Abstract
Over a 3-year period, 67 patients of the Hospital of Pau (Pau, France), including 64 patients hospitalized in the adult intensive care unit (ICU), were colonized and/or infected by strains of Pseudomonas aeruginosa P12, resistant to all potentially active antibiotics except colistin. Most patients were mechanically ventilated and presented respiratory tract infections. Since cefepime and amikacin were the least inactive antibiotics by MIC determination, all ICU patients were treated with this combination, and most of them benefited. Cefepime-amikacin was found highly synergistic in vitro. Ribotyping and arbitrary primer-PCR analysis confirmed the presence of a single clonal isolate. Isoelectrofocusing revealed that the epidemic strain produced large amounts of the chromosomal cephalosporinase and an additional enzyme with a pI of 5.7, corresponding to PSE-1, as demonstrated by PCR and sequencing. Outer membrane protein profiles on sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed the absence of a ca. 46-kDa protein, likely to be OprD, and increased production of two ca. 49- and 50-kDa proteins, consistent with the outer membrane components of the efflux systems, MexAB-OprM and MexEF-OprN. Thus, we report here a nosocomial outbreak due to multiresistant P. aeruginosa P12 exhibiting at least four mechanisms of β-lactam resistance, i.e., production of the penicillinase PSE-1, overproduction of the chromosomal cephalosporinase, loss of OprD, and overexpression of efflux systems, associated with a better activity of cefepime than ceftazidime.
Pseudomonas aeruginosa, like many other nonfermenting gram-negative rods, is a saprophytic organism widespread in nature, particularly in moist environments (water, soil, plants, and sewage), and endowed with only weak pathogenic potential. However, because of its ability to survive on inert materials and its resistance to most antiseptics and antibiotics, P. aeruginosa has become an important and frequent nosocomial pathogen. Indeed, in hospitals, sinks, respiratory therapy equipment, and antiseptic or detergent solutions can act as reservoirs of P. aeruginosa. This organism is responsible for a wide range of hospital-acquired infections such as pneumonia, urinary tract infections, or bacteremia. Patients with impaired specific or nonspecific defense systems particularly tend to suffer from severe and even fatal infections caused by P. aeruginosa (9, 11, 23, 24). Cross-transmission from patient to patient may occur via the hands of the health care staff or through contaminated materials or reagents (2, 18, 21, 24). Thus, a number of outbreaks of nosocomial infections due to P. aeruginosa have been reported, especially in intensive care units (ICUs) (2, 11, 18, 24), burn wound units (21), and cancer centers (23).
One feature of P. aeruginosa is its high level of intrinsic resistance to a number of structurally unrelated antimicrobial agents. Indeed, the broad-spectrum resistance of this organism is largely due to a low outer-membrane permeability (32, 33) and to multidrug resistance (MDR) efflux systems (25, 26, 32, 35). Moreover, P. aeruginosa possesses an inducible chromosomally encoded AmpC cephalosporinase belonging to Ambler class C (6). This enzyme, expressed at low levels, confers resistance to aminopenicillins and to narrow-spectrum and expanded-spectrum cephalosporins. Actually, the only antimicrobial agents effective against P. aeruginosa are some β-lactams (ticarcillin, piperacillin, aztreonam, imipenem, cefsulodin, cefoperazone, and ceftazidime), most recent aminoglycosides, fluoroquinolones, fosfomycin, and colistin. In addition, acquired resistance are particularly frequent in P. aeruginosa, and the difficulty of finding an effective treatment increases the mortality and the morbidity of P. aeruginosa-induced infections in hospitalized patients. The combination of ceftazidime plus amikacin is considered the first-choice antipseudomonal chemotherapy (22). Even the “fourth-generation” cephalosporins, cefpirome and cefepime, which have a higher degree of activity against gram-negative organisms, are less efficient in vitro than ceftazidime on P. aeruginosa (40).
Over a 3-year period, 67 patients at the Hospital of Pau, including 64 patients hospitalized in the adult ICU, were colonized and/or infected by strains of P. aeruginosa belonging to the same serotype, P12, and exhibiting the same antibiotype, i.e., resistant to all potentially active antibiotics except colistin. Since the least inactive antimicrobial agents were cefepime and amikacin, all ICU patients were treated with the combination of these two drugs, with apparent benefit for many of them. In this study, clinical cases were retrospectively analyzed, and the existence of an outbreak was demonstrated by molecular typing of the strains. The efficacy of cefepime-amikacin therapy observed in vivo was confirmed in vitro, and the mechanisms of β-lactam resistance in the epidemic strain were investigated.
MATERIALS AND METHODS
Hospital and ICU characteristics.
The hospital of the city of Pau in France is a 740-bed general hospital, including an adult ICU (10 beds), a pediatric ICU (11 beds), seven surgical units (162 beds), and 17 medical wards (557 beds). The hospital is composed of three separate buildings. The 447-bed block where the adult ICU is located also includes the surgical units and 10 medical wards. At the time of the outbreak, the ICU consisted of a single room, without separate cubicles. Clinical cases were retrospectively analyzed to elicit patients' characteristics, diseases, treatment, and evolution.
Bacterial strains and culture conditions.
Between June 1995 and March 1998, 202 multiresistant P. aeruginosa P12 strains were collected from 67 patients (1 to 15 strains isolated per patient, over a period varying from 1 day to 16 months). They were recovered from respiratory samples (n = 91), urine (n = 39), stools (n = 37), ear, nose, or throat samples (n = 10), surgical wounds (n = 6), bedsores (n = 6), pus (n = 5), bile samples (n = 2), cerebrospinal fluid (n = 1), skin (n = 1), and urethral (n = 1) and vaginal (n = 1) samples. Among the 202 strains, 145 came from patients in the adult ICU. All isolates were identified and serotyped by conventional methods (19). The wild-type strain P. aeruginosa P12 CIP 33359, obtained from the “Collection de l'Institut Pasteur,” was the reference strain for MIC determination, ribotyping, arbitrary primer (AP)-PCR assays, and outer membrane analysis. P. aeruginosa RPL11 (a gift of G. Paul), which produces the PSE-1 β-lactamase, was used as control for β-lactamase isoelectric focusing and gene amplification (17). All bacterial strains were either routinely cultured at 37°C on Mueller-Hinton (MH) agar (Diagnostics Pasteur, Marnes la Coquette, France) or grown in Luria broth (LB) (Gibco BRL, Cergy Pontoise, France) or Trypticase soy broth (TSB) (Diagnostics Pasteur) supplemented with KNO3 (4 mg/ml) and incubated at 37°C with aeration.
Antibiotic susceptibility testing.
Antibiotic susceptibility was determined for the 202 isolates by a standard agar diffusion method (7) using 20 disks (Diagnostics Pasteur). MICs were determined by a standard agar dilution method (7) for 26 selected strains isolated from different patients and distributed over the outbreak period; MICs were interpreted according to national guidelines (7). The in vitro effect of the combination of cefepime plus amikacin was evaluated for three selected strains (strain 459 [representing the onset of the outbreak], strain 507 [middle], and strain 521 [end]) by the checkerboard and the time-kill curve methods (10). The checkerboard technique was performed in MH agar with concentration ranges of 512 to 1 mg/liter for cefepime, and 512 to 0.5 mg/liter for amikacin. Time-kill studies were carried out in MH broth at the MIC, 1/2 of the MIC, and 1/4 of the MIC of cefepime and amikacin. Each antibiotic was tested alone and in combination at the same concentration. At time zero and after 2, 4, 6, and 24 h of incubation at 37°C, viable cells were enumerated by serial 10-fold dilutions plated on antibiotic-free MH agar.
β-Lactamase extraction and isoelectric focusing.
β-Lactamases of 12 representative strains were released by ultrasonic treatment, and their pIs were determined by isoelectric focusing on a pH 3.5 to 10 ampholin polyacrylamide gel as described by Matthew et al. (31). Enzyme activities were detected by the iodine procedure in a gel (1) using benzylpenicillin (75 mg/liter) as the substrate. β-Lactamases of known pIs, PSE-1 (pI 5.7), CARB-3 (pI 5.75), PSE-4 (pI 5.3), and SHV-3 (pI 7.0), were used as pI markers.
Preparation of outer membrane proteins.
Cultures of P. aeruginosa were grown overnight at 37°C in 5 ml of LB medium and then diluted 100-fold into fresh medium. Bacterial cells were incubated for 5 h with shaking at 37°C to yield late-logarithmic-phase cells. Outer membrane proteins of the three selected strains (strains 459, 507, and 521) were extracted by the method of Hosaka et al. (16), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Coomassie blue.
DNA methods, PCR, and sequencing.
Total DNA was extracted from the three selected strains by the method of Hall et al. (14), and plasmid DNA was obtained using an alkaline-lysis extraction method (39). The blaPSE-1 and mexR (37) coding regions were amplified with the primers sets PSE-1a–PSE-1b and MexR1–MexR2, respectively (Table 1), under standard PCR conditions (39). The amplicons were revealed by electrophoresis on a 1.5% agarose gel and subsequent exposure to UV light in the presence of ethidium bromide. For sequencing purposes, the PCR products were purified through S400 spin columns (Pharmacia, Orsay, France). Both strands were used as templates in a single-cycle reaction by using the dideoxy-chain termination method with the dRhodamine Dye Terminator Kit (Perkin-Elmer, Courtaboeuf, France) and primers PSE-1b, PSE-1c, PSE-1d and PSE-1e and MexR1–MexR2 (Table 1) for the blaPSE-1 and mexR coding regions, respectively. The cycle reaction consisted of 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. Sequence reaction products were precipitated and separated by electrophoresis on a 6% polyacrylamide gel containing 7 M urea and 1×Tris-borate buffer (TBE). Sequences were analyzed with an ABI 377 automatic sequencer (Perkin-Elmer) using the Sequencing Analysis software. The sequences were compared to references and to each other by using the Sequence Navigator software.
TABLE 1.
Oligonucleotides used as primers for PCR and sequencing of blaPSE-1 and mexR genes
| Primers | Sequence (5′ to 3′) | Position (nt)a | Lb (nt) |
|---|---|---|---|
| PSE-1a | GTGTGACAATCAAAATTATG | 115–134 | 20 |
| PSE-1b | GACTGTGATGTATAAACGTC | 986–1005 | 20 |
| PSE-1c | AATTAGAGTCTTGGATGGTG | 747–766 | 20 |
| PSE-1d | TTGATTGTTCACCATCCAAG | 756–775 | 20 |
| PSE-1e | CGCAACTATGACTACAAGTG | 502–521 | 20 |
| MexR1 | AACCAATGAACTACCCCGTG | 272–291 | 20 |
| MexR2 | ATCCTCAAGCGGTTGCGCGG | 695–714 | 20 |
Ribotyping.
Total DNA of the 15 representative strains was digested with the restriction enzyme NruI (Gibco BRL). Fragments of digested DNA were separated by electrophoresis on a 0.8% agarose gel and transferred to Hybond-N nylon filters (Schleicher and Schuell, Ecquevilly, France) by a vacuum blotting system (Appligene, Illkirch, France). After Southern blotting, ribotyping was performed with a digoxigenin (DIG)-labeled probe consisting of the purified plasmid DNA, pKK3535, a pBR322 derivative containing the rrnB ribosomal operon of Escherichia coli (4). The hybrids were detected by an immunoenzymatic method (DIG-DNA labeling and detection kit; Boehringer, Mannheim, Germany).
AP-PCR analysis.
For the 15 strains studied by ribotyping, AP-PCR assays were performed with primer ERIC2 (38) under PCR standard conditions. After a first cycle of denaturation for 10 min at 94°C, the 45 subsequent cycles of amplification consisted of denaturation for 1 min at 94°C, annealing for 1 min at 42°C, and extension for 1 min at 72°C, with a final extension step for 10 min at 72°C. The amplification products were analyzed by electrophoresis of 10-μl samples on 1.5% agarose gels in the presence of ethidium bromide.
RESULTS
Clinical cases and treatment.
From June 1995 through March 1998, 67 patients hospitalized in the Hospital of Pau were colonized and/or infected by multiresistant strains of P. aeruginosa serotype P12. The two index cases came from the neurosurgical unit of a private clinic: one of these patients was admitted in neurology and rapidly died of P. aeruginosa-related meningitis, and the other was hospitalized in the adult ICU due to vascular brain damage. This patient presented a pulmonary sample positive for multiresistant P. aeruginosa P12 on the day of admission to the ICU. Thereafter, 65 patients were found to harbor phenotypically identical strains. Among them, 63 were initially admitted to the adult ICU, 1 stayed in the pulmonary medicine unit, and 1 stayed in the hematology ward. The latter two wards had previously accommodated one or several infected ICU patients. Indeed, among the 64 patients initially admitted to the adult ICU, 45 were transferred to other wards of the hospital in the course of their hospitalization. The 67 patients, 23 females and 44 males, ranged in age from 17 to 85 years (mean, 57.5 years). The length of exposure before known infection or colonization varied from 1 day to 75 days (mean, 18.4 days). Of the 56 ICU patients who were mechanically ventilated, 44 developed a nosocomial windpipe-bronchial or pulmonary infection and 2 presented with sinusitis due to the multiresistant P. aeruginosa P12. Seven patients who had urinary catheters developed nosocomial urinary tract infections caused by the same pathogen. The temporal relationship between the colonized and/or infected patients showed the presence of at least one infected and/or colonized patient in the ICU during the epidemic period (data not shown). After identification and antibiotic susceptibility testing of the multiresistant P. aeruginosa P12, all patients were treated intravenously with the combination of cefepime at 6 g/day and amikacin at 15 mg/kg of body weight day. The length of treatment extended from 14 to 20 days for the pulmonary infections. In addition, 23 patients harboring the multiresistant P. aeruginosa P12 in their feces were treated orally with colistin plus neomycin for 6 days. Among the 64 ICU patients treated, 44 recovered and 20 died: the infection was not related to death in 12 cases but represented an aggravating factor in 7 cases and was responsible for death in 1 case. The latter patient was a 38-year-old woman admitted for a suicide attempt with drugs; she died of inhalative pneumonia caused by P. aeruginosa.
During the epidemiological investigations, 52 sites in the environment of the ICU were sampled, including water (n = 9), bathroom installations (n = 27), air (n = 4), doors (n = 3), hand lotion (n = 1), and the hands of health care staff (n = 8). P. aeruginosa was found in 8 of the 52 environmental sites, consisting of 1 water and 7 sanitary samples, but the epidemic P. aeruginosa P12 was never detected.
Antibiotic susceptibility testing.
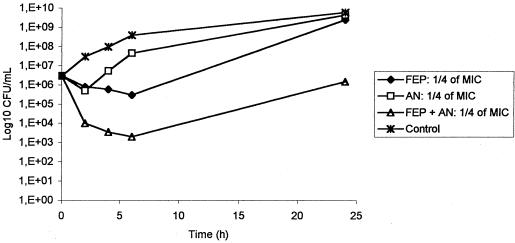
By the disk diffusion method, all the 202 P. aeruginosa P12 isolates were insensitive to all potentially active antibiotics except colistin. However, they had intermediate susceptibility to cefepime, and amikacin gave a small inhibition zone, in contrast with the other aminoglycosides. MICs for 26 representative strains (Table 2) confirmed these data and, in particular, confirmed that cefepime (mode MIC, 32 mg/liter) was slightly more active than ceftazidime and imipenem (mode MICs, 64 mg/liter). Moreover, the combination of cefepime plus amikacin was found highly synergistic in vitro. Indeed, by the checkerboard method, the fractionary inhibitory concentration indexes (ΣFIC) at the point of maximal effectiveness were equal to 0.375 for the three tested strains, (synergy: ΣFIC ≤0.5). The time-kill curves showed that the combination of cefepime and amikacin was significantly more active (difference of >1 log10 CFU/ml) than amikacin, the most active drug alone, e.g., at 1/4 of the MIC in Fig. 1.
TABLE 2.
Antimicrobial susceptibilities of 26 selected multiresistant strains of P. aeruginosa P12 isolated during the outbreak and the P. aeruginosa P12 reference strain (CIP 33359)
| Antimicrobiol agent | MIC (mg/liter) for:
|
MIC breakpointa (mg/liter) | ||
|---|---|---|---|---|
| Multiresistant P12 strains
|
CIP 33359 | |||
| Range | Mode | |||
| Ticarcillin | >512 | >512 | 16 | ≤16–>64 |
| Ticarcillin + clavulanic acid | ≥512 | >512 | 16 | ≤16–>64 |
| Piperacillin | 256–>512 | >512 | 4 | ≤16–>64 |
| Piperacillin + tazobactam | 256–>512 | 512 | 4 | ≤16–>64 |
| Cefoperazone | 256–512 | 256 | 8 | ≤4–>32 |
| Cefsulodin | 128–256 | 128 | 2 | ≤8–>32 |
| Cefotaxime | >512 | >512 | 16 | ≤4–>32 |
| Ceftazidime | 32–128 | 64 | 2 | ≤4–>32 |
| Cefepime | 32–64 | 32 | 1 | ≤4–>32 |
| Cefpirome | 64–128 | 64 | 2 | ≤4–>32 |
| Aztreonam | 16–128 | 32 | 4 | ≤4–>32 |
| Imipenem | 32–64 | 64 | 2 | ≤4–>8 |
| Gentamicin | 8–64 | 32 | 2 | ≤4–>8 |
| Tobramycin | 128–256 | 128 | 1 | ≤4–>8 |
| Amikacin | 32–128 | 64 | 8 | ≤8–>16 |
| Netilmicin | >512 | >512 | 8 | ≤4–>8 |
| Fosfomycin | >512 | >512 | 8 | >32 |
| Ciprofloxacin | 32–128 | 64 | 0.5 | ≤1–>2 |
According to reference 7.
FIG. 1.
In vitro killing of one clinical isolate of multiresistant P. aeruginosa (strain 521) by cefepime (1/4 of the MIC = 8 mg/liter) and amikacin (1/4 of the MIC = 16 mg/liter), alone and in combination. FEP, cefepime; AN; amikacin.
β-Lactamase characterization.
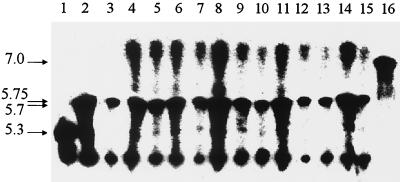
The 12 representative strains were analyzed for their β-lactamase contents. Two enzymes were present in these isolates. One of these gave a strong band at a pI of >7 and was likely to be the derepressed chromosomally encoded cephalosporinase of P. aeruginosa. The other comigrated with the PSE-1 reference enzyme, which exhibits a pI of 5.7 (Fig. 2). Total DNA amplification with primers PSE-1a and PSE-1b gave a fragment with the expected size of 890 bp, and nucleotide sequence analysis revealed the presence of the blaPSE-1 gene, with T at position 610 and C at position 845, allowing definitive distinction between PSE-1, CARB-3, and PSE-4 (17).
FIG. 2.
Analytical isoelectric focusing of β-lactamases. Lanes 1, 2, 3, and 16; β-lactamases with known pIs, PSE-4 (pI 5.3), CARB-3 (pI 5.75), PSE-1 (pI 5.7), and SHV-3 (pI 7.0), respectively; lanes 4 to 15; 12 representative clinical strains of multiresistant P. aeruginosa P12. Enzyme activities were revealed by the iodometric method using benzylpenicillin as the substrate.
Analysis of outer membrane proteins.
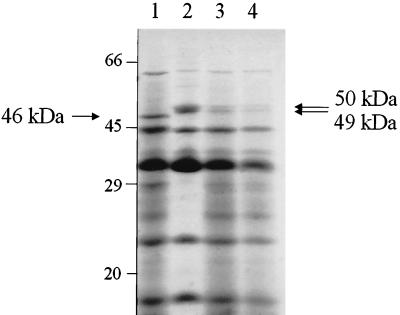
The outer membrane protein patterns of the three representative strains (strains 459, 507, and 521) were analyzed by SDS-PAGE and compared to the profile of the susceptible reference strain, P. aeruginosa P12 CIP 33359 (Fig. 3). This comparison revealed two differences: a ca. 46-kDa protein, consistent with the porin D2, was lacking in the clinical strains, and in contrast, there was increased production of two proteins of about 49 to 50 kDa, likely to correspond to the outer membrane proteins (OprM, OprJ, or OprN) associated with the antibiotic efflux systems of P. aeruginosa.
FIG. 3.
Electrophoretic analysis of the outer membrane proteins of multiresistant P. aeruginosa P12. Lane 1; the susceptible strain CIP 33359; lane 2; strain 459; lane 3; strain 507; lane 4; strain 521. The migration positions of standard proteins are shown on the left (in kilodaltons).
To assess whether the overproduction of MexAB-OprM could be associated with a mutation located in the mexR repressor gene, the mexR coding regions of the three selected strains were amplified and sequenced. The three sequences were identical and differed from the mexR gene described by Poole et al. (37) by four silent mutations at positions 264 (C→T), 327 (G→A), 384 (G→A), and 411 (G→A) and by a base substitution at position 377 (T→A) which converts an arginine to a glutamine at position 126 in the MexR protein.
Plasmid content.
Plasmid extraction carried out with the 15 isolates analyzed for ribotype and AP-PCR profile revealed the presence of a unique high-molecular-weight plasmid.
Ribotyping and AP-PCR.
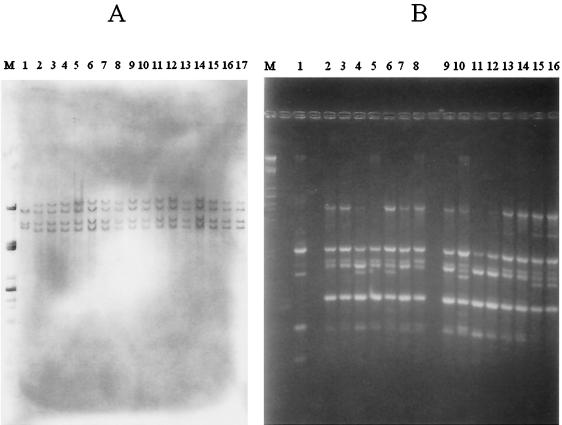
Ribotyping of the 15 representative strains, using chromosomal DNA digested with NruI and plasmid pKK3535 as the probe, gave identical patterns, with the same number of bands of identical sizes (Fig. 4A). This ribotype consisted of four bands ranging in size from ca. 14.5 to 6.6 kb. It differed from the profile of the reference strain, CIP 33359, by three bands. Moreover, AP-PCR analysis performed with the ERIC2 primer also yielded a similar pattern for the 15 representative strains. This AP-PCR profile contained five to six major bands (Fig. 4B) ranging in size from ca. 2.15 to 0.65 kb, and a major band of ca. 0.95 kb was present in five of the analyzed patterns. In contrast, the reference strain, CIP 33359, gave a pattern with four bands of sizes different from those of the epidemic strain profile.
FIG. 4.
(A) Ribotype profiles of NruI-digested total DNA from P. aeruginosa isolates detected with the digoxigenin-labeled pKK3535 plasmid as the probe. Lane M, DNA of λ phage digested with PstI; lane 1, ribotype profile of CIP 33359; lanes 2 to 17, ribotype patterns of 16 multiresistant P. aeruginosa P12 isolates. (B) AP-PCR patterns of P. aeruginosa isolates with primer ERIC2. Lane M, DNA of λ phage digested with PstI; lane 1, AP-PCR profile of CIP 33359; lanes 2 to 16, AP-PCR patterns of 15 epidemiologically related isolates
DISCUSSION
Over a 3-year period, a number of infections and/or colonizations associated with strains of P. aeruginosa exhibiting the same multiresistant antibiotype and serotype P12 were observed at the Hospital of Pau, suggesting the occurrence of an outbreak. In the course of this study, representative strains were shown to exhibit other identical characteristics, i.e., β-lactamase content, outer membrane profile, and plasmid pattern. Finally, the clonal origin of the multiresistant strains of P. aeruginosa P12 was confirmed by determining chromosomal markers. Ribotyping showed that 15 representative clinical strains were identical to each other and different from the control strain. Ribotyping is a highly specific and reproducible typing method (13). However, the ribotype of the strains studied consisted of only four bands. Thus, an additional method, AP-PCR, was used. The AP-PCR profiles of the 15 representative clinical strains were composed of five to six identical bands and differed from that of the control strain. The additional band of ca. 0.95 kb probably reflects a genetic divergence by mutation and/or DNA transfer rather than a strain differentiation. In addition, three of these representative strains were further shown to carry identical mutations in the mexR gene. Thus, all multiresistant strains of P. aeruginosa P12 examined in this study were considered to be a single epidemic strain. P12 is one of the most common serotypes of P. aeruginosa found in French hospitals and is frequently associated with resistance to β-lactams, aminoglycosides, and fluoroquinolones, but not usually with resistance to fosfomycin (3).
The route by which the epidemic strain was introduced into the hospital seems to be the admission of two infected patients coming from a private clinic. Since the first patient, admitted in neurology, died rapidly, the other patient, who had pneumonia caused by the clonal P. aeruginosa and stayed 10 weeks in the ICU, probably represents the real origin of the outbreak. Once the epidemic strain was introduced in 1995, a 3-year outbreak developed in the adult ICU. Indeed, 64 of the 67 patients implicated in this outbreak were in or came from the adult ICU. The transfer of the ICU patients into other units of the hospital accounts for the occurrence of the clonal strain in these units. However, secondary dissemination within and among these wards remained limited. The absence of P. aeruginosa P12 in the environmental samples and the presence of at least one infected and/or colonized patient in the ICU during the outbreak period suggest that the spread of the epidemic strain was related to cross-transmission between patients rather than to contamination from a single reservoir. Outbreaks caused by P. aeruginosa are often associated with probable or demonstrated cross-transmission via the hands of health care workers (42). Most epidemic isolates (45%) came from respiratory specimens and were associated with respiratory tract infections in mechanically ventilated patients (78.6%). Indeed, mechanical ventilation is the main risk factor for acquiring nosocomial respiratory tract infections, particularly in ICUs (11, 24). Receipt of nebulized medications, invasive procedures, and exposure to contaminated hospital water supplies and medications are additional risk factors (2, 9, 18, 21).
All the ICU patients infected by the clonal strain of P. aeruginosa were treated with the combination of cefepime plus amikacin, rather than with the classic ceftazidime-amikacin combination (22) because, by MIC determination, cefepime appeared to be more active than ceftazidime. Moreover, the imipenem resistance complicated the choice of an antipseudomonal treatment. The efficacy of the cefepime-amikacin combination was subsequently verified in vitro by the checkerboard and time-kill curve methods. For most of the ICU patients, the treatment led to the elimination of the multiresistant pathogen, demonstrating the efficacy of the cefepime-amikacin combination in vivo. Finally, the outbreak was also brought under control by the reinforcement of control procedures, by architectural modification of the adult ICU, i.e., the construction of three separate rooms, limiting the person-to-person transmission of the epidemic strain, and by digestive decontamination of patients who presented gut colonization with the clonal P. aeruginosa.
Because cefepime is, in general, less active than ceftazidime against P. aeruginosa, the mechanisms of β-lactam resistance in the epidemic strain were investigated. Resistance to β-lactam antimicrobials is often due to plasmid-mediated β-lactamases (8, 27, 34). Most of the enzymes found in P. aeruginosa are carbenicillinases of the Pseudomonas-specific-enzyme type (PSE), oxacillinases (OXA, class D enzymes) or broad-spectrum penicillinases of the TEM or SHV type (class A β-lactamases). The PSE-1 enzyme produced by our clonal strain of P. aeruginosa conveys a particularly high level of resistance to carboxy- and ureido penicillins but does not affect ceftazidime susceptibility. In the present case, the blaPSE-1 gene seems to be chromosomally located since, despite the presence of a plasmid, all attempts to transfer ampicillin or ticarcillin resistance by conjugation or transformation failed.
The most common mechanism of acquired resistance to broad-spectrum cephalosporins is the overproduction of the species-specific cephalosporinase (8), due to mutations in the regulatory region (29). In our epidemic strain of P. aeruginosa, the derepressed chromosomally encoded cephalosporinase may account for the resistance to all antipseudomonal β-lactams except imipenem. Cefepime, like ceftazidime, is poorly hydrolyzed by this cephalosporinase, even when overproduced. However, cefepime, in contrast with ceftazidime, may remain active against derepressed mutants, because it has an accelerated penetration through the outer membrane, allowing rapid binding to the targets (15). The activity of penicillins is not restored by the β-lactamase inhibitors, since they have little or no effect on PSE-1 and the chromosomal cephalosporinase.
Imipenem resistance in P. aeruginosa has been shown to be mainly related to decreased permeability due to the loss of the imipenem-specific porin D2 (OprD) (41), coupled with weak hydrolysis by the periplasmic cephalosporinase (5, 28). Analysis of outer membrane proteins by SDS-PAGE showed the absence of a band of ca. 46 kDa in the clonal P. aeruginosa, consistent with the loss of OprD. This, together with the overproduction of the chromosomal cephalosporinase, may explain the high level of imipenem resistance in the epidemic strain.
Active efflux pumps are now recognized to play a major role in nonenzymatic mechanisms of acquired β-lactam resistance in P. aeruginosa. Three major inducible MDR efflux systems that extrude antibiotics with different substrate specificities have been described for P. aeruginosa: MexAB-OprM, MexCD-OprJ, and MexEF-OprN. Indeed, while MexAB-OprM extrudes most β-lactams except imipenem (25, 26), MexCD-OprJ extrudes only cefepime and cefpirome (12, 30, 36); MexEF-OprN does not directly contribute to β-lactam efflux (20). Separation of the outer membrane proteins by SDS-PAGE revealed the presence in the epidemic P. aeruginosa strain of two thin bands of ca. 49 to 50 kDa that were absent in the reference strain, CIP 33359. Unfortunately, the positions of these bands are not helpful in distinguishing between the outer membrane components of the three efflux pump systems of P. aeruginosa, because of their similar mobilities on SDS. However, the better activity of cefepime than ceftazidime suggests an overproduction of MexAB-OprM rather than MexCD-OprJ. Moreover, the molecular masses of OprM (49 kDa) and OprN (50 kDa) are closer than that of OprJ (54 kDa) to those of the bands detected in the clonal strain profile. Thus, the MexAB-OprM and MexEF-OprN efflux systems are likely to be overproduced in this strain. Overexpression of the mexA-mexB-oprM operon is usually associated with mutations in the mexR repressor gene. Different alterations have been reported, such as frameshift mutations or amino acid substitutions. In our outbreak strain, an amino acid change at position 126 in MexR may account for the overproduction of the MexAB-OprM efflux system, allowing bacterial cells to reach higher levels of resistance to a wide range of substrate antibiotics, especially ticarcillin, ceftazidime, and aztreonam.
In conclusion, we report here a nosocomial outbreak due to a clonal multiresistant P. aeruginosa P12 strain in an adult ICU, mainly associated with respiratory tract infections in mechanically ventilated patients. The epidemic strain exhibited at least four mechanisms of β-lactam resistance: production of the penicillinase PSE-1, loss of OprD, and overproduction of the chromosomal cephalosporinase and of two Mex efflux systems. The latter two mechanisms likely explain the better activity of cefepime than ceftazidime. Both successful treatment with a cefepime-amikacin combination and hygiene measures contributed to the elimination of the epidemic strain from the adult ICU of the Hospital of Pau.
REFERENCES
- 1.Barthelemy M, Guionie M, Labia R. Beta-lactamases: determination of their isoelectric points. Antimicrob Agents Chemother. 1978;13:695–698. doi: 10.1128/aac.13.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becks V E, Lorenzoni N M. Pseudomonas aeruginosa outbreak in a neonatal intensive care unit: a possible link to contaminated hand lotion. Am J Infect Control. 1995;23:396–398. doi: 10.1016/0196-6553(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 3.Bert F, Lambert-Zechovsky N. Comparative distribution of resistance patterns and serotypes in Pseudomonas aeruginosa isolates from intensive care units and other wards. J Antimicrob Chemother. 1996;37:809–813. doi: 10.1093/jac/37.4.809. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 5.Buscher K H, Cullmann W, Dick W, Opferkuch W. Imipenem resistance in Pseudomonas aeruginosa resulting from diminished expression of an outer membrane protein. Antimicrob Agents Chemother. 1987;31:703–708. doi: 10.1128/aac.31.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carret, G., J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, J. Sirot, and C. J. Soussy. 2000. Comité de l'antibiogramme de la société française de microbiologie: communiqué. [Online.] http://www.sfm.asso.fr. [PubMed]
- 8.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the U.K. in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 9.Cobben N A, Drent M, Jonkers M, Wouters E F, Vaneechoutte M, Stobberingh E E. Outbreak of severe Pseudomonas aeruginosa respiratory infections due to contaminated nebulizers. J Hosp Infect. 1996;33:63–70. doi: 10.1016/s0195-6701(96)90030-4. [DOI] [PubMed] [Google Scholar]
- 10.Eliopoulos G M, Moellering R C. Antimicrobial combinations. In: Lorian V, editor. Antibiotics and laboratory medicine. 4th ed. Baltimore, Md: Williams and Wilkins; 1996. pp. 330–396. [Google Scholar]
- 11.File T M, Jr, Tan J S, Thomson R B, Jr, Stephens C, Thompson P. An outbreak of Pseudomonas aeruginosa ventilator-associated respiratory infections due to contaminated food coloring dye—further evidence of the significance of gastric colonization preceding nosocomial pneumonia. Infect Control Hosp Epidemiol. 1995;16:417–418. doi: 10.1086/647141. [DOI] [PubMed] [Google Scholar]
- 12.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in δmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grattard F, Pozzetto B, Ros A, Gaudin O G. Differentiation of Pseudomonas aeruginosa strains by ribotyping: high discriminatory power by using a single restriction endonuclease. J Med Microbiol. 1994;40:275–281. doi: 10.1099/00222615-40-4-275. [DOI] [PubMed] [Google Scholar]
- 14.Hall L M, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock R E, Bellido F. Factors involved in the enhanced efficacy against gram-negative bacteria of fourth generation cephalosporins. J Antimicrob Chemother. 1992;29(Suppl. A):1–6. doi: 10.1093/jac/29.suppl_a.1. [DOI] [PubMed] [Google Scholar]
- 16.Hosaka M, Gotoh N, Nishino T. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob Agents Chemother. 1995;39:1731–1735. doi: 10.1128/aac.39.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huovinen P, Jacoby G A. Sequence of the PSE-1 beta-lactamase gene. Antimicrob Agents Chemother. 1991;35:2428–2430. doi: 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumaa P, Chattopadhyay B. Outbreak of gentamicin, ciprofloxacin-resistant Pseudomonas aeruginosa in an intensive care unit, traced to contaminated quivers. J Hosp Infect. 1994;28:209–218. doi: 10.1016/0195-6701(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 19.Kiska D L, Gilligan P H. Pseudomonas. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 517–525. [Google Scholar]
- 20.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 21.Kolmos H J, Thuesen B, Nielsen S V, Lohmann M, Kristoffersen K, Rosdahl V T. Outbreak of infection in a burns unit due to Pseudomonas aeruginosa originating from contaminated tubing used for irrigation of patients. J Hosp Infect. 1993;24:11–21. doi: 10.1016/0195-6701(93)90085-e. [DOI] [PubMed] [Google Scholar]
- 22.Korvick J A, Yu V L. Antimicrobial agent therapy for Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:2167–2172. doi: 10.1128/aac.35.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krcmery V, Trupl J. Nosocomial outbreak of meropenem resistant Pseudomonas aeruginosa infections in a cancer centre. J Hosp Infect. 1994;26:69–71. doi: 10.1016/0195-6701(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 24.Labarca J A, Pegues D A, Wagar E A, Hindler J A, Bruckner D A. Something's rotten: a nosocomial outbreak of malodorous Pseudomonas aeruginosa. Clin Infect Dis. 1998;26:1440–1446. doi: 10.1086/516370. [DOI] [PubMed] [Google Scholar]
- 25.Li X Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livermore D M. β-Lactamases of Pseudomonas aeruginosa. Antibiot Chemother. 1991;44:215–220. doi: 10.1159/000420317. [DOI] [PubMed] [Google Scholar]
- 28.Livermore D M. Interplay of impermeability and chromosomal beta-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2046–2048. doi: 10.1128/aac.36.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodge J M, Minchin S D, Piddock L J, Busby J W. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC beta-lactamase. Biochem J. 1990;272:627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda N, Gotoh N, Ohya S, Nishino T. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:909–913. doi: 10.1128/aac.40.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 32.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido H. Role of permeability barriers in resistance to beta-lactam antibiotics. Pharmacol Ther. 1985;27:197–231. doi: 10.1016/0163-7258(85)90069-5. [DOI] [PubMed] [Google Scholar]
- 34.Nordmann P, Guibert M. Extended-spectrum beta-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998;42:128–131. doi: 10.1093/jac/42.2.128. [DOI] [PubMed] [Google Scholar]
- 35.Poole K. Bacterial multidrug resistance—emphasis on efflux mechanisms and Pseudomonas aeruginosa. J Antimicrob Chemother. 1994;34:453–456. doi: 10.1093/jac/34.4.453. [DOI] [PubMed] [Google Scholar]
- 36.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 37.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renders N, Romling Y, Verbrugh H, van Belkum A. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J Clin Microbiol. 1996;34:3190–3195. doi: 10.1128/jcm.34.12.3190-3195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Thornsberry C, Brown S D, Yee Y C, Bouchillon S K, Marler J K, Rich T. In vitro activity of cefepime and other antimicrobials: survey of European isolates. J Antimicrob Chemother. 1993;32(Suppl. B):31–53. doi: 10.1093/jac/32.suppl_b.31. [DOI] [PubMed] [Google Scholar]
- 41.Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widmer A F, Wenzel R P, Trilla A, Bale M J, Jones R N, Doebbeling B N. Outbreak of Pseudomonas aeruginosa infections in a surgical intensive care unit: probable transmission via hands of a health care worker. Clin Infect Dis. 1993;16:372–376. doi: 10.1093/clind/16.3.372. [DOI] [PubMed] [Google Scholar]