Abstract
Synthetic glucocorticoids (GCs) have been widely used in the treatment of a broad range of inflammatory diseases, but their clinic use is limited by undesired side effects such as metabolic disorders, osteoporosis, skin and muscle atrophies, mood disorders and hypothalamic-pituitary-adrenal (HPA) axis suppression. Selective glucocorticoid receptor modulators (SGRMs) are expected to have promising anti-inflammatory efficacy but with fewer side effects caused by GCs. Here, we reported HT-15, a prospective SGRM discovered by structure-based virtual screening (VS) and bioassays. HT-15 can selectively act on the NF-κB/AP1-mediated transrepression function of glucocorticoid receptor (GR) and repress the expression of pro-inflammation cytokines (i.e., IL-1β, IL-6, COX-2, and CCL-2) as effectively as dexamethasone (Dex). Compared with Dex, HT-15 shows less transactivation potency that is associated with the main adverse effects of synthetic GCs, and no cross activities with other nuclear receptors. Furthermore, HT-15 exhibits very weak inhibition on the ratio of OPG/RANKL. Therefore, it may reduce the side effects induced by normal GCs. The bioactive compound HT-15 can serve as a starting point for the development of novel therapeutics for high dose or long-term anti-inflammatory treatment.
Keywords: glucocorticoid receptor, anti-inflammation, SGRMs, transrepression
Introduction
Glucocorticoid receptor (GR), a ligand activated transcription factor, belongs to steroid hormone receptor family that also includes androgen receptor (AR), progesterone receptor (PR), mineralocorticoid receptor (MR), and estrogen receptor (ER). GR is widely expressed in the body and plays important roles in the regulation of a wide range of vital biological processes, such as development, metabolism, inflammation, and stress response [1, 2]. In the resting state, the GR monomer predominantly resides in the cell cytoplasm as a part of a chaperone complex. Upon ligand binding, GR undergoes a conformational change, dissociates from heat shock protein 90 (HSP90), and translocates into the nucleus and selectively binds to the conserved GC response elements (GREs) to regulate downstream genes. GR homodimer can bind to positive GREs to activate the transcription of target genes, also known as “transactivation” [3–5]. GR can also bind to negative GREs (nGREs) [6, 7] or interact with other transcription factors through a manner called “tethering” to repress the transcription of target genes, also known as “transrepression” [8, 9]. However, recent studies suggest that the “tethering” alone is not sufficient for GR anti-inflammatory effects and potent anti-inflammatory effects additionally require a direct binding of GR to the recognition motif of transcription factors activator protein-1 (AP-1) or nuclear factor-kappa B (NF-κB) [10–12]. GR recognizes a cryptic response element within the promoter of NF-κB and represses the expression of the inflammatory cytokines such as IL-8 and CCL-2 [13]. It has been confirmed that the anti-inflammatory effects of GR mostly attribute to its ability to inhibit downstream effectors such as the AP-1 and NF-κB of proinflammatory signaling pathways [14]. In addition, GCs can also exert rapid anti-inflammatory effects by non-genomic pathways [15].
Synthetic glucocorticoids (GCs), such as dexamethasone (Dex), are among the most widely prescribed drugs for the treatment of inflammatory and autoimmune diseases [16]. Recently, Dex was recommended for use in COVID-19 patients with severe respiratory symptoms in a press release named “Randomized Evaluation of COVID-19 Therapy” (RECOVERY) for a comprehensive trial in the United Kingdom [17]. However, long-term use of traditional synthetic GR agonists often causes severe adverse effects such as glucose intolerance, hypertension, muscle wasting and osteoporosis [18, 19]. Early clinical studies indicated that the adverse effects of GCs are predominantly associated with its transactivation activity because a series of genes activated via the transactivation pathway are involved in metabolic and endocrine functions [19]. However, it has also been reported that, through the transactivation mechanism, GR can induce the expression of anti-inflammatory proteins, such as glucocorticoid-induced leucine zipper (GILZ), MAPK phosphatase-1 (MKP-1) and annexin-1 (Anx-1) [20]. In addition, higher doses of GCs can bind to MR, resulting in increased sodium level and decreased potassium level [15, 21]. In general, discovery of GR ligands with anti-inflammatory function but less transactivation-mediated side effect is highly desirable. It is believed that selective glucocorticoid receptor modulators (SGRMs, also termed as “dissociated ligands”) have the potential to dissociate the promising anti-inflammatory efficacy from adverse effects through differential DNA binding and coregulator recruitment [18, 22]. During the past decades, several synthetic nonsteroidal SGRMs have been reported based on the transactivation/transrepression dissociation mechanism, such as mapracorat [23], CpdA [24], ZK 216348 [25], AL-438 [26] and AZD9567 [22]. These compounds exhibit GC-like anti-inflammatory activities and less GC-induced undesirable effects. Especially, Azd9567 had entered a phase IIa trial for rheumatoid arthritis (in patients with type 2 diabetes mellitus) in October 2020 (NCT04556760) [27]. However, so far, no SGRM has been approved for clinical use. Therefore, it is still quite urgent to develop novel SGRMs for inflammatory disorders.
Virtual screening is a powerful computational approach for the identification of lead compounds with novel structural scaffolds [28, 29]. In this study, a method of structure-based virtual screening (SBVS) combined bioassays was employed to discover novel SGRMs. By screening the ChemDiv chemical library, 54 potential compounds were identified and submitted to bioassays. HT-15, a new-scaffold SGRM with overall better activity and safety profile, was finally identified. Compared with Dex, HT-15 shows less transactivation potency that is associated with the main adverse effects of synthetic GCs, and no cross activities with other NRs. Furthermore, molecular dynamics (MD) simulations and free energy calculations were used to explore the GR LBD/ HT-15 interaction to guide future optimization of HT-15.
Materials and methods
Structure-based virtual screening
The crystal structure of the GR ligand binding domain (GR LBD) in complex with nonsteroidal glucocorticoid receptor modulators (PDB entry: 5G5W [30]) was retrieved from Protein Data Bank (http://www.rcsb.org) and used as the template for docking-based virtual screening. The crystal structure was prepared using the Protein Preparation Wizard in Schrödinger software [31]. The water and 1,2-ethanediol molecules were deleted. The protein grid box for docking was generated by enclosing the residues in a box with a size of 15 Å × 15 Å × 15 Å centered on the native ligand using the Receptor Grid Generation module with the default settings. Before molecule docking, the small molecules in the ChemDiv chemical library were prepared using the LigPrep module in Schrödinger, and filtered by the Lipinski’s rule-of-five [32] and Oprea’s [33] rules. The remaining 770,709 molecules were docked into the prepared structure by using the Glide module, and the binding energies were scored and ranked by the Glide SP scoring mode. The 1000 top-ranked compounds were clustered based on the 2D similarities (Tanimoto coefficients) of the MACCS fingerprints. Then, the binding poses of the clustered compounds were carefully checked. Finally, 54 potential compounds were purchased for subsequent bioassays.
Molecular dynamics (MD) simulations
The structure of the GR LBD bound with HT-15 predicted by molecular docking was used as initial conformation for the MD simulations. The GR agonist Dex was obtained from the crystal structures of the complexes (PDB entry: 1M2Z). Then, the crystal structures of 1M2Z was superimposed onto that of 5G5W, and Dex was extracted and merged into 5G5W to construct the structure of the GR LBD bound with Dex. The AM1-BCC atomic partial charges for the ligand were assigned with the antechamber program in the AMBER 18 package [34]. The ff14SB and GAFF2 force fields were assigned for the protein and ligand, respectively. The protein-ligand complex was solvated into a TIP3P water box with a distance of 10 Å extended from any solute atom. Two sodium ions were added to neutralize the whole systems.
Three phases of minimizations were performed to remove unfavorable contacts for the prepared system. First, the water molecules and counter ions were optimized by 1000 steps of steepest descent and 2000 steps of conjugated gradient minimizations with the other atoms restrained by 50 kcal·mol−1·Å−2. Then, the protein atoms were restrained by a 10 kcal·mol−1·Å−2 force constant and the other atoms were minimized by 1000 steps of steepest descent and 2000 steps of conjugated gradient minimizations. Finally, the whole system was relaxed without any restraint for 1000 steps of steepest descent and 2000 steps of conjugated gradient minimizations. Next, the system was gradually heated to 300 K over a period of 30 ps under the NVT ensemble, and equilibrated for 110 ps in the NPT (P = 1 atm, T = 300 K) ensemble. Finally, 500 ns MD simulations were carried out in the NPT (T = 300 K and P = 1 atm) ensemble with the PMEMD program [35]. The SHAKE algorithm [36] was used to constrain the covalent bonds. The length of the production simulation was 500 ns with a time step of 2 fs, and the conformations were saved per 10 ps.
The root-mean-square deviation (RMSD) and distance analyses were carried out by using the cpptraj module in AmberTools18. The principal component analysis (PCA) was carried out by using the NMWiz module in VMD [37] based on the coordinates of all the Cα atoms in the trajectory.
MM/GBSA free energy decomposition
The interactions between each residue in the GR LBD and each ligand (HT-15 or Dex) were analyzed using the MM/GBSA free energy decomposition analysis [38–41] applied in the MMPBSA.py script as implemented in AmberTools18. The binding interaction for each residue-inhibitor pair includes three terms: van der Waals contribution (ΔGvdw), electrostatic contribution (ΔGele) and desolvation contribution (ΔGsol) (Eq. (1)). The electrostatic desolvation energy (ΔGGB) was estimated by using the GB model based on the parameters developed by Onufriev et al. (igb = 2) [42]. The non-polar contribution of desolvation (ΔGSA) was measured by the solvent accessible surface area (SASA).
| 1 |
All the energy components were calculated using the 100 snapshots extracted from the last 50 ns MD trajectory.
Cell culture and materials
RAW264.7, HeLa, 3T3, HePG2, MG-63, PC3, 293T and A549 cell lines were purchased from the Cell Bank of the Chinese Academy of Science Type Culture Collection (China). Cells were cultured according to the supplier’s instructions.
LPS from Escherichia coli O55:B5 (Sigma-Aldrich, USA; # L-2637) and recombinant human tumor necrosis factor-alpha (TNF-ɑ) (Sangon Biotech, China; # C600021) were used to induce inflammation. Progesterone, aldosterone, PMA, Azd9567 and Dex were purchased from MCE (Medchem Express, USA), and and all the tested compounds were bought from Chemdiv and the compounds are listed in Supplementary Table S1. pGL4.36[luc2P MMTV Hygro] (GenBank accession no. FJ773214) was purchased from Promega. pCMV-GR11 (Addgene, USA; # 89105), pCMV-hAR (Addgene, USA; # 89078), and pcDNA3-PRB (Addgene, USA; # 89130) were gifts from Elizabeth Wilson. pNF-κB-luc (Beyotime Biotechnology, China; #D2206) was purchased from Beyotime. pCMV-hMR was constructed by cloning MR fragment into the plasmid of pCMV-hAR by the technology of seamless cloning (Vazyme, China; # C112-1). AP-1-luc was constructed by cloning five copies of AP-1 promoter into the BmtI and BglII sites of pNF-κB-luc. ARR3tk promoter was cloned into the HindIII and XhoI sites of PGL4.18 vector (GenBank accession no. DQ188838) (Promega, USA).
Transactivation assay in agonist mode (TAag)
The agonistic activity of the tested compounds toward GR was determined in HeLa cells stably transfected with pGL4.36[luc2P MMTV Hygro] by measuring the firefly luciferase signal. Briefly, MMTV-Luc-expressed cells were cultured in 5% charcoal stripped serum (CSS) DMEM medium with 1 × 104 cells/well in 96-well plates overnight. Then cells were treated with the gradient concentrations of the tested compounds for 18 h and their luciferase activities were measured with One-Lumi™ Firefly Luciferase Assay Kit (Beyotime Biotechnology, China; # RG055M). Bioluminescence was measured with Synergy H1 (BioTek; USA).
Transactivation assay in antagonist mode (TAantag)
In antagonist mode, the assay was run as above except that the cells were treated by gradient concentrations (0–50 μM) of the tested compounds with 100 nM Dex. The control wells were included on each plate to define 0% inhibition (DMSO) and 100% inhibition (1 μM mifepristone) of the Dex response.
| 2 |
Transrepression assay
HeLa cells were cultured in 5% CSS DMEM media in 96-well plates for 24 h. Then cells were transfected with 71 ng of GRɑ, 5 ng of Renilla and 24 ng of NF-κB-Luc or 5 × AP-1-Luc by lip3000 transfection reagent for 24 h. For transrepression, cells were treated with either TNF-α at 5 ng/mL (for NF-κB reporter) or PMA at 1 ng/mL (for AP-1 reporter), together with the indicated compounds. Cells were harvested by addition of 1× Passive Lysis Buffer (Promega, USA; # E1910), and luciferase activity was analyzed by the Dual-Glo Luciferase system (Promega, USA; # E1910). The control wells with DMSO and 10 μM Dex were included on each plate to define the 100% and 0% effects of NF-κB or AP-1 transcriptional activity, respectively.
| 3 |
In vitro GR ligand binding assay
The binding of the tested compounds toward GR was assessed with the LanthaScreen® TR-FRET GR Competitive Binding Assay (Invitrogen Life Technologies; USA). The data were plotted as a standard competition curve by GraphPad Prism 6. The values presented in Fig. 1a were normalized and the DMSO group was defined as 100% effect.
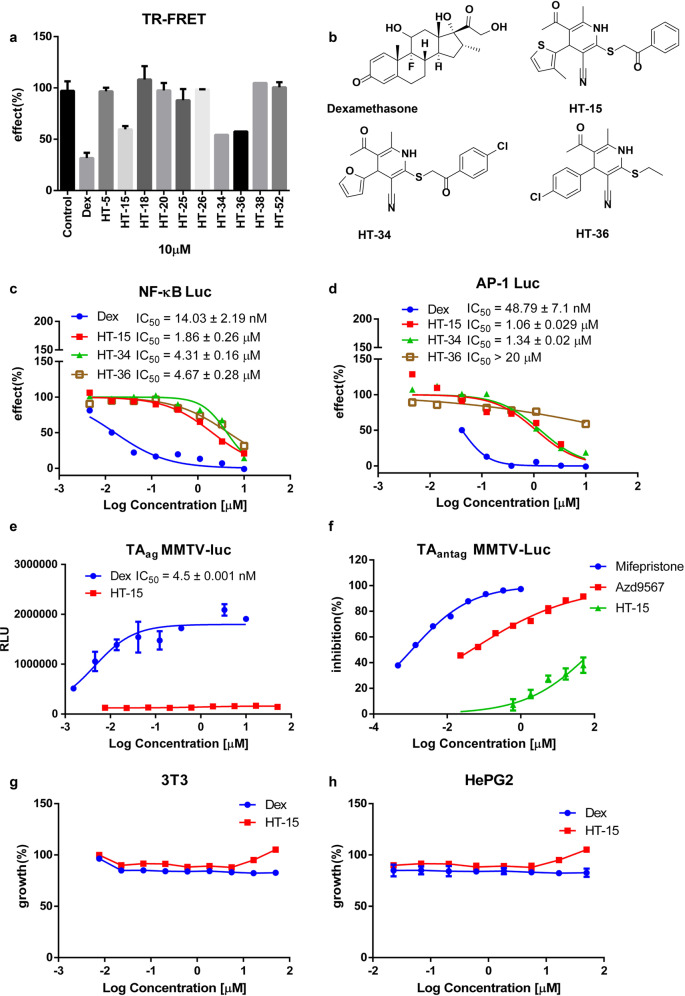
Fig. 1. HT-15 is a potent GR modulator.
a The relative GR competitive binding activity of the potent compounds (10 µM) was analyzed using the LanthaScreen TR-FRET GR competitive binding assay (n = 3). b The chemical structures of Dex, HT-15, HT-34, and HT-36. c, d The transrepression activity of the compounds on the NF-κB and AP-1 signaling pathway (n = 4). e The evaluation of the HT-15 transactivation activity in the MMTV-Luc reporter assay (n = 4). f The inhibition activities of mifepristone, Azd9567, and HT-15 against Dex in the TAantag MMTV-Luc reporter assay (n = 4). g, h The toxicity of HT-15 on 3T3 and HepG2 cell lines (n = 3).
Cell cytotoxicity assay
We evaluated the toxicity of the compounds purchased from Chemdiv. At first, 8 × 104 RAW264.7 cells were seeded in 96 plates and incubated overnight. Then cells were treated with 25 μM compounds for 24 h. The cell growth was evaluated with MTT assay. Furthermore, the inherent toxicity of the hit was evaluated with 3T3 and HepG2 cells. Cells were seeded in DMEM media at a density of 5 × 103 cells per well and treated with indicated tested compounds (0–50 μM) for 48 h. The cell viability was evaluated as previously described [43].
Real-time quantitative PCR
1 × 106 RAW264.7 cells or 5 × 105 A549 cells were seeded in 12 well plates and cultured in 3% CSS DMEM media. After 24 h, the RAW264.7 cells were stimulated with 20 ng/mL LPS and 10 μM tested compounds. A549 cells were stimulated with 1 ng/mL PMA and 10 μM tested compounds. Eighteen hours later, total RNA was isolated from cells using the EZ-10 DNAaway RNA Mini-Preps Kit (Sangon Biotech, China) and cDNA was generated using the Hifair® III 1st Strand cDNA Synthesis SuperMix (YEASEN, China). Diluted cDNA was mixed with the forward primer, reverse primer, SYBR Green PCR Master Mix (YEASEN, China), and RNase-free water. Analysis of mRNA expression was carried out using the Applied Biosystems QuantStudio 3. The threshold cycles (Ct) for the control (GAPDH) and gene of interest were determined. All the samples were normalized to the level of GAPDH and the relative mRNA levels were calculated by the 2−∆∆Ct method. All primers are listed in Supplementary Table S2.
OPG and RANKL analysis
MG-63 cells were plated into 6-well plates at a concentration of 5 × 104 per well. After 24 h, cells were treated with the indicated test compounds and cultured for other 24 h. The RNA was isolated and analyzed by qPCR as above.
The selectivity on other nuclear receptors
PR and MR
PC3 and 293T cells were cultured in 3% CSS DMEM media with 1 × 104 cells/well in 96-well plates for 24 h, respectively. For PR assay, PC3 cells were transfected with 50 ng hPR, 50 ng ARR3tk-luciferase and 5 ng Renilla per well by lip3000 transfection reagent for 24 h. For MR activity, 293T cells were transfected with 71 ng pCMV-hMR, 24 ng pGL4.36[luc2P MMTV Hygro] and 5 ng Renilla per well by lip3000 transfection reagent for 24 h. For PR selectivity, cells were treated with 10 nM progesterone or indicated compounds and cultured for 16 h. Then the luciferase activity was measured as described above. For MR selectivity, cells were treated with 10 nM aldosterone or indicated compounds and cultured for 16 h. Then the luciferase activity was measured as described above.
AR
To investigate the agonist activity of the selected compounds on AR, the eGFP AR transcriptional activity was performed as previously described [43]. Briefly, For AR selectivity, cells were treated with 10 nM DHT or indicated compounds and cultured for 72 h. Then the fluorescence was measured by Synergy H1 (BioTek, USA; excitation, 485 nm; emission, 535 nm). Control wells with DMSO or hormone at 10 nM were included on each plate to define the 0% and 100% activation effects, respectively. Raw data were transformed to % activation with the following equation:
| 4 |
GR knockdown
SiRNA knockdown assays were performed to evaluate whether the compounds exerted anti-inflammation activity via the GR signaling pathway. The targeting siRNA for GR knockdown and non-targeting control (SiRNA NC or SiNC) were purchased from Biomics Biotech (Nantong; China). A549 cells were seeded to 60% confluency and transfected with 50 nM siRNA oligonucleotides using Lipofectamine3000 (Thermo, USA). After transfection with siRNA for 48 h, cells were treated with vehicle, 20 ng/mL TNF-ɑ with or without the tested compounds for 24 h. And then the total RNA was extracted as described as above. Q-PCR technology and Western blot technology were used to detect the mRNA and protein levels of GR, respectively. The anti-GR antibody (# 5153) was from Cell Signaling Technology.
The SiGR oligonucleotides were used:
SiGR-F: 5’-AAGCUUUCCUGGAGCAAAUAU-3’
SiGR-R: 5’-AUAUUUGCUCCAGGAAAGCUU-3’
RNA-seq
RAW264.7 cells were treated with 10 μM Dex or HT-15 combined with 20 ng/mL LPS overnight. Each treatment was duplicated. On the following day, RNA samples were isolated with triazole reagent and sequenced by the Hiseq-PE150 (Novogene, Beijing, China). The raw expression data were processed and normalized as we reported previously [44].
First, the mouse reference genome GRCm38 was downloaded from the Ensembl web site (https://www.ensembl.org). Four FASTQ files containing the paired-end sequence reads were aligned to the mouse reference genome using HISAT2 (version 2.1.0) [45]. The output SAM (sequencing alignment/map) files were converted to BAM (binary alignment/map) files and sorted using SAMtools (version 1.9) [46]. Next, HTSeq (version 2.0.) [47] counts for each gene how many aligned reads overlap its exons via referring to the mouse genome annotation file GRCm38.102.gtf downloaded from Ensembl website (https://www.ensembl.org). These counts were then used to determine the differentially expressed genes (DEGs) using the DESeq2 method [48]. Only the reads mapping unambiguously to a single gene were counted, whereas the reads aligned to multiple positions or overlapping with more than one gene were discarded. DEGs were defined as the genes whose false discovery rate (FDR) adjusted P value < 0.1 and |Log2fold change| > 1. To identify biological processes and pathways that are significantly enriched by DEGs, the gene list was analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.8) [49].
Confocal microscopy
A549 cells were seeded on sterile coverslips in 5% CSS DMEM media and incubated for 24 h, and then the cells were treated with the 10 μM tested compounds for 2 h. After aspiration of the media, cells were fixed in 4% paraformaldehyde and incubated with anti-GR antibody overnight. An Alexa-488 conjugated goat-anti rabbit lgG (Cell Signaling Technology, USA; # 4412) diluted at 1:1000 was used as the secondary antibody. Cell nuclei DNA was visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining (Beyotime, Shanghai; China). Images were recorded at 60 magnification using the Nikon A1R confocal spinning disk microscope (NiKon, Japan).
Data and statistical analysis
Statistical analyses were performed with the GraphPad Prism software v 6.0. The results were presented as the mean ± SD unless stated otherwise. The number of replicates was 5 or 6 per group for each data set. A two-tailed Student’s t test was utilized to evaluate the differences of two groups. The differences were analyzed with one-way analysis of variance (ANOVA), followed by Dunnett’s post hoc test between more than two groups. Some results were normalized to the control to avoid unwanted sources of variation. *(P < 0.05) was defined as significant between groups; ns (P > 0.05) was not significant.
Results and discussion
Evaluation of candidate compounds
The crystal structure of the GR LBD (PDB entry: 5G5W) was used as the template for docking-based virtual screening, and 54 potential GR modulators were obtained. Then, the inherent toxicity of these compounds toward RAW264.7 cells was evaluated at 25 μM, and the compounds that inhibited the growth rate of RAW264.7 cells lower than 30% were considered to have no significant inhibitory effect (Supplementary Fig. S1).
Most GR agonists such as Dex and prednisone exert their anti-inflammatory effects through the GR signaling pathway. The stimulated GR reduces the expression of certain inflammatory cytokines by inhibiting other pathways mediated by certain transcription factors such as AP-1 and NF-κB [16, 50]. At first, we evaluated the transrepression activities of the candidate compounds by the NF-κB-dependent luciferase assays. We co-transfected the full-length GR, NF-κB-luc and Renilla plasmids into HeLa cells and stimulated the cells with human TNF-α and 10 μM tested compounds to evaluate their activities in vitro. A total of 10 compounds reached ~50% inhibition compared with the TNF-α control group (Supplementary Fig. S2). Then, the 10 compounds were submitted to the GR competitive binding assay at a concentration of 10 μM, and three hits (HT-15, HT-34 and HT-36) were discovered to bind to GR (Fig. 1a, b). Afterwards, two cell-based reporter systems were used to examine the repression of the NF-κB-dependent transcription and AP-1-dependent transcription of the hits. The transrepression effect of 10 μM Dex was defined as 100% inhibition in both systems. In the NF-κB reporter assay, HT-15 showed a lower potency relative to Dex. The IC50 values of HT-15 and Dex were 1.86 ± 0.26 μM and 14.03 ± 2.19 nM, respectively (Fig. 1c). In the AP-1 reporter assay, the repression IC50 values of HT-15 and Dex were 1.06 ± 0.029 μM and 48.79 ± 7.1 nM, respectively, which are consistent with the results of the NF-κB reporter assay (Fig. 1d). The transrepression activities of HT-34 and HT-36 were lower than that of HT-15. Therefore, HT-15 showed the best anti-inflammatory potency among the three hits and was selected for further analysis.
It has been widely acknowledged that the side effects of GCs are mainly associated with the transactivation activity of GR [51]. Therefore, we established a stable MMTV-Luc expressing cell line to further evaluate the effect of HT-15 on the GR transactivation activity. In this assay, HT-15 did not show any potency while the EC50 of Dex was 4.5 ± 0.001 nM (TAag mode, Fig. 1e). In addition, in the mode of TAantag which measures the inhibition on the Dex-induced transactivation, HT-15 exhibited inconsiderable effect compared with the GR antagonist mifepristone and a reported SGRM Azd9567 (Fig. 1f). In conclusion, HT-15 demonstrated a better profile on the transactivation activity of GR, indicating that it probably induces less adverse effects. Furthermore, HT-15 did not inhibit the proliferation of NIH-3T3 cells, HepG2 cells, A549 cells, and MG-63 cells (Fig. 1g, h, Supplementary Figs. S3–4), and showed moderate effect on the growth of HeLa cells, Chang cells, and GES-1 cells (Supplementary Figs. S5–7) only under high concentrations above 10 μM, exhibiting a relatively safe profile.
HT-15 targets GR
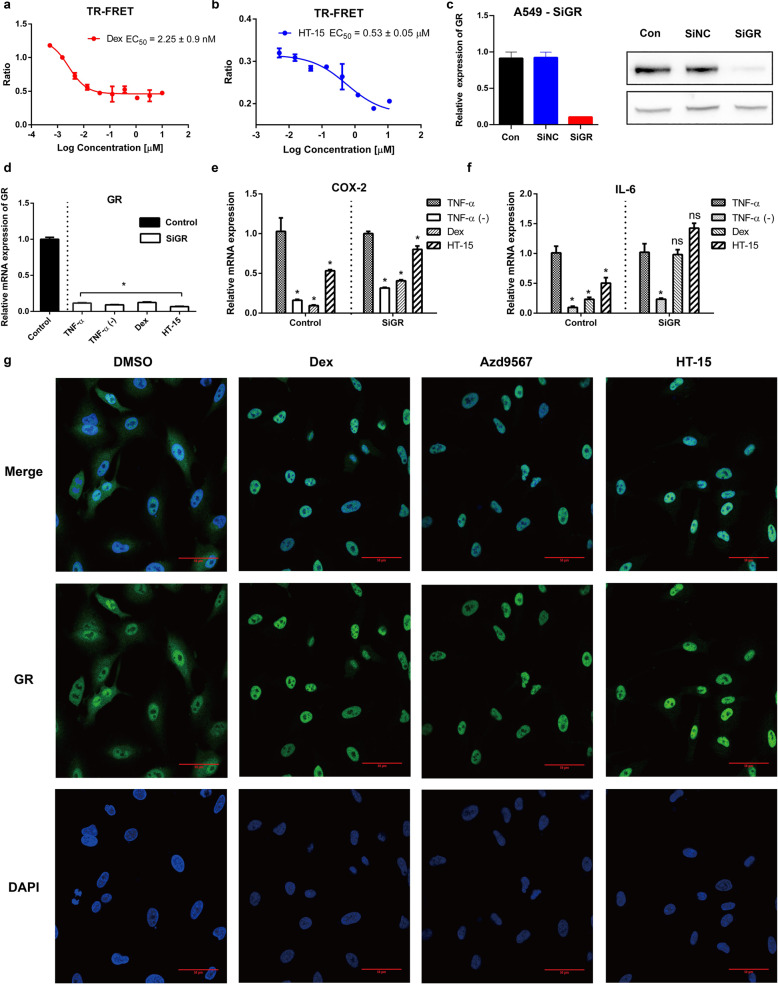
We investigated the affinity of HT-15 to GR with the LanthaScreen® TR-FRET GR Competitive Binding Assay at gradient concentrations. In the in vitro binding assay, HT-15 did display binding affinity with GR but relatively lower than Dex (EC50 HT-15 = 0.53 ± 0.05 μM and EC50 Dex = 2.25 ± 0.9 nM, Fig. 2a, b). To analyze whether the anti-inflammatory activity of HT-15 is truly related to GR, we knocked down the endogenous GR in A549 lung epithelial cells by small interfering RNA (SiRNA) (Fig. 2c, d) [52]. After the knockdown of the endogenous GR (A549-SiGR), COX-2 was downregulated by HT-15 and Dex, but the inhibitory effect was not as strong as the control group in which GR was normally expressed (Fig. 2e). The relative RNA expression of IL-6 was not suppressed by either Dex or HT-15 compared with the control group (Fig. 2f). In general, our results confirm that the anti-inflammatory effect of HT-15 is mainly associated with the GR signaling pathway.
Fig. 2. HT-15 targets GR.
a, b The LanthaScreen TR-FRET GR competitive binding assays of Dex and HT-15 (n = 4). c SiGR were transfected into A549 cells. After 48 h, the GR knockdown efficiency was determined with Q-PCR and Western blot (n = 5). d After knockdown, the mRNA expression levels of GR (n = 6). e, f The mRNA expression levels of IL-6 and COX-2 in normal A549 cells or in siRNA-treated control groups. Values were presented as mean ± SD. *P < 0.05 versus TNF-ɑ stimulated group (n = 6 in each group). g Effect of the compounds on GR nuclear localization. A549 cells were treated with the tested compounds (10 μM, 2 h) to visualize the nuclear accumulation of GR. (scale bar = 50 μm).
In addition, a true GR ligand is supposed to efficiently promote the GR nuclear translocation in which the translocation of GR into the nucleus is a key step for GCs to exert their anti-inflammatory effects [53]. Similar to Azd9567 and Dex, HT-15 promoted the translocation of GR from the cytoplasm to the nucleus (Fig. 2g). Taken together, HT-15 is a promising non-steroidal GR modulator.
HT-15 exhibited anti-inflammation activity through GR signaling pathway
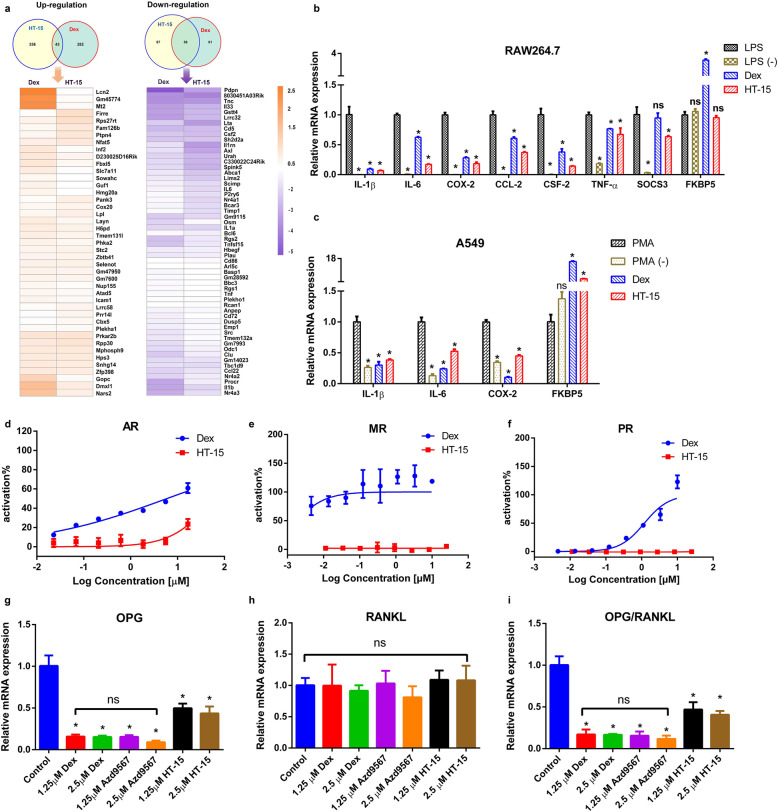
The anti-inflammatory effects of GCs are partially mediated via the inhibition of a vast number of pro-inflammatory cytokines, such as IL-1β, IL-6 and cyclooxygenase-2 (COX-2), by the transrepression mechanism or so-called tethering mechanism [54]. Macrophage cells play a key role in the initiation of inflammation response. Therefore, we conducted a transcriptome analysis with mouse macrophage RAW264.7 cells. To examine the overall anti-inflammatory activity of HT-15, the gene regulation pattern of HT-15 was profiled with Dex in parallel. The numbers of the repressed genes regulated by HT-15 and Dex are 143 and 117 respectively, and 56 are commonly repressed by both. Whereas, less than 15% induced genes are commonly induced by HT-15 and Dex (Fig. 3a). A pathway analysis of the 56 common genes regulated by HT-15 and Dex showed that the three most repressed pathways are TNF-α signaling pathway (Kegg pathway # mmu04668), hematopoietic cell lineage (Kegg pathway # mmu04060) and cytokine-cytokine receptor interaction (Kegg pathway # mmu04060) (Supplementary Table S3). Similar to Dex, HT-15 showed strong repression for a number of pro-inflammatory cytokines such as IL-1β, IL-6, IL-33 and TNF-α [12, 54]. The expression of pro-inflammatory cytokines triggers inflammatory response, and the inhibition of the cytokines is extremely important in immune regulation in patients with allergy, asthma, and other inflammatory diseases. In addition, in contrast to Dex, HT-15 was effective on the repression of cytokine signaling-3 (SOCS3) in macrophages. It has been reported that SOCS3 was upregulated in macrophages after infection and inflammation in different myeloid and lymphoid cell populations as well as in diverse non-hematopoietic cells. Furthermore, SOCS3 inhibited the TGFβ1/Smad3 signaling pathway, leading to enhanced LPS responses in macrophages [55]. The down-modulation of the SOCS3 expression might be effective in preventing the development of severe inflammatory diseases. We then validated the results by assessing the effect of HT-15 on the repression of proinflammatory cytokines by quantitative PCR, such as IL-1β, IL-6, COX-2, CSF-2, TNF-α, CCL-2, and SOCS3 (Fig. 3b). As expected, HT-15 repressed the LPS-induced transcription of a variety of proinflammatory genes such as IL-1β, TNF-α and COX-2, as efficiently as Dex. Moreover, HT-15 had a much higher repression activity on IL-6, CCL-2 and CSF-2 than Dex. Specially, SOCS3 was downregulated only by HT-15 but not Dex in RAW264.7 cells. Then the repression effects of HT-15 on IL-1β, IL-6 and COX-2 were also assessed in PMA-stimulated A549 cells, further confirming that HT-15 did inhibit the expression of pro-inflammatory cytokines (Fig. 3c). In addition, how FKBP5 is affected by HT-15 was also analyzed. FKBP5 is one of prominent GR upregulated genes, and it functions as a co-chaperone of GR, reduces the GCs-binding affinity to GR and decreases GR signaling capacity [12, 56]. We found that HT-15 did not induce the expression of FKBP5 in RAW264.7 while enhanced the expression of FKBP5 in A549 cells (Fig. 3b, c). The difference may be determined by the cell type-specific effects of GCs on the regulation of genes and signaling pathways [57]. Taken together, HT-15 exerted potent anti-inflammatory effects in vitro through the GR signaling pathway.
Fig. 3. HT-15 exerts anti-inflammation effects in vitro and potentially reduces the side effects caused by GCs.
a Venn diagrams of genes regulated by HT-15 and Dex in LPS (20 ng/mL) stimulated RAW264.7 cells and the gene expression profile of commonly induced or repressed genes by both HT-15 and Dex (n = 3). b Validation of the repressed genes (IL-1β, IL-6, TNF-ɑ, CCL-2, CSF-2, SOCS3, and COX-2) of HT-15 by Q-PCR and the effects of HT-15 on FKBP5 mRNA expression in RAW264.7 cells (n = 5). Values are presented as mean ± SD. *P < 0.05 versus LPS stimulated group. c The effect of HT-15 on the mRNA expressions of IL-1β, IL-6, COX-2 and FKBP5 in PMA-stimulated A549 cells (n = 5). Values are presented as mean ± SD. *P < 0.05 versus PMA-stimulated group. d–f An examination of the off-target effects by the reporter assays for AR, PR and MR (n = 4). g–i The influence of HT-15 on the mRNA expressions of OPG and RANKL (n = 5). *P < 0.05 versus DMSO treated group.
HT-15 reduced the side effects caused by GCs
As a transcription factor, GR belongs to the traditional steroid nuclear receptors (NRs), which also includes AR, PR, ER and MR. The activation of other NRs, such as PR and MR, is the major off-target effect caused by GCs due to their high structural similarity to GR [19, 21]. Therefore, we evaluated the off-target activity of HT-15 at various concentrations on AR, PR and MR. HT-15 displayed negligible activation effects on the three NRs, while Dex showed strong activation effects on the three NRs (Fig. 3d–f). Apparently, HT-15 has excellent selectivity for GR over the other three NRs, which would help to reduce the side effects caused by off-target.
The loss of bone is a well-known and unwanted phenomenon in the GCs treatment of patients with pulmonary, rheumatologic, autoimmune and hematopoietic diseases. Though the precise mechanism of GCs-induced bone loss remains unclear, a well-accepted theory is the imbalance between bone resorption and formation. Receptor activator of nuclear factor-κB ligand (RANKL) binds to receptor activator of nuclear factor-κB (RANK) to activate NF-κB and nuclear factor of activated T cells c1 (NFATC1), which stimulates osteoclast differentiation. The interaction between RANKL and RANK plays a pivotal role in the progress of osteoclast differentiation and function. Osteoprotegerin (OPG), a decoy receptor for RANKL, competitively disrupts the interaction between RANKL and RANK, and then inhibits osteoclast genesis and increases bone density in vivo. OPG is always regarded as a viable biomarker for bone formation rate [58, 59]. The OPG/RANKL ratio can be considered as an essential marker of osteoclast activation status, and the elevation of OPG/RANKL is favorable for bone formation. To evaluate the potential effect of HT-15 on bone, the mRNA expressions of OPG and RANKL were examined in human MG-63 cells and the OPG/RANKL ratio was determined. Though the mRNA expression of OPG was repressed by all the tested compounds, the expression level of the HT-15-treated groups was relatively higher than those of the Dex- and Azd9567-treated groups (Fig. 3g). The RANKL expression levels did not have significant difference between the groups (Fig. 3h). In addition, the results of OPG/RANKL were similar to those of OPG (Fig. 3i). In our assay, all the tested compounds showed repression on OPG, but the inhibitory effect of HT-15 is much weaker than those of Dex and Azd9567. Our experimental data manifest that HT-15 has reduced side effects on bone formation.
Binding mode analysis of HT-15 to GR LBD
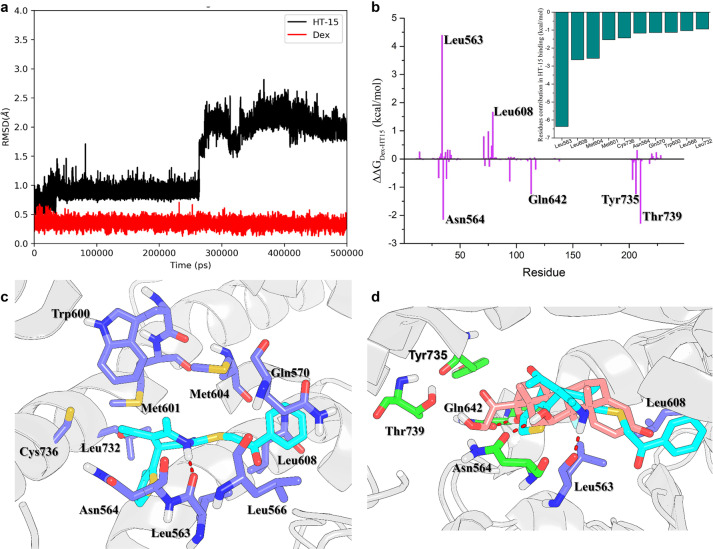
Our experiments confirmed that HT-15 has potent anti-inflammatory effects and reduced side effects in vitro. To understand the molecular basis of HT-15, MD simulations were performed to investigate the interactions between the GR LBD and Dex or HT-15. The RMSDs of HT-15 tend to converge after ~300 ns, while those of Dex are quite stable during the whole simulations with the RMSD fluctuations within 0.5 Å (Fig. 4a). The above results indicate that HT-15 is slightly more unstable in the ligand binding site of the GR LBD than Dex.
Fig. 4. The MD simulations and key residues in the GR LBD for the binding of HT-15.
a The time evolution of the RMSDs of the heavy atoms of HT-15 and Dex. b The energy differences between Dex and HT-15. The small figure is the 10 top-ranked residues in the GR LBD responsible for the binding of HT-15 predicted by MM/GBSA. c The structural analysis of the 10 top-ranked residues to the binding of HT-15. d Alignment of the representative structures of Dex (pink) and HT-15 (cyan) bound to the GR LBD. The residues that form stronger interaction with Dex than with HT-15 are highlighted in green. The residues that form stronger interaction with HT-15 than with Dex are highlighted in violet.
The snapshots from 450 to 500 ns were extracted from the MD trajectory for the subsequent structural and energetic analyses. As shown in Fig. 4b, the 10 top-ranked residues for the binding of HT-15 to the GR LBD predicted by the per-residue MM/GBSA free energy decomposition are Leu563, Leu608, Met604, Met601, Cys736, Asn564, Gln570, Trp600, Leu566 and Leu732. The structural analysis indicates the residue Leu563 forms hydrogen bond interactions with HT-15 to stabilize the binding of HT-15 (Fig. 4c). The benzene group of HT-15 forms hydrophobic interactions with Leu608, Met604 and Leu566.
To further characterize the binding difference between Dex and HT-15, the binding spectra of Dex and HT-15 were compared (ΔΔG = ΔGDex – ΔGHT-15). The positive values indicate that these residues form stronger interactions with HT-15 than Dex, while the negative values indicate that these residues form stronger interactions with Dex than HT-15 (Fig. 4b). It can be observed that most residues have similar energetic contributions to Dex and HT-15. However, the contributions of the residues of Leu563, Asn564, Leu608, Gln642, Tyr735 and Thr739 to Dex and HT-15 are quite different. The structural analysis shows that Dex can reach into the ligand binding site deeper than HT-15, and therefore the residues of Asn564, Gln642, Tyr735 and Thr739 in the deep ligand binding site can form stronger interactions with Dex than with HT-15 (Fig. 4d). In addition, the binding of HT-15 causes H12 to swing inward, and the volume of AF2 becomes smaller, which is not conducive to TIF2 binding (Supplementary Fig. S8) [60]. According to the above energetic and structural analyses, HT-15 bound in the shallower pocket will reduce the binding affinity with the GR LBD, but its side effects will be reduced.
Conclusion
For the therapy of several inflammatory and immune diseases, GCs maintains the dominance in the clinical application. However, multiple undesirable side effects of GCs limited their long-term use. Relevant therapeutic human monoclonal antibodies have been developed, but inconvenient for dosing and unfriendly in expense. Developing novel dissociated ligands of GR is probably an effective solution. In this work, we described a novel SGRM HT-15 discovered by virtual screening and bioassays. As expected, HT-15 was capable of dissociating transactivation from transrepression without cell toxicity. And HT-15 exhibited obvious binding affinity to the ligand binding pocket of GR. The predicted binding mode of HT-15 and GR LBD reveals that the hydrophobic interactions and the hydrogen bond interactions (residues Leu563 and Gln642) play crucial roles in stabilizing the binding of HT-15 to GR LBD. Besides, the GR knockdown assay and GR translocation assay further prove that the bioactivities of HT-15 are associated with GR. In addition, the inhibition on pro-inflammatory cytokines and chemokines has been shown to be an effective strategy for the treatment of diseases related to inflammation. It is observed in the gene expression analysis that the three most repressed pathways by HT-15 and Dex are the TNF-ɑ signaling pathway, hematopoietic cell lineage, and cytokine-cytokine receptor interaction. Especially, TNF-ɑ signaling pathway and cytokine-cytokine receptor interaction play crucial roles in the cellular inflammatory response [54, 61]. Further analysis shows that HT-15 has strong inhibition on the expression of pro-inflammatory cytokines such as IL-1β, IL-6, TNF-ɑ, and COX-2. Furthermore, HT-15 displays no cross-activity with AR, PR, or MR, and much weaker inhibition on OPG than Dex and Azd9567. In summary, HT-15 owns considerable anti-inflammation activity and meanwhile causes less adverse effects compared to Dex and Azd9567, which is a novel “dissociated” GR modulator and deserves further chemical modification and investigation.
Supplementary information
Acknowledgements
This study was supported by the Key R&D Program of Zhejiang Province (2020C03010), National Natural Science Foundation of China (81773632, 21907084), Zhejiang Provincial Natural Science Foundation of China (LZ19H300001), and the Key R&D Program of Zhejiang Province (2019C02024).
Author contributions
DL initiated and supervised the research. JPP and XPH designed the experiments, analyzed the experimental data and prepared the original draft. YXW, JJW, JNL, XC, XWW, XYW, LLZ, CS, FZ, QJW, and LX helped performing part of biological assays or the analysis of data. DL and TJH reviewed and edited the manuscript. All authors have critically revised the manuscript and approved its final version.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jin-ping Pang, Xue-ping Hu
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00855-6.
References
- 1.Krasselt M, Baerwald C. The current relevance and use of prednisone in rheumatoid arthritis. Expert Rev Clin Immunol. 2014;10:557–71. doi: 10.1586/1744666X.2014.904746. [DOI] [PubMed] [Google Scholar]
- 2.Hillier SG. Diamonds are forever: the cortisone legacy. J Endocrinol. 2007;195:1–6. doi: 10.1677/JOE-07-0309. [DOI] [PubMed] [Google Scholar]
- 3.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–10. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–86. doi: 10.1016/0092-8674(91)90370-E. [DOI] [PubMed] [Google Scholar]
- 5.Imai E, Stromstedt PE, Quinn PG, Carlstedt-Duke J, Gustafsson JA, Granner DK. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990;10:4712–9. doi: 10.1128/mcb.10.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20:53–8. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratman D, Vanden Berghe W, Dejager L, Libert C, Tavernier J, Beck IM, et al. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol Cell Endocrinol. 2013;380:41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Nixon M, Andrew R, Chapman KE. It takes two to tango: dimerisation of glucocorticoid receptor and its anti-inflammatory functions. Steroids. 2013;78:59–68. doi: 10.1016/j.steroids.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Escoter-Torres L, Greulich F, Quagliarini F, Wierer M, Uhlenhaut NH. Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 2020;48:8393–407. doi: 10.1093/nar/gkaa565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49:158–71. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weikum ER, de Vera IMS, Nwachukwu JC, Hudson WH, Nettles KW, Kojetin DJ, et al. Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes. Nucleic Acids Res. 2017;45:8596–608. doi: 10.1093/nar/gkx509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson WH, de Vera IMS, Nwachukwu JC, Weikum ER, Herbst AG, Yang Q, et al. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements. Nat Commun. 2018;9:1337. doi: 10.1038/s41467-018-03780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bosscher K, Haegeman G. Minireview: latest perspectives on antiinflammatory actions of glucocorticoids. Mol Endocrinol. 2009;23:281–91. doi: 10.1210/me.2008-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol. 2017;18:159–74. doi: 10.1038/nrm.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci. 1998;94:557–72. doi: 10.1042/cs0940557. [DOI] [PubMed] [Google Scholar]
- 17.Horby PW, Landray MJ, Mafham M, Bell JL, Linsell L, Staplin N, et al. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396:1345–52. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark AR, Belvisi MG. Maps and legends: The quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther. 2012;134:54–67. doi: 10.1016/j.pharmthera.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther. 2002;96:23–43. doi: 10.1016/S0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 20.Eddleston J, Herschbach J, Wagelie-Steffen AL, Christiansen SC, Zuraw BL. The anti-inflammatory effect of glucocorticoids is mediated by glucocorticoid-induced leucine zipper in epithelial cells. J Allergy Clin Immun. 2007;119:115–22. doi: 10.1016/j.jaci.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Frey FJ, Odermatt A, Frey BM. Glucocorticoid-mediated mineralocorticoid receptor activation and hypertension. Curr Opin Nephrol Hy. 2004;13:451–8. doi: 10.1097/01.mnh.0000133976.32559.b0. [DOI] [PubMed] [Google Scholar]
- 22.Ripa L, Edman K, Dearman M, Edenro G, Hendrickx R, Ullah V, et al. Discovery of a novel oral glucocorticoid receptor modulator (AZD9567) with improved side effect profile. J Med Chem. 2018;61:1785–99. doi: 10.1021/acs.jmedchem.7b01690. [DOI] [PubMed] [Google Scholar]
- 23.Shafiee A, Bucolo C, Budzynski E, Ward KW, Lopez FJ. In vivo ocular efficacy profile of mapracorat, a novel selective glucocorticoid receptor agonist, in rabbit models of ocular disease. Invest Ophth Vis Sci. 2011;52:1422–30. doi: 10.1167/iovs.10-5598. [DOI] [PubMed] [Google Scholar]
- 24.De Bosscher K, Vanden Berghe W, Beck IME, Van Molle W, Hennuyer N, Hapgood J, et al. A fully dissociated compound of plant origin for inflammatory gene repression. Proc Natl Acad Sci USA. 2005;102:15827–32. doi: 10.1073/pnas.0505554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittel S, Schacke H, Rehwinkel H, Docke W, Asadullah K, Zugel U, et al. The differential anti-inflammatory potential of SEGRA ZK 216348 and prednisolone relies on T cell apoptosis rather than modulation of dendritic cell activity. Exp Dermatol. 2009;18:305. [Google Scholar]
- 26.Owen HC, Miner JN, Ahmed SF, Farquharson C. The growth plate sparing effects of the selective glucocorticoid receptor modulator, AL-438. Mol Cell Endocrinol. 2007;264:164–70. doi: 10.1016/j.mce.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Myrback TH, Prothon S, Edman K, Leander J, Hashemi M, Dearman M, et al. Effects of a selective glucocorticoid receptor modulator (AZD9567) versus prednisolone in healthy volunteers: two phase 1, single-blind, randomised controlled trials. Lancet Rheumatol. 2020;2:E31–E41. doi: 10.1016/S2665-9913(19)30103-1. [DOI] [PubMed] [Google Scholar]
- 28.Onnis V, Kinsella GK, Carta G, Jagoe WN, Price T, Williams DC, et al. Virtual screening for the identification of novel nonsteroidal glucocorticoid modulators. J Med Chem. 2010;53:3065–74. doi: 10.1021/jm901452y. [DOI] [PubMed] [Google Scholar]
- 29.Hu X, Chai X, Wang X, Duan M, Pang J, Fu W, et al. Advances in the computational development of androgen receptor antagonists. Drug Discov Today. 2020;25:1453–61. doi: 10.1016/j.drudis.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Hemmerling M, Edman K, Lepisto M, Eriksson A, Ivanova S, Dahmen J, et al. Discovery of indazole ethers as novel, potent, non-steroidal glucocorticoid receptor modulators. Bioorg Med Chem Lett. 2016;26:5741–8. doi: 10.1016/j.bmcl.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 31.Schrödinger LLC NY, NY, USA, 2019.
- 32.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8:255–63. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Lee TS, Cerutti DS, Mermelstein D, Lin C, LeGrand S, Giese TJ, et al. GPU-accelerated molecular dynamics and free energy methods in Amber18: performance enhancements and new features. J Chem Inf Model. 2018;58:2043–50. doi: 10.1021/acs.jcim.8b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. generalized born. J Chem Theory Comput. 2012;8:1542–55. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambrakos SG, Boris JP, Oran ES, Chandrasekhar I, Nagumo M. A modified shake algorithm for maintaining rigid bonds in molecular-dynamics simulations of large molecules. J Comput Phys. 1989;85:473–86. doi: 10.1016/0021-9991(89)90160-5. [DOI] [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph Model. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem. 2011;32:866–77. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H, Duan L, Chen F, Liu H, Wang Z, Pan P, et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys Chem Chem Phys. 2018;20:14450–60. doi: 10.1039/C7CP07623A. [DOI] [PubMed] [Google Scholar]
- 40.Wang E, Weng G, Sun H, Du H, Zhu F, Chen F, et al. Assessing the performance of the MM/PBSA and MM/GBSA methods. 10. Impacts of enhanced sampling and variable dielectric model on protein-protein Interactions. Phys Chem Chem Phys. 2019;21:18958–69. doi: 10.1039/C9CP04096J. [DOI] [PubMed] [Google Scholar]
- 41.Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH, et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. 2019;119:9478–508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 42.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–94. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 43.Zhou W, Duan M, Fu W, Pang J, Tang Q, Sun H, et al. Discovery of novel androgen receptor ligands by structure-based virtual screening and bioassays. Genomics Proteom Bioinforma. 2018;16:416–27. doi: 10.1016/j.gpb.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang JP, Shen C, Zhou WF, Wang YX, Shan LH, Chai X, et al. Discovery of novel antagonists targeting the DNA binding domain of androgen receptor by integrated docking-based virtual screening and bioassays. Acta Pharmacol Sin. 2022;43:229–39. doi: 10.1038/s41401-021-00632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Landmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belvisi MG, Brown TJ, Wicks S, Foster ML. New glucocorticosteroids with an improved therapeutic ratio? Pulm Pharmacol Ther. 2001;14:221–7. doi: 10.1006/pupt.2001.0284. [DOI] [PubMed] [Google Scholar]
- 51.Austin RJH, Maschera B, Walker A, Fairbairn L, Meldrum E, Farrow SN, et al. Mometasone furoate is a less specific glucocorticoid than fluticasone propionate. Eur Respir J. 2002;20:1386–92. doi: 10.1183/09031936.02.02472001. [DOI] [PubMed] [Google Scholar]
- 52.He YZ, Xu Y, Zhang CH, Gao X, Dykema KJ, Martin KR, et al. Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory response. Sci Signal. 2011;4:ra44. doi: 10.1126/scisignal.2001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaaf MJM, Lewis-Tuffin LJ, Cidlowski JA. Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol Endocrinol. 2005;19:1501–15. doi: 10.1210/me.2005-0050. [DOI] [PubMed] [Google Scholar]
- 54.Akdis M, Burgler S, Crameri R, Eiwegger T, Fujita H, Gomez E, et al. Interleukins, from 1 to 37, and interferon-gamma: Receptors, functions, and roles in diseases. J Allergy Clin Immun. 2011;127:701–U317. doi: 10.1016/j.jaci.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Zhang YL, Yu YZ, Yang XA, Cao XT. SOCS3 promotes TLR4 response in macrophages by feedback inhibiting TGF-beta 1/Smad3 signaling. Mol Immunol. 2008;45:1405–13. doi: 10.1016/j.molimm.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 56.Van Zuiden M, Geuze E. Willemen HLDM, Vermetten E, Maas M, Amarouchi K, et al. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: a prospective study. Biol Psychiat. 2012;71:309–16. doi: 10.1016/j.biopsych.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 57.Franco LM, Gadkari M, Howe KN, Sun J, Kardava L, Kumar P, et al. Immune regulation by glucocorticoids can be linked to cell type-dependent transcriptional responses. J Exp Med. 2019;216:384–406. doi: 10.1084/jem.20180595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kramer HH, Hofbauer LC, Szalay G, Breimhorst M, Eberle T, Zieschang K, et al. Osteoprotegerin: a new biomarker for impaired bone metabolism in complex regional pain syndrome? Pain. 2014;155:889–95. doi: 10.1016/j.pain.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 59.Van Campenhout A, Golledge J. Osteoprotegerin, vascular calcification and atherosclerosis. Atherosclerosis. 2009;204:321–9. doi: 10.1016/j.atherosclerosis.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu X, Pang J, Zhang J, Shen C, Chai X, Wang E, et al. Discovery of novel GR ligands toward druggable GR antagonist conformations identified by MD simulations and Markov state model analysis. Adv Sci (Weinh). 2021; e2102435. 10.1002/advs.202102435 [DOI] [PMC free article] [PubMed]
- 61.Pietras EM. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130:1693–8. doi: 10.1182/blood-2017-06-780882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.