Abstract
During the hepatitis B virus lifecycle, 120 copies of homodimeric capsid protein assemble around a copy of reverse transcriptase and viral RNA and go on to produce an infectious virion. Assembly needs to be tightly regulated by protein conformational change to ensure symmetry, fidelity and reproducibility. Here we show that structures at the intradimer interface regulate conformational changes at the distal interdimer interface and so regulate assembly. A pair of interacting charged residues, D78 from each monomer, conspicuously located at the top of a four-helix bundle that forms the intradimer interface, were mutated to serine to disrupt communication between the two monomers. The mutation slowed assembly and destabilized dimer to thermal and chemical denaturation. Mutant dimers showed evidence of transient partial unfolding based on appearance of new proteolytically-sensitive sites. Though mutant dimer was less stable, the resulting capsids were as stable as wildtype, based on assembly and thermal denaturation studies. Cryo-EM image reconstructions of capsid indicated that the subunits adopted an “open” state more usually associated with free dimer and that the spike tips were either disordered or highly flexible. Molecular dynamics simulations provide mechanistic explanations for these results, suggesting that D78 stabilizes helix 4a, which forms part of the intradimer interface, by capping its N-terminus and hydrogen-bonding to nearby residues, whereas the D78S mutation disrupts these interactions, leading to partial unwinding of helix 4a. This in turn weakens the connection from helix 4 and the intradimer interface to helix 5, which forms the interdimer interface.
Graphical Abstract
Introduction
Virus capsids are protein complexes that contain and protect the viral genome. Typically, capsids are built from hundreds of identical subunits with ordered symmetries [1, 2]. For about half of known virus families, capsids have icosahedral symmetry. Assembly of subunits into a symmetrical capsids usually proceeds with high fidelity both in vivo for virus reproduction and in vitro for capsid assembly with purified subunits [3–8]. We propose that each subunit undergoes programmed changes to accommodate close packing of subunits during assembly and that the overall assembly process must be tightly regulated.
The core protein of Hepatitis B virus (HBV), which causes chronic infection in more than 250 million people [9], is a well-established model for studying the regulation of capsid assembly. In the HBV viral lifecycle, 120 copies of core protein (Cp) dimer assemble an icosahedral T=4 capsid while packaging a copy of viral RNA and reverse transcriptase to form an immature core [10]; within this core, the reverse transcriptase synthesizes the circular dsDNA of the mature infectious virion. In addition to infectious nucleic acid-filled particles, about 90% of HBV capsids in vivo are empty [11], indicating that nucleation may be spontaneous and that assembly does not require nucleic acid. HBV Cp can also form T=3 capsids with 90 dimers per capsid [12]. There is a translational interest in HBV assembly, as assembly agonists have been developed that can precipitously reduce virus production; some of these agonists are now in clinical trials.
The full length Cp has 183 amino acids and can be divided into the assembly domain (residues 1–149, also known as Cp149) and the nucleic acid binding domain (residues 150–183) [13]. HBV capsid assembly has been investigated intensively in vitro [14]. Purified Cp149 dimers can self-assemble into capsids in response to ionic strength, and those assembled capsids look indistinguishable compared to capsids isolated from cell culture [15, 16]. Simulations, bulk experiments (SAXS, light scattering, and size exclusion chromatography), and single molecule experiments (charge-detection mass spectrometry and resistive-pulse sensing) reveal that the self-assembly process has sigmoidal assembly kinetics [17–22]. During the lag phase, a steady state pipeline of intermediates is established; after the lag, capsids accumulate rapidly until the reaction equilibrates [18, 19]. Subunits make weak dimer-dimer interactions, approximately −3 to −5 kcal/mol per contact; these weak interactions can hold a capsid together because each Cp dimer is tetravalent [23, 24]. During assembly, weak interactions minimize the possibility of mis-assembly by allowing poorly bound subunits to dissociate and re-assemble correctly.
Structures of Cp149 capsid and dimer have shown systematic modulation of intradimer and interdimer interfaces [25, 26]. Comparison of dimer and capsid suggest how changes at the intradimer interface propagate toward the interdimer interface, affecting assembly pathways and products. For example, naturally occurred mutations, F97L or I97L, located at the intradimer interface, led to secretion of immature virions in cell cultures [27] and more aggressive assembly in vitro [28]. It was suggested that such mutation increases the flexibility of the intradimer interface [28]. Conversely, crosslinking the intradimer interface by a disulfide restrains the movement of the intradimer interface and results in slower assembly and less stable capsids [29]. These results suggest an allosteric connection between the intradimer and interdimer interfaces.
In this study, we investigated the regulatory function of the intradimer interface, focusing on a pair of aspartates, D78 from each monomer in the dimer, that are in surprisingly close proximity at the intradimer spike tip (Figure 1). Our initial hypothesis was that removing the potentially repulsive D78-D78 interaction would favor a more assembly-active conformation for Cp149 dimers, leading to enhanced capsid assembly. However, Cp149-D78S led to slower assembly with no change in capsid stability, destabilization of dimer structure, and partial disruption of a pair of helices contributing to the dimer interface. In summary, our results suggest that integrated secondary and quaternary structures of the intradimer interface are essential to regulated assembly to produce correct capsids.
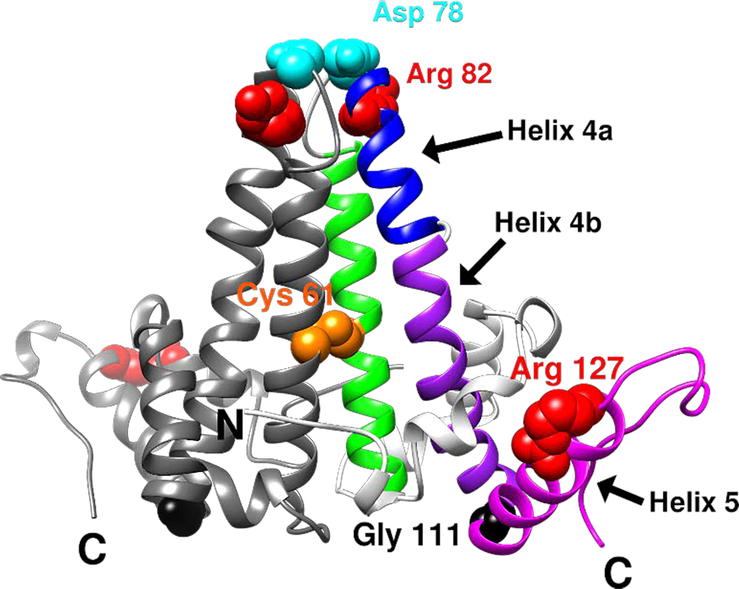
Figure 1.
Ribbon representation of an HBV Cp149 dimer (PDB: 1QGT) and key residues discussed in this study. One half-dimer is shown in gray and the other is divided into different regions by colors: helix 4 is divided into helix 4a (blue) and 4b (purple). The interdimer interface is formed by interactions between helix 5 (magenta) from one dimer and the loop at the end of helix 5 from a neighboring dimer. Key residues are shown in space-filled representation: Cys61 in orange, Asp78 in cyan, Gly111 in black, Arg82 and Arg127 in red.
Results
D78S mutation slows down capsid assembly under low ionic strength.
Despite its position distal to the interdimer interface, D78 is part of helix 4, which is linked by G111 to helix 5, which forms the interdimer interface. Thus, movement of helix 4 could directly impact the adjustment of helix 5 in assembly. We hypothesized that the conserved D78, and the predicted D78-D78 repulsion at the intradimer interface, have regulatory functions in capsid assembly.
Structural comparison suggests that the spike tip of Cp149 exhibits a more open intradimer interface as free dimer [26] than when bound in a capsid [25]. It was hypothesized that conformational changes can allosterically regulate capsid assembly. Interference with conformational changes would affect the assembly process. Selzer et al. showed that modification of the intradimer interface of Cp149 dimer, specifically via a C61-C61 crosslink, can limit conformational changes and impede assembly [29]. To further investigate the regulatory function of the intradimer interface, we focused on the D78-D78 residue pair, which may include a repulsive interaction that relates to openness of the dimer spike tip (Figure 1). D78, located at the N-terminal end of helix 4a at the intradimer interface, is conserved in human HBV genotypes (A-D and F-H) [29]. Mutation of D78 inhibited packing of the genome and formation of capsids [30, 31], suggesting the importance of D78 or the D78-D78 interaction.
To test our hypothesis, D78 was mutated to serine, replacing the charged aspartate with a similarly sized polar sidechain. We then studied whether the D78S mutation affected capsid assembly in vitro. First, we compared the assembly kinetics of Cp149 and mutant Cp149-D78S in response to increasing concentrations of NaCl (Figure 2a, b) using 90° light scattering, in which capsid assembly is proportional to the increase in light scatter. At relatively low ionic strength (200 mM and 300 mM NaCl), Cp149-D78S assembly (Figure 2b) was much slower than Cp149 (Figure 2a). However, the difference became negligible at high ionic strength (500 mM and 750 mM NaCl). This result was contrary to expectations based on our initial hypothesis: eliminating D78-D78 electrostatic repulsion was expected to decrease sensitivity of assembly to ionic strength.
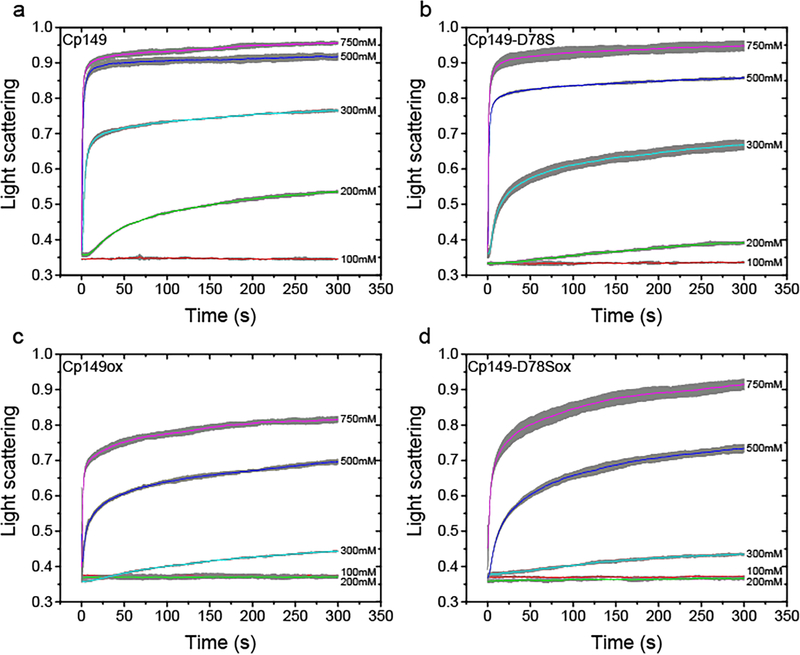
Figure 2.
Light scattering of assembly shows that D78S mutation and dimer oxidation impede capsid assembly. (a and b) Reduced Cp149 and Cp149-D78S dimers assemble with different kinetics in an ionic strength dependent manner. Under lower ionic strength (200 and 300 mM NaCl), reduced Cp149-D78S assembles slower than reduced Cp149. Higher ionic strength (500 mM and 750 mM NaCl) minimizes the assembly differences. (c and d) When dimers are oxidized by C61-C61 disulfides crosslinking the four helix bundles, there is little difference between the assembly of Cp149 and Cp149-D78S in all ionic strengths tested. The averaged signal of three repeats are shown as colored lines with error bars in grey.
The sigmoidal kinetics of HBV capsid assembly provides additional details of assembly energetics [18, 19, 32]. As we are examining a bulk reaction with many nuclei, the lag phase is proportional to the time of formation of a steady state of intermediates for capsid completion [19], which can be used to estimate the average rate for addition of each subunit to the growing capsid, the elongation rate [33]. The lag phase of each reaction was determined by extrapolating the steepest part of the assembly curve to the baseline using a linear regression, as described by Hagan et al [33]. We observed that Cp149-D78S lag phase was three times longer than the lag for Cp149 under 200 mM NaCl (Figures 2a, b and Figure S1). The difference became smaller as ionic strength increased. These results indicate that Cp149-D78S must overcome a higher energetic barrier to participate in assembly.
C61-C61 crosslink in the intradimer interface silences the D78S mutation.
To test whether the D78S mutation modulated the intradimer interface, we forced both Cp149 and Cp149-D78S into a similar intradimer conformation via the C61-C61 crosslink (Figure 1). Under the same assembly conditions, crosslinked Cp149 and Cp149-D78S (Cp149ox, Cp149-D78Sox) assembled much slower than reduced Cp149 and Cp149-D78S (compare Figures 2a and c or b and d), as previously observed [29]. However, despite the distinct differences between their reduced states (Figures 2a, b), Cp149ox and Cp149-D78Sox showed similar assembly kinetics (Figures 2c, d). Cp149ox and Cp149-D78Sox had similar lag times and thus assembly rates at 500 mM and 750 mM NaCl (Figure S1). These results imply the regulatory function of D78 through the intradimer interface.
D78S does not change capsid stability but does lead to a preference for T=3 capsids.
The D78S mutation affected assembly kinetics. We anticipated it would also change assembly thermodynamics. From SEC experiments, we determined the pseudo-critical concentrations of Cp149 to be 2.83 μM and Cp149-D78S to be 3.54 μM (Figure S2). The corresponding interdimer interaction energy per contact, ΔGcontact, was calculated to be −3.56 kcal/mol for Cp149 and −3.50 kcal/mol for Cp149-D78S [23]. Thus, the D78S mutation minimally affects the dimer-dimer association energy, despite the obvious effect on assembly kinetics.
Of note, the elution peaks of Cp149 and Cp149-D78S capsids are distinctly different (Figure S3a). D78S assembly has a broader capsid peak with a shoulder at 10 mL, indicating smaller assembly products, possibly T=3 capsids (Figure S3a, red line). To investigate this difference, both assembly reactions were resolved using linear sucrose gradients [34]. We observed that Cp149-D78S showed more of a distinct slow-sedimenting band than Cp149 at the position expected for T=3 capsids (Figure S3b). Both bands of the Cp149-D78S assembly were extracted and examined by negative stain electron microscopy. Micrographs for the upper band were dominated by smaller, 32 nm particles, while 36 nm T=4 capsids were the dominant species in the lower band (Figure S4). Thus, Cp149-D78S favors a larger proportion of T=3 capsids. Oxidized dimers also assemble more T=3 capsids [29]. Both results show that modification of the intradimer interface affects the geometry of the assembly products.
D78S mutation decreases Cp149-D78S dimer stability.
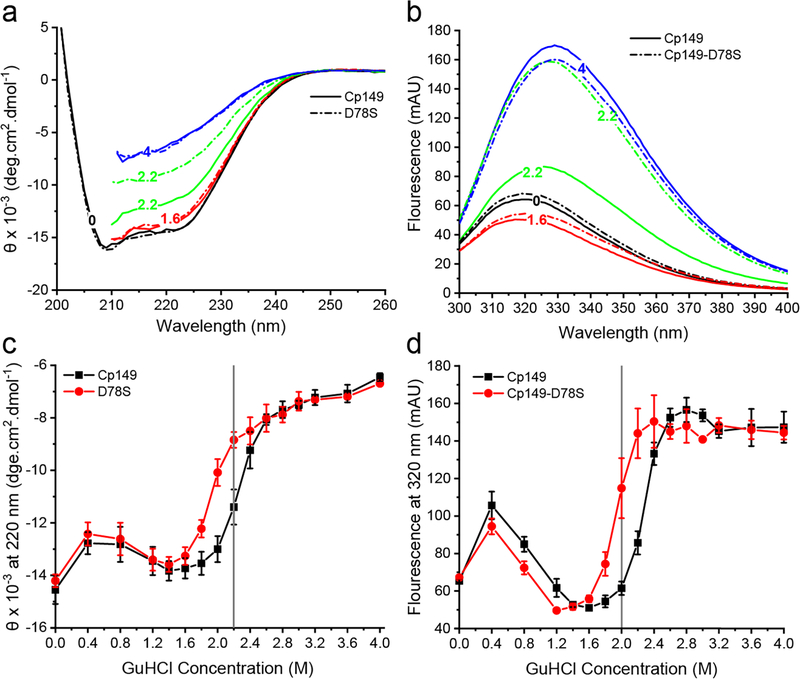
Cp149-D78S dimers assembled slower than Cp149 dimers, possibly because the D78S mutation alters dimer structure in a way that hinders conformational adjustments during assembly. To test this hypothesis, we examined the stability of secondary structure of Cp149 and Cp149-D78S dimers to GuHCl. HBV Cp149 dimers are helix-rich [25]. As shown in Figure 3a, Cp149 and Cp149-D78S showed similar circular dichroism (CD) spectra at low concentrations of GuHCl (0–1.6 M), typical for helix-rich structures. With increasing concentrations of GuHCl, spectra for both dimers indicated increasing random coil (Figure 3a). Plotting the CD amplitude at 220 nm against the GuHCl concentrations revealed that Cp149-D78S denatured earlier than Cp149 (Figure 3c). The same samples were examined for changes in tryptophan fluorescence emission. Denaturation of Cp149 dimers leads to a red shift in the emission spectrum [35]. Similar to the CD results, Cp149 and Cp149-D78S maintained their structures below 1.6 M GuHCl (Figure 3b); a small increase in fluorescence at 0.4 M GuHCl corresponds to ionic strength-induced assembly. Cp149-D78S denatured earlier at lower GuHCl (Figure 3d). These results show that although the D78S mutation does not affect the native helical structure, it decreases stability to chemical denaturation.
Figure 3.
The D78S mutation reduces dimer stability to GuHCl denaturation. (a) Circular Dichroism (CD) and (b) tryptophan fluorescence emission of Cp149 (solid line) and Cp149-D78S (dashed line) show dimer unfolding in GuHCl. (c) CD amplitude at 220 nm and (d) fluorescence at 320 nm plotted against GuHCl concentration show that Cp149-D78S (red) has a lower GuHCl threshold for denaturation than Cp149 (black).
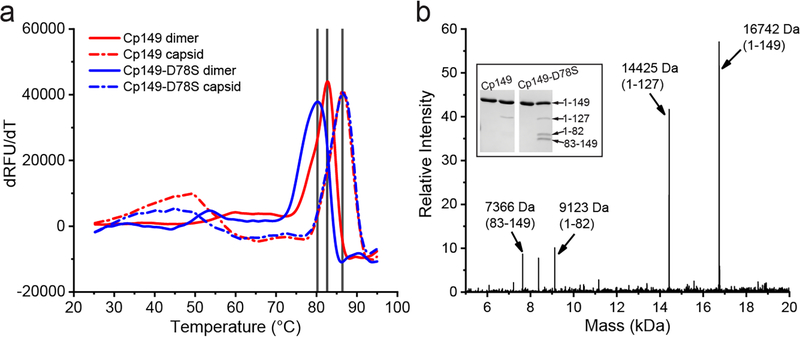
We further examined the stability of Cp149 and Cp140-D78S in dimer and capsid states using a protein thermal shift assay. As shown in Figure 4a, Cp149 had a denaturation Tm around 82.7°C (solid red). With the D78S mutation, the Tm dropped to 80°C (solid blue), consistent with decreased structural stability to GuHCl denaturation. In contrast to dimers, both capsids (dash lines) showed similar denaturation Tm values of 86°C, indicating that structural disruption arising from the D78S mutation is compensated for by the stability of capsid quaternary structure.
Figure 4.
The Cp149-D78S mutant is less resistant than Cp149 to thermal denaturation and proteolytic attack in the absence of denaturants. (a) Protein thermal shift assay shows that Cp149-D78S dimer (blue solid line) has a lower melting temperature (Tm) than Cp149 dimer (red solid line); while Cp149-D78S and Cp149 capsids have the same Tm (dashed lines), suggesting capsid quaternary structure overcomes the destabilization caused by the D78S mutation. (b) The Cp149-D78S mutation introduces a new trypsinolysis site, R82, presumably by destabilizing the α-helix near the spike tips (Inset). Mass spectrometry analysis of the trypsin-treated Cp149-D78S identifies the digestion site.
To pinpoint local changes in structure and/or stability, dimers were examined for sensitivity to proteolysis. In Cp149, Arg127 is essentially the only residue cleaved by trypsin [36]. As shown by SDS-PAGE, a band corresponding to residues 1–127 is seen after mild trypsin treatment. Under the same conditions, trypsin treatment of Cp149-D78S led to two new bands (Figure 4b) identified by LC-MS as evidence of cleavage at a new site, Arg82 (Figure 1). This result suggests that Arg82, protected by the secondary structure of helix 4a in Cp149, is at least temporally unfolded and accessible to trypsin cleavage.
D78S changes the conformation of spike tips on the capsid.
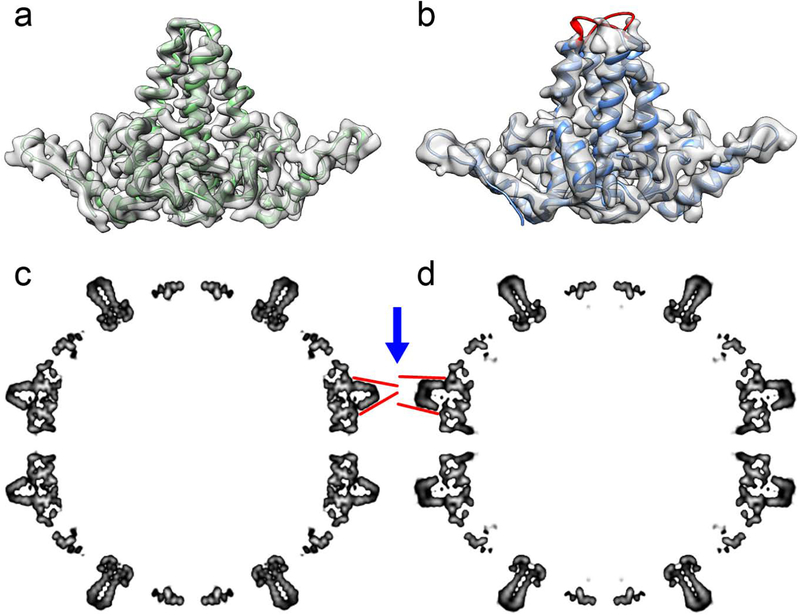
To visualize structural changes in Cp149 and Cp149-D78S, their T=4 capsid structures were determined by Cryo-EM reconstruction. The D78S capsid was reconstructed to 4.6 Å resolution, and Cp149 capsid to 3.6 Å using RELION 3.0 [37] (Table S1). The interdimer interface of both capsids showed no obvious differences, consistent with the similarity of the dimer contact energies. However, there are clear differences between the spike tips (Figure 5a, b). The Cp149 dimer map has continuous density covering AB and CD dimers at a 2σ contour level that allow reliable building of a complete molecular model (Figure 5a). However, the Cp149-D78S density map is disordered around the spike tips, specifically for residues 77–81 (Figure 5b), suggesting that this region of Cp149-D78S capsids is very flexible. To identify major structural differences between capsids, we compared the central sections of their density maps after low-pass filtering to 8 Å to exclude unnecessary details (Figure 5c, d). This view shows how the AB dimers (blue arrow) in Cp149 and Cp149-D78S differ in their conformations. In Cp149-D78S, the two copies of helix 4 that make up the intradimer interface are more nearly parallel than in wildtype Cp149. Helix 3 pairs from CD dimers, also in plane in Figure 5, are nearly identical in Cp149 and C149-D78S. Helix 4 from C149-D78S CD dimers is similar in conformation to helix 4 from the AB dimers, but cannot be seen in the thin plane of density shown in Figure 5.
Figure 5.
Cryo-EM analysis of Cp149 and Cp149-D78S capsids. The density maps of AB dimers isolated from Cp149 (a) and Cp149-D78S (b) capsids compared with determined dimer models. (a) The AB dimer model fits well to the Cp149 density map. (b) The D78S model captures most of the dimer structure, with missing density for the spike tips (labelled in red). (c, d) The central sections of the low-pass filtered capsid maps, highlighting the AB dimers. The spikes of AB dimers in the Cp149-D78S capsid (d) have a wide-open conformation compared to the equivalent spikes in the wild type Cp149 capsid map (c). The red lines accentuate the differences in the spikes. Similar differences are present in CD dimers, but they are not evident in this view.
D78S impairs refolding of helix 4a and diminishes sodium affinity.
Cryo-EM reconstruction indicates that the C149-D78S spike tips are extremely flexible; biochemical experiments indicate that the effects of the mutation on assembly are attenuated at high ionic strength. To investigate structural changes induced in the spike tip by D78S, all-atom molecular dynamics simulations were performed for both Cp149 and Cp149-D78S dimers under low (150 mM NaCl) and high (750 mM NaCl) ionic strength conditions using NAMD 2.13 [38] (Table S2). Systems were sampled for 500 ns. Both fully helical and partially helical states of helix 4a were represented in the initial models used for simulations (Figure 6a).
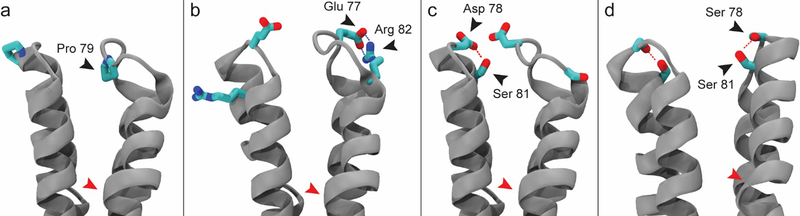
Figure 6.
Molecular dynamics simulations reveal variations in secondary structure and stabilizing interactions in dimer spike tips. (a) The left half-dimer exhibits the fully helical spike tip conformation. The right half-dimer exhibits the partially helical state, where Pro79 to Arg82 of helix 4a are unfolded. (b) A salt bridge between Glu77 and Asp82 can stabilize the partial helical state and impair the ability of helix 4a to recover. (c) Asp78 stabilizes helix 4a by capping its N-terminus and by forming a strong hydrogen bond with Ser81. (d) Ser78 can fulfill the role of N-cap but does not consistently hydrogen bond with Ser81. Hydrogen atoms omitted for clarity. Location of the spike hinge indicated by red arrow.
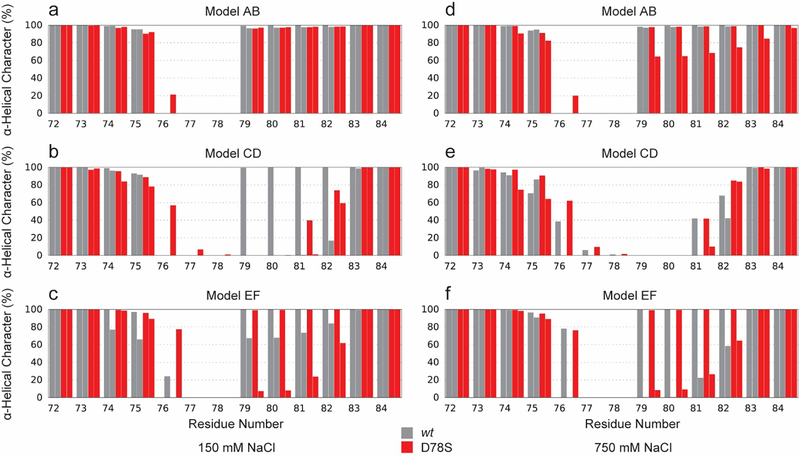
For Cp149 at low ionic strength, residues 79–82 of helix 4a either remained helical or refolded to be helical in all cases, except one (Figure S5a, blue). In the single case where recovery did not occur, a salt bridge was observed between Glu77 and Arg82 (Figure 6b and S5a, red), which impaired recovery of the helix by stabilizing the partially helical state. This salt bridge had not been previously suspected based on experimental structures. For Cp149-D78S at low ionic strength, and both Cp149 and Cp149-D78S at high ionic strength, models initiated with helix 4a in a partially helical state never successfully recovered secondary structure even in the absence of the Glu77-Arg82 salt bridge (Figure S5b–d). With the combination of the D78S mutation and high ionic strength, one initially helical spike tip was seen to unfold to adopt a partially helical state (Figure S5d). Increased concentration of salt can play a role in disrupting intramolecular hydrogen bonds. Average helicity for all spike tip residues observed during simulations is given in Figure 7.
Figure 7.
Average helicity of spike tip residues over 500 ns of simulation. Cp149 shown in gray, Cp149-D78S shown in red. Secondary structure assigned by STRIDE [42]. Initial conformations for models AB and CD were from the respective quasi-equivalent dimers from a T=4 capsid (2G33 [43]). Model EF is from the crystal structure of an assembly incompetent dimer (3KXS [26]).
Simulation results reveal that Asp78 is more effective than Ser78 at stabilizing helix 4a. In addition to forming an anionic cap on the N-terminal end of the helix with backbone amides of Ala80 and Ser81 (the N-cap [39]), Asp78 can form a strong, consistent hydrogen bond with the Ser81 sidechain (Figure 6c and S5a, black). While Ser78 can partially fulfill the role of N-cap, an auxiliary hydrogen bond with Ser81 is only observed transiently (Figure 6d and S5b, black). Further, the negative charge of Asp78 leads to notable sodium occupancy and specific interactions with sodium ions around the spike tip, which are not observed with the D78S mutation or under conditions of high ionic strength (Figure S6 and Movie S1).
Loss of helicity in helix 4a leads to a more open conformation in the spike tip.
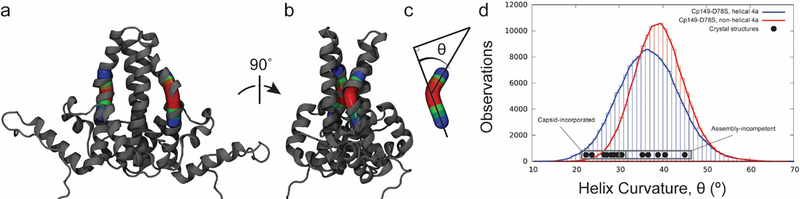
Cp149 contains a hinge region that divides the intradimer four-helix bundle into the chassis subdomain and the spike tip region [26]; glycine residues at positions 63 and 94 define the hinge (Figure 8a, b). Hinge behavior was characterized by measuring helix 4 curvature over residues 88–98, with the apex of curvature (hinge vertex) identified at Val93 (Figure 8c). MD simulations indicate that non-helical secondary structure in helix 4a shifts the distribution of hinge states to increased curvature (Figure 8d), which results in a more open conformation in the spike. For Cp149 and Cp149-D78S at 150 mM ionic strength, distributions of hinge curvature were centered at 36° for the three half-dimers that maintained helical secondary structure throughout simulation. For the three Cp149-D78S half-dimers that remained partially helical, the distribution narrowed and shifted to 39°. This shift of 3° per half-dimer corresponds to opening the spike tip by 2 Å.
Figure 8.
Helix curvature along the dimer hinge. (a-b) Hinges were abstracted using Bendix [44], fitting Bendices through residues 88–98. (c) The angle of helix curvature captures the deviation from linearity. The hinge vertex is at Val93. (d) The distribution of hinge angles is larger by 3° for half-dimers with helical (blue) versus partial helical (red) secondary structure in helix 4a. A shift of 3° per half-dimer corresponds to widening the spike tip opening by 2 Å. Curvature angles measured for dimers in crystal structures (PDB: 1QGT, 2G33, and 3KXS) are indicated (black).
Discussion
In the HBV lifecycle, capsid assembly must be regulated to prevent mis-assembly and to allow formation of RNA-filled immature virions [10]. Even so, 90% of released virions are empty capsids [11], indicating that assembly may be triggered spontaneously. Hagan et al. showed that allosteric activation of assembly and induced fit of subunits make assembly robust over a broad range of conditions [33]. Structural comparison of HBV free dimer and capsid-incorporated dimer revealed evidence of allostery and/or induced fit: structural changes of interdimer contacts and an open to closed transition of the intradimer interface during assembly [25, 26, 31]. We suggest assembly is regulated by such conformational change from ensembles of inactive states (predominantly opened conformation, e.g. Figure 8b) to ensembles of active states (predominantly closed conformation with flexibility for conformational adjustments, e.g. Figure 5a). Here, we observe that a point-mutation in the spike tip of the Cp intradimer interface inhibits assembly, which involves a completely different region of Cp. This indicates a conformational connection between two different interfaces involved in capsid assembly. Complementary to our observations, Patterson and co-workers observe that a mutation near the end of helix 5, Y132A, modulates the hydrogen bonding pattern in helix 4a, even in free dimer [40]. Because we paradoxically observe substantial structural changes and structural conservation, our data suggest flexible components with shifts in ensembles of conformations, consistent with the entropic description of allostery outlined by Hilser and co-workers [41].
The D78S mutation at the spike tip led to dimer destabilization and structural deformation. Although D78S did not affect the dimer helical structure overall, it substantially reduced dimer stability to chemical and thermal denaturation (Figure 3 and 4a). Proteolysis analysis revealed a new digestion site, Arg82, within the Cp149-D78S mutant (Figure 4b), indicating sporadic partial unfolding of helix 4a which is required for attack by trypsin [36]. Cryo-EM reconstruction of Cp149 and Cp149-D78S capsids showed that the mutant capsid has an open conformation for the intradimer interface and deteriorated density at the spike tips (Figure 5). In simulations, the D78S mutation decreased the stability of helix 4a by removing D78 as an N-cap and as a hydrogen bond acceptor (Figure 6 and S5). Such changes also appeared to decrease the interaction between the spike tip and ions that screen the charge-rich spike tip (Figure S6). The loss of helical secondary structure in helix 4a can lead to a more open conformation of the spike tip (Figure 6b and 8d), where a newly formed loop at the top of the spike protrudes into the spike opening, leading to steric clash and influencing the dimer hinge to increase its curvature to accommodate the change in secondary structure. In summary, biochemically and structurally we observed that D78S destabilizes helix 4a. In simulations, we observed new preferred conformations due to the loss of necessary hydrogen bonds to constrain the tight helical structure. We suggest that the ensemble of conformations available to the intradimer interface shifted to those compatible with a more open conformation due to the D78S mutation.
Due to the change in the ensemble of preferred conformations, the mutant dimers became less assembly active in response to ionic strength (Figure 2). Under low ionic strength conditions (200 mM and 300 mM NaCl), Cp149-D78S showed slower assembly, while the difference disappeared under high ionic strength conditions. Detailed analysis of the assembly kinetics revealed that the D78S mutant has a longer lag phase, meaning a slower on rate. We also note that the C-terminal end of helix 4 is connected by a single residue, G111, to helix 5, which forms a large part of the interdimer interface. These observations suggest that at low ionic strength the D78S mutation impairs conformational change from inactive to active states, presumably opened to closed states, and signal transduction from helix 4 to helix 5 for building nuclei and intermediates. Simulations paint a consistent picture at high ionic strength (Figure S6), the electrostatic nature of Asp78 was Coulombically screened and became negligibly distinct from Ser78. It is also possible that high ionic strength strengthens the hydrophobic interactions between monomers to overcome the conformational barrier. Crosslinking C61-C61 overcomes the effects of disrupting the spike tip, presumably by forcing a closed conformation. Though C61-C61 attenuates assembly kinetics [29], crosslinked Cp149 and Cp149-D78S showed similar assembly kinetics (Figure 2c and 2d). Such results support our hypothesis that the D78S-induced conformational changes at the intradimer interface modulates assembly, which involves a distal set of interfaces.
It is striking that the D78S mutation slows assembly and destabilizes dimer but does not destabilize capsid in terms of pseudo-critical concentration (Figure S2) and thermal stability (Figure 4a). Therefore, the energy barrier to adopting the assembly-active state is higher for D78S but the interdimer contacts are, as observed, unchanged.
MD simulations of Cp149 and Cp149-D78S dimers reveal that Asp78 can function as a capping residue for helix 4a [39]. With the D78S mutation, partially helical structures failed to recover helical conformation (Figure 6 and S5). Although Ser78 can form a hydrogen bond with Ser81, as Asp78 does, the transient nature of the hydrogen bond and the limited ability to cap the N-terminal end of helix 4a provide insufficient stabilization. Conversely, Cp149, which has Asp78, showed greater propensity to recover helix. This correlation suggests that during assembly, helix 4a at the intradimer interface plays a critical role for orchestrating conformational movements.
In summary, the D78S mutation impairs the conformational change necessary for assembly. In Cp149, helix 4 can be divided into two small helices, helix 4a and helix 4b, a division marked by a helix breaking residue, Gly94. However, in Cp149-D78S, dimers preferentially adopt an open conformation where helix 4a has a propensity to partially unwind (Figure 7 and 8) and dimers assemble with slower kinetics. We suggest that the defective helix 4a inadequately transduces conformational change at the intradimer interface to the interdimer interface via helix 5. This effect can lead to slower ionic strength induced assembly. Altogether, this emphasizes the importance of the intradimer interface in allosteric regulation of capsid assembly.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01-AI144022 to AZ and NSF grant MCB-2027096, funded in part by Delaware EPSCoR, to JAH-P. Some electron microscopy was supported by a grant from the Indiana CTSI to AZ. We gratefully acknowledge the use of the Indiana University Physical Biochemistry Instrumentation Facility and the Indiana University Electron Microscopy Center. Molecular dynamics components of this research are part of the Blue Waters sustained-petascale computing project, which is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois. Blue Waters is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications. The authors acknowledge use of Blue Waters through allocation awards to JAH-P.
Footnotes
Supporting Information
Supporting information includes text, figures, and movies: Experimental materials and methods; Figure S1 shows early phase of assembly kinetics. Figure S2 shows the pseudo-critical concentrations of assembly for mutant and wildtype proteins. Figures S3 and S4 show characterization of T=3 capsids. Figures S5, S6 and Movie S1 show the molecular dynamics simulations of the dimer interfaces. Table S1 provides details of cryo-EM data collection and reconstruction. Table S2 summarizes the conditions tested by molecular dynamics simulations. This material is available free of charge via the internet at http://pubs.acs.org
References
- 1.Crick FHC and Watson JD, The structure of small viruses. Nature, 1956. 177: p. 473–475. [DOI] [PubMed] [Google Scholar]
- 2.Caspar DLD and Klug A, Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol, 1962. 27: p. 1–24. [DOI] [PubMed] [Google Scholar]
- 3.Bancroft JB, Hills GJ, and Markham R, A study of the self-assembly process in a small spherical virus. Formation of organized structures from protein subunits in vitro. Virology, 1967. 31(2): p. 354–79. [DOI] [PubMed] [Google Scholar]
- 4.Adolph KW and Butler PJG, Reassembly of a spherical virus in mild conditions. Nature, 1975. 255: p. 737–738. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay S, et al. , In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J Virol, 2002. 76(21): p. 11128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlotnick A, et al. , Mechanism of capsid assembly for an icosahedral plant virus. Virology, 2000. 277(2): p. 450–6. [DOI] [PubMed] [Google Scholar]
- 7.Conway JF, et al. , Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature, 1997. 386: p. 91–94. [DOI] [PubMed] [Google Scholar]
- 8.Zlotnick A and Mukhopadhyay S, Virus assembly, allostery and antivirals. Trends Microbiol, 2011. 19(1): p. 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razavi-Shearer D and Razavi H, Global prevalence of hepatitis B virus infection and prevention of mother-to-child transmission - Authors’ reply. Lancet Gastroenterol Hepatol, 2018. 3(9): p. 599. [DOI] [PubMed] [Google Scholar]
- 10.Bartenschlager R and Schaller H, Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J., 1992. 11: p. 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ning X, et al. , Secretion of genome-free hepatitis B virus--single strand blocking model for virion morphogenesis of para-retrovirus. PLoS pathogens, 2011. 7(9): p. e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowther RA, et al. , Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell, 1994. 77(6): p. 943–50. [DOI] [PubMed] [Google Scholar]
- 13.Birnbaum F and Nassal M, Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J.Virol., 1990. 64: p. 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingfield PT, et al. , Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry, 1995. 34(15): p. 4919–32. [DOI] [PubMed] [Google Scholar]
- 15.Kenney JM, et al. , Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure, 1995. 3: p. 1009–1019. [DOI] [PubMed] [Google Scholar]
- 16.Zlotnick A, et al. , Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry, 1996. 35(23): p. 7412–21. [DOI] [PubMed] [Google Scholar]
- 17.Zlotnick A, Theoretical aspects of virus capsid assembly. J Mol Recognit, 2005. 18(6): p. 479–90. [DOI] [PubMed] [Google Scholar]
- 18.Hagan MF and Chandler D, Dynamic pathways for viral capsid assembly. Biophys J, 2006. 91(1): p. 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katen SP and Zlotnick A, Thermodynamics of Virus Capsid Assembly. Methods in Enz., 2009. 455: p. 395–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutomski CA, et al. , Hepatitis B Virus Capsid Completion Occurs through Error Correction. J Am Chem Soc, 2017. 139(46): p. 16932–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asor R, et al. , Assembly Reactions of Hepatitis B Capsid Protein into Capsid Nanoparticles Follow a Narrow Path through a Complex Reaction Landscape. ACS Nano, 2019. 13(7): p. 7610–7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, et al. , Characterization of Virus Capsids and Their Assembly Intermediates by Multicycle Resistive-Pulse Sensing with Four Pores in Series. Anal Chem, 2018. 90(12): p. 7267–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceres P and Zlotnick A, Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry, 2002. 41(39): p. 11525–31. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JM, et al. , Regulating self-assembly of spherical oligomers. Nano Lett, 2005. 5(4): p. 765–70. [DOI] [PubMed] [Google Scholar]
- 25.Wynne SA, Crowther RA, and Leslie AGW, The crystal structure of the human hepatitis B virus capsid. Mol Cell, 1999. 3(6): p. 771–780. [DOI] [PubMed] [Google Scholar]
- 26.Packianathan C, et al. , Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol, 2010. 84(3): p. 1607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan TT, et al. , The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J Virol, 1999. 73(7): p. 5731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceres P, Stray SJ, and Zlotnick A, Hepatitis B virus capsid assembly is enhanced by naturally occurring mutation F97L. J Virol, 2004. 78(17): p. 9538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selzer L, Katen SP, and Zlotnick A, The hepatitis B virus core protein intradimer interface modulates capsid assembly and stability. Biochemistry, 2014. 53(34): p. 5496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koschel M, Thomssen R, and Bruss V, Extensive mutagenesis of the hepatitis B virus core gene and mapping of mutations that allow capsid formation. J Virol, 1999. 73(3): p. 2153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponsel D and Bruss V, Mapping of Amino Acid Side Chains on the Surface of Hepatitis B Virus Capsids Required for Envelopment and Virion Formation. Journal of Virology, 2003. 77(1): p. 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endres D and Zlotnick A, Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys J, 2002. 83(2): p. 1217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagan MF and Elrad OM, Understanding the concentration dependence of viral capsid assembly kinetics--the origin of the lag time and identifying the critical nucleus size. Biophys J, 2010. 98(6): p. 1065–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zlotnick A, et al. , Separation and crystallization of T = 3 and T = 4 icosahedral complexes of the hepatitis B virus core protein. Acta Crystallogr D Biol Crystallogr, 1999. 55(Pt 3): p. 717–20. [DOI] [PubMed] [Google Scholar]
- 35.Singh S and Zlotnick A, Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem, 2003. 278(20): p. 18249–55. [DOI] [PubMed] [Google Scholar]
- 36.Hilmer JK, Zlotnick A, and Bothner B, Conformational equilibria and rates of localized motion within hepatitis B virus capsids. J Mol Biol, 2008. 375(2): p. 581–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zivanov J, et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips JC, et al. , Scalable molecular dynamics with NAMD. J Comput Chem, 2005. 26(16): p. 1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson JS and Richardson DC, Amino acid preferences for specific locations at the ends of alpha helices. Science, 1988. 240(4859): p. 1648–52. [DOI] [PubMed] [Google Scholar]
- 40.Patterson A, et al. , Capsid protein dynamics regulate Hepatitis B virus assembly through an allosertic network. Submitted, 2020.
- 41.Hilser VJ and Thompson EB, Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A, 2007. 104(20): p. 8311–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frishman D and Argos P, Knowledge-based protein secondary structure assignment. Proteins, 1995. 23(4): p. 566–79. [DOI] [PubMed] [Google Scholar]
- 43.Bourne CR, Finn MG, and Zlotnick A, Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol, 2006. 80(22): p. 11055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahl ACE, Chavent M, and Sansom MSP, Bendix: intuitive helix geometry analysis and abstraction. Bioinformatics, 2012. 28(16): p. 2193–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.