ABSTRACT
In summer 2019, widespread occurrence of crown gall disease caused by Agrobacterium spp. was observed on commercially grown ornamental plants in southern Iran. Beside agrobacteria, pale yellow-pigmented Gram-negative strains resembling the members of Xanthomonas were also associated with crown gall tissues on weeping fig (Ficus benjamina) and Amaranthus sp. plants. The purpose of the present study was to characterize the crown gall-associated Xanthomonas strains using plant inoculation assays, molecular-phylogenetic analyses, and comparative genomics approaches. Pathogenicity tests showed that the Xanthomonas strains did not induce disease symptoms on their host of isolation. However, the strains induced hypersensitive reaction on tobacco, geranium, melon, squash, and tomato leaves via leaf infiltration. Multilocus sequence analysis suggested that the strains belong to clade IA of Xanthomonas, phylogenetically close to Xanthomonas translucens, X. theicola, and X. hyacinthi. Average nucleotide identity and digital DNA-DNA hybridization values between the whole-genome sequences of the strains isolated in this study and reference Xanthomonas strains are far below the accepted thresholds for the definition of prokaryotic species, signifying that these strains could be defined as two new species within clade IA of Xanthomonas. Comparative genomics showed that the strains isolated from crown gall tissues are genetically distinct from X. translucens, as almost all the type III secretion system genes and type III effectors are lacking in the former group. The data obtained in this study provide novel insight into the breadth of genetic diversity of crown gall-associated bacteria and pave the way for research on gall-associated Xanthomonas-plant interactions.
IMPORTANCE Tumorigenic agrobacteria—members of the bacterial family Rhizobiaceae—cause crown gall and hairy root diseases on a broad range of plant species. These bacteria are responsible for economic losses in nurseries of important fruit trees and ornamental plants. The microclimate of crown gall and their accompanying microorganisms has rarely been studied for the microbial diversity and population dynamics of gall-associated bacteria. Here, we employed a series of biochemical tests, pathogenicity assays, and molecular-phylogenetic analyses, supplemented with comparative genomics, to elucidate the biological features, taxonomic position, and genomic repertories of five crown gall-associated Xanthomonas strains isolated from weeping fig and Amaranthus sp. plants in Iran. The strains investigated in this study induced hypersensitive reactions (HR) on geranium, melon, squash, tobacco, and tomato leaves, while they were nonpathogenic on their host of isolation. Phylogenetic analyses and whole-genome-sequence-based average nucleotide identity (ANI)/digital DNA-DNA hybridization (dDDH) calculations suggested that the Xanthomonas strains isolated from crown gall tissues belong to two taxonomically unique clades closely related to the clade IA species of the genus, i.e., X. translucens, X. hyacinthi, and X. theicola.
KEYWORDS: Agrobacterium sp., Amaranthus sp., clade I Xanthomonas, Ficus benjamina, T3SS, weeping fig
INTRODUCTION
Crown gall, a bacterial plant disease caused by tumorigenic agrobacteria (i.e., Agrobacterium spp., Allorhizobium spp., and Rhizobium spp.), is one of the economically most important constraints in commercial nurseries and the ornamental-plant industry (1). Crown gall symptoms include overgrowth and hyperplasia in the young tissues of plants due to infection with agrobacterial strains harboring the tumor-inducing (Ti) plasmid. Transfer of the T-DNA fragment of the Ti plasmid into the plant cells transforms them to synthesize plant hormones, i.e., indole-3-acetic acid (IAA; auxin) and cytokinin, and initiates gall formation. The imbalanced status of phytohormones inside the plant tissues leads to overgrowth and gall formation at the site of infection (2). Genetically transformed plant cells within the gall tissue are also responsible for synthesizing strain-specific nutrient sources called opines to be used by the gall-inducing pathogen. The nutrient-rich environment of crown gall tissue is a sink of amino acids and sugars or organic acids that can also be used as nutrients for the growth of several bacterial groups other than the pathogenic agrobacteria. Hence, gall tissue is a suitable environment to be colonized by epiphytic and endophytic populations of bacterial species (3).
During the past decade, severe outbreaks of crown and stem gall diseases were observed in commercial nurseries and ornamental-plant greenhouses in southern Iran. Several agrobacterial strains were isolated from symptomatic plants and investigated using molecular-phylogenetic approaches. As a result, dozens of Agrobacterium spp. and Allorhizobium species strains which were isolated from gall tissues were shown to cause the same symptoms on the host plants under greenhouse conditions (4). However, we have simultaneously isolated a number of atypical bacterial strains from the gall issues of weeping fig (Ficus benjamina) and Amaranthus sp. plants in the Fars province of southern Iran. Preliminary examinations, i.e., Gram reaction, colony color, and morphology on culture media, revealed that the gall-associated atypical bacterial strains resemble members of the genus Xanthomonas (5).
Xanthomonas constitutes a group of plant-associated or plant-pathogenic bacterial strains capable of infecting taxonomically diverse plant species. These bacteria cause devastating diseases on hundreds of annual crops, fruit trees and ornamental species, including economically important crops within the families Cucurbitaceae, Fabaceae, Poaceae, Rosaceae, and Solanaceae (6, 7). Currently, the genus comprises 31 validly described species, some of which have several subspecies and pathovars (8). Plant-pathogenic xanthomonads use a type III secretion system (T3SS) to translocate a cocktail of different effector proteins into host plant cells, referred to as type III effectors (T3Es). These T3Es are generally classified into transcription activator-like effectors (TALEs) and non-TALEs, with the latter group also being known as Xanthomonas outer proteins (Xops). Nucleic acid sequence-based phylogenetic analyses suggest that xanthomonads can be divided into two distinct clades; clade I includes the economically important small-grain-cereal pathogen Xanthomonas translucens, as well as a number of less important species, such as X. albilineans, X. hyacinthi, and X. theicola (9), whereas clade II xanthomonads include most of the well-studied plant pathogens, i.e., X. campestris, X. oryzae, and X. arboricola, along with the former X. axonopodis species complex, which includes X. citri and X. euvesicatoria (6, 10).
The purposes of the present study were to identify and characterize the Xanthomonas-like bacterial strains isolated from crown gall tissues of weeping fig and Amaranthus sp. plants in Iran. With this aim, we conducted a series of inoculation assays on the host of isolation as well as a number of taxonomically diverse plant species under greenhouse conditions. Multilocus sequence analysis (MLSA) of four housekeeping genes was also conducted to determine the phylogenetic position of the strains within the Xanthomonas species. Furthermore, whole-genome-sequence-based comparisons and comparative genomics were used to elucidate the exact taxonomic position, genomic features, and virulence-associated repertories of the strains. Taken together, phenotypic and molecular-phylogenetic analyses revealed that the strains associated with weeping fig and Amaranthus sp. crown gall tissues belong to two taxonomically unique clades closely related to the clade IA species of the genus, i.e., X. translucens, X. hyacinthi, and X. theicola.
RESULTS
Microbiological features of the bacterial strains.
Five bacterial strains (Table 1) which were morphologically distinct from the crown gall pathogenic agrobacteria were isolated from crown gall tissues of weeping fig and Amaranthus sp. plants in Shiraz (Iran) in 2019. Pale yellow, domed, circular colonies 1 to 2 mm in diameter were observed on general culture media, i.e., nutrient agar (NA) and yeast-extract peptone glucose agar (YPGA), after 48 to 72 h of incubation. All the strains were Gram negative, oxidase negative, and catalase positive and had oxidative metabolism. All the strains had amylolytic and pectolytic activity and were able to hydrolyze esculin, Tween 20/80, gelatin, and casein. They were able to utilize cellobiose, maltose, inositol, galactose, fructose, arabinose, xylose, and sorbitol. They were negative for utilization of raffinose, while all the reference strains of xanthomonads were positive for this feature. The strains produced pale yellow and mucoid colonies on yeast extract-dextrose-calcium carbonate (YDC) agar medium, characteristic of Xanthomonas spp. Based on the morphological features and results of biochemical tests, the bacterial strains were suspected to belong to the genus Xanthomonas.
TABLE 1.
Biochemical characteristics and pathogenicity of the Xanthomonas strains investigated in this studya
| Strain | Pectolytic activity | Amylolytic activity | Utilization of: |
Virulence on: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raffinose | Mannitol | Barley | Common bean | Cucumber | Maize | Melon | Squash | Sunflower | Tomato | Watermelon | Wheat | Tobacco | Geranium | |||
| Xanthomonas sp. strain FX1 | +b | + | − | + | − | − | − | − | * | * | − | * | − | − | * | * |
| Xanthomonas sp. strain FX2 | + | + | − | + | − | − | − | − | * | * | − | * | − | − | * | * |
| Xanthomonas sp. strain FX3 | + | + | − | + | − | − | − | − | * | * | − | * | − | − | * | * |
| Xanthomonas sp. strain FX4 | + | + | − | + | − | − | − | − | * | * | − | * | − | − | * | * |
| Xanthomonas sp. strain AmX2 | + | + | − | + | − | − | − | − | * | * | − | * | − | − | * | * |
| X. translucens pv. translucens ICMP 5752T | − | + | + | + | + | ND | ND | ND | ND | * | ND | * | ND | − | ND | ND |
| X. translucens pv. undulosa ICMP 11055 | − | + | + | + | + | ND | ND | ND | ND | ND | ND | ND | ND | + | ND | ND |
| X. arboricola pv. pruni ICMP 17186 | − | + | + | − | ND | ND | ND | ND | ND | ND | ND | + | ND | ND | + | + |
| X. arboricola pv. juglandis CFPB 8608 | − | + | + | − | ND | ND | ND | ND | ND | ND | ND | + | ND | ND | + | + |
| X. citri subsp. citri SU261 | − | + | + | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| X. euvesicatoria pv. euvesicatoria ICMP 22075 | − | − | + | + | ND | ND | − | ND | ND | ND | ND | + | ND | ND | ND | ND |
| X. gardneri ICMP 16689 | − | − | + | + | ND | ND | − | ND | ND | ND | ND | + | ND | ND | ND | ND |
| X alfalfae subsp. alfalfa Xa119 | − | + | + | + | ND | + | ND | ND | ND | ND | ND | − | ND | ND | + | ND |
ND, not determined; *, induced an HR on these plants.
+, indicates positive; −, indicates negative results.
Pathogenicity and plant colonization of the strains.
The bacterial strains were evaluated for pathogenicity on their host of isolation as well as a set of taxonomically diverse annual crops, vegetables, and weeds using three inoculation techniques, i.e., spraying, leaf infiltration, and needle prick. All the plant species inoculated with spraying and needle prick techniques remained asymptomatic until 20 days postinoculation (dpi). Thus, the gall-associated Xanthomonas-like bacteria did not induce symptoms on the plants when they were inoculated alone. However, when the strains isolated in this study were coinoculated on sunflower plantlets along with the Agrobacterium larrymoorei strain Ficamol, the former group did not affect the pathogenicity of crown gall pathogen, and Agrobacterium-induced crown gall symptoms were observed on the inoculated plants 20 to 30 dpi (Fig. 1A to C). In the leaf infiltration method, a hypersensitivity reaction (HR), i.e., necrotic areas with a blackish brown appearance, was observed at the site of inoculation on squash (Fig. 1D), melon (Fig. 1E), and tomato (Fig. 1F) as well as tobacco and geranium leaves 36 to 48 hpi. The same HR symptoms were observed on tomato and squash leaves infiltrated with X. translucens pv. translucens ICMP 5752T (Fig. 1G and H; Table 1). Reisolation of the bacterial strains from crown gall tissues of the plants which were coinoculated with A. larrymoorei and Xanthomonas sp. revealed a lower proportion of Xanthomonas colonies than Agrobacterium colonies on culture media (data not shown).
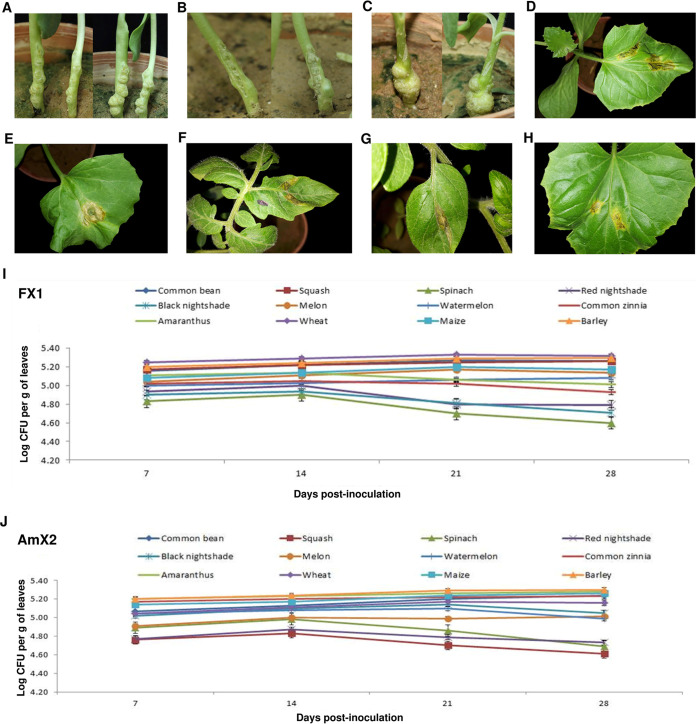
FIG 1.
Crown gall symptoms induced on sunflower plants inoculated with Agrobacterium larrymoorei strain Ficamol alone (A), Ficamol plus Xanthomonas strain FX1 (B), and Ficamol plus Xanthomonas strain AmX2 (C) under greenhouse conditions 14 (left) and 21 (right) days postinoculation. Leaf infiltration of squash (D), melon (E), and tomato (F) plants using bacterial suspension of the strain FX1 led to a hypersensitive reaction (HR) (necrotic areas with a blackish brown appearance) at the site of inoculation 36 to 48 hpi. The same symptoms were observed when tomato (G) and squash (H) leaves were infiltrated with X. translucens pv. translucens ICMP 5752T as control. In planta bacterial population sizes of Xanthomonas strains FX1 (I) and AmX2 (J) on 12 annual crops and weed species under greenhouse conditions, recorded and presented in log CFU/g of the leaf tissue at 7, 14, 21, and 28 days postinoculation. Wheat and spinach plants showed the highest and lowest population of the strains, respectively, in all the time frames. Error bars indicate standard deviations.
Furthermore, in order to provide precise insight into the biology of the strains isolated in this study, in planta growth of strains AmX2 and FX1 was investigated on 12 plant species, i.e., Amaranthus sp., barley, black nightshade (Solanum nigrum), red nightshade (Solanum villosum), common bean, common zinnia (Zinnia elegans), maize, melon, spinach (Spinacia oleracea L. cv. Hudson), squash, watermelon, and wheat, under greenhouse conditions. Population sizes of the inoculated bacterial strains were determined 7, 14, 21, and 28 dpi and presented in log CFU/g of the leaf tissue (Fig. 1I and J). On the seventh day of inoculation, populations of both FX1 and AmX2 strains were smaller on spinach, black nightshade, and red nightshade than on the other plant species, while the largest populations of FX1 and AmX2 were observed on wheat and barley leaves, respectively. The same patterns were observed 14, 21, and 28 dpi; populations of both strains subsided over this period of time on spinach, black nightshade, and red nightshade to the lowest level at 28 dpi. On the other hand, no significant variation was observed in the population count of the strains on barley, common bean, maize, squash, and wheat over time. The highest population over the period of 28 days was observed on wheat plants inoculated with strain FX1 (Fig. 1I and J).
Molecular-phylogenetic analyses.
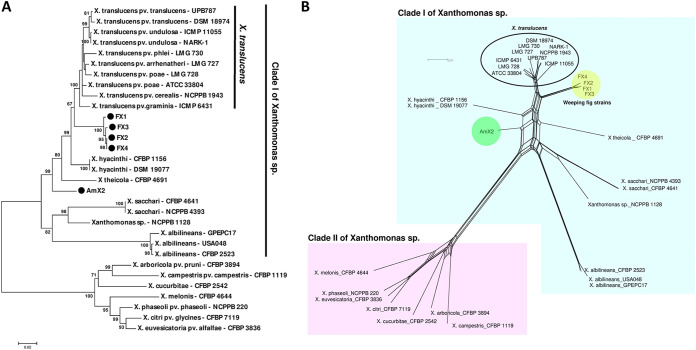
The five Xanthomonas-like strains were subjected to a set of molecular-phylogenetic analyses to elucidate their precise taxonomic position. Xanthomonas-specific PCR primers Xc-lip-F2 and Xc-lip-R2 directed the amplification of a 777-bp DNA fragment in all the strains, confirming their identity as members of this genus. However, none of the species-specific PCR primers designed for detection of different plant-pathogenic Xanthomonas species/pathovars (see Table S1 in the supplemental material) could amplify the expected DNA fragment, leaving the species status of the strains undetermined. A BLAST search on the NCBI GenBank database using the individual sequences of four housekeeping genes, i.e., atpD, efp, gyrB, and rpoD, revealed that the five strains had variable sequence similarity (93 to 99% identity) to different species within clade I of Xanthomonas, i.e., X. translucens, X. hyacinthi, X. theicola, and X. sacchari (data not shown). MLSA-based phylogenetic investigations using the concatenated sequences of the four genes showed that the strains isolated from weeping fig were clustered in a monophyletic clade close to the small-grain-cereal pathogen X. translucens, while strain AmX2, isolated from Amaranthus sp., clustered distinctly from all the species within clade I (Fig. 2A). Because maximum-likelihood phylogeny can present conflicting topologies, a phylogenetic network was generated using the NeighborNet method (11) for the concatenated data set of sequences (Fig. 2B). The phylogenetic network confirmed divergence of the weeping fig strains from the X. translucens species complex, showing that the two groups were clustered in separate clades. The closest species to strain AmX2 was X. hyacinthi, while the two clades were too distinct to be considered members of the same species (Fig. 2B).
FIG 2.
Phylogeny of crown gall-associated Xanthomonas strains isolated in this study based on the concatenated sequences of four housekeeping genes, i.e., atpD, efp, gyrB, and rpoD, using tree-based maximum-likelihood (A) and network-based NeighborNet (B) analyses. The five Xanthomonas strains isolated from weeping fig and Amaranthus sp. were divided into two clades, where the weeping fig strains were closely related to but still distinct from the X. translucens species complex. The Amaranthus sp. strain was distinct from all the validly described species with clade I of Xanthomonas. Further, parallel lines/branches within the nodes of the NeighborNet network indicate possible recombination between the connected strains.
Genome sequencing and overall genome relatedness.
Considering the MLSA results, three representative strains, i.e., FX1, FX4, and AmX2, were subjected to whole-genome sequencing using the Illumina HiSeq X platform and assembled using the SPAdes genome assembler (12). Quality assessment of the genome assembly and annotation using the BUSCO online service (13) confirmed the accuracy and completeness of all three genomes compared to the complete genome sequence of X. translucens DSM 18974. Details of the genome quality assessment criteria are shown in Table S2A. The genome sizes of the strains were 5,383 kbp (in 152 contigs [FX1]), 5,383 kbp (in 89 contigs [FX4]), and 5,069 kbp (in 75 contigs [AmX2]), with the GC content ranging from 68.8% to 69.8%, which is within the typical range of clade I Xanthomonas strains. Based on the annotation obtained with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (14), strains FX1, FX4, and AmX2 contain a total of 4,502, 4,468, and 4,170 coding genes, respectively, with 40, 68, and 41 pseudogenes, respectively. Genomic features of the strains sequenced in this study in comparison to the phylogenetically closely related species are shown in Tables S3 and S4.
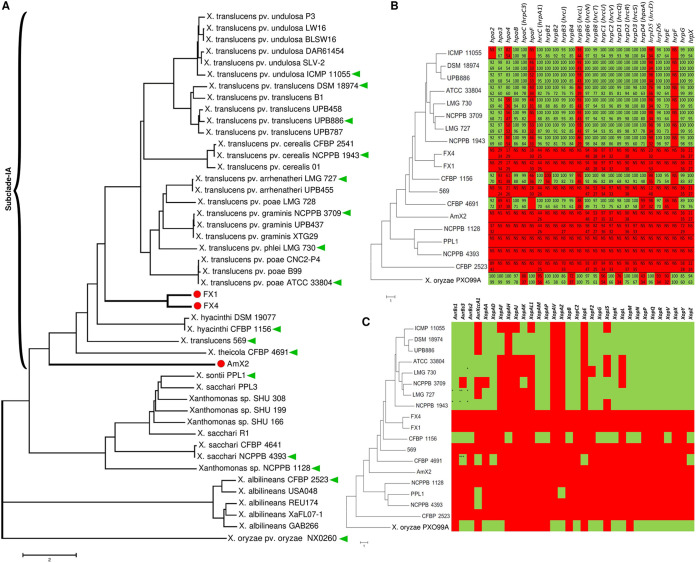
A genome-based distance matrix calculator (ANI/AAI-Matrix) was used to conduct average nucleotide identity (ANI) distance clustering of genome sequences obtained in this study, first for all 32 reference strains of Xanthomonas spp. and then for the representative species of clade I xanthomonads available in the NCBI GenBank database. The resulted neighbor-joining tree revealed distinct clustering of strains FX1 and FX4 and the other clade IA xanthomonads. The former strains were clustered in a monophyletic clade closely related to but still distinct from the X. translucens species complex. The strain AmX2 was also distinct from the closest neighbor species, i.e., X. hyacinthi (Fig. 3A). In order to estimate DNA similarity indexes between the strains sequenced in this study and all Xanthomonas species, ANI values were calculated using three online services—JSpeciesWS, ANI Calculator, and OrthoANIu (15–17) (http://enve-omics.ce.gatech.edu/g-matrix/)—and the ultimate ANI value was obtained via combination and averaging of the results generated by the three online services.
FIG 3.
Average nucleotide identity (ANI)-based neighbor-joining distance clustering tree of all available clade I Xanthomonas strains constructed using the ANI/AAI-Matrix genome-based distance matrix calculator (A). The clade I xanthomonads were divided into two subclades (9), where strains FX1, FX4, and AmX2, which were sequenced in this study, were clustered in subclade IA. Predicted type III secretion system (T3SS) (B) proteins and Xanthomonas outer proteins (Xops or non-TALEs) (C) in the three Xanthomonas strains sequenced in this study were compared to a representative set of clade I Xanthomonas genomes retrieved from the NCBI GenBank. (A) Red circles indicate the three strains sequenced in this study; green triangles indicate the strains included in the comparative genomics analyses. (B and C) Green and red squares indicate the presence and absence of the respective genes, respectively, while the numbers inside the squares (B) indicate the query coverage (top numbers) and sequence similarity (bottom numbers) indices in the BLASTp search. NS, no significant similarity was found in BLASTp.
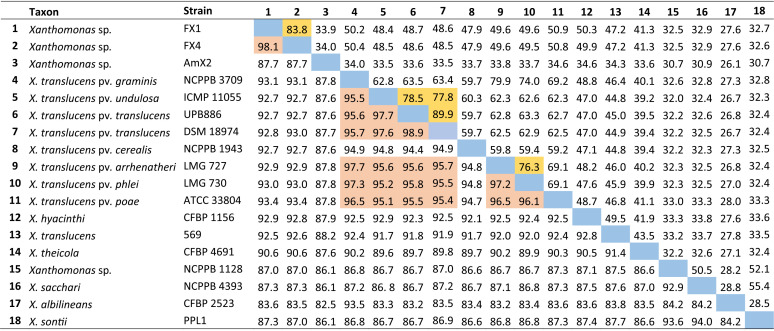
According to the MLSA results, which confirmed taxonomic position of the strains isolated in this study in clade I of Xanthomonas spp., type strains of all clade I Xanthomonas species were included in DNA similarity analyses. The resulting data showed that the strains FX1 and FX4 had only 83.6% to 93.1% sequence identity with the type strains of previously described Xanthomonas species, while the ANI value between strain AmX2 and the other Xanthomonas species was less than 90.0% (Table 2). On the other hand, the digital DNA-DNA hybridization (dDDH) value (18) among the strains sequenced in this study and those of the reference strains was consistently lower than 60.0%. These ANI and dDDH values are far below the accepted threshold for prokaryotic species definition, i.e., ≤95% and ≤70% for ANI and dDDH, respectively, suggesting that strains FX1, FX4, and AmX2 belong to two new species within clade IA of Xanthomonas. While strains FX1 and FX4 belong to the same species, strain AmX2 could be considered another standalone species distinct from all validly described Xanthomonas spp. (Fig. 3A). A formal comprehensive polyphasic taxonomic study is warranted to further refine the position of these strains within the genus.
TABLE 2.
ANI and dDDH values among the representative set of clade I Xanthomonas, including the strains sequenced in this studya
A combination of ANI and dDDH indices was used to designate a taxonomic status for a given phylogenetic clade; strains FX1 and FX4 were suspected to belong to a novel species, while strain AmX2 could be considered another novel species. Highlighted squares indicate ANI (below the diagonal) and dDDH (above the diagonal) values higher than the accepted threshold for the definition of prokaryotic species.
Comparative genomics.
It has been shown that clade I Xanthomonas species possess high genetic diversity in terms of the T3SS, T3Es, and TALEs. Genome-informed investigation of Xanthomonas spp. would provide valuable information on the virulence repertories, pathogenicity mechanisms, and host adaptation of the strains (9). Furthermore, genomic investigation of the Xanthomonas strains associated with crown gall tissues would decipher their potential unique capabilities that support their survival in the gall environment. Significant variations were observed among the clade 1 xanthomonads in their T3SS repertories (Fig. 3B). When a 60% cutoff index was used for both query coverage and sequence similarity criteria, no T3SS gene was detected in the three strains sequenced in this study in BLASTp-based investigations (http://www.ncbi.nlm.nih.gov/blast/). However, a number of suspected sequences with high query coverage but low sequence similarity were predicted in strains AmX2, FX1, and FX4 (Fig. 3B). For instance, a sequence similar to hrpB6 (94% query coverage and 48% sequence similarity) was predicted in AmX2, FX1, and FX4. hrpB6 is a T3SS ATPase in X. euvesicatoria (gene ID: 61777304). Furthermore, hrpC2 (originally described in X. euvesicatoria; accession number P80150), which belongs to the FHIPEP (flagella/HR/invasion protein export pore) family, was detected in all the three strains with 97% query coverage and 32% similarity. The T3SS pattern in the strains AmX2, FX1, and FX4 was similar to those of X. translucens strain 569, Xanthomonas sp. strain NCPPB 1128, “X. sontii” strain PPL1, X. sacchari strain NCPPB 4393, and X. albilineans strain CFBP 2523, while members of the X. translucens species complex carried most of the T3SS genes (Fig. 3B).
As for the Xanthomonas outer proteins (Xops or non-TALEs), in silico analyses revealed that the three strains sequenced in this study encode none of the Xops included in this investigation. The same pattern was observed in X. translucens 569, Xanthomonas sp. strain NCPPB 1128, and X. albilineans CFBP 2523, which was significantly different from that observed in the X. translucens species complex (Fig. 3C). The protein XopAV was absent in all the clade I xanthomonads investigated in this study, while XopAH, XopE, and XopAZ were present only in X. hyacinthi CFBP 1156, X. theicola CFBP 4691, and “X. sontii” strain PPL1, respectively. In order to visualize the T3SS and Xop repertories of the strains sequenced in this study, BRIG (19) was implemented in whole-genome-based comparisons of strains FX1, FX4, and AmX2 using X. theicola strain CFBP 4691 and X. translucens pv. translucens strain DSM 18974 as reference genomes (Fig. S1). Considering the satisfactory completeness of the genome sequencing and assembly of the three gall-associated strains (Table S2A), circular mapping against the reference genomes CFBP 4691 (Fig. S1A) and DSM 18974 (Fig. S1B) confirmed the absence of the T3SS and xop genes in the three sequences obtained in this study.
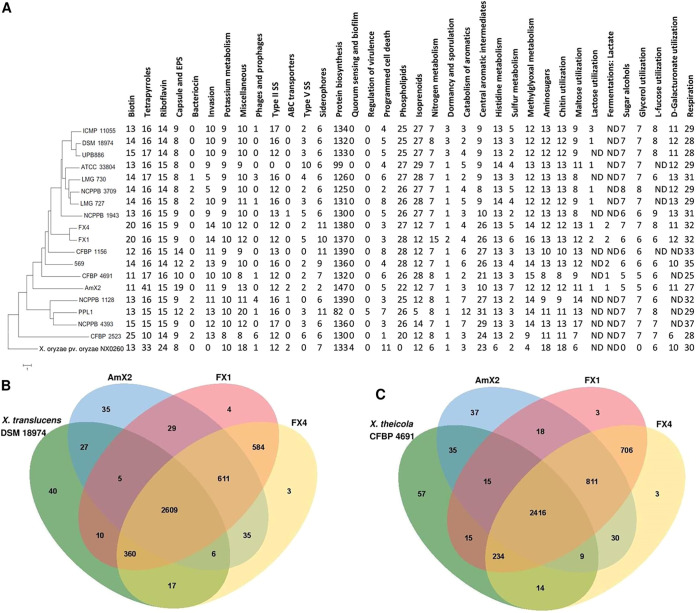
Comparative genomics using the data provided by the RAST (20) and SEED-Viewer (21) databases revealed several lines of evidences highlighting differences in metabolic networks and subsystem profiles between the three strains sequenced in this study and those of the clade I xanthomonads, as well as the reference strain X. oryzae pv. oryzae PXO99A (Fig. 4A). For instance, the number of subsystems that are responsible for biotin biosynthesis, invasion and intracellular resistance, siderophore production, protein biosynthesis, nitrogen metabolism, central aromatic intermediate metabolism, sulfur metabolism, methylglyoxal metabolism, potassium metabolism, maltose and maltodextrin utilization, respiration, and the miscellaneous subsystems in strains FX1 and FX4 was higher than in the members of their neighbor clade X. translucens. The number of subsystems responsible for tetrapyrroles, capsular and extracellular polysaccharides, ABC transporters, protein biosynthesis, and sulfur metabolism in the strain AmX2 was higher than in the other genomes investigated. In the genome of X. oryzae pv. oryzae, which belongs to clade II of the xanthomonads, numbers of subsystems related to riboflavin biosynthesis, T3SS, amino sugars, chitin and N-acetylglucosamine utilization, programmed cell death, and toxin-antitoxin systems were significantly higher than in the clade I xanthomonads.
FIG 4.
Genomic characteristics of three Xanthomonas strains sequenced in this study compared to a set of representative clade I xanthomonads calculated using the online annotating service RAST (A). Venn diagram constructed using the OrthoVenn online service showing the shared gene families (orthologous clusters) among the three strains sequenced in this study and the two reference genomes from X. translucens pv. translucens DSM 18974 (B) and X. theicola CFBP 4691 (C).
Orthologous gene clusters were determined using the OrthoVenn online service (22) through four-versus-four designations of the strains, where one of the clade I strains DSM 18974 and CFBP 4691 was used as the reference genome in each analysis (Fig. 4B and C). When X. translucens strain DSM 18974 was the reference, the three strains sequenced in this study shared 611 proteins besides the 2,609 proteins which were shared among all four strains. Strains AmX2, FX1, and FX4 possessed 35, 4, and 3 unique proteins, respectively, when DSM 18974 was designated the reference genome. Furthermore, FX1 and FX4 shared 584 proteins with each other while these strains shared 29 and 35 proteins, respectively, with AmX2, indicating closer genomic repertories of FX1 and FX4. On the other hand, when X. theicola CFBP 4691 was the reference genome, the three strains sequenced in this study showed 811 shared proteins along with 2,416 proteins which were shared within all four genomes. Strains AmX2, FX1, and FX4 possessed 37, 3, and 3 unique proteins, respectively, when CFBP 4691 was considered the reference genome (Fig. 4B and C).
The representative genomes of clade I xanthomonads along with the three genomes obtained in this study were screened using the web-based tool BAGEL4 (23) for the presence of hypothetical bacteriocin-encoding genes/clusters and antibiotic peptides. Zoocin A was detected in strains FX1, FX4, and AmX2 as well as all the other clade I xanthomonads except X. theicola CFBP 4691. The sactipeptide class of microbial products was detected in FX1 but not in FX4 and AmX2. Interestingly, rhodanodin, which is a lasso peptide produced by Rhodanobacter thiooxydans, was detected in AmX2 but not in any other Xanthomonas genome evaluated in this study (Table S5). The PHASTER online service (24) was used to detect prophage sequences within the bacterial genomes. Escherichia phage 500465-1 was predicted in the genome of FX1, FX4, and AmX2 as well as a number of reference strains. The Escherichia phage 500465-1 was the only prophage predicted in FX1 and FX4, but strain AmX2 had the Ralstonia phage phiRSA1 in its genome along with X. translucens pv. undulosa strain ICMP 11055. Table S6 shows the list of prophage sequences in the clade I xanthomonads that were missing in the strains sequenced in this study. However, it should be noted that due to the draft nature of the genomes, it is possible that more phage sequences are present in these strains but were not identified in the present study due to unresolved repetitive sequence.
No integrative plasmid (episome) was detected using the PlasmidFinder online service (25) in the genomes of strains in this study. Visualization of genomic islands in the whole-genome sequences of the three Xanthomonas sp. strains obtained in this study against the genomes of two reference strains, X. theicola CFBP 4691 and X. translucens pv. translucens DSM 18974, is shown in Fig. S2. The distribution pattern of the main genomic islands in all the three strains sequenced in this study remained constant regardless of the reference genome used for prediction. When strain CFBP 4691 was considered the reference genome, two dominant genomic islands were predicted in FX1, located between 0.9Mbp and 1.0Mbp and between 2.5Mbp and 2.6Mbp. The two islands were predicted when DSM 18974 was used as the reference, although the positions of the islands were slightly different (1.5Mbp to 1.6Mbp and 2.8Mbp to 2.9Mbp). Similarly, a genomic island was constantly predicted in FX4 when both the reference genomes were used, while the position of the island changed from 2.1Mbp to 2.2Mbp and 2.9Mbp to 3.0Mbp when CFBP 4691 and DSM 18974 were used as reference genomes, respectively (Fig. S2). The same pattern was observed in strain AmX2, where a dominant genomic island was predicted at 1.6Mbp to 1.7bp and 2.3Mbp to 2.4Mbp when CFBP 4691 and DSM 18974 were used as reference genomes, respectively. These results indicate different patterns of genomic island distribution among the strains sequenced in this study despite the fact that they belong to the same species, i.e., FX1 and FX4 (Fig. S2).
DISCUSSION
Five Xanthomonas sp. strains have accidentally been isolated from crown gall tissues along with several Agrobacterium strains, while the latter organisms were the main causal agent of the disease. In this study, we employed a series of biochemical tests, pathogenicity assays, and MLSA-based phylogenetic analyses on five crown gall-associated Xanthomonas strains isolated from weeping fig and Amaranthus sp. plants in Iran. Subsequently, three representative strains, i.e., FX1, FX4, and Amx2, were subjected to genome sequencing and comparative genomics to elucidate their biological features, taxonomic positions, and genomic repertories. The strains investigated in this study induced a hypersensitive reaction (HR) on geranium, melon, squash, tobacco, and tomato leaves, while they were nonpathogenic on their host of isolation. MLSA-based phylogenetic analyses accomplished with whole-genome sequence-based ANI/dDDH calculations suggested that the Xanthomonas strains isolated from crown gall tissues could be considered two novel species within clade I of Xanthomonas.
No Xanthomonas pathogen has so far been described as colonizing Amaranthus sp., while two distinct xanthomonads infecting Ficus spp. plants are described in the literature (26, 27): angular leaf spot of ornamental Ficus spp. species caused by X. campestris pv. fici (28, 29) and angular leaf spot of pipal (Ficus religiosa) caused by a taxonomically undetermined Xanthomonas species. In fact, the latter pathogen (pipal pathogen) has originally been designated a standalone species and described as Xanthomonas fici in India (27). However, Young et al. (30) noted that the name “X. fici Jindal & Patel 1972” is illegitimate because it is antedated by “X. fici Cavara 1905.” On the other hand, the causal agent of the bacterial blight of Ficus elastica in Florida (USA) was identified as X. axonopodis based on the nucleotide sequence of its 16S rRNA gene (31). The strains isolated in this study had no phylogenetic similarity to those of the previously described species infecting Ficus sp. plants. Our strains were clustered within the clade I of Xanthomonas, while the above-mentioned species—originally described by Young et al. (32) and Parkinson et al. (33)—are members of clade II of the genus.
A physiologically active crown gall produces specific nutrients (opines) utilizable by microorganisms, providing a rich niche for plant-associated bacteria to colonize, grow, and form complex ecological interactions (34–37). The microbial communities involved in opine ecology are not limited to Agrobacterium members. Complex bacterial communities of many species coexist in crown gall tissues (38). During the past few decades, various bacterial species, i.e., Pseudomonas spp. and coryneform bacteria, were isolated from crown gall tissues and possess the capability of competing for opines as growth substrates (34). Hence, agrobacteria, either in the soil or in association with healthy or diseased plants, are likely in environments that require mechanisms to compete with other microbes for resources (39). The bacterial type VI secretion system (T6SS) delivers effector proteins in a contact-dependent manner to antagonize other bacteria and provides a quantifiable competitive advantage over accompanying susceptible counterparts. It has been noted that only a fraction of sequenced Agrobacterium tumefaciens strains carry T6SS-encoding loci. Wu et al. (39) stated that in some cases, all the members of a genomospecies appear to lack T6SS-encoding loci, whereas in other genomospecies, T6SS loci are polymorphic in terms of presence and/or absence. Since we did not generate whole-genome sequences of the agrobacterial strains isolated alongside xanthomonads, the presence and putative role of T6SS in their interaction remain undetermined.
The genome of Pseudomonas kilonensis strain 1855-344, which can catabolize octopine, was recently shown to contain an octopine-catabolic operon named ooxAB (40). Successful colonization and long-term persistence of Agrobacterium strains on/in crown gall tissue depend partly on competition for opine nutritive resources, either between the agrobacteria or among other microorganisms, such as pseudomonads, Sinorhizobium spp., coryneform bacteria, or even fungal species such as Cylindrocarpon heteronema and Fusarium solani (36, 37). It was recently shown that the microbial community of natural grapevine (Vitis vinifera) crown galls caused by Allorhizobium vitis contains more than 150 bacterial species, among which members of Pseudomonas, Sphingomonas, and Erwinia were more common (38). Furthermore, crown galls can harbor bacteria belonging to Corynebacterium or Arthrobacter (37). The abundance of A. vitis in grapevine crown gall is positively correlated with the abundance of Novosphingobium sp., Xanthomonas sp., and members of Microbacteriaceae, suggesting the presence of a microbial hub in the crown gall environment (3). The co-occurrence of Xanthomonas with A. vitis on crown gall tissues can be explained by the presence of high concentrations of lignin, cellulose, N-glycosylated proteins, and other cell wall precursors as well as cell wall degradation products found in bark on developing gall tissue (3, 41, 42). BLAST-based in silico analyses using the sequences of ooxA, ooxB, and ooxAB against the three genome sequences obtained in this study revealed the presence of these genes in the three Xanthomonas strains. As detailed in Table S2B, sequences of DNA fragments with 66 to 98% query coverage and 25 to 27% sequence similarity to ooxA, ooxB, and ooxAB genes were detected in the genomes of all three strains. These observations suggest the potential acquisition of opine catabolism genes by gall-associated xanthomonads, which needs further in-depth investigation.
Plant inoculation assays showed that the crown gall-associated Xanthomonas strains were nonpathogenic on the foliage parts of their host plant and the other plant species evaluated via spray inoculation. They also did not affect the virulence and aggressiveness of their Agrobacterium counterparts with which they were isolated from gall tissues. However, when the strains were inoculated onto geranium, melon, squash, tobacco, and tomato via leaf infiltration, an HR was observed 36 to 48 hpi. A similar pattern was observed in Gram-positive actinobacteria, where Curtobacterium flaccumfaciens strains isolated from asymptomatic solanaceous vegetables, i.e., eggplant, pepper, and tomato, were pathogenic on leguminous crops, i.e., common bean, cowpea, and mung bean, but not on solanaceous plants (43, 44). A high level of in planta (either epiphytic or endophytic) populations of the gall-associated Xanthomonas strains on barley and wheat 28 dpi revealed their potential ability to persist on small grain cereals. The gall-associated strains are taxonomically closely related to the X. translucens species complex, which infects gramineous plants and includes economically important pathogens around the globe. However, neither spray inoculation nor leaf infiltration of the three gall-associated Xanthomonas strains induced symptoms on wheat plants under greenhouse conditions (data not shown). Hence, these gall-associated strains could epiphytically persist on nonhost plants, i.e., small-grain cereals and seed materials, interfering with the quarantine inspection and pathogen survey procedure.
Epiphyte growth and persistence of different xanthomonads on nonhost plants have frequently been reported (45, 46). For instance, X. arboricola pv. pruni and X. citri subsp. citri, pathogens of stone fruits and citrus, respectively, multiplied on red nightshade, black nightshade, bindweed, Chenopodium, common bean, and wheat up to 20 dpi under greenhouse conditions. Interestingly, X. arboricola pv. pruni successfully proliferated on both lemon and sweet lemon up to 140 dpi, attaining a population density even higher than that of X. citri subsp. citri (47). Xanthomonas euvesicatoria pv. euvesicatoria strains isolated from pepper were able to colonize nightshade and common bean (48, 49). All these observations highlight the role of nonhost plants in the epidemiology of bacterial plant pathogens, which needs further attention, especially when it comes to decision-making for management of the corresponding diseases under natural field conditions.
Virulence of most plant-pathogenic xanthomonads is attributed to the function of the T3SS and a set of transcription activator-like effectors (TALEs) and non-TALES or T3Es/Xops (7, 50). Comparative genomics revealed that the crown gall-associated Xanthomonas strains might be adapted to a nonpathogenic lifestyle, which is reflected by the lack of pathogenicity gene clusters present in the pathogenic members of clade I (Fig. 3B and C). In spite of the absence of all T3SS and T3E repertories in the three strains sequenced in this study, the induction of HR phenotype warrants further exploration. The same results were observed by Jacobs et al. (51), who reported that X. cannabis strains NCPPB 3753 and NCPPB 2877 isolated from cannabis (Cannabis sativa L.) as well as the Xanthomonas maliensis strain 97M isolated from rice lacked the Hrp T3SS and did not contain any of the known T3Es while stile inducing HR on nonhost plants. Further, it has been shown that nonpathogenic Pseudomonas strains lacking a T3SS are common leaf colonizers of healthy plants and grow as well as or better than other Pseudomonas syringae strains on nonhost species without causing disease (52). These nonpathogenic Pseudomonas strains also typically fail to induce an HR on tobacco, unlike the Xanthomonas strains isolated in this study.
An in-depth comparative analysis with the other clade I xanthomonads' genomes allowed us to obtain more precise insight underlying genetic diversity of these bacteria. Subsystem analyses using RAST (20) highlighted differences in the genetic contents of clade I xanthomonads, especially regarding the differences between the cereal pathogen X. translucens and the strains isolated in this study. For instance, the number of subsystems that are responsible for biotin biosynthesis, invasion and intracellular resistance, siderophore production, protein biosynthesis, nitrogen metabolism, central aromatic intermediates metabolism, sulfur metabolism, methylglyoxal metabolism, potassium metabolism, maltose and maltodextrin utilization, respiration, and the miscellaneous subsystems in the strains FX1 and FX4 was higher than in the members of their neighbor clade X. translucens. Furthermore, prophage sequences were missing in the strains sequenced in this study, but since these are draft genomes, it is possible that more phage sequences (which are often repetitive) are present in these strains but were not identified in the present study due to unresolved repetitive sequence.
The strains sequenced in this study are phylogenetically closely related to the tea (Camellia sinensis) pathogen X. theicola and small-grain-cereal pathogen X. translucens, all belonging to clade I of the genus, also known as early-branching xanthomonads (9, 32, 33). Providentially, high-quality complete genome sequences of representative strains from both these clade IA pathogens are available in the NCBI GenBank database, allowing us to obtain precise comparative genomics among the early-branching group of xanthomonads (33). Recently, a complete genome sequence of the type strain of X. theicola CFBP 4691 was published, aiming to better understand the biology and evolution of the clade 1 xanthomonads (9). Further, Shah et al. (53) conducted a comprehensive phylogenomics and comparative genomics analysis using the complete genome sequences of 14 X. translucens strains and several other xanthomonads as reference genomes. Draft genome sequences of the three gall-associated Xanthomonas strains revealed their unique phylogenetic position, suggesting that they should be described as two novel species (Fig. 3A). Furthermore, strain 569, which is labeled as X. translucens, should not be included within this complex species, as the ANI values between this strain and the other members of X. translucens were consistently below 93%, suggesting that strain 569 cannot be considered X. translucens. Together, our observations indicate that a formal and comprehensive taxonomic study is warranted to address the last two taxonomic issues within X. translucens and further refine the classification of this complex species. While the phylogenetic position of strains FX1, FX4, and AmX2 was clarified in these analyses, only further investigations using a larger collection of strains will shed light on the genetic content, biological characteristics, and taxonomic status of crown gall-associated members of Xanthomonas.
MATERIALS AND METHODS
Phenotypic characterization of the strains.
Five yellow-pigmented Gram-negative bacterial strains (FX1 to FX4 and AmX2) (Table 1) resembling the members of Xanthomonas were isolated from crown gall tissues along with dozens of Agrobacterium strains in southern Iran (4, 54, 55). The bacterial strains were resuspended in sterile distilled water (SDW) and were stored at 4°C for further use and in 15% glycerol at −70°C for long-term purposes. All purified bacterial strains were subjected to standard biochemical and physiological tests (5). Gram reaction, oxidase and catalase activity, aerobic/anaerobic growth (O/F), and colony characteristics on YDC medium were determined. Furthermore, the strains were evaluated for their capacity to degrade starch and pectate. Type/reference strains of “Agrobacterium fabrum” (C58), X. euvesicatoria pv. euvesicatoria (ICMP 109T), X. euvesicatoria pv. perforans (ICMP 16690T), X. translucens pv. undulosa (ICMP 11055), X. translucens pv. translucens (DSM 18974T), and X. arboricola pv. pruni (ICMP 17186) were used as controls in all phenotypic and biochemical tests. The biochemical tests were repeated twice.
Plant inoculation assays.
The hypersensitive reaction (HR) was evaluated on tobacco (Nicotiana tabacum cv. Turkish) and geranium (Pelargonium graveolens) leaves using the bacterial suspension from a 48-h-old culture on NA medium at a concentration of 108 CFU/ml. The five strains were evaluated for their pathogenicity on their host of isolation, i.e., weeping fig and Amaranthus sp., as well as a set of taxonomically diverse annual crops, vegetables, and weeds, i.e., barley (Hordeum vulgare cv. Behrokh, https://link.springer.com/article/10.1007/s11032-019-1026-z, https://doi.org/10.1111/ppl.13102), common bean (Phaseolus vulgaris cv. Sadri), cucumber (Cucumis sativus cv. RS2N-NADA), maize (Zea mays cv. KSC703), melon (Cucumis melo cv. Nagham), squash (Cucurbita pepo cv. Elena), sunflower (Helianthus annuus cv. Armavirski), tomato (Solanum lycopersicum cv. Sunseed 6189), watermelon (Citrullus lanatus cv. B32), and wheat (Triticum aestivum cv. Behrokh, https://link.springer.com/article/10.1007/s11032-019-1026-z, https://doi.org/10.1111/ppl.13102). Plant growth conditions and inoculum preparation were as described previously (56), while inoculation was performed using spray inoculation, leaf infiltration, and needle prick techniques as described by Mafakheri et al. (4). The strains were evaluated either alone or in combination with the tumorigenic Agrobacterium sp. strain Ficamol, with which they were simultaneously isolated from crown gall tissues. Reference strains of the above-mentioned xanthomonads were used as positive controls, while SDW was used as the negative control. Koch’s postulates were accomplished by reisolating the inoculated strains on YPGA medium from the inoculated plants showing symptoms. Confirmation of the identity of the reisolated bacteria was performed using the Gram reaction and colony morphology on YDC medium as described above.
In planta growth of the strains AmX2 and FX1 was investigated on 12 plant species using the procedure detailed previously (47, 56). Population size of the inoculated bacterial strains was evaluated on each of the plant species at 7, 14, 21, and 28 dpi. In brief, three subsamples from the lower, middle, and upper portions of the canopy were collected from each inoculated plant species. The leaves were combined, the weight of each leaf sample was adjusted to 2 g, and the plant tissues were macerated using a sterile mortar and pestle in 40 ml of SDW. After appropriate serial dilutions (10−1 to 10−4), 10 μl of the resulted bacterial suspension was plated on YPGA medium (in 9-cm-diameter plates). All bacterial colonies were counted 4 days postincubation using a colony counter (CX-300; Gallenkamp, England) and the bacterial population sizes were expressed as log CFU/g of fresh leaf tissues as described previously (42). All the experiments were repeated at least once.
Molecular-phylogenetic analyses.
In order to determine the precise taxonomic and phylogenetic position of the strains, they were subjected to PCR tests using the Xanthomonas-specific PCR primers Xc-lip-F2 and Xc-lip-R2 as well as several species-specific primers, as detailed in Table S1. Furthermore, multilocus sequence analysis (MLSA) using the sequences of four housekeeping genes, i.e., atpD, efp, gyrB, and rpoD, was performed on all five Xanthomonas strains using the procedure described by Khojasteh et al. (57) (Table S1). Bacterial DNA extraction, PCR parameters, and sequencing procedure were the same as described previously (57). Subsequently, sequences of these four genes in a collection of reference Xanthomonas sp. strains were retrieved from the GenBank databases and included in the phylogenetic analyses. The sequences were concatenated following the alphabetic order of the genes, and a phylogenetic tree was constructed using the maximum-likelihood method with MEGA7 software (58). The model of evolution for maximum-likelihood analysis was determined using the Modeltest tab in MEGA7, and the phylogenetic tree was constructed with bootstrapping (1,000 replications). Furthermore, SplitsTree version 4.14.4 was used to construct the NeighborNet network of the concatenated sequences of the four genes using the same set of sequences (11).
Whole-genome sequencing and annotation.
Based on the MLSA results, the weeping fig strains FX1 and FX4 and the Amaranthus sp. strain AmX2 were subjected to whole-genome sequencing to further refine the taxonomic status and genomic repertoires of the strains. Total DNA of the strains was extracted using NucleoSpin microbial DNA extraction kit (Macherey-Nagel, Germany). DNA libraries were obtained with a Nextera XT DNA library prep kit (Illumina, USA). Paired-end sequencing (2 × 150 bp) was performed on an Illumina HiSeq X platform. Reads were demultiplexed using BaseSpace (Illumina). Paired-reads quality filtering and trimming were performed with the bbduk program (59). Adaptor trimming was performed using Trimmomatic (Galaxy version 0.38.1) implemented on the Galaxy web server (60). The read quality was assessed with FastQC (Galaxy version 0.72+galaxy1; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and de novo sequence assembly was performed with the SPAdes genome assembler (Galaxy version 3.12.0+galaxy1) (12) using the “careful correction” option (–careful) and automatically chosen k-mer values. Short (<200-bp) and very-low-coverage (<2-fold) contigs were excluded from the final assembly. Quantitative assessment of genome assembly and annotation completeness was evaluated using BUSCO online software (13). Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline with default settings (14). Additionally, the RAST server (20) was used for fully automated annotation of the bacterial genomes. Total numbers of protein-coding genes, RNA genes, and pseudogenes were determined for all the genomes.
Whole-genome-sequence-based comparisons and comparative genomics.
MLSA-based phylogenetic analyses revealed the gall-associated strains belong to clade I of Xanthomonas; hence; genomic data for a representative set of 42 clade I xanthomonad strains were retrieved from the NCBI GenBank database and subjected to phylogenetic analyses. Average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values were calculated among all the type/reference strains of Xanthomonas species using the procedure described previously (53, 61). ANI was estimated using both one-versus-one and all-versus-all strategies via different algorithms, i.e., JSpeciesWS (15), ANI Calculator (16), and OrthoANIu (17). Additionally, the Genome-to-Genome Distance Calculator (GGDC, v2.1) online service was used to calculate dDDH values among the strains (18). ANI-based all-versus-all distance matrix and the resulting neighbor-joining tree were generated using the ANI/AAI-Matrix genome-based distance matrix calculator (16) (http://enve-omics.ce.gatech.edu/g-matrix/). A combination of ANI and dDDH indices was used to assign “stand-alone species” taxonomic status to a given taxon. When both ANI and dDDH values were below the accepted threshold, i.e., ≤95% and ≤70% for ANI and dDDH, respectively, the corresponding strain was considered a potential novel species (62).
Based on the results obtained from the phylogenetic analyses, the three strains sequenced in this study were subjected to comparative genomics against several clade I Xanthomonas species, including the closest reference strains, X. theicola CFBP 4691 and X. translucens pv. translucens DSM 18974 (9, 63). Both tBLASTn- and BLASTp-based investigations (http://www.ncbi.nlm.nih.gov/blast/) were performed to decipher the T3SS repertories and Xanthomonas outer proteins (Xops) of the strains sequenced in this study in comparison to the other clade I xanthomonads. Sequences of a set of 26 T3SS and 32 Xops were retrieved from the complete genome sequences of X. translucens pv. translucens DSM 18974, X. theicola CFBP 4691, and X. oryzae pv. oryzae PXO99A and included in the BLAST searches. Amino acid sequences (BLASTp) were used in all the analyses, while nucleotide sequences (tBLASTn) were also implemented in the investigations for further confirmation. For both the BLASTp and tBLASTn, an E value of <1e−10 with 60% query coverage and 60% sequence similarity was considered the cutoff criterion, as recommended previously (64).
Genome collinearity analyses provide valuable insight into the genome arrangement and gene distribution in bacteria (65, 66). Thus, pairwise genome collinearity alignment of the three strains was performed and visualized with BRIG 0.95 using both X. theicola CFBP 4691 and X. translucens pv. translucens DSM 18974 as reference genomes (19). The annotation obtained by RAST (20) was used to reconstruct metabolic networks and subsystems. Given that the annotation procedure might affect the outcome of such comparative genomics analyses, we used the genome sequences that all annotated using the same technique as shown in Table S4. Subsequently, the genomes were transferred to the comparative environment of SEED-Viewer for comparative genomics analyses (21). The online services PlasmidFinder 2.0 (25), PHASTER (24), and BAGEL4 (23) were used to screen the genomes for the presence of integrative plasmids/episomes, prophage sequences, and bacteriocin-encoding genes/clusters, respectively. Further, genome-wide comparisons and visualization of orthologous clusters were performed using the online service OrthoVenn (22). IslandViewer 4 was used for the identification and visualization of genomic islands in the three strains sequenced in this study using X. theicola CFBP 4691 and X. translucens pv. translucens DSM 18974 as reference genomes (67).
Data availability.
The nucleotide sequences obtained in this study were deposited into the GenBank database under the following accession numbers: atpD, MW222614 to MW222618; efp, MW222619 to MW222623; gyrB, MW222624 to MW222628; and rpoD, MW222629 to MW222633. Whole-genome sequences of the three strains were also deposited into the NCBI GenBank database under the following accession numbers: AmX2, JAFIWC000000000 (BioProject no. PRJNA700118); FX1, JAFJNT000000000 (BioProject no. PRJNA700116); and FX4, JAFIWB000000000 (BioProject no. PRJNA700117). Furthermore, pure cultures of the strains have been deposited in the CIRM-CFBP culture collection (French Collection for Plant-associated Bacteria) and assigned the following accession numbers: AmX2, CFBP 8902; FX1, CFBP 8700, FX2, CFBP 8701, FX3, CFBP 8702, and FX4, CFBP 8703.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cécile Dutrieux and Audrey Lathus at CIRM-CFBP for preservation of the bacterial strains.
E.O. conceived and designed the study with assistance from H.M. and S.Z. H.M. and S.Z. carried out the experiments with assistance from S.M.T., M.S.H., N.K., and I.D. E.O. analyzed and interpreted the data with assistance from T.R., H.M., and R.K. Technical support was provided by P.P. and M.F.-L.S. E.O. prepared the paper with assistance from H.M. All the authors revised the final version of the manuscript.
This article is based upon work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology). The work of N.K. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Project number 429677233. I.D. acknowledges the Ministry of Education, Science and Technological Development of the Republic of Serbia and INTERREG ADRION OIS-AIR (Open Innovation System of the Adriatic-Ionian Region) proof-of-concept project for the financial support of his research (contracts 451-03-9/2021-14/200178 and OIS-AIR 77). H.M. and S.Z. benefited from a sabbatical grant provided by the Iranian Ministry of Science and Technology for a 6-month fellowship at the University of Tehran, Iran. The work of E.O. was funded by University of Tehran, Iran.
Footnotes
Supplemental material is available online only.
Contributor Information
Nemanja Kuzmanović, Email: nemanja.kuzmanovic@julius-kuehn.de.
Ebrahim Osdaghi, Email: eosdaghi@ut.ac.ir.
Lindsey Price Burbank, USDA—San Joaquin Valley Agricultural Sciences Center.
REFERENCES
- 1.Kuzmanović N, Puławska J, Hao L, Burr TJ. 2018. The ecology of Agrobacterium vitis and management of crown gall disease in vineyards. Curr Top Microbiol Immunol 418:15–53. doi: 10.1007/82_2018_85. [DOI] [PubMed] [Google Scholar]
- 2.González-Mula A, Lang J, Grandclément C, Naquin D, Ahmar M, Soulère L, Queneau Y, Dessaux Y, Faure D. 2018. Lifestyle of the biotroph Agrobacterium tumefaciens in the ecological niche constructed on its host plant. New Phytol 219:350–362. doi: 10.1111/nph.15164. [DOI] [PubMed] [Google Scholar]
- 3.Gan HM, Szegedi E, Fersi R, Chebil S, Kovács L, Kawaguchi A, Hudson AO, Burr TJ, Savka MA. 2019. Insight into the microbial co-occurrence and diversity of 73 grapevine (Vitis vinifera) crown galls collected across the Northern hemisphere. Front Microbiol 10:1896. doi: 10.3389/fmicb.2019.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mafakheri H, Taghavi SM, Puławska J, de Lajudie P, Lassalle F, Osdaghi E. 2019. Two novel genomospecies in the Agrobacterium tumefaciens species complex associated with rose crown gall. Phytopathology 109:1859–1868. doi: 10.1094/PHYTO-05-19-0178-R. [DOI] [PubMed] [Google Scholar]
- 5.Schaad NW, Jones JB, Chun W. 2001. Laboratory guide for the identification of plant pathogenic bacteria, 3rd ed. APS Press, St. Paul, MN. [Google Scholar]
- 6.Jacques M-A, Arlat M, Boulanger A, Boureau T, Carrère S, Cesbron S, Chen NWG, Cociancich S, Darrasse A, Denancé N, Fischer-Le Saux M, Gagnevin L, Koebnik R, Lauber E, Noël LD, Pieretti I, Portier P, Pruvost O, Rieux A, Robène I, Royer M, Szurek B, Verdier V, Vernière C. 2016. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu Rev Phytopathol 54:163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 7.Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB. 2020. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat Rev Microbiol 18:415–427. doi: 10.1038/s41579-020-0361-8. [DOI] [PubMed] [Google Scholar]
- 8.Catara V, Cubero J, Pothier JF, Bosis E, Bragard C, Đermić E, Holeva MC, Jacques M-A, Petter F, Pruvost O, Robène I, Studholme DJ, Tavares F, Vicente JG, Koebnik R, Costa J. 2021. Trends in molecular diagnosis and diversity studies for phytosanitary regulated Xanthomonas. Microorganisms 9:862. doi: 10.3390/microorganisms9040862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koebnik R, Burokiene D, Bragard C, Chang C, Saux MF-L, Kölliker R, Lang JM, Leach JE, Luna EK, Portier P, Sagia A, Ziegle J, Cohen SP, Jacobs JM. 2021. The complete genome sequence of Xanthomonas theicola, the causal agent of canker on tea plants, reveals novel secretion systems in clade 1 xanthomonads. Phytopathology® 111:611–616. doi: 10.1094/PHYTO-07-20-0273-SC. [DOI] [PubMed] [Google Scholar]
- 10.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, De Vos P, Cottyn B. 2016. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 11.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 12.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 14.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-R LM, Konstantinidis KT. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 17.Yoon SH, Ha SM, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 18.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:1–14. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alikhan NF, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Coleman-Derr D, Chen G, Gu YQ. 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jong A, van Hijum SA, Bijlsma JJ, Kok J, Kuipers OP. 2006. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res 34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavara E. 1905. Bacteriosi del fico. Atti Accad Gioenia Sci Nat Catania 14:1–17. [Google Scholar]
- 27.Jindal JK, Patel PN. 1972. Angular leaf spot of pipal (Ficus religiosa) due to Xanthomonas fici sp. nov. Indian Phytopathol 25:164–166. [Google Scholar]
- 28.Duff J. 1991. Angular leaf spot of ornamental Ficus spp. due to Xanthomonas campestris pv. fici. Austral Plant Pathol 20:1–2. doi: 10.1071/APP9910001. [DOI] [Google Scholar]
- 29.Dye DW, Lelliott RA, Buchanan R, & Gibbons N. 1974. Genus II. Xanthomonas Dowson 1939, 187. In Bergey's Manual of Determinative Bacteriology, 89. [Google Scholar]
- 30.Young JM, Dye DW, Bradbury JF, Panagopoulos CG, Robbs CF. 1978. A proposed nomenclature and classification for plant pathogenic bacteria. N Z J Agric Res 21:153–177. doi: 10.1080/00288233.1978.10427397. [DOI] [Google Scholar]
- 31.Campoverde EV, Palmateer AJ. 2016. Bacterial blight of Ficus elastica caused by Xanthomonas. UF/IFAS Extension. https://edis.ifas.ufl.edu/pdffiles/PP/PP30500.pdf.
- 32.Young JM, Park DC, Shearman HM, Fargier E. 2008. A multilocus sequence analysis of the genus Xanthomonas. Syst Appl Microbiol 31:366–377. doi: 10.1016/j.syapm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson N, Cowie C, Heeney J, Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol 59:264–274. doi: 10.1099/ijs.0.65825-0. [DOI] [PubMed] [Google Scholar]
- 34.Bell CR, Cummings NE, Canfield ML, Moore LW. 1990. Competition of octopine-catabolizing Pseudomonas spp. and octopine-type Agrobacterium tumefaciens for octopine in chemostats. Appl Environ Microbiol 56:2840–2846. doi: 10.1128/aem.56.9.2840-2846.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barton IS, Fuqua C, Platt TG. 2018. Ecological and evolutionary dynamics of a model facultative pathogen: Agrobacterium and crown gall disease of plants. Environ Microbiol 20:16–29. doi: 10.1111/1462-2920.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dessaux Y, Faure D. 2018. Niche construction and exploitation by Agrobacterium: how to survive and face competition in soil and plant habitats. Curr Top Microbiol Immunol 418:55–86. doi: 10.1007/82_2018_83. [DOI] [PubMed] [Google Scholar]
- 37.Meyer T, Thiour-Mauprivez C, Wisniewski-Dyé F, Kerzaon I, Comte G, Vial L, Lavire C. 2019. Ecological conditions and molecular determinants involved in agrobacterium lifestyle in tumors. Front Plant Sci 10:978. doi: 10.3389/fpls.2019.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faist H, Keller A, Hentschel U, Deeken R. 2016. Grapevine (Vitis vinifera) crown galls host distinct microbiota. Appl Environ Microbiol 82:5542–5552. doi: 10.1128/AEM.01131-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu CF, Smith DA, Lai EM, Chang JH. 2018. The Agrobacterium type VI secretion system: a contractile nanomachine for interbacterial competition. Curr Top Microbiol Immunol 418:215–231. doi: 10.1007/82_2018_99. [DOI] [PubMed] [Google Scholar]
- 40.Eng WWH, Gan HM, Gan HY, Hudson AO, Savka MA. 2015. Whole-genome sequence and annotation of octopine-utilizing Pseudomonas kilonensis (previously P. fluorescens) strain 1855-344. Genome Announc 3:e00463-15. doi: 10.1128/genomeA.00463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulanger A, Zischek C, Lautier M, Jamet S, Rival P, Carrère S, Arlat M, Lauber E. 2014. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. mBio 5:e01527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osdaghi E, Taghavi SM, Hamzehzarghani H, Fazliarab A, Harveson RM, Tegli S, Lamichhane JR. 2018. Epiphytic Curtobacterium flaccumfaciens strains isolated from symptomless solanaceous vegetables are pathogenic on leguminous but not on solanaceous plants. Plant Pathol 67:388–398. doi: 10.1111/ppa.12730. [DOI] [Google Scholar]
- 44.Osdaghi E, Taghavi SM, Calamai S, Biancalani C, Cerboneschi M, Tegli S, Harveson RM. 2018. Phenotypic and molecular-phylogenetic analysis provide novel insights into the diversity of Curtobacterium flaccumfaciens. Phytopathology 108:1154–1164. doi: 10.1094/PHYTO-12-17-0420-R. [DOI] [PubMed] [Google Scholar]
- 45.Schaad NW, Dianes JC. 1981. Cruciferous weeds as sources of inoculum of Xanthomonas campestris in black rot of crucifers. Phytopathology 71:1215–1220. doi: 10.1094/Phyto-71-1215. [DOI] [Google Scholar]
- 46.Thompson DC, Schaad NW, Forster RL. 1989. New perennial hosts of epiphytic populations of Xanthomonas campestris pv. translucens. Phytopathology 79:1168. [Google Scholar]
- 47.Zarei S, Taghavi SM, Hamzehzarghani H, Osdaghi E, Lamichhane JR. 2018. Epiphytic growth of Xanthomonas arboricola and Xanthomonas citri on non‐host plants. Plant Pathol 67:660–670. doi: 10.1111/ppa.12769. [DOI] [Google Scholar]
- 48.Osdaghi E, Taghavi SM, Hamzehzarghani H, Lamichhane JR. 2016. Occurrence and Characterization of the bacterial spot pathogen Xanthomonas euvesicatoria on pepper in Iran. J Phytopathology 164:722–734. doi: 10.1111/jph.12493. [DOI] [Google Scholar]
- 49.Osdaghi E, Sharma A, Goss EM, Abrahamian P, Newberry EA, Potnis N, Carvalho R, Choudhary M, Paret ML, Timilsina S, Vallad GE, Jones JB. 2021. A centenary for bacterial spot of tomato and pepper. Mol Plant Pathol 22:1500–1519. doi: 10.1111/mpp.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dia NC, Morinière L, Cottyn B, Bernal E, Jacobs JM, Koebnik R, Osdaghi E, Potnis N, Pothier JF. 2022. Xanthomonas hortorum – beyond gardens: current taxonomy, genomics, and virulence repertoires. Molecular Plant Pathology, In Press. doi: 10.1111/mpp.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs JM, Pesce C, Lefeuvre P, Koebnik R. 2015. Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front Plant Sci 6:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohr TJ, Liu H, Yan S, Morris CE, Castillo JA, Jelenska J, Vinatzer BA. 2008. Naturally occurring nonpathogenic isolates of the plant pathogen Pseudomonas syringae lack a type III secretion system and effector gene orthologues. J Bacteriol 190:2858–2870. doi: 10.1128/JB.01757-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah SMA, Khojasteh M, Wang Q, Taghavi SM, Xu Z, Khodaygan P, Zou L, Mohammadikhah S, Chen C, Osdaghi E. 2021. Genomics-enabled novel insight into the pathovar-specific population structure of the bacterial leaf streak pathogen Xanthomonas translucens in small grain cereals. Front Microbiol 12:674952. doi: 10.3389/fmicb.2021.674952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mafakheri H, Taghavi SM, Banihashemi Z, Osdaghi E, Lamichhane JR. 2017. Pathogenicity, host range and phylogenetic position of Agrobacterium species associated with sugar beet crown gall outbreaks in Southern Iran. Eur J Plant Pathol 147:721–730. doi: 10.1007/s10658-016-1034-3. [DOI] [Google Scholar]
- 55.Mafakheri H, Taghavi SM, Zarei S, Kuzmanović N, Osdaghi E. 2021. Occurrence of crown gall disease on Japanese spindle (Euonymus japonicas var. Green Rocket) caused by Agrobacterium rosae in Iran. Plant Dis. Epub ahead of print. doi: 10.1094/PDIS-03-21-0580-PDN. [DOI] [Google Scholar]
- 56.Osdaghi E, Ansari M, Taghavi SM, Zarei S, Koebnik R, Lamichhane JR. 2018. Pathogenicity and phylogenetic analysis of Clavibacter michiganensis strains associated with tomato plants in Iran. Plant Pathol 67:957–970. doi: 10.1111/ppa.12801. [DOI] [Google Scholar]
- 57.Khojasteh M, Taghavi SM, Khodaygan P, Hamzehzarghani H, Chen G, Bragard C, Koebnik R, Osdaghi E. 2019. Molecular typing reveals high genetic diversity of Xanthomonas translucens strains infecting small-grain cereals in Iran. Appl Environ Microbiol 85:e01518-19. doi: 10.1128/AEM.01518-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bushnell B, Rood J, Singer E. 2017. BBMerge–Accurate paired shotgun read merging via overlap. PloS One. 12:e0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osdaghi E, Rahimi T, Taghavi SM, Ansari M, Zarei S, Portier P, Briand M, Jacques MA. 2020. Comparative genomics and phylogenetic analyses suggest several novel species within the genus Clavibacter, including nonpathogenic tomato-associated strains. Appl Environ Microbiol 86:e02873-19. doi: 10.1128/AEM.02873-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 63.Jaenicke S, Bunk B, Wibberg D, Spröer C, Hersemann L, Blom J, Winkler A, Schatschneider S, Albaum SP, Kölliker R, Goesmann A, Pühler A, Overmann J, Vorhölter F-J. 2016. Complete genome sequence of the barley pathogen Xanthomonas translucens pv. translucens DSM 18974T (ATCC 19319T). Genome Announc 4:e01334-16. doi: 10.1128/genomeA.01334-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pearson WR. 2013. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinformatics 42:3.1.1–3.1.8. doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darling AE, Mau B, Perna NT. 2010. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov D, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL, Simon Fraser University Research Computing Group. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00577-21_Supp_1_seq7.pdf, PDF file, 4.9 MB (5MB, pdf)
Data Availability Statement
The nucleotide sequences obtained in this study were deposited into the GenBank database under the following accession numbers: atpD, MW222614 to MW222618; efp, MW222619 to MW222623; gyrB, MW222624 to MW222628; and rpoD, MW222629 to MW222633. Whole-genome sequences of the three strains were also deposited into the NCBI GenBank database under the following accession numbers: AmX2, JAFIWC000000000 (BioProject no. PRJNA700118); FX1, JAFJNT000000000 (BioProject no. PRJNA700116); and FX4, JAFIWB000000000 (BioProject no. PRJNA700117). Furthermore, pure cultures of the strains have been deposited in the CIRM-CFBP culture collection (French Collection for Plant-associated Bacteria) and assigned the following accession numbers: AmX2, CFBP 8902; FX1, CFBP 8700, FX2, CFBP 8701, FX3, CFBP 8702, and FX4, CFBP 8703.