ABSTRACT
Dickeya zeae is a worldwide destructive pathogen that causes soft rot diseases on various hosts such as rice, maize, banana, and potato. The strain JZL7 we recently isolated from clivia represents the first monocot-specific D. zeae and also has reduced pathogenicity compared to that of other D. zeae strains (e.g., EC1 and MS2). To elucidate the molecular mechanisms underlying its more restricted host range and weakened pathogenicity, we sequenced the complete genome of JZL7 and performed comparative genomic and functional analyses of JZL7 and other D. zeae strains. We found that, while having the largest genome among D. zeae strains, JZL7 lost almost the entire type III secretion system (T3SS), which is a key component of the virulence suite of many bacterial pathogens. Importantly, the deletion of T3SS in MS2 substantially diminished the expression of most type III secreted effectors (T3SEs) and MS2’s pathogenicity on both dicots and monocots. Moreover, although JZL7 and MS2 share almost the same repertoire of cell wall-degrading enzymes (CWDEs), we found broad reduction in the production of CWDEs and expression levels of CWDE genes in JZL7. The lower expression of CWDEs, pectin lyases in particular, would probably make it difficult for JZL7 to break down the cell wall of dicots, which is rich in pectin. Together, our results suggest that the loss of T3SS and reduced CWDE activity together might have contributed to the host specificity and virulence of JZL7. Our findings also shed light on the pathogenic mechanism of Dickeya and other soft rot Pectobacteriaceae species in general.
IMPORTANCE Dickeya zeae is an important, aggressive bacterial phytopathogen that can cause severe diseases in many crops and ornamental plants, thus leading to substantial economic losses. Strains from different sources showed significant diversity in their natural hosts, suggesting complicated evolution history and pathogenic mechanisms. However, molecular mechanisms that cause the differences in the host range of D. zeae strains remain poorly understood. This study carried out genomic and functional dissections of JZL7, a D. zeae strain with restricted host range, and revealed type III secretion system (T3SS) and cell wall-degrading enzymes (CWDEs) as two major factors contributing to the host range and virulence of D. zeae, which will provide a valuable reference for the exploration of pathogenic mechanisms in other bacteria and present new insights for the control of bacterial soft rot diseases on crops.
KEYWORDS: Dickeya zeae, host range, comparative genomics, virulence, T3SS, T3SEs, CWDEs
INTRODUCTION
Dickeya bacteria usually cause soft rot disease on many economically important crops and ornamental plants all over the world. As a highly diverse group, the genus contains 12 species according to current classification criteria, namely, D. chrysanthemi, D. dadantii, D. dianthicola, D. paradisiaca, D. zeae, D. solani, D. aquatica, D. fangzhongdai, D. poaceaephila, D. lacustris, D. undicola, and D. oryzae (1–8). Among them, D. zeae was isolated mostly from plants in tropical and subtropical regions, especially in Southeast Asian countries (9), suggesting its preference to high temperature and humidity. On the other hand, D. solani was usually isolated from potato in temperate and frigid zones, especially across Europe (10, 11), with a wider temperature range for growth and disease development (12).
Although isolated mainly from monocotyledonous hosts in nature, D. zeae is so far known to infect 24 types of dicots and 23 types of monocots, indicating a broad host range (9). In recent years, D. zeae has caused considerable losses in crop yields in China, especially on rice and banana (13–16). At the same time, D. zeae strains from diverse sources also display considerable variations in their pathogenesis. For instance, the strains EC1 (isolated from rice) and MS2/MS3 (isolated from banana) showed different virulence on various monocot and dicots plants and exhibited 2-fold difference in their ability to inhibit rice seed germination (9, 17). Furthermore, consistent with these phenotypic variations, there are also key genomic differences between EC1 and MS2/MS3, including the zms gene cluster encoding the phytotoxic zeamines, one of the most important virulence factors unique to and shared by rice strains isolated from geographically distant regions (9, 18, 19), the biosynthetic gene cluster (C1O30_RS04995 to C1O30_RS05185) encoding a novel phytotoxin in MS2, and the pipR and pipA that are present only in MS1 and MS2 (17).
Importantly, we have recently isolated from the ornamental plant Clivia miniata three D. zeae strains which are, for the first time, found to specifically infect monocots (9). These strains were pathogenic on nearly all tested monocots (e.g., rice, banana, clivia), although their virulence was usually lower than that of other D. zeae strains, and could not infect any of the nine dicots tested (e.g., potato, tomato, radish, cabbage) (9). A distinctive characteristic of these clivia-isolated strains is the remarkably lower activity of cell wall-degrading enzymes (CWDEs) compared with that of other D. zeae (9). However, the molecular basis of the dramatic alteration in host range remains largely unknown.
To investigate determinants of the host range of D. zeae, we sequenced the genome of JZL7, one of the monocot-specific strains isolated from C. miniata, and performed comparative genomic as well as functional analyses of JZL7 and other D. zeae strains that infect both monocots and dicots. The findings of this study will facilitate our understanding of the nature and the evolution of the pathogenicity mechanisms of D. zeae.
RESULTS AND DISCUSSION
General genomic features of D. zeae strain JZL7.
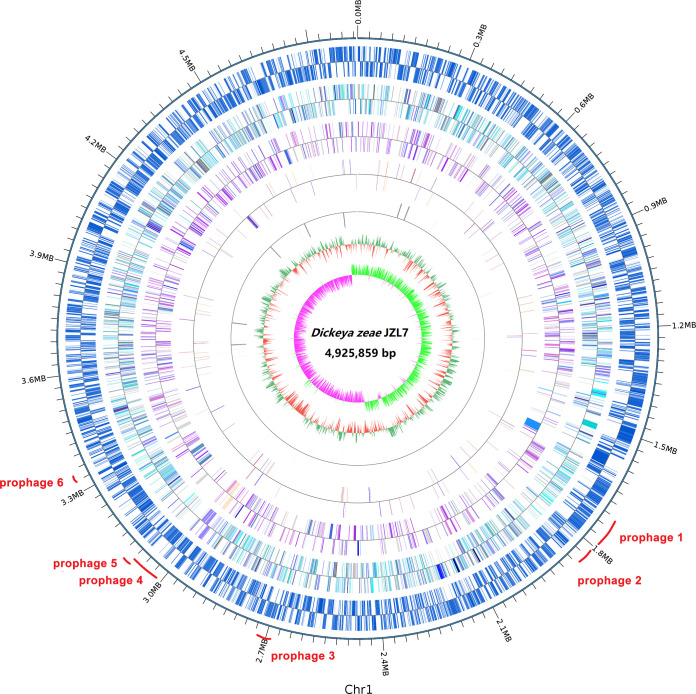
The genome of D. zeae strain JZL7 was sequenced on both the PacBio RSII and the Illumina HiSeq X Ten platforms, producing 196,305 long sequencing reads (N50: 8,071 bp; mean read length: 5.356 Kb; total size: 1.051 Gb) and 5.44 million short sequencing reads (PE150; total size: 1.789 Gb), respectively. A hybrid de novo assembly using both the short and long sequencing reads resulted in a high-quality and gapless assembly consisting of a single circular chromosome of 4,925,859 bp in size; no plasmids were detected (Fig. 1). BUSCO evaluation of the JZL7 genome detected 438 of the 440 genes (99.5%) in the enterobacterales_odb10 data set, suggesting that the assembly is highly complete. The JZL7 genome contains 4,268 protein-coding genes, 75 tRNA genes, and 22 rRNA genes which belong to seven complete rRNA loci, including the unusual 16S-23S-5S-5S operon previously observed in other Dickeya genomes (Table 1) (18). JZL7 has both the largest genome and the largest repertoire of protein-coding genes among all sequenced D. zeae strains; it contains 167 more genes than Ech586, which has the second largest genome, and 443 more genes than EC1, which has the smallest genome (Table 1). On the other hand, the G+C content of JZL7 genome (53.68%) is highly similar to that of the other D. zeae genomes (53.34% to 53.65%) (Table 1).
FIG 1.
Circular visualization of genome characteristics of D. zeae strain JZL7. The circles from outermost to innermost indicate the distributions of protein-coding genes, COG annotations, KEGG annotations, GO annotations, rRNAs, GC ratio, and GC skew, respectively.
TABLE 1.
Genomic features of the six D. zeae strains with complete genome
| Features | JZL7 | MS2 | EC1 | EC2 | CE1 | Ech586 |
|---|---|---|---|---|---|---|
| BUSCO (complete) | 99.5% | 99.5% | 99.8% | 99.8% | 99.5% | 99.5% |
| No. of replicons | 1 | 1 | 1 | 1 | 1 | 1 |
| Size (bp) | 4,925,859 | 4,740,052 | 4,532,364 | 4,575,125 | 4,714,731 | 4,818,394 |
| G+C content (%) | 53.68 | 53.45 | 53.43 | 53.34 | 53.65 | 53.64 |
| Genes | 4,372 | 4,171 | 3,947 | 3,985 | 4,143 | 4,205 |
| Protein-coding genes | 4,268 | 4,068 | 3,825 | 3,879 | 4,037 | 4,101 |
| rRNAs | 22 | 22 | 22 | 22 | 22 | 22 |
| tRNA | 75 | 75 | 88 | 75 | 75 | 76 |
| ncRNA | 3 | 2 | 8 | 5 | 5 | 2 |
| Pseudo genes | 112 | 50 | 65 | 74 | 71 | 64 |
| Transposases | 47 | 48 | 38 | 59 | 25 | 22 |
| Prophage | ||||||
| Region | 9 | 3 | 4 | 4 | 4 | 3 |
| Gene | 250 | 68 | 160 | 63 | 108 | 87 |
| Genomic island | ||||||
| Region | 35 | 24 | 21 | 25 | 19 | 30 |
| Gene | 657 | 355 | 364 | 357 | 337 | 346 |
Genomic differences between JZL7 and other D. zeae strains.
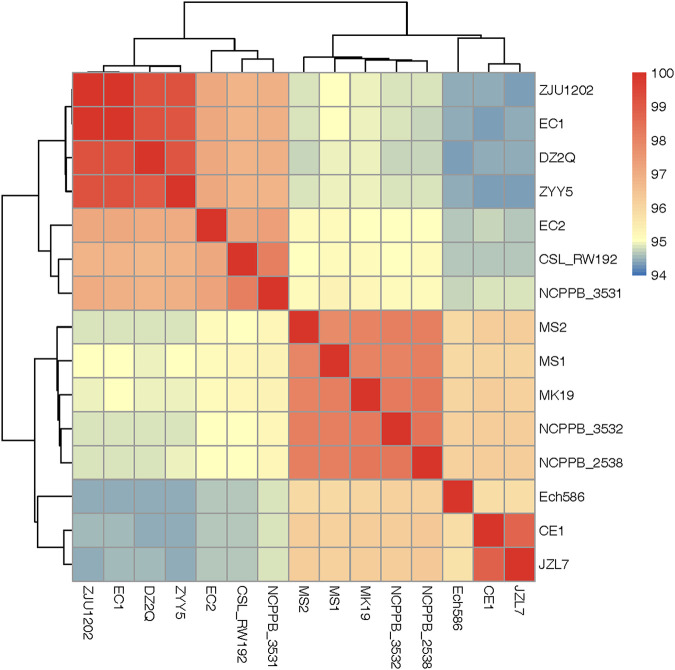
To determine the evolution relationship between JZL7 and other D. zeae strains, all sequenced D. zeae and the closely related D. oryzae ZYY5 (8) genomes were used for phylogenetic analysis based on the 3,110 single-copy core genes (present in all strains). Results showed that the 15 D. zeae strains can be divided into two clusters (Fig. 2); the average nucleotide identity (ANI) values are above 95% for all pairwise comparisons within each cluster but below 95% for nearly all comparisons between clusters (Fig. 2). The first cluster consists of seven strains, including DZ2Q, EC1, EC2, ZJU1202, and ZYY5, the five rice-pathogenic strains, as well as CSL_RW192 and NCPPB_3531. Our results thus corroborate the recent proposal by Wang et al. (8) to classify this clade as a novel species, Dickeya oryzae.
FIG 2.
ANI analyses of 15 D. zeae and D. oryzae strains. Based on their pairwise ANI values, all strains were clustered into two well-separated groups corresponding to D. zeae (type strain: NCPPB 2538) and D. oryzae (type strain: ZYY5), respectively. JZL7 belongs to the D. zeae group and the ANI values between JZL7 and all other D. zeae strains are greater than 95%.
The second clade contains JZL7, CE1, and Ech586, as well as five other strains that are highly similar to each other (ANI values of >97.85%), namely, MS1, MS2, MK19, NCPPB_2538, and NCPPB_3532 (Fig. 2). We have recently shown that EC1 (first clade) and MS2 (second clade) are pathogenic toward both monocot and dicot hosts, whereas JZL7 (second clade) can infect only monocots (9). Taken together, our results suggest that the broad host range may have been the ancestral state to both D. zeae and D. oryzae, while JZL7 became specialized on monocots during recent evolution.
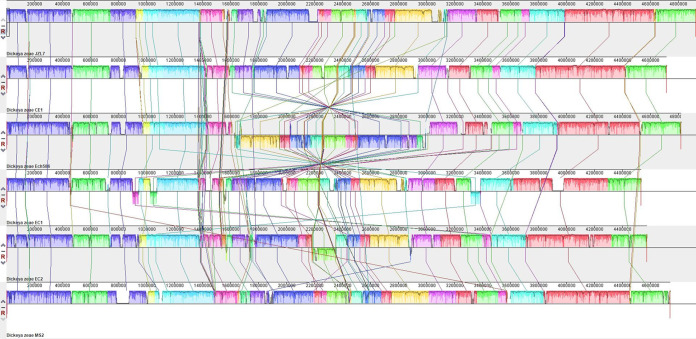
To characterize the genomic differences between JZL7 and other D. zeae and D. oryzae strains that might underlie their divergent host specificities (Table S1), we first constructed whole-genome alignment between JZL7, CE1, Ech586, EC1, EC2, and MS2, all of which have completely sequenced genomes. Results showed that the six strains are largely conserved in their overall genome structures, with the only exceptions being a large inversion of ∼1.4 Mb in the genome of Ech586 and a few smaller translocations in the genomes of EC1 and EC2 (Fig. 3). At the same time, insertions/deletions up to several tens of kilobases are frequently observed and distributed throughout the six genomes (Fig. 3).
FIG 3.
Whole-genome comparison between Dickeya strains JZL7, CE1, Ech586, EC1, EC2, and MS2. The genome structures are largely conserved among the strains, with only a few large-scale rearrangements and much more frequent small-scale insertions and deletions.
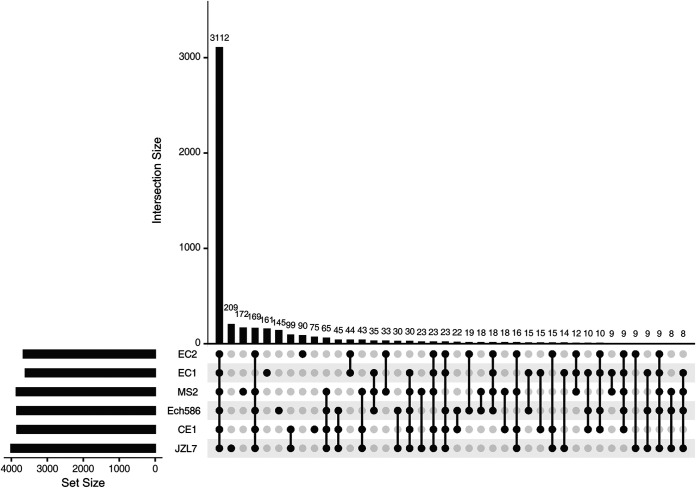
We then performed pangenome analyses on the six strains to further examine differences in their gene contents. In total, 4,966 distinct gene families were identified, the majority of which (3,112 families; 62.67%) are shared by all six strains (Fig. 4), indicating a relatively stable core genome. On the other hand, 209 ortholog group family genes are JZL7 specific (Table S4), whereas 9 gene families are absent in JZL7 but shared by all the other five strains, which may contain genes that are critical for the pathogenicity to dicots.
FIG 4.
A diagram UpSet plot showing the results of pangenome analysis of JZL7, CE1, Ech586, EC1, EC2, and MS2.
Among the unique genes in the JZL7 genome, an additional set of type IV secretion system (T4SS) that consists of 14 trb genes was detected (Table S5). This extra T4SS in JZL7 shares high levels of nucleotide sequence similarity to the T4SS-encoding gene sets in Ralstonia solanacearum strains FQY_4 and GMI1000, Pseudomonas aeruginosa F30658, and Pectobacterium carotovorum subsp. brasiliense strain SX309. T4SS usually functions in the translocation of nucleic acids or proteins from donor to recipient by conjugation and DNA release/uptake (20). The two sets of T4SSs encoded in the JZL7 genome (Table S5) may have contributed to the abundant strain-specific genes of JZL7 (Fig. 1 and 3, Tables S6 and S7). However, given that strain JZL7 has more restricted host range than the strains EC1 and MS2 (9), we conjectured that this extra T4SS is not important for the pathogenesis of D. zeae.
The loss of T3SS in JZL7 partially explains its restricted host range.
The more restricted host range of JZL7 (Table S1) might be due to the loss of some vital determinants of virulence or host specificity during its evolution. Accordingly, we found in our pangenome analysis 85 gene families that are shared by both EC1 and MS2 but absent in JZL7 (Fig. 4, Table S8), including a set of 30 genes that are colocalized in the genome; they comprise the majority of the dsp gene cluster, which encodes multiple T3SS secreted proteins (e.g., DspE, HrpZ, and HrpW) as well as their chaperone (DspF), and the hrp/hrc gene cluster, which encodes a T3SS (Fig. 5, Table S8). The hecA-hecB genes between the dsp and hrp/hrc clusters, however, were conserved in JZL7; they encode the type V secretion system (T5SS), which contributes to the adherence to hosts (21). Exactly the same organization in this genomic region was found in CE1, the closest relative of JZL7 (Fig. 5). Interestingly, the D. oryzae strain EC2 also lost its dsp and hrp/hrc clusters, but it has a substantially different set of remaining genes.
FIG 5.
Gene arrangements of T3SS in genomes of strains Ech586, EC1, MS2, JZL7, CE1, and EC2.
T3SS forms a syringe-like structure to directly translocate type III secreted effectors (T3SEs) from bacterial cells into host cells and is considered a key determinant for virulence in many bacteria. It has been reported in a number of plant bacterial pathogens (e.g., Erwinia pyrifoliae, Ps. syringae, and Xanthomonas campestris pv. campestris) that the loss of the T3SS would severely reduce the elicitation of host defenses and thus lead to attenuated disease symptoms (22–24). Moreover, T3SS was demonstrated to be required for the full virulence of D. dadantii 3937 (25, 26).
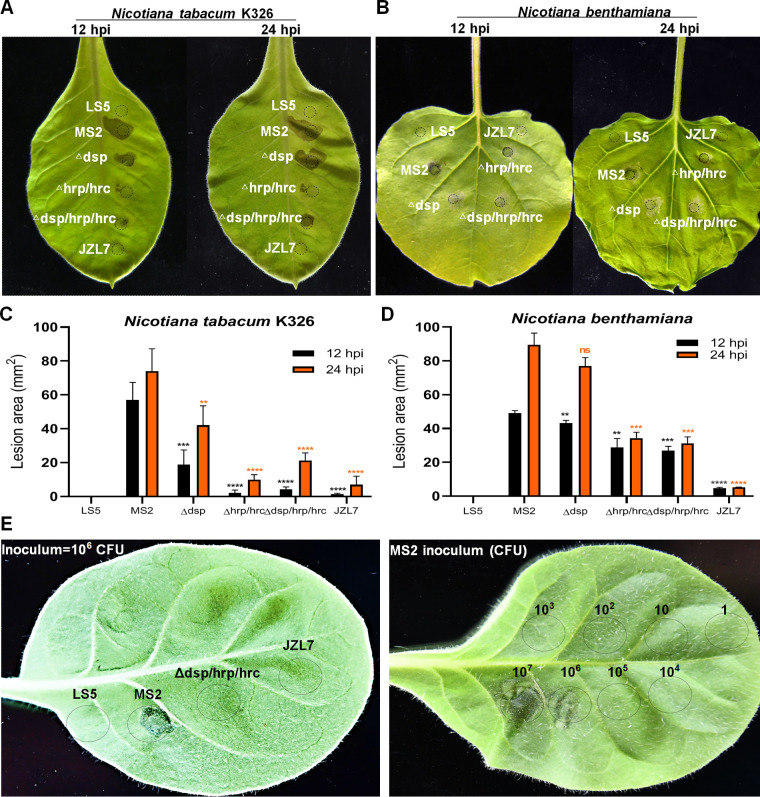
To evaluate whether strain JZL7 could elicit hypersensitive response (HR) as strain MS2 harboring T3SS, we performed HR assay on Nicotiana tabacum K326 (27) and Nicotiana benthamiana (28). Results indicated that strain JZL7 could not cause HR on either of the tobacco leaves, the same as the negative-control LS5 medium, whereas strain MS2 elicited a typical HR quickly in the early stage of inoculation (12 hpi) (Fig. 6). To further verify the importance of T3SS, we respectively deleted the dsp cluster, the hrp/hrc cluster, and both in MS2. HR induction of Δdsp was substantially reduced in the early stage of inoculation compared with that of wild-type MS2, but little difference was found between them after 24 hpi (Fig. 6), suggesting that the T3SEs DspE, HrpZ, HrpW, and HrpK function in the initial infection of plant hosts. The T3SS structure deletion mutant Δhrp/hrc almost could not induce HR both in the early and later stages of infection, while the whole T3SS deletion mutant Δdsp/hrp/hrc significantly attenuated the HR reaction at 12 hpi but recovered part of it at 24 hpi (Fig. 6). HR is a form of programmed cell death (PCD) at the site of pathogen infection commonly controlled by interactions between pathogen avirulence gene products and plant resistance genes. Although the T3SS has been deleted from strain MS2, some avirulence genes, like the known avrL (C1O30_RS14255), are still present in the genome; thus, reduced HR could be observed by mutant infiltration. Furthermore, the components of the T3SS and the T3SEs are not the only pathogen-associated molecular patterns (PAMPs) that plant defenses are known to recognize. Flagellin, peptidoglycan, and lipopolysaccharides also contribute to triggering host innate immune response through recognizing by plant pattern recognition receptors (PRRs). To quantify the function of the T3SS, we infiltrated tobacco leaves with controlled inoculum size. Results showed that the threshold of 106 MS2 CFU elicited visible HR at 12 h (Fig. 6E), which may help distinguish between true HR and other types of PCD.
FIG 6.
The HR of the T3SS mutants of MS2 and strain JZL7 on Nicotiana tabacum K326 (A, C, and E) and N. benthamiana (B and D) after 12 and 24 hpi. Bacterial cultures (OD600 of 1.0 in LS5 medium) were dipped by a puncher (5 mm), pressed on the back of tobacco leaves, and incubated at 28°C. The area of HR lesions was measured using Image J 1.52a after 12 and 24 h. **, P < 0.001; ***, P < 0.0001; ****, P < 0.0001; ns, not significant (Student’s t test, n = 3 independent experiments). (E) Quantitative determination of the T3SS functions in hypersensitive response indicated that the threshold of 106 CFU of MS2 elicits HR at 12 h.
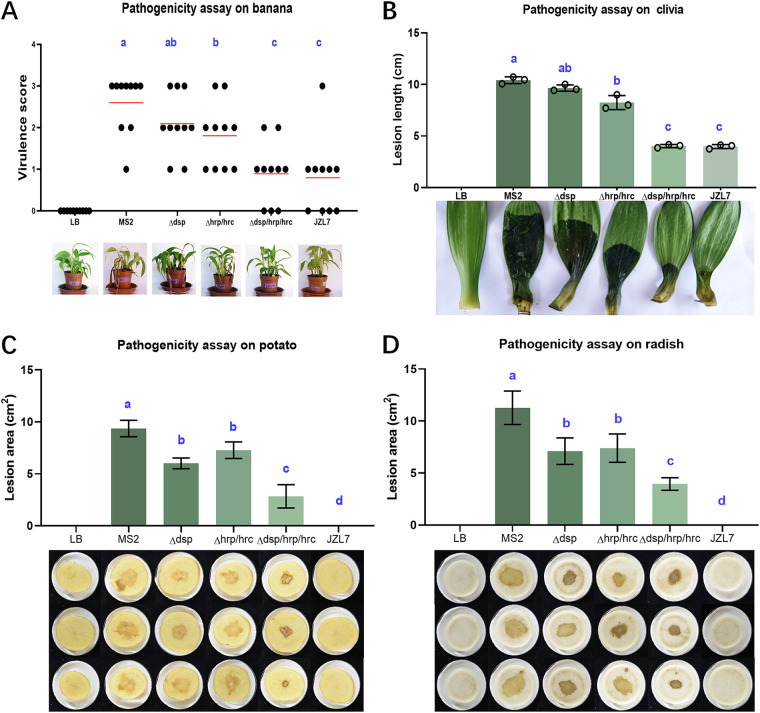
To further investigate the role of T3SS in the pathogenicity and host specificity of D. zeae pathogens, we tested the pathogenicity of Δdsp, Δhrp/hrc, and Δdsp/hrp/hrc mutants in strain MS2 on monocotyledonous banana stems and clivia leaves, as well as dicotyledonous potato and radish slices. We found that the Δhrp/hrc showed 30% reduced virulence on banana and clivia seedlings (Fig. 7A and B) and 22% and 24% reduced virulence on potato and radish slices, respectively, compared with wild-type MS2 (Fig. 7C and D). The Δdsp mutant similarly showed lower (35% on potato and 36% on radish) virulence on dicots, whereas its virulence on monocots was 20% reduced on banana seedlings and 7% reduced on clivia seedlings, not significantly different from that of MS2. Notably, the deletion of the whole T3SS (Δdsp/hrp/hrc) dramatically attenuated the virulence of MS2 by more than 60% on monocots (Fig. 7A and B), to a level that is comparable to that of JZL7, and by more than 65% on dicots (Fig. 7C and D). Altogether, our results indicate that T3SS has a major role in the virulence of D. zeae. Previous studies have revealed that tissue maceration requires type II secretion (T2SS) of CWDEs and does not involve T3SS (18). Our study revealed the contribution of T3SS to the development of soft rot symptoms, which probably results from the action by the T3SEs regulated by HrpL, as shown in Fig. S1. Unlike JZL7, however, the Δdsp/hrp/hrc mutant of MS2 still exhibited considerable virulence on dicots, suggesting that there are likely other factors contributing to the host specificity of D. zeae pathogens in addition to T3SS.
FIG 7.
The virulence of the T3SS mutants of MS2 and strain JZL7 on monocotyledonous and dicotyledonous plants. (A) Strain inoculation on banana seedlings. Every 100 μL of bacterial culture (OD600 of 1.2 in LB medium) was injected into the basal stem and incubated at 30°C with 12-h alternating light-dark cycles for 7 days. The disease was assessed using the virulence scoring method described previously (17, 80). Red lines indicate the mean virulence score of the strains. (B) Strain inoculation on clivia leaves. Every 100 μL of bacterial culture was injected into the base of clivia leaf. (C and D) Strain inoculation on potato and radish slices. Bacterial cells of 2 μL were applied to the center of the tuber slices. Same volume of LB medium was inoculated as a negative control. All plants were kept in a growth chamber with controlled conditions of 28 ± 2°C, 75% ± 15% relative humidity and 24-h white light (7,350 lx) illumination until symptoms appeared. Visible macerate areas on clivia and potato and radish slices were measured using Image J 1.52a. Statistical analysis was performed on each group of data, and significantly different values (ANOVA P < 0.05) are indicated by different letters.
HrpL regulates most predicted T3SEs in MS2.
In addition to the syringe structure of T3SS, genes in the dsp and hrp/hrc clusters also encode multiple transcription regulators and effectors. We generated single-gene knockout mutants for selected regulator (HrpL HrpS, HrpX, and HrpY) or effector (DspE, HrpK, HrpN, HrpW, and HrpZ) coding genes in MS2 and tested their pathogenicity on potato slices. Results showed that three regulator gene mutants (ΔhrpL, ΔhrpX, and ΔhrpY) and three effector gene mutants (ΔhrpK, ΔhrpN, and ΔhrpZ) exhibited significantly reduced virulence compared with that of MS2 wild type (Fig. S1). In particular, the ΔhrpL mutant showed the most dramatic reduction in virulence, which is comparable to the knockout of the entire T3SS (Δdsp/hrp/hrc) (Fig. S1).
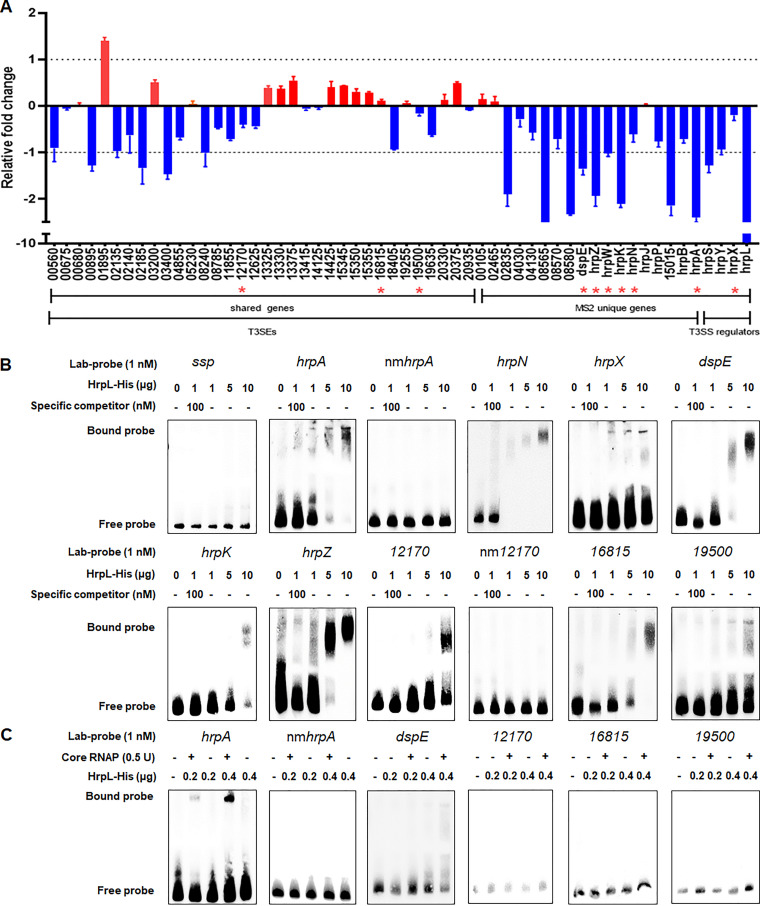
HrpL is a sigma factor activating T3SS at different levels, including gene transcription, mRNA stability, and enzymatic activity in many plant-pathogenic bacteria, such as Erwinia, Pseudomonas, Ralstonia, and Xanthomonas (25, 29–34). In Dickeya bacteria, however, only one effector, DspE, has been characterized alongside two harpins, HrpN and HrpW (35, 36). To investigate the regulatory role of HrpL, we first used a combination of four state-of-the-art bioinformatic tools (i.e., Bastion3, BEAN2, DeepT3, and pEffect) to identify candidate T3SEs in the genomes of MS2 and JZL7. As a result, we predicted 34 T3SEs that are present in both strains, as well as 16 and 10 T3SEs that are unique to MS2 and JZL7, respectively (Table 2). Given that hrpL is absent in JZL7, we measured the expression of all T3SE genes in MS2 wild type and ΔhrpL mutant using quantitative reverse transcriptase PCR (qRT-PCR). Results showed that the expression of 5 shared T3SE and 8 MS2-unique T3SE genes was significantly downregulated in the ΔhrpL mutant, including dspE, hrpA, hrpK, hrpS, hrpW, and hrpZ, which are all members of the dsp and hrp/hrc clusters (Fig. 8A). Chi-square tests on the fold change values of shared versus unique T3SEs showed that HrpL regulates the expression of the MS2-unique T3SE genes more strongly than the shared ones.
TABLE 2.
T3SE candidates predicted in the genomes of Dickeya zeae MS2 and JZL7
| Accession no. of: |
NCBI accession no. | Product | Predicted hrp box (GGAACC-Nx-CCACNNA) | |
|---|---|---|---|---|
| MS2 | JZL7 | |||
| C1O30_RS00560 | JZL000100 | WP_013319992.1 | Hypothetical protein VfmS | |
| C1O30_RS00675 | JZL000138 | WP_102800815.1 | Hypothetical protein | |
| C1O30_RS00680 | JZL000139 | WP_102800816.1 | Hypothetical protein | |
| C1O30_RS00895 | JZL000183 | WP_102800840.1 | Murein hydrolase activator EnvC | |
| C1O30_RS01895 | JZL000383 | WP_102800904.1 | Type I-E CRISPR-associated protein Cse2/CasB | |
| C1O30_RS02135 | JZL000448 | WP_102800932.1 | Hypothetical protein | |
| C1O30_RS02140 | JZL000451 | WP_102800933.1 | Hypothetical protein | |
| C1O30_RS02185 | JZL000460 | WP_102800940.1 | Hypothetical protein | |
| C1O30_RS03200 | JZL000666 | WP_102801017.1 | Glycine dehydrogenase GcvP | |
| C1O30_RS03400 | JZL000710 | WP_019846222.1 | Hypothetical protein | |
| C1O30_RS04855 | JZL000906 | WP_019844323.1 | LOG family protein | |
| C1O30_RS05230 | JZL000962 | WP_023639126.1 | Hypothetical protein | |
| C1O30_RS08240 | JZL001538 | WP_102801528.1 | Alginate lyase family protein | |
| C1O30_RS08785 | JZL001698 | WP_023639967.1 | Hypothetical protein | |
| C1O30_RS11855 | JZL002390 | WP_012884524.1 | DUF1852 domain-containing protein | |
| C1O30_RS12170 | JZL002462 | WP_102801891.1 | Hypothetical protein | −241 GGAACT-N69-CCACCGA −133 |
| C1O30_RS12625 | JZL002555 | WP_102801925.1 | Anhydro-N-acetylmuramic acid kinase AnmK | |
| C1O30_RS13325 | JZL002697 | WP_019843821.1 | Flagellin FliC | |
| C1O30_RS13330 | JZL002698 | WP_102801989.1 | Flagellar filament capping protein FliD | |
| C1O30_RS13375 | JZL002707 | WP_102801991.1 | Flagellar hook-length control protein FliK | |
| C1O30_RS13415 | JZL002715 | WP_102801993.1 | Flagellar hook-filament junction protein FlgL | |
| C1O30_RS14125 | JZL002985 | WP_102802039.1 | Flagella biosynthesis regulator Flk | |
| C1O30_RS14425 | JZL003042 | WP_023640291.1 | General secretion pathway protein GspB | |
| C1O30_RS15345 | JZL003254 | WP_028085677.1 | Pectate lyase PelA | |
| C1O30_RS15350 | JZL003255 | WP_102802153.1 | Pectate lyase PelE | |
| C1O30_RS15355 | JZL003256 | WP_102802154.1 | Pectate lyase PelD | |
| C1O30_RS16815 | JZL003523 | WP_102802268.1 | Hypothetical protein | −590 GGAACA-N56-GCACCAA −522 |
| C1O30_RS18405 | JZL003848 | WP_102802410.1 | Hypothetical protein | |
| C1O30_RS19255 | JZL004042 | WP_102802473.1 | SPOR domain-containing protein | |
| C1O30_RS19500 | JZL004096 | WP_023640946.1 | Hypothetical protein | +22 GGAACG-N37-TCACCAA +71 |
| C1O30_RS19635 | JZL004123 | WP_102802502.1 | Uroporphyrinogen-III C-methyltransferase HemX | |
| C1O30_RS20330 | JZL004262 | WP_102802565.1 | Four-carbon acid sugar kinase family protein | |
| C1O30_RS20375 | JZL004272 | WP_102802574.1 | Cell envelope biogenesis protein TolA | |
| C1O30_RS20935 | JZL004396 | WP_038906084.1 | Der GTPase-activating protein YihI | |
| C1O30_RS00105 | WP_102800757.1 | Hypothetical protein | ||
| C1O30_RS02465 | WP_102800962.1 | Hypothetical protein | ||
| C1O30_RS02835 | WP_038915541.1 | Pectate lyase Pnl | ||
| C1O30_RS04030 | WP_102801114.1 | Hypothetical protein | ||
| C1O30_RS04130 | WP_102801119.1 | TonB family protein | ||
| C1O30_RS08565 | WP_102801570.1 | Hypothetical protein | ||
| C1O30_RS08570 | WP_102801571.1 | Hypothetical protein | ||
| C1O30_RS08580 | WP_102801572.1 | Hypothetical protein | ||
| C1O30_RS11100 | WP_102801796.1 | Type III secretion system effector DspE | −82 GGAACC-N15-CCACTCA −55 | |
| C1O30_RS11105 | WP_102801797.1 | Type III effector protein HrpZ | −110 GGAACC-N16-TCACTCA −82 | |
| C1O30_RS11110 | WP_102801798.1 | Pectate lyase HrpW | −98 GGAACT-N15-GTACTCA −71 | |
| C1O30_RS11125 | WP_102801801.1 | Type III effector HrpK | −70 GGAACT-N15-CCACTCA −43 | |
| C1O30_RS11560 | WP_023639571.1 | Harpin HrpN | −125 GGAACC-N15-TCACTCA −98 | |
| C1O30_RS11660 | WP_102801841.1 | Type III secretion system gatekeeper HrpJ | −73 GGAACC-N15-CCACATA −46 | |
| C1O30_RS11685 | WP_102801846.1 | Type III secretion protein HrpP | ||
| C1O30_RS15015 | WP_158653989.1 | Hypothetical protein | ||
| JZL000137 | Hypothetical protein | |||
| JZL000140 | Hypothetical protein | |||
| JZL000450 | Hypothetical protein | |||
| JZL001636 | Hypothetical protein | |||
| JZL001650 | Hypothetical protein | |||
| JZL001967 | Hypothetical protein | |||
| JZL002766 | Hypothetical protein | |||
| JZL002871 | Hypothetical protein | |||
| JZL002874 | Hypothetical protein | |||
| JZL002884 | Hypothetical protein | |||
FIG 8.
The T3SEs regulated by the HrpL regulator. (A) qRT-PCR of the predicted T3SEs in the genome of MS2. The expression of the predicted T3SE encoding genes (listed in Table 2), including 34 genes shared in both MS2 and JZL7 genomes and 15 genes present uniquely in the MS2 genome, as well as the regulatory genes hrpX, hrpY, hrpS, and hrpL, were measured by qRT-PCR. Expression of the housekeeping gene atpD was used as a reference. The y axis indicates the values log2(fold change of ΔhrpL mutant relative to wild-type MS2). Red bar indicates expression levels higher in the mutant, while blue indicates those lower in the mutant. Red stars indicate the target genes whose promoters were verified to be interacted by HrpL protein in the panel below. (B) EMSA of the T3SEs with predicted hrp box. The promoter DNA fragments of the genes C1O30_RS12170, C1O30_RS16815, C1O30_RS19500, dspE, hrpZ, hrpK, and hrpN and the known HrpL-regulated gene hrpA, which contain the predicted hrp box in Table 2, were amplified and labeled by biotin and then performed for EMSA with different concentrations of the expressed and purified HrpL protein. Fragments without hrp box (non GGAACC-Nx-CCACNNA motif) in the hrpA and C1O30_RS12170 promoters, designated nmhrpA and nm12170, respectively, were amplified and used for EMSA reaction to confirm the importance of the presence of hrp box. For specific competition, a 100 nM unlabeled DNA fragment was incubated with 1 μg HrpL protein for 15 min before addition of a 25 nM labeled DNA fragments. (C) E. coli core RNAP (0.5 U) was incubated on ice for 20 min with 0.2 or 0.4 μg of purified HrpL protein. The remaining steps were as described above.
Previous studies in Er. amylovora, Pantoea stewartii, and Ps. syringae have characterized a conserved binding motif for HrpL, which is called the “hrp box” (GGAACC/T-N15/16-C/T/GCACNNA) (32, 33, 36). We analyzed the promoter sequences of all predicted T3SE genes and found the typical hrp box in six of them, including the known effector DspE (36), two harpins, HrpN and HrpW, secreted through the T3SS (31), two putative effectors, HrpZ and HrpK, and a T3SS gatekeeper, HrpJ (Table 2). Three other candidate T3SE genes, C1O30_RS12170, C1O30_RS16815, and C1O30_RS19500, encoding hypothetical proteins, also contain the hrp box in their promoters, except the distances between the two conserved modules are abnormally long (69, 56, and 37 nucleotides, respectively) (Table 2). Additionally, 400 ng of HrpL was demonstrated to bind to the promoter of hrpA encoding a T3SS substrate in D. dadantii 3937 with the help of core RNA polymerase (RNAP) (34). In MS2, HrpA was not predicted as a T3SE but contains a conserved hrp box in its promoter (GGAACC-N15-CTACTTA). To verify the affinity of the HrpL to the above-mentioned hrp box-containing promoters, we carried out electrophoretic mobility shift assays (EMSAs) and found that a high concentration (5 μg) of HrpL could directly bind to the promoters of hrpA, hrpN, hrpZ, hrpK, dspE, hrpX, C1O30_RS12170, C1O30_RS16815, and C1O30_RS19500 in an hrp box-dependent manner without adding core RNAP in vitro (Fig. 8B). This is similar to the phenomenon observed in Bacillus subtilis primary sigma σA, which by itself is able to interact with promoter DNA at the concentration of 10 μM without the assistance from core RNAP (37). The observed binding of HrpL in high concentrations to the promoters of C1O30_RS12170, C1O30_RS16815, and C1O30_RS19500 containing unusual hrp-boxes in the absence of core RNAP is not likely to be biologically relevant or evidence of function, since these atypical hrp-boxes are not functional HrpL-dependent promoters, and the expression of these three genes is not significantly affected by HrpL (Fig. 8A). To verify this, low concentrations of HrpL in addition to RNAP were also used to test the affinity to these three promoters with atypical hrp boxes. No bound band was observed for the atypical promoters, while bound bands were formed by incubation of 0.2 μg HrpL protein with hrpA promoter, and 0.4 μg HrpL protein with dspE promoter, with RNAP (Fig. 8C).
The expression of CWDE genes was broadly reduced in JZL7.
Plant cell wall is a major barrier for the invasion of pathogenic bacteria and is composed of mainly cellulose, hemicellulose, and pectin polymers. To breach this barrier, D. zeae bacteria produce a full set of cellulases (Cels), pectinases (Pels), polygalacturonases (Pehs), and proteases (Prts), along with a glucuronoxylanase (XynA) and two rhamnogalacturonanate lyases (RhiE1/2) (Table 3). Our previous study showed that JZL strains produced significantly smaller amounts of all four types of CWDEs compared to EC1 and MS2 when they were all cultivated at the same, relatively high cell density (optical density at 600 nm [OD600] = 1.8) (9).
TABLE 3.
Genes encoding the CWDEs in genomes of Dickeya zeae
| Gene | Accession no. of: |
Identity between MS2 and JZL7 | |||
|---|---|---|---|---|---|
| EC1 | Ech586 | MS2 | JZL7 | ||
| celZ | W909_12595 | Dd586_1489 | C1O30_RS13580 | JZL002780 | 96.94 |
| celY | W909_19635 | Dd586_4057 | C1O30_ RS20545 | JZL004313 | 97.07 |
| bglA | W909_01965 | Dd586_0376 | C1O30_ RS02015 | JZL000410 | 97.56 |
| bgxA | W909_07850 | Dd586_2493 | C1O30_ RS08765 | JZL001697 | 95.72 |
| bglB | W909_11710 | Dd586_1660 | C1O30_ RS12705 | JZL002602 | 95.14 |
| nagZ | W909_12020 | Dd586_1600 | C1O30_ RS13010 | JZL002663 | 94.38 |
| bglC | W909_16350 | JZL003611 | |||
| bglD | W909_16355 | JZL003612 | |||
| celH | W909_16395 | Dd586_3407 | C1O30_ RS17155 | JZL003621 | 97.89 |
| lfaA | W909_11990 | Dd586_1606 | C1O30_ RS12980 | JZL002657 | 95.71 |
| pnl | W909_02890 | C1O30_ RS02835 | |||
| pelN | W909_08805 | Dd586_2245 | C1O30_ RS09885 | JZL001947 | 97.72 |
| pelL | W909_12600 | Dd586_1488 | C1O30_ RS13585 | JZL002781 | 96.36 |
| pelI | W909_14095 | Dd586_2937 | C1O30_ RS14580 | JZL003127 | 84.96 |
| pelA | W909_14860 | Dd586_3083 | C1O30_ RS15345 | JZL003289 | 96.11 |
| pelE | W909_14870 | Dd586_3084 | C1O30_ RS15350 | JZL003291 | 96.50 |
| pelD | W909_14875 | Dd586_3085 | C1O30_ RS15355 | JZL003292 | 97.84 |
| pelC | W909_18430 | Dd586_3788 | C1O30_ RS19205 | JZL004051 | 96.63 |
| pelB | W909_18435 | Dd586_3789 | C1O30_ RS19210 | JZL004052 | 97.61 |
| pelZ | W909_18440 | Dd586_3790 | C1O30_ RS19215 | JZL004053 | 92.89 |
| pelW | W909_10230 | Dd586_1962 | C1O30_ RS11330 | JZL002317 | 98.16 |
| pelX | W909_20195 | Dd586_4161 | C1O30_ RS21105 | JZL004439 | 97.03 |
| paeX | W909_10260 | Dd586_1956 | C1O30_ RS11360 | JZL002324 | 95.61 |
| paeY | W909_14880 | Dd586_3086 | C1O30_ RS15360 | JZL003293 | 91.08 |
| pemA | W909_14885 | Dd586_3087 | C1O30_ RS15365 | JZL003294 | 97.07 |
| pehN | C1O30_ RS05710 | JZL001060 | 98.69 | ||
| pehK | W909_15935 | Dd586_3319 | C1O30_ RS16710 | JZL003524 | 96.93 |
| pehX | W909_19025 | Dd586_3904 | C1O30_ RS19785 | JZL004171 | 96.91 |
| rhiE1 | C1O30_ RS04480 | JZL000839 | 98.00 | ||
| rhiE2 | W909_09610 | Dd586_2097 | C1O30_ RS10600 | JZL002175 | 97.29 |
| xynA | W909_10005 | Dd586_2011 | C1O30_ RS11050 | JZL002266 | 94.12 |
| prtX | W909_09760 | Dd586_2059 | C1O30_ RS10785 | JZL002216 | 95.49 |
| prtC | W909_09765 | Dd586_2058 | C1O30_ RS10790 | JZL002217 | 97.71 |
| prtB | W909_09770 | Dd586_2057 | C1O30_ RS10795 | JZL002218 | 96.53 |
| prtG | W909_09795 | Dd586_2052 | C1O30_ RS10820 | JZL002223 | 96.30 |
| prtF | W909_09775 | Dd586_2056 | C1O30_ RS10800 | JZL002219 | 97.51 |
| prtE | W909_09780 | Dd586_2055 | C1O30_ RS10805 | JZL002220 | 97.09 |
| prtD | W909_09785 | Dd586_2054 | C1O30_ RS10810 | JZL002221 | 96.89 |
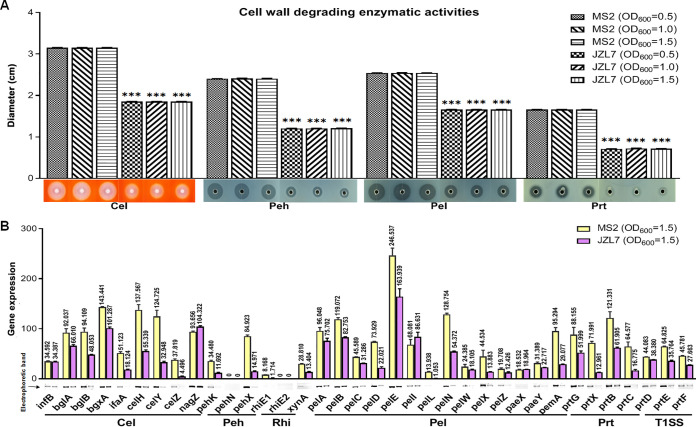
In this study, we further compared the CWDE activities of strains JZL7 and MS2 at three different cell densities (i.e., OD600 of 0.5, 1.0, and 1.5) and observed a similar pattern that JZL7 produced a considerably smaller amount of CWDEs (Cels: 41.3%; Pehs: 50%; Pels: 34.6%; Prts: 57.2%) than MS2 at all densities (Fig. 9A). The lower CWDE activities of JZL7 might be due to either a much smaller repertoire of CWDE encoding genes or a greatly reduced expression of the same set of genes. Our genome comparison revealed a highly conserved set of 34 CWDE genes shared by both JZL7 and MS2 (84.96% to 98.69% DNA sequence identity) and very few strain-specific genes, including two beta-glucosidase-encoding genes (bglC and bglD) in JZL7 and one pectin lyase-encoding gene (pnl) in MS2 (Table 3). Interestingly, recent studies showed that pnl was drastically upregulated during early disease development in Pectobacterium carotovorum but not so in D. dadantii (38). Here, in D. zeae, we found that neither the complementation of pnlMS2 in JZL7 nor the deletion of pnl in MS2 altered the virulence of respective strains on dicotyledonous tissues (i.e., potato and radish slices) (Fig. S2). We then measured the expression of the shared CWDE genes in the two strains using semiquantitative PCR. Results showed that the ratios (JZL7 versus MS2) of cumulative expression levels of genes encoding Cels, Pehs, RhiEs, XynA, Pels, and Prts are 0.56 (430.579:774.477), 0.22 (26.663:119.403), 0.21 (1.714:8.168), 0.47 (13.404:28.810), 0.62 (632.929:1,023.790), and 0.11 (143.640:1,346.054), respectively. In addition, Dickeya exports Prts via T1SS encoded by the prtD, prtE, and prtF genes after recognizing a C-terminal signal sequence in their substrates (18, 39, 40). The expression level of T1SS genes in strain JZL7 is 0.66 of those in strain MS2 (Fig. 9B). Furthermore, 15 of the 34 shared CWDE genes had significantly lower expression levels in JZL7 than in MS2, including lfaA, celH, celY, celZ, pehK, pehX, rhiE1, xynA, pelD, pelL, pelN, pelX, pemA, prtX, and prtC (Fig. 9B).
FIG 9.
Extracellular cell wall-degrading enzymes (CWDEs) produced by D. zeae strains MS2 and JZL7. (A) CWDE activities of strains MS2 and JZL at different cell concentrations. Samples of 20 μL bacterial cells (OD600 of 0.5, 1.0 and 1.5) were added to the assay plate wells (5 mm in diameter) and incubated at 28°C. Pel and Peh assay plates were treated with 1 M HCl after 14 h, respectively. Cel assay plate was stained with 0.1% Congo red for 15 min after 14 h and decolored twice with 1 M NaCl. Photos were taken of Prt assay plate after 24 h without any further treatment. (B) RT-PCR of CWDE genes of strains MS2 and JZL (OD600 of 1.5). The reference gene infB (coding transfer initiation factor 2) was used to equilibrate the concentration of cDNA samples. The expression of genes was determined by measuring the signal intensity of the bands (under the x axis) using Image Lab software (Bio-Rad, USA). Experiments were repeated three times in triplicate, and the mean data above the bars indicate the signal intensity of RT-PCR bands.
The broadly reduced expression of CWDE genes observed here was consistent with the aforementioned lower CWDE activities in JZL7 and may provide a basis for the strain’s lack of virulence on dicots. Specifically, monocots and dicots differ in the composition and structure of their primary cell walls; the former contain mainly cellulose and rarely pectin, while the latter consist of both (41). In this study, we showed that several pectin lyase-encoding genes were either lost (e.g., pnl) or expressed at remarkably lower levels (e.g., pelD, pelL, pelN, pelX, and pemA) in JZL7 (Fig. 9B). The resulting reduction in Pel activity might considerably compromise the ability of JZL7 to degrade cell wall and invade dicot hosts, which, together with the loss of T3SS, might abolish the pathogenicity of JZL7 on dicots. To verify whether the significantly lower expression of CWDEs is due to the lower expression of some important regulators controlling CWDE activity, we measured the expression of the genes encoding CWDE regulators, such as Fis, SlyA, VfmE, PecS, PecT, and KdgR, in both MS2 and JZL7 by RT-PCR. Results showed that none of these genes was differentially expressed between MS2 and JZL7 (Fig. S3).
Other 13 genes were newly found to encode pathogenicity-related proteins in D. zeae MS2.
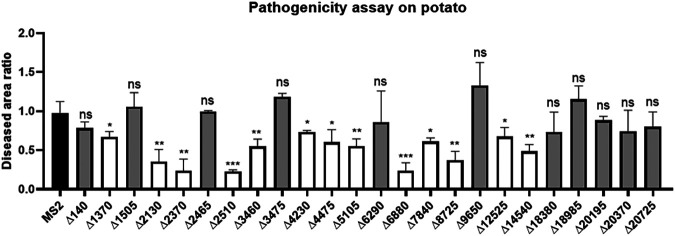
In order to identify additional factors related to the virulence of D. zeae pathogens on dicots, we obtained knockout mutants in MS2 for most of the genes absent in JZL7 but shared by MS2 and EC1 (apart from the T3SS and T3SE genes, which have already been examined above; Table S8). Pathogenicity tests showed that deletion of C1O30_RS01370-01390, C1O30_RS02130, C1O30_RS02370, C1O30_RS02510, C1O30_RS03460, C1O30_RS04230, C1O30_RS04475, C1O30_RS05105, C1O30_RS06880, C1O30_RS07840, C1O30_RS08725, C1O30_RS12525, and C1O30_RS14540 significantly reduced the virulence of MS2 on potato slices (Fig. 10). Among them, C1O30_RS01370-1390, C1O30_RS02510, C1O30_RS03460, C1O30_RS04230, C1O30_RS04475, C1O30_RS05105, C1O30_RS07840, C1O30_RS08725, C1O30_RS12525, and C1O30_RS14540, respectively, carry a dimethyl sulfoxide reductase anchor subunit gene cluster (dmsC, dmsB, dmsA, dmsD, and a SDR family oxidoreductase) and encode a GNAT family N-acetyltransferase, LysE family transporter, a PAS domain-containing protein (helix-turn-helix transcriptional regulator), an SDR family oxidoreductase, an HAMP domain-containing protein, an FMN-binding negative transcriptional regulator, a glycosyltransferase, DUF2335 domain-containing protein, and a diguanylate cyclase, while all the others encode hypothetical proteins (Table S8). All these genes were first reported to be involved in Dickeya pathogenesis and, thus, represent prominent targets of future studies.
FIG 10.
Maceration tests of other D. zeae MS2-unique gene mutants on potato slices. The data present the means of three replicates, and error bars represent the standard deviation. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Student’s t test).
Recently, an saxA gene encoding isothiocyanate hydrolase was found to enable a potato-specific Pectobacterium parmentieri strain to increase the ability to macerate Arabidopsis, suggesting its significant role in defining the host range (42). We searched the homologs of this interesting gene in the genomes of D. zeae and found 4 and 6 copies of saxA, respectively, present in the genomes of MS2 and JZL7. Thus, we ruled out the possibility of SaxA as a host range determinant.
Pathogenic mechanism of Dickeya and other soft rot Pectobacteriaceae.
In this study, we sequenced the genome of D. zeae JZL7 and carried out in-depth comparative and functional analyses with other D. zeae strains. The results not only provide insights into the unique pathogenicity and host specificity of JZL7 but also help to better understand the pathogenic mechanism of soft rot Pectobacteriaceae pathogens (e.g, Dickeya, Erwinia, and Pectobacterium) in general.
Xanthomonas, Ps. syringae, and many other bacterial phytopathogens adopt the “stealth” model of pathogenicity whereby they make use of an extensive battery of type III secreted effector proteins and phytotoxins for successful infection. These effectors act to suppress or manipulate host defenses as the bacterial population grows to numbers that are sufficient to induce disease symptoms (43–47). Additionally, many studies have demonstrated that T3SEs could alter the physiology of host cells in a way that is beneficial for the pathogens. In Xanthomonas, a mutation in the T3SS impairs the ability to inject T3SEs in the host plant and, as a consequence, abolishes pathogenicity and multiplication in planta (48). Among pathogenicity determinants shown to display heterogeneous distribution between strains are T3SEs. Furthermore, in Xanthomonas and Ps. syringae, diversity in T3SE repertoires from different hosts revealed determinants of host specificity (45, 49, 50).
Soft rot Pectobacteriaceae pathogens also possess in their genomes the molecular machineries required for the stealth mode of infection, such as T3SS, T4SS, phytotoxins, and so on. Functional studies have also demonstrated the importance of T3SS in soft rot Pectobacteriaceae (SRP) pathogens. For instance, T3SS is important for the full virulence of D. dadantii 3937 (25, 26) and required for pellicle formation and cell aggregation (51, 52). However, T3SS or T3SE has not been reported in SRP as a determinant of host range, and the role of T3SS in the virulence of necrotrophic SRP and other bacterial pathogens appears quite complex.
In our study, we knocked out the whole T3SS gene clusters dsp/hrp/hrc in D. zeae MS2, which resulted in significantly reduced virulence on both monocotyledonous and dicotyledonous hosts (Fig. 7), demonstrating its role as an important virulence factor in the course of disease. However, Δdsp/hrp/hrc was not virulence free on either monocots or dicots, suggesting that T3SS alone does not determine host range in D. zeae. In addition, Er. pyrifoliae, Pe. carotovorum, Pectobacterium wasabiae, and Ps. syringae strains lacking a functional T3SS have been reported to infect plants (22, 23, 53, 54). In Ps. syringae, nonpathogenic isolates are separate from the pathogenic ones due to the deficiency of T3SS and T3SEs (23). In contrast, T3SS-deficient Pe. wasabiae strains are still virulent (53). More strikingly, human-pathogenic Ps. aeruginosa isolates from patients with chronic lung infections are typically T3SS deficient, even though 90% of environmental isolates carry a T3SS (55). These T3SS-deficient pathogens may have alternative ways of modifying the host plant at the initiation of infection.
It has often been assumed that the mechanisms of infection used by SRP are distinctly different from those used by other bacteria like Xanthomonas and Ps. syringae (56). In addition to the above-mentioned stealth model, SRP pathogens also share a “brute-force” model of pathogenicity; they produce many CWDEs to physically attack the plant cell walls and their surrounding apoplast, which thus promotes soft rotting (45, 57). It is unquestionable that brute force has made the SRP highly successful pathogens, and the CWDEs are critical factors in both the pathogenicity and host range of SRP. We compared the concentrated enzymatic activities of soft rot D. zeae MS2 and JZL7 and found that broadly reduced expression of CWDE genes might be a major cause for the significantly lower virulence of JZL7 on dicots (Fig. 9). Importantly, the brute-force and stealth models seem to operate in parallel in SRP pathogens, as the production of CWDEs remained unaffected in the T3SS mutants of MS2 (Fig. S4), which might explain their remaining soft rot symptoms on potato and radish slices (Fig. 7C and D).
Bacterial phytopathogens employ various approaches to infect and kill their host(s). Plant-pathogen interaction is a multifaceted process, mediated by the pathogen- and plant-derived molecules. Secreted as well as translocated molecules, derived from the pathogens, are the key factors, which determine their pathogenicity and allow successful colonization of pathogens on the host. On the other hand, plant-derived molecules are involved in the recognition of pathogen and triggering of the defense response. In the future, to learn more about the latent stage of Dickeya infection and host range, we should consider factors including both CWDEs and T3SEs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium. D. zeae and its derivatives strains were cultivated at 28°C in LB medium, minimal medium (MM), or LS5 medium (58, 59). Antibiotics were added to the media at the following final concentrations when required: streptomycin 50 µg/mL, kanamycin 50 µg/mL, tetracycline 15 μg/mL, and ampicillin 100 μg/mL.
Genome sequencing and assembly.
Genomic DNA was extracted from D. zeae strain JZL7 in LB medium culture using MasterPure DNA purification kit (EPICENTRE Biotechnologies, Madison), which was then subjected to quality control by agarose gel electrophoresis and quantified by Qubit. The genome sequencing was performed at Health Time Gene (Shenzhen, China) using both the PacBio RS II and the Illumina Hiseq X Ten platforms.
For the PacBio sequencing, genomic DNA was treated into fragments in 10 kb by g-TUBE first. The fragments were damage-repaired and end-repaired. Both sides of the DNA fragments were, respectively, connected with hairpin adapter to get a dumbbell structure, which is known as SMRTbell. After annealing, the SMRTbell was fixed at the bottom of the ZWM polymerase and was sequenced last. Adapter trimming and quality filtering were performed by using fastp (version 0.20.0) (60) with default parameters.
For the Illumina sequencing, genomic DNA was randomly broken, and 350 bp fragments were purified and end-repaired using T4 DNA polymerase, Klenow DNA polymerase, and T4 PNK. The fragments were added with an “A” base at the 3′ terminal and ligated with the adaptor with a “T” base at its terminal. The library was sequenced on the Hiseq X Ten platform to generate PE150 reads. A hybrid genome assembly with both the PacBio long-reads and Illumina short-reads was carried out using Unicycler v0.4.7 (default parameters) (61).
Genome annotation.
The genome annotation of JZL7 was carried out using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v2019-11-25 (standalone version) (62). Functional annotations of all proteins predicted in JZL7 were conducted using eggNOG-mapper v1.0.3 (for GO) (63), KofamKOALA v2020-08-04 (for KEGG) (64), and COGsoft v201204 (for COG) (65). The genome and annotated protein sequences of other Dickeya strains analyzed in this study were all obtained from the NCBI RefSeq and GenBank databases (Table S2). Genomic islands and prophages were predicted using the webservers of IslandViewer 4 (http://www.pathogenomics.sfu.ca/islandviewer/) (66) and PHASTER (http://phaster.ca/) (67), respectively. Genes encoding type I to VI secretion system-related proteins were identified using TXSScan v1.0.2 (68). Type III secreted effectors (T3SEs) were predicted by using a combination of four bioinformatics approaches, including the webserver of Bastion3 (https://bastion3.erc.monash.edu/) (69) and standalone versions of BEAN2 v2.0 (70), DeepT3 (71), and pEffect (72); a protein is classified as T3SE only if it is predicted by at least three of the four approaches. The circular visualization of JZL7 genome characteristics was created by Circos (73). Prediction of hrp box was performed by searching the promoter sequences (2,000 bp upstream the start codon) of the candidate T3SE-coding genes in MS2 genome using the following regular expressions: GGAAC[CT][ATCG]{1,100}[CGT][CT]AC[ATG][CT]A (forward) and T[AG][ATC]GT[AG][AG][AGCT]{1,100}[AG]GTTCC (reverse).
Comparative genomic analyses.
Pairwise average nucleotide identity (ANI) values between all D. zeae and D. oryzae strains were calculated using pyani v0.2.10 (with the “-m ANIb” option) (74). The whole-genome alignment of CE1, EC1, EC2, Ech586, JZL7, and MS2 was constructed using Mauve v2.4.0 (75). The pangenome analysis of EC1, JZL7, and MS2 was carried out using OrthoFinder v2.4.0 (76). To build the phylogenetic tree, we generated multiple DNA sequence alignment for each of the 3,110 single-copy core genes (present in all strains) using MAFFT v7.453 (77). All single-gene alignments were then concatenated, and a phylogenetic inference was conducted using IQ-TREE v1.6.12 (78). Reliability of the inferred topology was assessed by ultrafast bootstrap with 1,000 replicates.
Generation of in-frame deletion mutants.
For generation of in-frame deletion mutants of D. zeae MS2, triparental mating was performed using the methods described previously (15). For instance, to knock out the hrp/hrc gene cluster of T3SS in strain MS2, the upstream and downstream fragments of hrp/hrc gene cluster were amplified using the primer pairs of hrp/hrc-1 accompanied with hrp/hrc-2, and hrp/hrc-3 accompanied with hrp/hrc-4, respectively (Table S3), and purified with AxyPrep DNA gel extraction kit (Axygen Biotech Co., Hangzhou, China). The fragments were ligated with the BamHI and Spe digested suicide plasmid pKNG101 using ClonExpress MultiS kit (Vazyme Biotech Co., Nanjing, China) and transformed into E. coli CC118 competent cells. In triparental mating, the donor and receptor cells were mixed with the helper strain E. coli RK2013 (grown in LB medium containing kanamycin) in a ratio of 2:1:1 on LB plate and incubated at 28°C overnight. The transformants were grown on minimal medium (MM) agar plates (13) containing streptomycin sulfate. Single colony culture was then spread on MM agar plate containing 5% sucrose to exclude the suicide plasmid. The resultant deletion mutants were confirmed by PCR using the detection primer pair of hrp/hrc-F and hrp/hrc-R and DNA sequencing. In the same way, we correspondingly deleted the important T3SS transcriptional regulation genes hrpX, hrpY, hrpS, and hrpL and the candidate T3SE genes dspE, hrpZ, hrpW, hrpK, and hrpN, the pectin lyase-encoding gene pnl, and, compared to JZL7, some genes specific to MS2. In addition, the Δdsp/hrp/hrc mutant was also generated by deleting the dsp gene cluster based on the obtained Δhrp/hrc mutant. The primers are listed in Table S3.
Measurement of bacterial growth curves.
Bacterial strains were grown in LB medium overnight at 28°C, adjusted to an OD600 of 1.5, and diluted into fresh LB and LS5 medium in a 1:100 ratio, respectively (9). Aliquots of 500-μL dilutions were transferred into 2.0-mL tubes. Bacteria were grown with shaking at 200 rpm under 28°C, and cell density was measured every 2 h. The experiment was repeated three times in triplicate.
HR assay.
Bacteria were grown in LS5 medium (pH 5.5) (59) until they reached an OD600 of 1.0. Leaves of Nicotiana tabacum variant K326 (27) and N. benthamiana (28) were, respectively, inoculated on the back by pressing a sterilized puncher (5 mm) dipped with bacterial culture and incubated at 28°C. The area of HR lesions on leaves was measured using Image J 1.52a 12 and 24 h postinoculation (hpi) (79). To quantify the function of the T3SS, we infiltrated leaves of N. tabacum K326 with 100 μL of MS2 bacterial dilutions in LS5 medium (OD600 of 0.5, containing approximately 108 CFU/mL), and 100 μL each of LS5 medium, Δdsp/hrp/hrc, and JZL7 (107 CFU/mL) was also infiltrated as a negative control. Each assay was repeated three times in triplicate.
Pathogenicity tests on monocotyledonous and dicotyledonous plants.
Strains were cultured to the logarithmic phase until an OD600 of 1.2 was reached in LB medium, and pathogenicity tests were carried out on monocotyledonous and dicotyledonous hosts. For monocots, every 100 μL of bacterial culture was injected into the basal stem of banana and the base of clivia leaf. For dicotyledonous plants, potato and radish tubers were surface-sterilized with 70% ethanol, cut evenly about 5 mm in thickness, and then placed onto moistened filter paper in a tray. Bacterial cells of 2 μL were applied to the center of the tuber slices. All plants were kept in a growth chamber with controlled conditions of 28 ± 2°C, 75% ± 15% relative humidity, and 24 h white light (7,350 lx) illumination until symptoms appeared, except that the bananas were incubated at 30°C with 12-h alternating light-dark cycles for 7 days and then disease was assessed using a modified virulence scoring (17, 80). Same volume of LB medium was inoculated as a negative control. Visible macerate areas on clivia leaves and potato and radish slices were measured using Image J 1.52a. Each assay was repeated three times in triplicate.
RNA purification.
The bacterial cultures used for RNA extraction were the ones described below for CWDE activity measurement. Since the same strain shared basically the same enzymatic activity in different cell densities (OD600 of 0.5, 1.0, and 1.5), the bacterial cultures of strains MS2 and JZL7 at an OD600 of 1.5 were selected for RNA extraction. On the other hand, to determine whether the predicted T3SE expression is induced by the HrpL regulator, we grew strains MS2 and ΔhrpL in LS5 medium (pH 5.5) (59) for RNA extraction until an OD600 of 1.0 was reached. The RNA was extracted using the SV total RNA isolated system kit (Promega, Madison, WI, USA), further purified using the RNA clean kit (Qiagen, Hilden, Germany), and treated with DNase I to degrade any possible DNA contamination. Quantity of RNA was first measured using a NanoDrop 2000c (Thermo Fisher Scientific, MA, USA), and the integrity of RNA was detected by agarose gel electrophoresis.
RT-PCR analysis.
For each RNA sample for CWDE activity measurement, two dilutions (5 and 50 ng) were reverse transcribed into cDNAs using FastKing gDNA dispelling RT mix (Tiangen Biotech, Co., Ltd., Beijing, China), and the concentration of each resultant cDNA was quantitatively equilibrated according to the expression quantities of reference gene infB of strains MS2 and JZL7. PCR was performed using the primers listed in Table S3, and gene expression (signal intensity) of each gene was determined using the software Image Lab (Bio-Rad, USA). The experiment was repeated three times in triplicate.
qRT-PCR analysis.
In qRT-PCR analysis, an aliquot of 300 ng RNA sample was used for genomic DNA elimination and cDNA synthesis by a FastKing RT kit (with gDNase) (Tiangen Biotech, Co., Ltd., Beijing, China) following the manufacturer’s protocol. The housekeeping gene atpD was used as a reference. The PCR efficiency of each gene was determined using five DNA standards at different concentrations (10, 1, 0.1, 0.01, and 0.001 g/mL). The qRT-PCR was conducted on a Quantstudio 6 Flex system using ChamQ Universal SYBR qPCR master mix (Vazyme Biotech Co., Nanjing, China) with the following cycle profile: 1 cycle at 95°C for 30 s, followed by 40 cycles at 95°C for 10 s, 60°C for 30 s, and then 1 cycle at 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. The experiment was repeated three times, each time in triplicate. Data were analyzed using the 2−ΔΔCT method as described previously (81).
Protein expression and purification.
The open reading frame (ORF) that encodes HrpL protein was amplified from MS2 genomic DNA using primers pET32a-hrpL-F and pET32a-hrpL-R containing BamHI and HindIII restriction enzyme sites (Table S3). The PCR product was purified and ligated to the BamHI/HindIII-digested pET32a vector harboring a thioredoxin (TRX)-His6 tag at its N terminus. pET32a-hrpL was then transformed into the E. coli strain BL21(DE3) competent cells (TransGen Biotech Co., Beijing, China) and confirmed by sequencing. To express the HrpL protein, a single colony of BL21(pET32a-hrpL) was grown in LB medium containing 100 μg/μL of ampicillin overnight and transferred into 1 L of fresh LB medium in a 1:100 ratio to grow until reaching an OD600 of 0.6. Then, 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) was added to the culture to induce protein expression at 18°C for 16 h. Bacterial cells were harvested by centrifuging at 5,000 rpm at 4°C for 30 min and suspended in 20 mL of phosphate-buffered saline (PBS). The cells were lysed with sonication and centrifuged at 12,000 rpm at 4°C for 30 min. The supernatant was filtered with a 0.45-μm-pore filter (MilliporeSigma, St. Louis, MO, USA), and the filtrate was added into a Ni-nitrilotriacetic acid (NTA) column (Changzhou Smart-Life Sciences Biotechnology Co., Changzhou, China) that had been equilibrated with lysis buffer. After washing five times with lysis buffer, the column was eluted with 30 mL gradients of 10 to 500 mM imidazole prepared in wash buffer, respectively. Fractions were collected, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to verify the molecular weight of the target protein. The protein was concentrated by centrifugation (MilliporeSigma, St. Louis, MO, USA) at 4°C and then stored at −80°C.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assay (EMSA) was performed as described previously (58). DNA probes were PCR amplified using primers listed in Table S3. The purified PCR fragments were labeled by biotin using the biotin 3′ end DNA labeling kit (Thermo Fisher Scientific, MA, USA). Binding reactions were performed in a final volume of 10 μL using LightShift chemiluminescent EMSA kit following the manufacturer’s protocol (Thermo Fisher Scientific, MA, USA). For each reaction, 1 μL of 10× binding buffer, 0.5 μL of 1 μg/μL Poly, 1 nmol of labeled DNA fragments, and 1, 5, or 10 μg of HrpL protein placed in an ice bath were added together and incubated for 30 min at 20°C. The protein-DNA complexes and the unbound free DNA fragments were separated on 6% nondenaturing polyacrylamide (acrylamide/bisacrylamide 29: 1 [vol/vol]) gels using the electrophoresis buffer Tris-borate-EDTA (TBE) and were detected by chemiluminescence (Tanon, China). A 100-fold molar excess of unlabeled DNA fragment was incubated with HrpL protein for 15 min before addition of the labeled DNA fragments to verify specific interaction of the HrpL protein-DNA fragment. In the experiment of adding core RNA polymerase (RNAP), E. coli core RNAP (0.5 U) (New England Biolabs [Beijing] Ltd., Beijing, China) (82) was incubated on ice for 20 min with 200 or 400 ng of purified HrpL protein (34). The remaining steps were as described above.
Measurement of CWDE activities.
The activities of CWDEs were measured according to the methods described previously (9). Specifically, pectate lyase (Pel) assay medium (10 g/L polygalacturonic acid, 10 g/L yeast extract, 8 g/L agarose, 0.38 μM CaCl2, and 100 mM Tris-HCl [pH 8.5]), polygalacturonase (Peh) assay medium (5 g/L polygalacturonic acid, 2 g/L sucrose, 2 g/L (NH4)2SO4, and 15 g/L agar [pH 5.5]), cellulase (Cel) assay medium (1.0 g/L carboxymethyl ethyl cellulose, 3.8 g/L Na3PO4, and 8.0 g/L agarose [pH 7.0]), and protease (Prt) assay medium (10 g/L skimmed milk, 5 g/L Bacto tryptone, 2.5 g/L yeast extract, 5 g/L NaCl, and 15 g/L agar) were prepared, and 30 mL of each medium was added into a 10 by 10 cm square plate. Wells in 5 mm diameter were made, and 20 µL of bacterial cells (OD600 of 0.5, 1.0, or 1.5) was applied to the wells. Plates were incubated at 28°C until Pel and Peh assay plates were treated with 1 M HCl after 14 h, and Cel assay plate was stained with 0.1% Congo red for 15 min after 14 h and decolored with 1 M NaCl twice. The protease activity was measured without any further treatment after 24 h. The experiment was repeated three times with duplicates.
Introduction of pnl gene in strain JZL7.
To create the complementing plasmid, sequence of pnI open reading frame (ORF) with 19 bp before the start codon harboring a ribosome-binding site (RBS) was amplified and cloned from strain MS2 into the EcoRI/BamHI-digested pLAFR3 vector using pEASY-Uni seamless cloning and assembly kit (TransGen Biotech Co., Beijing, China) and transformed into E. coli DH5α competent cells. Plasmid construct was confirmed by DNA sequencing and introduced into strain JZL7 by conjugal triparental mating. The primers used here are listed in Table S3.
Statistical analysis.
All the experiments were repeated three times in duplicate or triplicate. GraphPad Prism 8.4.1 (GraphPad Software, San Diego, California) was used to performed unpaired two-tailed t test (83), and the data of D. zeae strain JZL7 were normalized to those of strain MS2. * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, and **** indicates P < 0.0001. Analysis of variance (ANOVA) was used between D. zeae MS2 and its mutants. The values are means of three replicates, and the error bars are standard deviations. Statistical analysis was performed on each group of data, and significantly different values (ANOVA P < 0.05) are indicated by different letters.
To investigate association between the frequency of fold change (>0.5/≤0.5) and that of unique/shared T3SEs of MS2, the Pearson χ2 statistic was computed (Pearson’s chi-square test, χ2 = 8.77, degrees of freedom [df] = 1, χ20.995 = 7.88, χ20.999 = 10.83, and χ20.995 < χ2 < χ20.999). The P value was less than 0.005.
Data availability.
Both the original genome sequencing data and the genome assembly were deposited in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA656647. The JZL7 genome sequence has been deposited in GenBank under accession number CP060263.1.
ACKNOWLEDGMENTS
This work was financially supported by grants from the Key-Area Research and Development Program of Guangdong Province (2020B0202090001 and 2018B020205003), the National Natural Science Foundation of China (31972230, 31901843), the Natural Science Foundation of Guangdong Province, China (2020A1515011534), the Guangzhou Basic Research Program (202102080613), the Foundation from the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-b201908), and the China Scholarship Council Fellowship Program Grant (202108440367).
J.Z. conceived the study, J.Z. and M.H. drafted the manuscript, M.H., Y.X., and M.L. performed the experiments, X.Z., C.L., and M.H. analyzed the genome sequences and experimental data, J.Z., X.Z., L.Z., G.L., and M.R.P. revised the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaofan Zhou, Email: xiaofan_zhou@scau.edu.cn.
Jianuan Zhou, Email: jianuanzhou@scau.edu.cn.
Beile Gao, South China Sea Institute of Oceanology, Chinese Academy of Sciences.
REFERENCES
- 1.Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, Gardan L. 2005. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol 55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- 2.Brady C, Cleenwerck I, Denman S, Venter SN, Rodriguez-Palenzuela P, Coutinho TA, De Vos P. 2012. Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int J Syst Evol Microbiol 62:1592–1602. doi: 10.1099/ijs.0.035055-0. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson N, De Vos P, Pirhonen M, Elphinstone JG. 2014. Dickeya aquatica sp nov., isolated from waterways. Int J Syst Evol Microbiol 64:2264–2266. doi: 10.1099/ijs.0.058693-0. [DOI] [PubMed] [Google Scholar]
- 4.Tian Y, Zhao Y, Yuan X, Yi J, Fan J, Xu Z, Hu B, De Boer SH, Li X. 2016. Dickeya fangzhongdai sp. nov., a plant-pathogenic bacterium isolated from pear trees (Pyrus pyrifolia). Int J Syst Evol Microbiol 66:2831–2835. doi: 10.1099/ijsem.0.001060. [DOI] [PubMed] [Google Scholar]
- 5.Duprey A, Taib N, Leonard S, Garin T, Flandrois JP, Nasser W, Brochier-Armanet C, Reverchon S. 2019. The phytopathogenic nature of Dickeya aquatica 174/2 and the dynamic early evolution of Dickeya pathogenicity. Environ Microbiol 21:2809–2835. doi: 10.1111/1462-2920.14627. [DOI] [PubMed] [Google Scholar]
- 6.Hugouvieux-Cotte-Pattat N, Jacot-Des-Combes C, Briolay J. 2019. Dickeya lacustris sp. nov., a water-living pectinolytic bacterium isolated from lakes in France. Int J Syst Evol Microbiol 69:721–726. doi: 10.1099/ijsem.0.003208. [DOI] [PubMed] [Google Scholar]
- 7.Oulghazi S, Pédron J, Cigna J, Lau YY, Moumni M, Van Gijsegem F, Chan KG, Faure D. 2019. Dickeya undicola sp. nov., a novel species for pectinolytic isolates from surface waters in Europe and Asia. Int J Syst Evol Microbiol 69:2440–2444. doi: 10.1099/ijsem.0.003497. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, He S, Guo H, Han J, Thin KK, Gao J, Wang Y, Zhang X. 2020. Dickeya oryzae sp. nov., isolated from the roots of rice. Int J Syst Evol Microbiol 70:4171–4178. doi: 10.1099/ijsem.0.004265. [DOI] [PubMed] [Google Scholar]
- 9.Hu M, Li JL, Chen RT, Li WJ, Feng LW, Shi L, Xue Y, Feng XY, Zhang LH, Zhou JN. 2018. Dickeya zeae strains isolated from rice, banana and clivia rot plants show great virulence differentials. BMC Microbiol 18:136. doi: 10.1186/s12866-018-1300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toth IK, van der Wolf JM, Saddler G, Lojkowska E, Hélias V, Pirhonen M, Tsror Lahkim L, Elphinstone JG. 2011. Dickeya species: an emerging problem for potato production in Europe. Plant Pathol 60:385–399. doi: 10.1111/j.1365-3059.2011.02427.x. [DOI] [Google Scholar]
- 11.van der Wolf JM, Nijhuis EH, Kowalewska MJ, Saddler GS, Parkinson N, Elphinstone JG, Pritchard L, Toth IK, Lojkowska E, Potrykus M, Waleron M, de Vos P, Cleenwerck I, Pirhonen M, Garlant L, Hélias V, Pothier JF, Pflüger V, Duffy B, Tsror L, Manulis S. 2014. Dickeya solani sp. nov., a pectinolytic plant pathogenic bacterium isolated from potato (Solanum tuberosum). Int J Syst Evol Microbiol 64:768–774. doi: 10.1099/ijs.0.052944-0. [DOI] [PubMed] [Google Scholar]
- 12.Golanowska M, Kielar J, Lojkowska E. 2017. The effect of temperature on the phenotypic features and the maceration ability of Dickeya solani strains isolated in Finland, Israel and Poland. Eur J Plant Pathol 147:803–817. doi: 10.1007/s10658-016-1044-1. [DOI] [Google Scholar]
- 13.Hussain MBBM, Zhang HB, Xu JL, Liu QG, Jiang ZD, Zhang LH. 2008. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J Bacteriol 190:1045–1053. doi: 10.1128/JB.01472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin BR, Shen HF, Pu XM, Tian XS, Zhao WJ, Zhu SF, Dong MM. 2010. First report of a soft rot of banana in Mainland China caused by a Dickeya sp. (Pectobacterium chrysanthemi). Plant Dis 94:640. doi: 10.1094/PDIS-94-5-0640C. [DOI] [PubMed] [Google Scholar]
- 15.Zhou JN, Zhang HB, Lv MF, Chen YF, Liao LS, Cheng YY, Liu SY, Chen SH, He F, Cui ZN, Jiang ZD, Chang CQ, Zhang LH. 2016. SlyA regulates phytotoxin production and virulence in Dickeya zeae EC1. Mol Plant Pathol 17:1398–1408. doi: 10.1111/mpp.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Shen H, Pu X, Lin B, Hu J. 2014. Identification of Dickeya zeae as a causal agent of bacterial soft rot in banana in China. Plant Dis 98:436–442. doi: 10.1094/PDIS-07-13-0711-RE. [DOI] [PubMed] [Google Scholar]
- 17.Feng LW, Schaefer AL, Hu M, Chen RY, Greenberg EP, Zhou JN. 2019. Identification of a virulence factor in the banana pathogen Dickeya zeae MS2. Appl Environ Microbiol 85:e01611-19. doi: 10.1128/AEM.01611-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou JN, Cheng YY, Lv MF, Liao LS, Chen YF, Gu YF, Liu SY, Jiang ZD, Xiong YY, Zhang LH. 2015. The complete genome sequence of Dickeya zeae EC1 reveals substantial divergence from other Dickeya strains and species. BMC Genomics 16:571. doi: 10.1186/s12864-015-1545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou JN, Zhang HB, Wu J, Liu QG, Xi PG, Lee J, Liao JL, Jiang ZD, Zhang LH. 2011. A novel multi-domain polyketide synthase is essential for zeamine antibiotics production and the virulence of Dickeya zeae. Mol Plant Microbe Interact 24:1156–1164. doi: 10.1094/MPMI-04-11-0087. [DOI] [PubMed] [Google Scholar]
- 20.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A 99:13142–13147. doi: 10.1073/pnas.202358699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jock S, Kim WS, Barny MA, Geider K. 2003. Molecular characterization of natural Erwinia pyrifoliae strains deficient in hypersensitive response. Appl Environ Microbiol 69:679–682. doi: 10.1128/AEM.69.1.679-682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohr TJ, Liu H, Yan S, Morris CE, Castillo JA, Jelenska J, Vinatzer BA. 2008. Naturally occurring nonpathogenic isolates of the plant pathogen Pseudomonas syringae lack a type III secretion system and effector gene orthologues. J Bacteriol 190:2858–2870. doi: 10.1128/JB.01757-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun WX, Liu LJ, Bent AF. 2011. Type III secretion-dependent host defence elicitation and type III secretion-independent growth within leaves by Xanthomonas campestris pv. campestris. Mol Plant Pathol 12:731–745. doi: 10.1111/j.1364-3703.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CH, Gavilanes-Ruiz M, Okinaka Y, Vedel R, Berthuy I, Boccara M, Chen J, Perna NT, Keen NT. 2002. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol Plant Microbe Interact 15:472–480. doi: 10.1094/MPMI.2002.15.5.472. [DOI] [PubMed] [Google Scholar]
- 26.Yap MN, Yang CH, Barak JD, Jahn CE, Charkowski AO. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J Bacteriol 187:639–648. doi: 10.1128/JB.187.2.639-648.2005. [DOI] [PMC free article] [PubMed]
- 27.Chen YM, Dong YH, Liang ZB, Zhang LH, Deng YZ. 2018. Enhanced vascular activity of a new chimeric promoter containing the full CaMV 35S promoter and the plant XYLOGEN PROTEIN 1 promoter. 3 Biotech 8:380. doi: 10.1007/s13205-018-1379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie JJ, Yin ZY, Li Z, Wu Y, Huang L. 2019. A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol 222:995–1011. doi: 10.1111/nph.15631. [DOI] [PubMed] [Google Scholar]
- 29.Oh CS, Kim JF, Beer SV. 2005. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol Plant Pathol 6:125–138. doi: 10.1111/j.1364-3703.2005.00269.x. [DOI] [PubMed] [Google Scholar]
- 30.Xie YP, Shao XL, Deng X. 2019. Regulation of type III secretion system in Pseudomonas syringae. Environ Microbiol 21:4465–4477. doi: 10.1111/1462-2920.14779. [DOI] [PubMed] [Google Scholar]
- 31.Choi MS, Kim W, Lee C, Oh CS. 2013. Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol Plant Microbe Interact 26:1115–1122. doi: 10.1094/MPMI-02-13-0050-CR. [DOI] [PubMed] [Google Scholar]
- 32.Wei ZM, Beer SV. 1995. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol 177:6201–6210. doi: 10.1128/jb.177.21.6201-6210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouts DE, Abramovitch RB, Alfano JR, Baldo AM, Buell CR, Cartinhour S, Chatterjee AK, D'Ascenzo M, Gwinn ML, Lazarowitz SG, Lin N-C, Martin GB, Rehm AH, Schneider DJ, van Dijk K, Tang X, Collmer A. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc Natl Acad Sci U S A 99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang SH, Peng QA, Zhang Q, Zou LF, Li Y, Robert C, Pritchard L, Liu H, Hovey R, Wang Q, Birch P, Toth IK, Yang CH. 2010. Genome-wide identification of HrpL-regulated genes in the necrotrophic phytopathogen Dickeya dadantii 3937. PLoS One 5:e13472. doi: 10.1371/journal.pone.0013472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glasner JD, Yang CH, Reverchon S, Hugouvieux-Cotte-Pattat N, Condemine G, Bohin JP, Van Gijsegem F, Yang SH, Franza T, Expert D, Plunkett G, Francisco MJS, Charkowski AO, Py B, Bell K, Rauscher L, Rodriguez-Palenzuela P, Toussaint A, Holeva MC, He SY, Douet V, Boccara M, Blanco C, Toth I, Anderson BD, Biehl BS, Mau B, Flynn SM, Barras F, Lindeberg M, Birch PRJ, Tsuyumu S, Shi XY, Hibbing M, Yap MN, Carpentier M, Dassa E, Umehara M, Kim JF, Rusch M, Soni P, Mayhew GF, Fouts DE, Gill SR, Blattner FR, Keen NT, Perna NT. 2011. Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J Bacteriol 193:2076–2077. doi: 10.1128/JB.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frederick RD, Ahmad M, Majerczak DR, Arroyo-Rodriguez AS, Manulis S, Coplin DL. 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol Plant Microbe Interact 14:1213–1222. doi: 10.1094/MPMI.2001.14.10.1213. [DOI] [PubMed] [Google Scholar]
- 37.Yeh HS, Chen TC, Liou KM, Hsu HT, Chung KM, Hsu LL, Chang BY. 2011. The core-independent promoter-specific interaction of primary sigma factor. Nucleic Acids Res 39:913–925. doi: 10.1093/nar/gkq911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellieny-Rabelo D, Tanui CK, Miguel N, Kwenda S, Shyntum DY, Moleleki LN. 2019. Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in Pectobacterium and Dickeya spp. Appl Environ Microbiol 85:e02050-18. doi: 10.1128/AEM.02050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palacios JL, Zaror I, Martinez P, Uribe F, Opazo P, Socias T, Gidekel M, Venegas A. 2001. Subset of hybrid eukaryotic proteins is exported by the type I secretion system of Erwinia chrysanthemi. J Bacteriol 183:1346–1358. doi: 10.1128/JB.183.4.1346-1358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delepelaire P, Wandersman C. 1990. Protein secretion in Gram-negative bacteria. The extracellular metalloprotease B from Erwinia chrysanthemi contains a C-terminal secretion signal analogous to that of Escherichia coli alpha-hemolysin. J Bio Chem 265:17118–17125. doi: 10.1016/S0021-9258(17)44877-0. [DOI] [PubMed] [Google Scholar]
- 41.Bailey RW, Monro JA, Pickmere SE, Chesson A. 1976. Herbage hemicellulose and its digestion by the ruminant, p 1–16. In Carbohydrate research in plants and animals. Landbouwhogeschool Wageningen, The Netherlands. [Google Scholar]
- 42.Van den Bosch TJM, Niemi O, Welte CU. 2020. Single gene enables plant pathogenic Pectobacterium to overcome host-specific chemical defence. Mol Plant Pathol 21:349–359. doi: 10.1111/mpp.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buttner D, Noel L, Thieme F, Bonas U. 2003. Genomic approaches in Xanthomonas campestris pv. vesicatoria allow fishing for virulence genes. J Biotechnol 106:203–214. doi: 10.1016/j.jbiotec.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M. 2004. Conserved features of type III secretion. Cell Microbiol 6:805–816. doi: 10.1111/j.1462-5822.2004.00432.x. [DOI] [PubMed] [Google Scholar]
- 45.Hajri A, Brin C, Hunault G, Lardeux F, Lemaire C, Manceau C, Boureau T, Poussier S. 2009. A «repertoire for repertoire» hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One 4:e6632. doi: 10.1371/journal.pone.0006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xin XF, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 47.Toth IK, Birch P. 2005. Rotting softly and stealthily. Curr Opin Plant Biol 8:424–429. doi: 10.1016/j.pbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Gürlebeck D, Thieme F, Bonas U. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J Plant Physiol 163:233–255. doi: 10.1016/j.jplph.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar SF, Gordon JS, Martin GB, Guttman DS. 2006. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174:1041–1056. doi: 10.1534/genetics.106.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz AR, Potnis N, Timilsina S, Wilson M, Patané J, Martins J, Jr, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White FF, Miller SA, Ritchie D, Goss E, Bart RS, Setubal JC, Jones JB, Staskawicz BJ. 2015. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol 6:535. doi: 10.3389/fmicb.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang S, Perna NT, Cooksey DA, Okinaka Y, Lindow SE, Ibekwe AM, Keen NT, Yang C-H. 2004. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol Plant Microbe Interact 17:999–1008. doi: 10.1094/MPMI.2004.17.9.999. [DOI] [PubMed] [Google Scholar]
- 52.Yap MN, Rojas CM, Yang CH, Charkowski AO. 2006. Harpin mediates cell aggregation in Erwinia chrysanthemi 3937. J Bacteriol 188:2280–2284. doi: 10.1128/JB.188.6.2280-2284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HS, Ma B, Perna NT, Charkowski AO. 2009. Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains. Appl Environ Microbiol 75:4539–4549. doi: 10.1128/AEM.01336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nykyri J, Niemi O, Koskinen P, Nokso-Koivisto J, Pasanen M, Broberg M, Plyusnin I, Törönen P, Holm L, Pirhonen M, Palva ET. 2012. Revised phylogeny and novel horizontally acquired virulence determinants of the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. PLoS Pathog 8:e1003013. doi: 10.1371/journal.ppat.1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, Bar-Meir M, Sullivan CL, McColley SA, Hauser AR. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 57.Robert-Baudouy J, Nasser W, Condemine G, Reverchon S, Schevchik V, Hugouvieux-Cotte-Pattat N. 2000. Pectic enzymes of Erwinia chrysanthemi: regulation and role in pathogenesis, p 221–268. In Stacey G, Keen N (ed), Plant microbe interactions. American Phytopathological Society Press, St. Paul, MN. [Google Scholar]
- 58.Lv MF, Hu M, Li P, Jiang ZD, Zhang LH, Zhou JN. 2019. A two-component regulatory system VfmIH modulates multiple virulence traits in Dickeya zeae. Mol Microbiol 111:1493–1509. doi: 10.1111/mmi.14233. [DOI] [PubMed] [Google Scholar]
- 59.Liao LS, Cheng YY, Liu SY, Zhou JN, An SW, Lv MF, Chen YF, Gu YF, Chen SH, Zhang LH. 2014. Production of novel antibiotics zeamines through optimizing Dickeya zeae fermentation conditions. PLoS One 9:e116047. doi: 10.1371/journal.pone.0116047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen SF, Zhou YQ, Chen YR, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, Mushegian A. 2010. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487. doi: 10.1093/bioinformatics/btq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FSL, Simon Fraser University Research Computing Group. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha EPC. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Li H, Yang B, Xie R, Marquez-Lago TT, Leier A, Hayashida M, Akutsu T, Zhang Y, Chou KC, Selkrig J, Zhou T, Song J, Lithgow T. 2019. Bastion3: a two-layer ensemble predictor of type III secreted effectors. Bioinformatics 35:2017–2028. doi: 10.1093/bioinformatics/bty914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong XB, Lu XT, Zhang ZD. 2015. BEAN 2.0: an integrated web resource for the identification and functional analysis of type III secreted effectors. Database (Oxford) 2015:bav064. doi: 10.1093/database/bav064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue L, Tang B, Chen W, Luo J. 2019. DeepT3: deep convolutional neural networks accurately identify Gram-negative bacteria type III secreted effectors using the N-terminal sequence. Bioinformatics 35:2051–2057. doi: 10.1093/bioinformatics/bty931. [DOI] [PubMed] [Google Scholar]
- 72.Goldberg T, Rost B, Bromberg Y. 2016. Computational prediction shines light on type III secretion origins. Sci Rep 6:34516. doi: 10.1038/srep34516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 75.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, Hu M, Xue Y, Chen X, Lu G, Zhang L, Zhou J. 2020. Screening, identification and efficacy evaluation of antagonistic bacteria for biocontrol of soft rot disease caused by Dickeya zeae. Microorganisms 8:697. doi: 10.3390/microorganisms8050697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 82.Yang L-Y, Yang L-C, Gan Y-L, Wang L, Zhao W-Z, He Y-Q, Jiang W, Jiang B-L, Tang J-L. 2018. Systematic functional analysis of sigma (σ) factors in the phytopathogen Xanthomonas campestris reveals novel roles in the regulation of virulence and viability. Front Microbiol 9:1749. doi: 10.3389/fmicb.2018.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaba N, Gaba S. 2020. Study of liver dysfunction in hyperemesis gravidarum. Cureus 12:e8709. doi: 10.7759/cureus.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01590-21_Supp_1_seq13.pdf, PDF file, 1.5 MB (1.5MB, pdf)
Data Availability Statement
Both the original genome sequencing data and the genome assembly were deposited in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA656647. The JZL7 genome sequence has been deposited in GenBank under accession number CP060263.1.