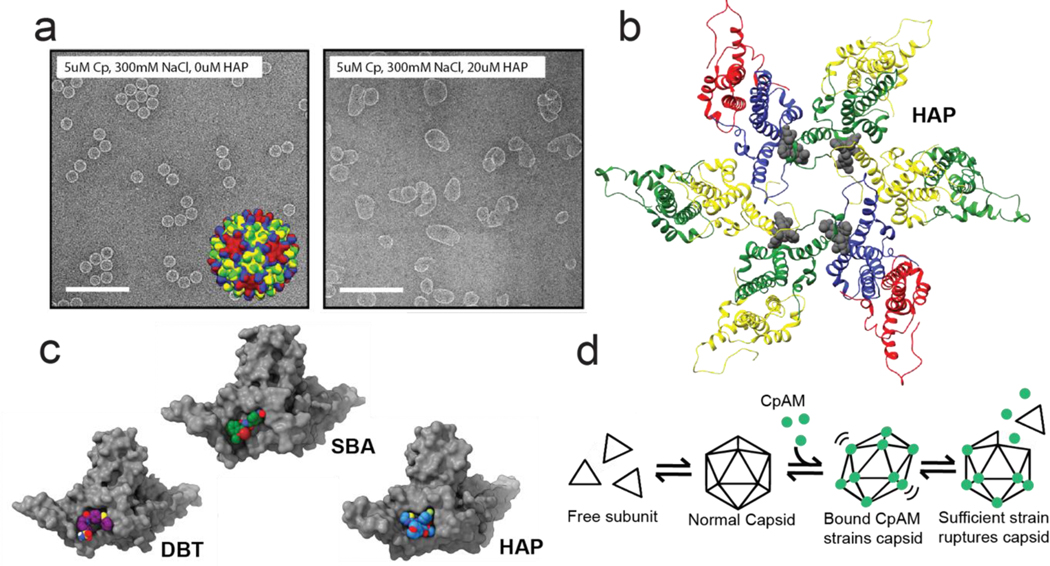
Figure 2. The effects of CpAM binding to HBV core protein.
(a) Electron micrographs of assembled HBV Cp dimer. (left panel) Virus-like particles are ~35nm icosahedra. Inset: an model of a T=4 capsid structure with chains colored according to quasi-equivalent environment. When assembled in the presence of a HAP (right panel), reaction products include large misassembled structures (40). (b) A hexamer fragment of a T=4 capsid structure in complex with a HAP (44). Coloring is the same as in the inset in the left panel of a, except with the HAP molecules colored gray. The HAP molecules bind at the interface between two adjacent subunits. (c) One site, multiple chemistries: a selection of CpAMs which target the interface between HBV capsid protein subunits. Despite occupying the same site, each molecule has a distinct binding mode, and associated phenotype (47, 49, ••52) (d) A simplified reaction schematic for how CpAMs disrupt pre-formed capsids. Increasing the per-contact association energy causes favorable, but modified pairwise orientation that induces global strain. Strained capsids will rupture to relieve strain (53). Components in this figure are extracted from or adapted from figures in references (40), (44), and (••52).
An important component to this mechanism is that CpAMs strengthen the association energy between subunits, often on the order of −1kcal/mol per-contact (42). This modest energy shift is amplified as each Cp dimer is tetravalent, and because assembly is already a steep energy gradient.
Crystal structures of core protein bound to CpAMs used pre-assembled capsids, co-crystalized with a HAP (43, 44) or a PPA (45). All three CpAM-containing capsids expanded by about 5% compared to ligand-free capsids, effectively an allosteric response to CpAM binding. For both chemotypes, the molecules bound to two of the four quasi-equivalent protein interfaces of the hexameric capsomers. The molecules bind in a pocket formed at the junction of two Cp dimers (Figure 2b, c). The dimers must be oligomerized to form a complete pocket, suggesting that subunits associate spontaneously in the absence of a CpAM, which can then insert into the pocket and raise the energy barrier for dissociation.
Capsid-CpAM structures have provided a clear view of the CpAM binding site(s) and the effect of CpAMs on quaternary structure, however capsids are not convenient for crystallographic studies. Dimers with a point mutation (Y132A) that removes a tyrosine from the dimer-dimer interface and renders the protein assembly-incompetent (46) will co-crystalize with CpAMs, packing as a planar lattice of hexamers with a CpAM bound at every inter-subunit interface (47). Using Y132A, structures have been solved of at least three HAPs (47–49), a sulfamoylbenzamide (49), and the antifungal derived CpAM Ciclopirox (50). All molecules bind at the inter-subunit interface, but each has a distinct binding pose. With assembly-active dimer, each of these molecules drives Cp assembly and therefore strengthens Cp-Cp interaction.
The effects of some CpAMs on capsid structure imply that the molecules not only interfere with the assembly process but can also disrupt intact capsids. This could have the effect of blocking deposition of viral DNA in the nucleus (51). We envision a single antiviral with antiviral capability at opposite ends of the viral lifecycle (release of nucleic acid to establish infection, packaging of nucleic acid during assembly). Evidence for structural disruption includes the capsid crystal structures, where drug binding causes faceting, becoming flatter at hexamers (where the molecules bind), and becoming more highly curved at the 5-fold vertices. Faceting occurs due to concerted changes in quaternary structure, so it was likely constrained due to the inter-dimer crosslinks that were necessary to get diffraction quality crystals. Using cryo-EM to visualize capsids with a bound fluorescently-labeled HAP CpAM, un-crosslinked capsids were irregular, with elliptical and asymmetrically faceted capsids visible (42). The mechanisms for disruption of preformed capsids were investigated further with another CpAM, a dibenzothiazapine (DBT), which both drives assembly and paradoxically causes pre-formed capsids to ultimately disassemble (••52). HAPs will also cause capsids to become strained, eventually rupturing and reassembling into larger flatter structures. These observations led to a model where CpAMs induce local deformations that propagate to global capsid strain, leading to eventual rupture (Figure 2d). The energetic cost of adopting a strained state, an unfavorable quaternary structure, is paid for at every protein interface where the binding energy of a CpAM strengthens the interaction. Essentially, capsid strain is an emergent property of local changes to the subunit interfaces.
HIV: successfully targeting a non-icosahedral virus
Human immunodeficiency virus (HIV) capsid assembly is a multistep process that starts with the Gag polyprotein (53). Starting from the N-terminus, most Gag polyproteins are comprised of matrix, capsid (CA), spacer peptide 1 (SP1), nucleocapsid, spacer peptide 2, and p6 domains. About 5% of HIV Gag translation products incorporate a ribosomal frameshift that appends the protease, polymerase, and integrase domains. Formation of a new virion starts with Gag localized to the plasma membrane bound to viral RNA (53). In nascent virions, Gag is arranged in a hexagonal lattice with numerous flaws, forming a partially complete spherical shell (54). Consistent with other examples of virus capsid proteins, Gag-Gag interaction (largely mediated by CA-CA association) is weak (55). Hexameric and pentameric capsomers are formed by CA N-terminal domains (CA-NTDs) which are supported by interdigitating C-terminal domains (CA-CTDs). Capsomers are crosslinked by dimerization of CA-CTDs. During or after budding, the viral protease becomes active and processes Gag into individual proteins. In the mature virus, about 1500–2000 CA proteins reorganize into a Fullerene cone, an arrangement of CA hexamers and pentamers (56, •57). After mature virus infects a new host cell, the fullerene cone is released, and its dissociation appears to play important roles in the transport and reverse transcription of the viral genome. Both the maturation and ultimate capsid dissociation steps have been targets for antiviral intervention.
Gag maturation begins with an ordered series of cleavages by the HIV protease (53). Typical transition state analogs are effective protease inhibitors. Maturation inhibitors, however, bind Gag, not the protease. Maturation inhibitors like the betulinic acid derivative Beviramat (BVM) or PF46396 specifically inhibit cleavage of the CA-SP1 peptide bond (58). Normal HIV assembly is sensitive to mutations near the CA-SP1 junction (59), implying that the mobility and structure(s) of this junction are restricted (60, 61). Remarkably, some PF46396-resistant mutants are drug-dependent for normal assembly (58, 62). The structure of a CA-CTD-SP1 hexamer with a bound BVM was determined to 2.9Å using micro-electron diffraction (Figure 3) (••63). The proteins are arranged with approximate 6-fold symmetry with a single BVM filling a pore along the oligomer axis. It is presumed that PF46396 binds near or at the BVM site (58). The CA-SP1 junction is alpha helical and located near the hexamer axis; its conformation is unlike the extended conformation of a CA-SP1 peptide bound to the protease, indicating that the sequence is cleaved in an unfolded state (64). Because BVM can support Gag assembly and stabilize Gag polymers (65), we can deduce that the small molecule is stabilizing the helical conformation. Of course, it may also act to block protease access to the scissile bond. Without proteolytic cleavage, the Gag polymer remains trapped as a low energy intermediate in HIV assembly. Maturation inhibitors buttress this stable state.
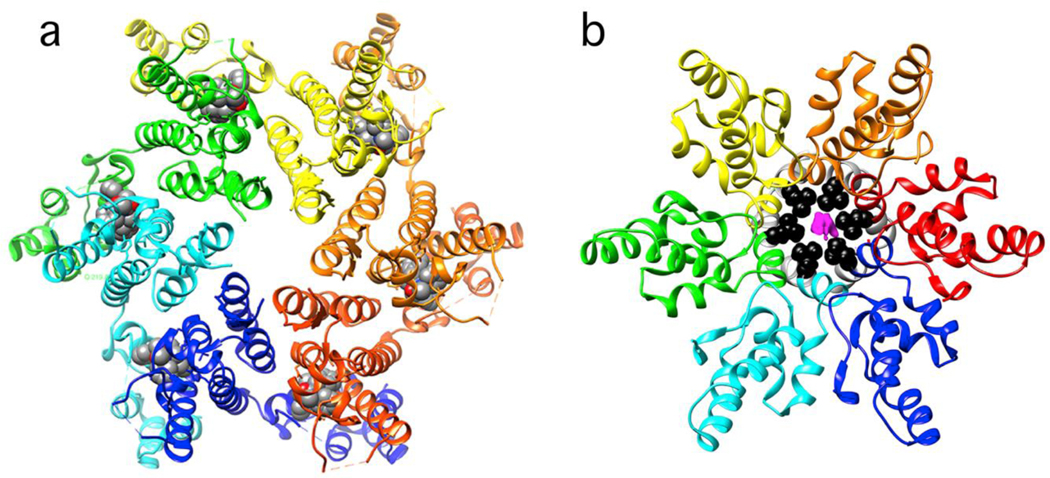
Figure 3. The HIV CA domain with hexamer-stabilizing small molecules.
(a) A ribbon diagram of a top view of a mature CA hexamer with bound P74 (PDB: 4u0e). The P74 molecule (space filling with carbons in gray) fits into a pocket largely formed by the NTD (top) and completed by the CTD from the neighboring subunit (CTDs are partially obscured by the NTD). (b) A CTD-SP1 construct representing a fragment of immature Gag with bound bevirimat (PDB: 6n3u). The CTD is on top. The last residue in the CTD, L363 (black), and SP1 (gray) are partially obscured. Electron density for bevirimat (magenta), contoured at 1.1 σ, fills a cylindrical gap in the ring of helices, presumably stabilizing them and blocking accessibility to protease.
At the other end of the viral lifecycle, uncoating is temporally linked to the processes of reverse transcription and nuclear entry, making it a critical step in initiating infection (53). Uncoating inhibitors stabilize interactions between CA subunits. The molecule PF74 is a much-studied tool compound that informs us of the mechanisms of more recently developed molecules (•66). Like DBT1 with HBV, PF74 has the effect of inducing dissolution of HIV capsid complexes (67). In the simplest case, from a thermodynamic perspective, this would require PF74 bind CA monomer more tightly than capsid: PF74 binds CA monomer with a KD of ~4000 nM and hexamers with a KD of 262nM, indicating that it stabilizes CA polymers (••68). Historically, efforts to identify molecules that inhibit assembly, binding monomer more tightly than oligomer, has led to candidates that only worked at very high concentrations and were, in the end, unsuccessful (69, 70). Structurally, it makes sense that PF74 binds more tightly to oligomer: PF74 binds an exposed pocket in the CA-NTD that is partially capped by the CTD from an adjacent monomer (••68) (Figure 3). The site is apparently modified by formation of a complex of hexamers: PF74 stabilizes cylinders of CA but induces dissociation of isolated viral cores. By analogy to studies with HBV and DBT1, we speculate that PF74 may stabilize local hexagonal conformations that lead to ruptures in an irregular conical HIV capsid. Global strain arises because virus capsids are an oligomeric network, where a conformational change at one point must be accommodated throughout the shell.
Conclusion
Capsid-directed antivirals remain in the early stages of development and will require tailoring the medicinal chemistry to each specific protein target. Nevertheless, core principles can be identified which generalize across viruses and across specific chemistries. Capsid-directed antivirals strengthen the association energy between subunits with the following implications:
assembly-directed drugs bind to oligomeric complexes tighter than to single subunits
dissociation of subunits becomes locally unfavorable
the rate and extent of assembly increases
free subunit is depleted
incomplete and defective particles are kinetically trapped
protein dynamics are modulated
In many cases, structural protein-directed molecules modify the preferred orientation of adjacent subunits:
assembly reactions can proceed off-path
reaction products include heterogenous mis-assemblies
pre-formed capsids experience a global strain
strained capsids may rupture
By preferentially binding subunit oligomer, assembly reactions are driven forward. A hypothetical capsid assembly inhibitor would have a much more demanding task: binding protein subunit very tightly, and with nearly equivalent stoichiometry. In contrast, a modest increase of the subunit interaction energy can be leveraged into a dramatic change in the reaction products, as seen with CpAMs driving HBV assembly to yield empty and deformed capsids. The proposed mechanism of picornavirus uncoating inhibitors and HIV maturation inhibitors also share this theme: picornavirus capsids are over-stabilized, preventing uncoating; HIV CA hexamers are over-stabilized at the expense of Gag cleavage and subsequent maturation.
A further consequence of using small molecules to strengthen the interface between two protein subunits is that the relative orientations of subunits are likely to be modified. This could be as simple as extra atoms forming a wedge, or it could be a more dynamic or allosteric shift in the conformational ensemble between two subunits. Finally, a combination of the two effects (increased local association energy and modified preferred orientation) can lead to circumstances where a capsid becomes globally strained and ruptures, as observed with DBT1 in HBV and PF74 in HIV.
Antiviral molecules directed against structural proteins facilitate action at a distance. They are assembly agonists, maturation inhibitors, modulators of capsid dynamics, inhibitors of nucleic acid release, and destabilizers of large complexes. In all cases evolution has optimized the structural protein interactions to the requirements of the viral lifecycle. Targeted modification of these interactions is disruptive to the virus. These principles provide a framework with which to approach further antiviral research.
Footnotes
Conflict of Interest
AZ acknowledges an interest in biotech companies involved in developing antivirals directed at HBV; these exercised no influence in the preparation of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- ••1. Tanner EJ, Liu HM, Oberste MS, Pallansch M, Collett MS, Kirkegaard K. 2014. Dominant drug targets suppress the emergence of antiviral resistance. Elife 3. This paper describes how viral capsids and other viral oligomers can remain susceptible to antiviral agents even when selective pressure leads to the appearance of resistant mutants. The presence of even a relatively small fraction of sensitive wild type protein can result in drug-sensitivity of the whole oligomer.
- 2.Zlotnick A. 1994. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol 241:59–67. [DOI] [PubMed] [Google Scholar]
- 3.Katen S, Zlotnick A. 2009. The thermodynamics of virus capsid assembly. Methods Enzymol 455:395–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston IG, Louis AA, Doye JP. 2010. Modelling the self-assembly of virus capsids. J Phys Condens Matter 22:104101. [DOI] [PubMed] [Google Scholar]
- ••5. Perlmutter JD, Hagan MF. 2015. Mechanisms of virus assembly. Annu Rev Phys Chem 66:217–39.25532951 This encyclopedic review describes the state of the art for modelling virus capsid assembly, touching on ordinary differential equation, Gillespie (stochastic event), and dynamic models. There are also substantive descriptions of means to analyze experimental data in light of model predictions.
- 6.Garmann RF, Comas-Garcia M, Knobler CM, Gelbart WM. 2016. Physical Principles in the SelfAssembly of a Simple Spherical Virus. Acc Chem Res 49:48–55. [DOI] [PubMed] [Google Scholar]
- 7.Zlotnick A. 2003. Are weak protein-protein interactions the general rule in capsid assembly? Virology 315:269–74. [DOI] [PubMed] [Google Scholar]
- 8.Singh S, Zlotnick A. 2003. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem 278:18249–55. [DOI] [PubMed] [Google Scholar]
- 9.Hagan MF, Elrad OM, Jack RL. 2011. Mechanisms of kinetic trapping in self-assembly and phase transformation. J Chem Phys 135:104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••10. Asor R, Selzer L, Schlicksup CJ, Zhao Z, Zlotnick A, Raviv U. 2019. Assembly Reactions of Hepatitis B Capsid Protein into Capsid Nanoparticles Follow a Narrow Path through a Complex Reaction Landscape. ACS Nano 13:7610–7626.31173689 Small angle X-ray scattering is used to analyze heterogeneous mixtures of complexes from equilibrated and trapped capsid assembly reactions. A library of model intermediates was generated to fit data to a sum of calculated scattering. Production of this library led to an estimate of 10^30 possible intermediates in the assembly of a 120-piece HBV capsid.
- 11.Garmann RF, Comas-Garcia M, Koay MS, Cornelissen JJ, Knobler CM, Gelbart WM. 2014. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J Virol 88:10472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagan MF, Zandi R. 2016. Recent advances in coarse-grained modeling of virus assembly. Curr Opin Virol 18:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykeman EC, Stockley PG, Twarock R. 2014. Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci U S A 111:5361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••14. Twarock R, Stockley PG. 2019. RNA-Mediated Virus Assembly: Mechanisms and Consequences for Viral Evolution and Therapy. Annu Rev Biophys 48:495–514.30951648 The role of specifically packaged RNA in stimulating and regulating assembly is described from the perspectives of experimental observation and a theoretical model based on possible arrays of “packaging sites” embedded in the RNA sequence.
- 15.Veesler D, Johnson JE. 2012. Virus maturation. Annu Rev Biophys 41:473–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Wang JC, Taylor MW, Zlotnick A. 2012. In vitro assembly of an empty picornavirus capsid follows a dodecahedral path. Journal of Virology 86:13062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero JR. 2001. Pleconaril: a novel antipicornaviral drug. Expert Opin Investig Drugs 10:369–79. [DOI] [PubMed] [Google Scholar]
- 19.Egorova A, Ekins S, Schmidtke M, Makarov V. 2019. Back to the future: Advances in development of broad-spectrum capsid-binding inhibitors of enteroviruses. Eur J Med Chem 178:606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20. Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286–93.3018924 A classic paper. The first demonstration of an antivral agent bound to viral structural proteins
- ••21. Abdelnabi R, Geraets JA, Ma Y, Mirabelli C, Flatt JW, Domanska A, Delang L, Jochmans D, Kumar TA, Jayaprakash V, Sinha BN, Leyssen P, Butcher SJ, Neyts J. 2019. A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses. PLoS Biol 17:e3000281. There are numerous examples now of drugs bound to the classic site described in reference 20. Here is a wholly new site with untapped potential to be a generalized target for enterovirus-directed antivirals.
- 22.Tsang SK, Danthi P, Chow M, Hogle JM. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J Mol Biol 296:335–40. [DOI] [PubMed] [Google Scholar]
- 23.Phelps DK, Post CB. 1995. A novel basis of capsid stabilization by antiviral compounds. J Mol Biol 254:544–51. [DOI] [PubMed] [Google Scholar]
- 24.Lewis JK, Bothner B, Smith TJ, Siuzdak G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci U S A 95:6774–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakeel S, Westerhuis BM, Domanska A, Koning RI, Matadeen R, Koster AJ, Bakker AQ, Beaumont T, Wolthers KC, Butcher SJ. 2016. Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis. Nat Commun 7:11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graber-Stiehl I. 2018. The silent epidemic killing more people than HIV, malaria or TB. Nature 564:24–26. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Liu K. 2017. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhason MS, Wang JC, Hagan MF, Zlotnick A. 2012. Differential assembly of Hepatitis B Virus core protein on single- and double-stranded nucleic acid suggest the dsDNA-filled core is springloaded. Virology 430:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng S, Gao L, Han X, Hu T, Hu Y, Liu H, Thomas AW, Yan Z, Yang S, Young JAT, Yun H, Zhu W, Shen HC. 2018. Discovery of Small Molecule Therapeutics for Treatment of Chronic HBV Infection. ACS Infect Dis 4:257–277. [DOI] [PubMed] [Google Scholar]
- 31.King RW, Ladner SK, Miller TJ, Zaifert K, Perni RB, Conway SC, Otto MJ. 1998. Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (−)beta-L-2’,3’-dideoxy-3’-thiacytidine. Antimicrob Agents Chemother 42:317986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaney WEt, Edwards R, Colledge D, Shaw T, Furman P, Painter G, Locarnini S. 2002. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrob Agents Chemother 46:305760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res 76:168–77. [DOI] [PubMed] [Google Scholar]
- 34.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893–6. [DOI] [PubMed] [Google Scholar]
- 35.Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2010. Trapping of Hepatitis B Virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 5:1125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hacker HJ, Deres K, Mildenberger M, Schroder CH. 2003. Antivirals interacting with hepatitis B virus core protein and core mutations may misdirect capsid assembly in a similar fashion. Biochem Pharmacol 66:2273–9. [DOI] [PubMed] [Google Scholar]
- 38.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. 2005. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A 102:8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Chirapu SR, Finn MG, Zlotnick A. 2013. Phase Diagrams Map the Properties of Antiviral Agents Directed against Hepatitis B Virus Core Assembly. Antimicrob Agents Chemother 57:1505–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •40. Kondylis P, Schlicksup CJ, Brunk NE, Zhou J, Zlotnick A, Jacobson SC. 2018. Competition between normative and drug-induced virus self-assembly observed with single-particle methods. JACS 141:1251–1260. HBV assembly in vivo is normally nucleated by viral RNA-reverse transcriptase or is spontaneous. This paper describes conditions where an assembly agonist competes with apontaneous nucleation.
- 41.Liu C, Fan G, Wang Z, Chen HS, Yin CC. 2017. Allosteric conformational changes of human HBV core protein transform its assembly. Sci Rep 7:1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlicksup CJ, Wang JC-Y, Francis S, Venkatakrishnan B, Turner WW, VanNieuwenhze M, Zlotnick A. 2018. Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. eLife 7:e31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourne CR, Finn MG, Zlotnick A. 2006. Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol 80:11055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatakrishnan B, Katen SP, Francis S, Chirapu S, Finn MG, Zlotnick A. 2016. Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket. J Virol 90:3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2013. Assembly-directed antivirals differentially bind quasi-equivalent pockets to modify Hepatitis B Virus capsid tertiary and quaternary structure. Structure 21:1406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packianathan C, Katen SP, Dann CE 3rd, Zlotnick A. 2010. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol 84:1607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, Espiritu C, Baydo R, Atkins K, Abendroth J, Liao G, Efimov A, Hartman G, Flores OA. 2015. High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci U S A 112:15196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu Z, Lin X, Zhou M, Liu Y, Zhu W, Chen W, Zhang W, Guo L, Liu H, Wu G, Huang M, Jiang M, Xu Z, Zhou Z, Qin N, Ren S, Qiu H, Zhong S, Zhang Y, Zhang Y, Wu X, Shi L, Shen F, Mao Y, Zhou X, Yang W, Wu JZ, Yang G, Mayweg AV, Shen HC, Tang G. 2016. Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors. J Med Chem 59:7651–66. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Z, Hu T, Zhou X, Wildum S, Garcia-Alcalde F, Xu Z, Wu D, Mao Y, Tian X, Zhou Y, Shen F, Zhang Z, Tang G, Najera I, Yang G, Shen HC, Young JA, Qin N. 2017. Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms. Sci Rep 7:42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang J-A, Kim S, Park M, Park H-J, Kim J-H, Park S, Hwang J-R, Kim Y-C, Kim YJ, Cho Y. 2019. Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly. Nature Commun 10:2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 2017. Capsid Assembly Modulators Have a Dual Mechanism of Action in Primary Human Hepatocytes Infected with Hepatitis B Virus. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••52. Schlicksup CJ, Laughlin P, Dunkelbarger S, Wang JC- Y, Zlotnick A. 2020. Local stabilization of subunit-subunit contacts causes global destabilization of Hepatitis B virus capsid. ACS Chem Biol 15:1708–17.32369333 Cryo-EM image reconstruction shows a small molecule binding to a local interface that distorts local geometry. This defect in capsid structure is proposed to propagate, leading to capsid rupture and ultimately dissociation.
- 53.Lee SK, Potempa M, Swanstrom R. 2012. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem 287:40867–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 517:505–8. [DOI] [PubMed] [Google Scholar]
- 56.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. 2013. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497:643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •57. Wang M, Quinn CM, Perilla JR, Zhang H, Shirra R Jr., Hou G, Byeon IJ, Suiter CL, Ablan S, Urano E, Nitz TJ, Aiken C, Freed EO, Zhang P, Schulten K, Gronenborn AM, Polenova T. 2017. Quenching protein dynamics interferes with HIV capsid maturation. Nat Commun 8:1779.29176596 A combination of solution results and state of the art molecular dynamics defines a mechanism for inhibiting virus maturation by stabilizing the immature state.
- 58.Ghimire D, Timilsina U, Srivastava TP, Gaur R. 2017. Insights into the activity of maturation inhibitor PF-46396 on HIV-1 clade C. Sci Rep 7:43711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datta SA, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. 2011. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J Virol 85:4111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Krausslich HG, Briggs JA. 2016. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353:506–8. [DOI] [PubMed] [Google Scholar]
- 61.Datta SA, Clark PK, Fan L, Ma B, Harvin DP, Sowder RC 2nd, Nussinov R, Wang YX, Rein A. 2016. Dimerization of the SP1 Region of HIV-1 Gag Induces a Helical Conformation and Association into Helical Bundles: Implications for Particle Assembly. J Virol 90:1773–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murgatroyd C, Pirrie L, Tran F, Smith TK, Westwood NJ, Adamson CS. 2016. Structure-Activity Relationships of the Human Immunodeficiency Virus Type 1 Maturation Inhibitor PF-46396. J Virol 90:8181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••63. Purdy MD, Shi D, Chrustowicz J, Hattne J, Gonen T, Yeager M. 2018. MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc Natl Acad Sci U S A 115:13258–13263.30530702 An elegant use of very small crystals and electron diffraction, along with clever biochemistry, to obtain the structure of a maturation inhibitor bound to a hexameric ring. The inhibitor presumably stabilizes the cleavage site from the partial unfolding needed to bind the protease.
- 64.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. 2002. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369–81. [DOI] [PubMed] [Google Scholar]
- 65.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •66.McArthur C, Gallazzi F, Quinn TP, Singh K. 2019. HIV Capsid Inhibitors Beyond PF74. Diseases 7. A well done review of PF74 and variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol 85:542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••68. Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. 2014. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A 111:18625–30.25518861 A crystal structure of a HIV CA hexamer shows how PF74 binds at the interface of the NTD of one subunit and the CTD of its neighbor. This structure explains how the small molecule can inhibit dissociation.
- 69.Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, Sehr P, Lewis J, Rey FA, Krausslich HG. 2008. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem 283:32024–33. [DOI] [PubMed] [Google Scholar]
- 70.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. 2005. The HIV-1 capsid protein Cterminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol 12:678–82. [DOI] [PubMed] [Google Scholar]