ABSTRACT
The nitrogen (N) cycle is of global importance, as N is an essential element and a limiting nutrient in terrestrial and aquatic ecosystems. Excessive anthropogenic N fertilizer usage threatens sensitive downstream aquatic ecosystems. Although freshwater lake sediments remove N through various microbially mediated processes, few studies have investigated the microbial communities involved. In an integrated biogeochemical and microbiological study on a eutrophic and oligotrophic lake, we estimated N removal rates from pore water concentration gradients in sediments. Simultaneously, the abundance of different microbial N transformation genes was investigated using metagenomics on a seasonal and spatial scale. We observed that contrasting nutrient concentrations in sediments were associated with distinct microbial community compositions and significant differences in abundances of various N transformation genes. For both characteristics, we observed a more pronounced spatial than seasonal variability within each lake. The eutrophic Lake Baldegg showed a higher denitrification potential with higher nosZ gene (N2O reductase) abundances and higher nirS:nirK (nitrite reductase) ratios, indicating a greater capacity for complete denitrification. Correspondingly, this lake had a higher N removal efficiency. The oligotrophic Lake Sarnen, in contrast, had a higher potential for nitrification. Specifically, it harbored a high abundance of Nitrospira, including some with the potential for comammox. Our results demonstrate that knowledge of the genomic N transformation potential is important for interpreting N process rates and understanding how the lacustrine sedimentary N cycle responds to variations in trophic conditions.
IMPORTANCE Anthropogenic nitrogen (N) inputs can lead to eutrophication in surface waters, especially in N-limited coastal ecosystems. Lakes effectively remove reactive N by transforming it to N2 through microbial denitrification or anammox. The rates and distributions of these microbial processes are affected by factors such as the amount and quality of settling organic material and nitrate concentrations. However, the microbial communities mediating these N transformation processes in freshwater lake sediments remain largely unknown. We provide the first seasonally and spatially resolved metagenomic analysis of the N cycle in sediments of two lakes with different trophic states. We show that lakes with different trophic states select for distinct communities of N-cycling microorganisms with contrasting functional potentials for N transformation.
KEYWORDS: metagenomics, microbial ecology, freshwater, pore water, DNRA, nitrification, denitrification, anammox, comammox, nitrogen transformation
INTRODUCTION
Nitrogen (N) is an essential nutrient, and microbes play central roles in the environmental N cycle. In most environments on earth, N-fixing microbes dominate the conversion of dinitrogen (N2) to reactive N (N compounds readily available for biological conversion; e.g., NO3−, NO2−, NH4+, and N2O). The different reactive N compounds are used as electron acceptors or donors in several microbial metabolic pathways. In addition, the availability of the essential nutrient N limits rates of primary production in many terrestrial and aquatic ecosystems, including coastal oceans (1–3). As a result, excessive anthropogenic reactive N input, e.g., from runoff of agricultural fertilizer, human wastewater, and fossil fuel combustion, increases biomass production and organic carbon loads in aquatic ecosystems and can lead to the occurrence of harmful algal blooms and anoxia (4, 5). Despite its small area, Switzerland is an essential headwater system of European rivers and a nonnegligible N source. N loads from atmospheric deposition (44 kt year−1), mineral fertilizers (52 kt year−1), and sewage (43 kt year−1) are about equally responsible for the N contamination of ecosystems in Switzerland (6). Atmospheric deposition rates on the Swiss Plateau, locally exceeding 40 kg of N ha−1 year−1, are among the highest in the world (6, 7). Today, Switzerland exports 63 kt year−1 of dissolved N via the Rivers Rhine and Rhone (an average from 1995 to 2013) (8). This load corresponds to 1.3% of the total N exports of Europe to the seas (4,761 kt year−1) (9).
Lakes have been identified as important N sinks that convert up to 90% of reactive N to less bioavailable N2 gas and, thus, substantially reduce N loads to oceans (10). N removal occurs through several microbially mediated processes, mainly within the oxic-anoxic transition zone. This zone extends across the first few millimeters of sediments in most lakes. The main N loss processes include denitrification and anaerobic ammonia oxidation (anammox), both of which produce N2 as an end product, as well as long-term burial of N-containing organic matter (sequestration) (11–17). Furthermore, the effectiveness of reactive N conversion to N2 depends on interactions with other N-cycling reactions that produce or compete for the same substrates. However, a thorough understanding of the environmental controls of these N removal processes is lacking despite being essential to understanding and predicting N removal by lakes, both today and under conditions of global change in the future.
Many different environmental drivers influence the reactive N transformation processes in lakes. These include NO3− concentrations, organic matter (OM) input, remineralization rates, and availability of electron acceptors besides NO3− [e.g., O2, SO42−, Fe(III), Mn(IV), and CO2] (1, 4, 10, 11, 16, 18–23). Most of these factors are intricately linked to the overall trophic status of lakes. For example, Finlay et al. (12) reported 7-fold higher N removal rates in eutrophic than in oligotrophic lakes. Others found that interlake variations in denitrification rates were positively correlated with water residence times and N loads (5, 24, 25).
There is a considerable lack of understanding about the controls on microbial communities involved in N transformation processes in lake sediments. Microbial ecology has made substantial progress in recent decades in identifying, characterizing, and quantifying key genes of microbial N-cycling enzymes (N transformation genes) in environmental systems using various molecular tools (15, 17, 26–29). This has improved understanding of the environmental controls of nitrification, complete ammonia oxidation (comammox), assimilatory and dissimilatory nitrate reduction to ammonia (ANRA and DNRA), denitrification, anammox, and N2 fixation (1, 14, 22, 27, 28, 30, 31). Metagenome sequencing allows for a more complete view of microbial N cycling by enabling the study of multiple N gene families and other pathways in environmental samples in parallel (32). Fierer et al. (33), for example, noted a significant change in the microbial community and its metabolic function (e.g., increase of DNA/RNA replication, electron transport, and protein metabolism) along a gradient of N availability established by fertilization with NH4NO3 in different soils. Nelson et al. (34) showed that soil C and N contents explained the number of detected N pathways in the soil samples. Furthermore, they distinguished between N cycling specialists, encoding only a few N transformation processes, and N cycling generalists, encoding all N transformation processes (34). Graf et al. (29) reported that partial denitrifiers with nirS (cytochrome cd1 variant for nitrite reductase) often carried genes that encode nitric oxide (norBC) and nitrous oxide (nosZ) reductases and, thus, have the potential to do the complete denitrification pathway. This was in contrast to denitrifiers that carry nirK (copper-binding variant of nitrite reductase), which showed less cooccurrence with norBC and nosZ (29). Another metagenomic study revealed atypical nosZ sequences in soils (35). Furthermore, metagenomic investigations have improved our understanding of the microbial N network in many stratified ecosystems with an oxic-anoxic transition zone, such as those found in open oceans or estuaries (1–3, 36). This indicates that the framework of metagenomics-based research also provides a promising approach for investigating freshwater sediment N cycling.
In contrast to soils, ocean, and wastewater systems, few metagenomic studies of the N cycle have focused on freshwater lake sediments. However, lake sediments are central sites of N transformation and of crucial importance in regulating continental N exports. Here, we investigate the potential influence of lake trophic state on the N cycling genetic potential based on lake sediments of the eutrophic Lake Baldegg and the oligotrophic Lake Sarnen over seasonal and spatial scales (Fig. 1). We investigate lateral, vertical, and temporal variations in N-cycling microbial communities using high-throughput 16S rRNA gene amplicon sequencing and compare these to pore water fluxes of NO3− and NH4+ recently reported for the same lakes by Müller et al. (20). We furthermore sequence shotgun metagenomes to provide a detailed view of N gene compositions and reveal the phylogenetic identities of microorganisms involved in N transformation reactions.
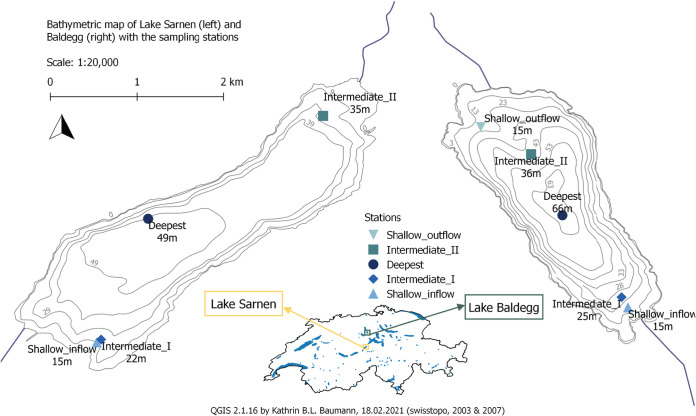
FIG 1.
Sampling locations in Lake Sarnen (SAR) (left, yellow) and Lake Baldegg (BAL) (right, green), sampled in 2018 in March, May, August, and September. Pore water (0.25-cm resolution) and 16S rRNA gene sequencing (0.5-cm resolution) data were measured in all sampling points. DNA from the top 3 cm was pooled for metagenome analysis of the deepest station (SAR and BAL), Shallow_inflow (SAR), and Shallow_outflow (BAL) in all months. Sediment traps were deployed at the deepest point of each lake (dark blue); see Müller et al. (20).
We hypothesize that the lakes’ contrasting N transformation processes are mediated by distinct microbial communities and that the genetic potential for microbial N cycling responds to environmental drivers such as the availability of different nutrients, including N species and OM inputs. Furthermore, we suggest that varying spatiotemporal pore water chemistry results in significant seasonal and spatial differences in the microbial N transformation potential and, thus, put into question the common practice of relying on single-core sampling to characterize whole lake ecosystems.
RESULTS
Microbial community composition and environmental drivers.
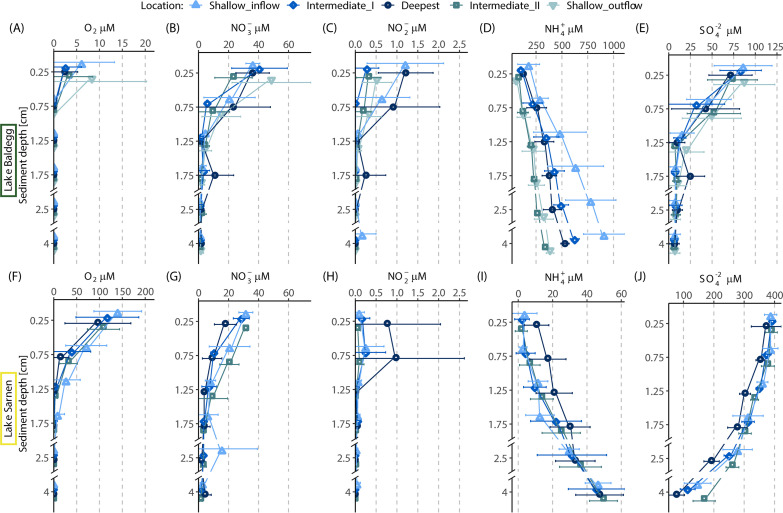
Sediment pore water profiles from both lakes showed pronounced gradients of electron acceptors and reduced compounds, typical of freshwater lake sediment underlying an oxygenated water column. The sediment O2 concentrations at the sediment-water interface were ∼20 times higher in Lake Sarnen (maximum, 217 μM) than Lake Baldegg (maximum, 25 μM) (Fig. 2A and F). O2 also penetrated up to 4 times deeper in Lake Sarnen (<2 cm) than in Lake Baldegg (<0.75 mm) (Fig. 2A and F). In both lakes, the highest O2 concentrations were measured at the shallower station. O2 concentrations decreased in both lakes over the course of the year, with an average of 63 μM in March and 0 μM in September in Lake Baldegg and 181 μM in March and 67 μM in September in Lake Sarnen. The NO3− levels in Lake Baldegg were slightly higher (maximum, 64 μM) than those of Lake Sarnen (maximum, 51 μM) (Fig. 2B and G). In Lake Sarnen, NO3− penetrates deeper (∼4 cm), whereas NO3− is depleted within the first 2 cm in Lake Baldegg. We did not observe any seasonal or spatial patterns for NO3− (Fig. 2B and G). NO2− concentrations generally remained below 2 μmol in both lakes (Fig. 2C and H). In Lake Baldegg, NO2− was highest in the surface sediment (mean, 0.21 μM, in contrast to Lake Sarnen, where NO2− peaked at 0.75 cm depth (mean, 0.13 μM) (Fig. 2C and H). NH4+ concentrations increased with sediment depth in both lakes, but in Lake Baldegg, concentrations (average, 337 μM) were over 10 times higher than those in Lake Sarnen (average, 21 μM) (Fig. 2D and I). The NH4+ concentration in Lake Baldegg decreased from inflow to outflow (Fig. 2D). In Lake Sarnen, the deepest station showed the highest NH4+ concentration in the upper sediment (Fig. 2I). Lake Sarnen had a higher concentration of SO42− than Lake Baldegg, with a mean SO42− concentration of 295 μM and 27.5 μM, respectively (Fig. 2E and J). The SO42− depletion curve in Lake Baldegg was steeper and stabilized at a low concentration below 2 cm sediment depth, in contrast to Lake Sarnen, where high SO42− concentrations were measured even at 4 cm sediment depth (Fig. 2E and J).
FIG 2.
(A to E) Pore water and oxygen profiles of Lake Baldegg from the five different sampling locations (Shallow_inflow, 15 m; Intermediate_I, 25 m; deepest, 66 m; Intermediate_II, 36 m; and Shallow_outflow, 15m) and Lake Sarnen (F to J) from the four different sampling locations (Shallow_inflow, 15 m; Intermediate_I, 22 m; deepest, 49 m; and Intermediate_II, 35 m). Each graph shows the seasonal means (points; the points from each sediment depth were shifted slightly to prevent overlap and to distinguish them better) with standard deviations (vertical error bars) for each location within the sediment depth for the following chemical parameters: O2 (A and F), NO3− (B and G), NO2− (C and H), NH4+ (D and I), and SO42− (E and J).
We observed distinct microbial community compositions in the two lakes. In Lake Sarnen, alpha diversity was significantly higher (Wilcoxon test, P < 0.001), with an observed total richness of 64,699 amplicon sequence variants (ASVs) (unfiltered) compared to Lake Baldegg, with 29,457 ASVs (see Fig. S1A and B in the supplemental material). The difference was robust to filtering out rare ASVs (taxa appearing less than five times in at least 10% of the samples), leaving 2,599 ASVs for Lake Baldegg and 6,124 ASVs for Lake Sarnen (Fig. S1A and B).
Overview of the alpha diversity measures (A and B) for the total microbial community composition based on the 16S rRNA of Lake Baldegg (green, 119 samples) and Lake Sarnen (yellow, 90 samples) and the significant differences between the alpha diversity of the two lakes. (C) Overview of the beta diversity measures for the whole microbial community composition based on the 16S rRNA of Lake Baldegg (green, 119 samples) and Lake Sarnen (yellow, 90 samples). All analyses were done in R with the vegan package by following the tutorial from the Küsel lab. Download FIG S1, EPS file, 1.4 MB (1.5MB, eps) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
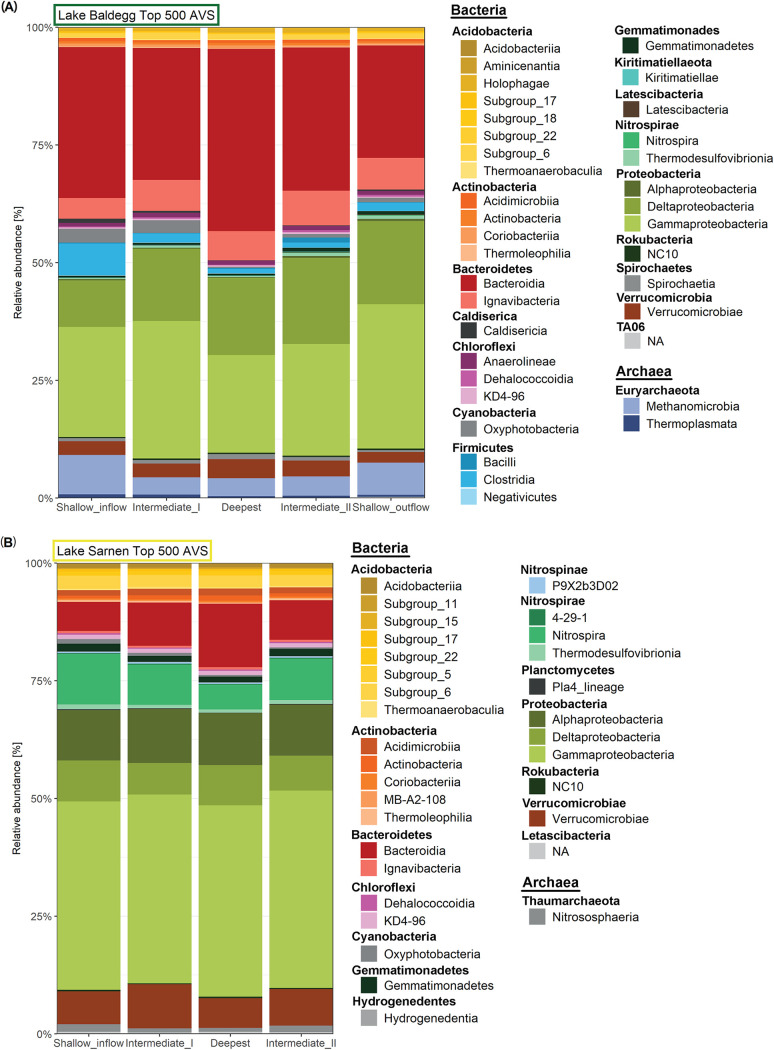
The most abundant ASVs (top 500 ASVs) in Lake Baldegg were dominated by representatives of the phyla Bacteroidetes (represented by the main classes Bacteroidia, 31%, and Ignavibacteria, 6%), Gamma (26%)- and Deltaproteobacteria (16%), Euryarchaeota (mainly Methanomicrobia, 5%), and Firmicutes (mainly Clostridia, 2%) (Fig. 3A). In Lake Sarnen, the phylum Proteobacteria was also abundant, with a higher contribution of Gamma- (41%) and Alphaproteobacteria (11%) than Lake Baldegg. The Bacteroidetes, primarily of the class Bacteroidia (9%), were three times less abundant than in Lake Baldegg. The third most abundant phyla in Lake Sarnen were Nitrospirae (mostly Nitrospira, 8%), Verrucomicrobiae (8%), and Acidobacteria (5%) (Fig. 3B), which were only a minor component in Lake Baldegg. In Lake Sarnen, Thaumarchaeota (<1%) was the only archaeal phylum represented.
FIG 3.
Microbial community composition of the top 500 ASVs summed at the class level in Lake Baldegg (top) and Lake Sarnen (bottom). Classes that belong to the same phylum are grouped together, and the phylum is written above each class group. The data shown are averaged over sediment depth and time for each station and separately for each lake.
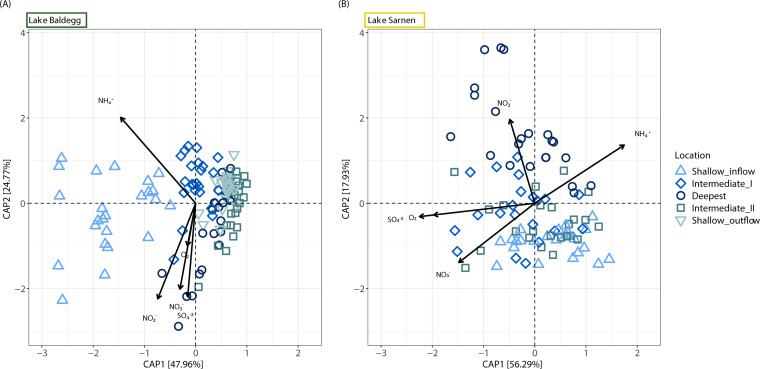
Constrained ordination by RDA confirmed that community composition was significantly different in the two lakes (permutational analysis of variance [PERMANOVA], P < 0.001, R2 = 0.59) (Fig. S1C). The analysis also showed that beta-diversity patterns in the two lakes differed considerably (Fig. 4). In Lake Baldegg, the first ordination axis (CAP1, explained variance of 48%) reflects the location, and the second component (CAP2, explained variance of 25%) explains the microbial community composition differences over sediment depth (Fig. 4A and Fig. S2A and C). The microbial community composition significantly differed by sampling station (PERMANOVA, P < 0.001, R2 = 0.44) in Lake Baldegg, with stations generally sorting along axis 1 from inflow to outflow (Fig. 4A). The influence of sediment depth and sampling month on the community composition was also significant (PERMANOVA, P < 0.001), although with much lower explanatory power (R2 = 0.06 months and R2 = 0.05 sediment depth). In contrast, for Lake Sarnen, the first component reflected the microbial community composition shift with sediment depth (CAP1, explained variance of 56%), and the second component reflected community compositional differences by location (CAP2, explained variance of 18%), indicating fewer differences between locations than in Lake Baldegg (Fig. 4B). The community composition varied significantly between sampling stations in Lake Sarnen (PERMANOVA, P < 0.001, R2 = 0.21). The deepest station of Lake Sarnen revealed the most distinct community composition compared to the three other locations, where the two intermediate stations showed the most similar microbial community composition (Fig. 4B). As in Lake Baldegg, the community differences by sediment depth and sampling month were significant (PERMANOVA, P < 0.001) but with lower explanatory power (R2 = 0.09 sediment depth and R2 = 0.05 months) (Fig. S2B and D). RDA indicated that NH4+ concentration displayed the highest correlation with the observed changes in community composition in both lakes’ deeper sediments (Fig. S2A and B). In contrast, the other four environmental drivers (O2, NO3−, NO2−, and SO42−) were drivers primarily for the microbial communities living in the surface sediment (Fig. S2A and B).
FIG 4.
Constrained ordination biplots (redundancy analysis [RDA] on weighted UniFrac distance) of microbial community composition and the different environmental drivers (O2, NO3−, NO2−, NH4+ and SO42−) for Lake Baldegg (A) and Lake Sarnen (B). The percentage of the variance explained for each component is labeled on the x and y axes. In Lake Baldegg, the two main components (CPA1 and CPA2) explained 72% of the variance in the microbial communities. In Lake Sarnen, the main components (CPA1 and CPA2) explain 74% of the variance in the microbial communities. The samples are colored and shaped by location.
Constrained ordination biplots (redundancy analysis [RDA] on weighted UniFrac distance) of microbial community composition and the different environmental drivers (O2, NO3−, NO2−, NH4+, and SO42−) for Lake Baldegg (A and C) and Lake Sarnen (B and D). The samples are colored by sediment depth (A and B) and month (C and D). All analyses were done in R with the vegan package by following the tutorial from the Küsel lab. Download FIG S2, EPS file, 0.1 MB (68.1KB, eps) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial nitrogen transformation potential and possible key players.
A total of 32,363 and 18,951 genes were indicative of N transformation processes in Lake Baldegg and Lake Sarnen, respectively (Table S2). Ordination of N transformation gene composition clearly distinguished lakes (PC1, 62%) and also locations (PC2, 8%) (Fig. 5C), whereas seasonal variations were comparatively minor (Fig. 5C). N gene abundance varied considerably, from 35,565 genes per million (gpm) for N degradation and biosynthesis in Lake Baldegg to 10 gpm for anammox in Lake Sarnen. The N gene abundance differed substantially between the lakes, especially for DNRA (Lake Baldegg, 7,638 gpm; Lake Sarnen, 4,716 gpm), N2 fixation (Lake Baldegg, 1,155 gpm; Lake Sarnen, 443 gpm), hydroxylamine reductase (hcp) (Lake Baldegg, 1,006 gpm; Lake Sarnen, 80 gpm), anammox (Lake Baldegg, 108 gpm; Lake Sarnen, 10 gpm), and nitrification (Lake Baldegg, 400 gpm; Lake Sarnen, 1,456 gpm) (Fig. 5A and Table S3). Denitrification (Lake Baldegg, 4,517 gpm; Lake Sarnen, 4,650 gpm) and nitrate reduction genes belonging to either DNRA or denitrification (Lake Baldegg, 4,347 gpm; Lake Sarnen, 4,336 gpm) occurred at similar abundances in both lakes (Fig. 5A and Table S3). We further analyzed methane-monooxygenase (pmoABC), a homologous gene family with high sequence similarity to amoABC. In both lakes, the pmoABC genes were less abundant than amoABC (Lake Baldegg, 105 gpm; Lake Sarnen, 379 gpm) (Table S3).
FIG 5.
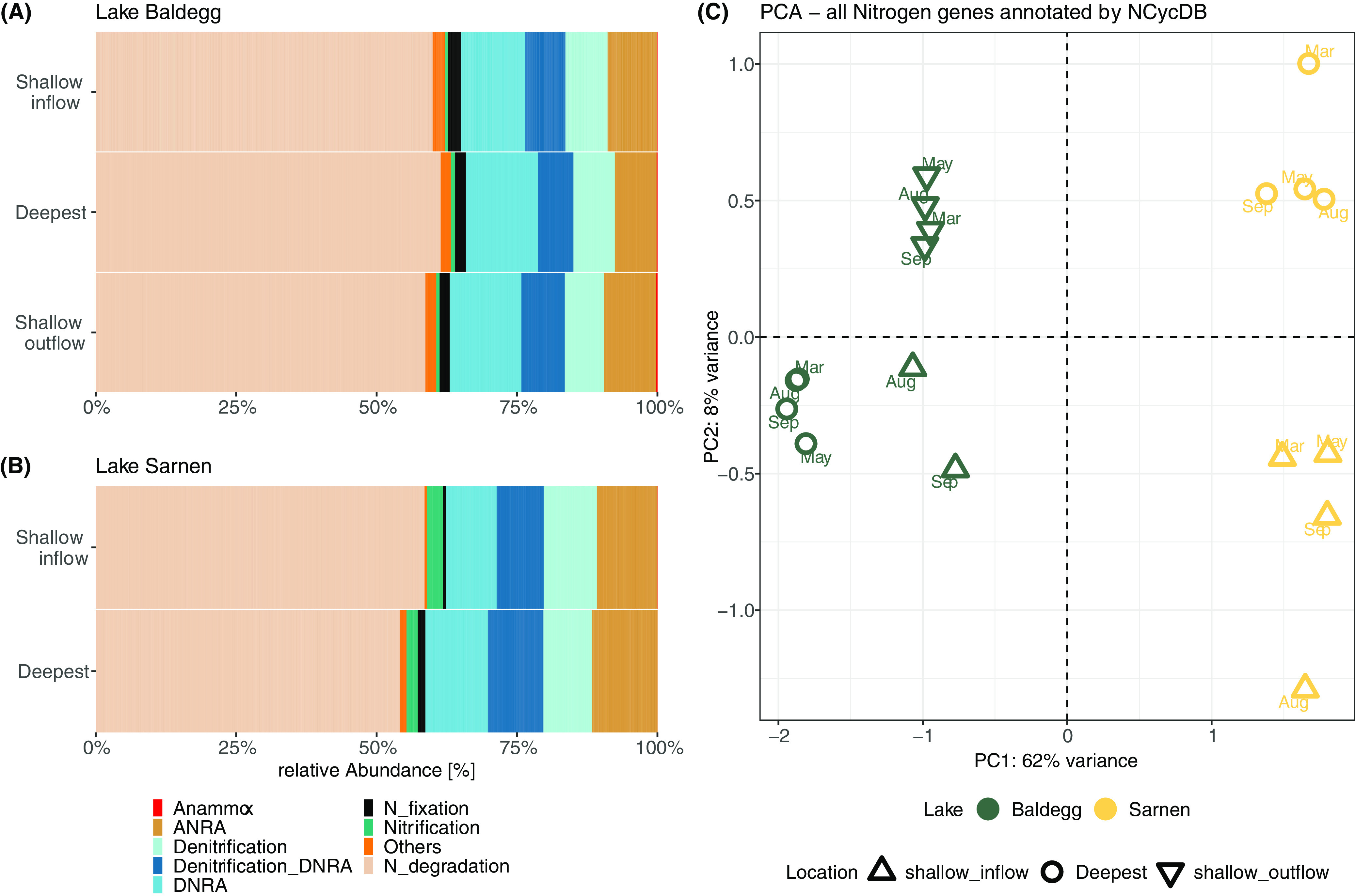
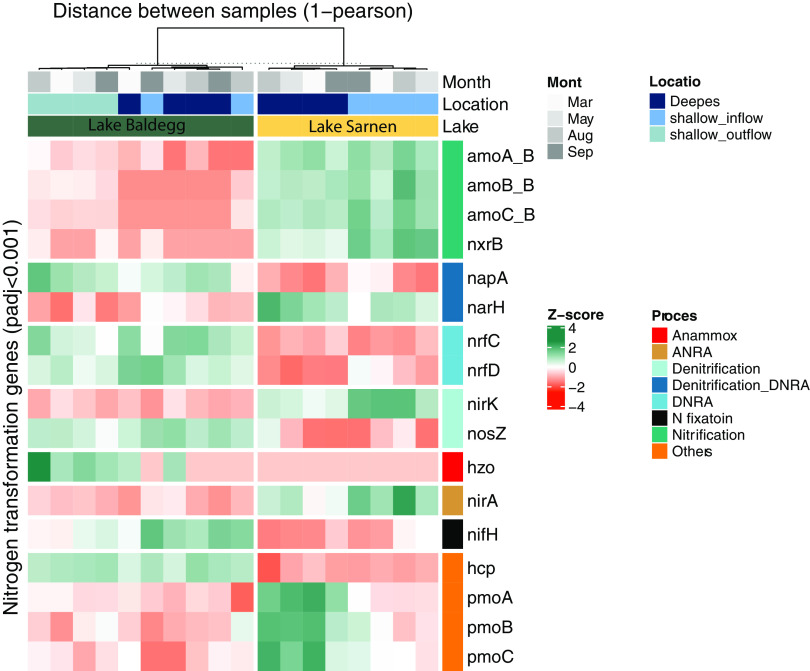
Nitrogen transformation gene distribution in Lake Baldegg (A) and Lake Sarnen (B) for each station summed over all seasons, grouped by the nitrogen transformation processes (Table 2). Nitrate reduction genes that cannot be assigned to either denitrification or DNRA are grouped together (Denitrification_DNRA). (C) The principal component analysis shows the nitrogen gene counts for each station, calculated with the DESeq2 package.
Sequencing overview for the metagenomic data, with the number of raw reads, raw bases (G, giga), GC content, number of contigs (assembled sequences), number of contigs aligned to NCycDB, and number of total N genes (count N genes from NCycDB pipline) per sample (1). Download Table S2, XLSX file, 0.01 MB (12.2KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of all nitrogen cycle genes found in Lake Baldegg and Lake Sarnen from the different locations and months. The numbers are the sum of gpm (genes per million) per sample. The genes are clustered in the N transformation process they belong to. Download Table S3, XLSX file, 0.01 MB (37.4KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
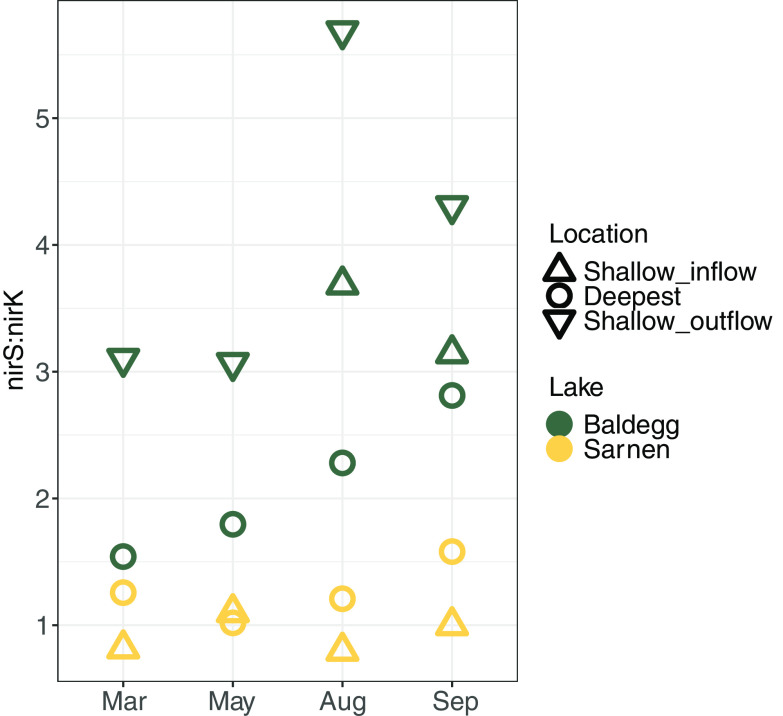
We found 17 genes involved in various N transformation pathways that had significantly different abundances between Lake Baldegg and Lake Sarnen (Deseq2, Wald test, adjusted P value [padj] of <0.001) (Fig. 6). In agreement with observations made for the microbial community composition (Fig. 4), we did not observe a distinct temporal clustering for N gene abundance data, but there was a clustering by sampling location evident in the heatmap (Fig. 6). We tested the spatial and temporal N transformation differences with DESeq2 using lakes plus station and lakes plus months as conditions. The results revealed 14 significantly different genes between the lakes and sampling location (P < 0.01) and 0 significantly different genes between the lakes and sampling time point (P < 0.01). Lake Sarnen had a significantly higher abundance of bacterial ammonia (amoABC_B) and nitrite-oxidizing (nxrB) genes than Lake Baldegg (Deseq2, Wald test, padj < 0.001), indicating a higher nitrification potential (Fig. 6). In Lake Baldegg, the shallow_outflow location showed a higher nitrification potential than the deep location (Fig. 6). Lake Baldegg had a significantly higher potential to reduce NO3− via periplasmic nitrate reductase (napA) (Deseq2, Wald test, padj < 0.001), whereas Lake Sarnen showed a significantly higher abundance for a different nitrate reductase gene, narH (Deseq2, Wald test, padj < 0.001) (Fig. 6). Two representative genes that encode NO2− reduction to NH4+ in the DNRA pathway (nrfC and nrfD) were significantly higher in Lake Baldegg than in Lake Sarnen (Deseq2, Wald test, padj < 0.001) (Fig. 6). Among genes that encode steps of denitrification, nirK was significantly more abundant in Lake Sarnen, especially in the shallow stations (Deseq2, Wald test, padj < 0.001), while nosZ was significantly more abundant in Lake Baldegg (Deseq2, Wald test, padj < 0.001). nirS had similar abundances in both lakes, resulting in a higher nirS:nirK ratio for Lake Baldegg than Lake Sarnen, especially in the shallower stations (Fig. 7). Anammox genes were significantly more abundant in Lake Baldegg (Deseq2, Wald test, padj < 0.001) and were highest in the shallow outflow location (Fig. 6). The gene encoding the assimilatory NO3− reduction to NO2− (nirA) was significantly more abundant in Lake Sarnen than Lake Baldegg (Deseq2, Wald test, padj < 0.001) (Fig. 6). In contrast, Lake Baldegg showed a significantly higher abundance of nifHK, which encodes nitrogenase, the central enzyme for N2 fixation. The N2 fixation potential was especially high in the deep and shallow inflow stations (Deseq2, Wald test, padj < 0.001) (Fig. 6). The hydroxylamine reductase gene hcp was significantly more abundant in Lake Baldegg, whereas the genes encoding methane oxidation (pmoABC) were significantly more abundant in Lake Sarnen (Deseq2, Wald test, padj < 0.001).
FIG 6.
Heatmap of nitrogen gene abundances showing significant differences between Lake Baldegg (green) and Lake Sarnen (yellow) (alpha = 0.001; padj, adjusted P value with Benjamini-Hochberg method from DESeq2) clustered by lake and station (column clustering distance calculated using Pearson from the ComplexHeatmap package) in the columns and nitrogen process in the rows. The z-score of abundances was calculated for each gene individually to emphasize the differences.
FIG 7.
nirS:nirK ratio calculated from gpm values per lake (Baldegg, green; Sarnen, yellow), location (shallow inflow, triangle; deepest, circle; shallow outflow, rotated triangle), and month.
About 47% of the significant N transformation genes could be taxonomically assigned to at least the class level. The results suggest that different key players dominate N cycling in each lake (Table 1 and Table S5). Overall, most represented among nitrogen-cycling taxa were members of the Gammaproteobacteria (2,950 gpm), followed by Deltaproteobacteria (1,462 gpm), Bacteroidetes (720 gpm), and Nitrospirae (638 gpm). The most abundant nitrification genes (nxrB and amoABC) were assigned to the nitrite-oxidizing bacteria (NOB) of the phylum Nitrospirae, with Lake Sarnen having a higher abundance (300 gpm) than Lake Baldegg (9 gpm) (Table 1). In contrast, the ammonia-oxidizing bacteria (AOB) of the order Nitrosomonadales (Gammaproteobacteria) were represented in both lakes. The nitrate and nitrite reductase genes from ANRA, DNRA, and Dentrification_DNRA were assigned to a phylogenetically diverse group of microbes (Table 1). In Lake Sarnen, the most abundant phyla carrying nirK were Nitrospirae (157 gpm) and Chloroflexi (113 gpm). In contrast, in Lake Baldegg, nirK sequences were mainly assigned to Gammaproteobacteria (86 gpm) and Bacteroidetes (52 gpm) (Table 1). The phylum most frequently assigned to nirS was Gammaproteobacteria, representing 448 gpm in Lake Baldegg and 211 gpm in Lake Sarnen, respectively. The most abundant phyla containing the nitrous oxide-reducing denitrification gene (nosZ) in Lake Baldegg and Lake Sarnen were Bacteroidetes (140 and 54 gpm) and Gammaproteobacteria (158 and 49 gpm). Nitrogenase genes (nifK) in Lake Baldegg were often assigned to Euryarchaeota (173 gpm).
TABLE 1.
Overview of the mapping rates of the N removal genes to the Kaiju database to reveal possible key players (top 12 phyla) for the significantly different gene families plus nirSa
| Organism(s) | Nitrification for amoABC, nxrB |
Denitrification (DNRA) |
DNRA for nrfCD |
Denitrification |
Anammox for hzo, hszA |
ANRA for nirA |
N_fixation for nifHK |
Others for hcp |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
napA
|
narH
|
nirK
|
nirS
|
nosZ
|
||||||||||||||||||
| BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | BAL | SAR | |
| Acidobacteria | 2 | 12 | 6 | 6 | 119 | 18 | 2 | 12 | 2 | 14 | 13 | 11 | 2 | 2 | 1 | 6 | 4 | |||||
| Actinobacteria | 5 | 6 | 7 | 13 | 4 | 6 | 8 | 2 | 1 | 4 | 3 | 3 | 6 | 1 | ||||||||
| Alphaproteobacteria b | 2 | 4 | 2 | 34 | 25 | 9 | 7 | 22 | 3 | 8 | 5 | 19 | 8 | 32 | 11 | 2 | 2 | |||||
| Bacteroidetes | 19 | 1 | 5 | 364 | 20 | 52 | 8 | 15 | 140 | 54 | 4 | 7 | 4 | 16 | 3 | 8 | ||||||
| Chlorobi | 1 | 106 | 11 | 10 | 6 | 22 | ||||||||||||||||
| Chloroflexi | 17 | 14 | 3 | 26 | 38 | 31 | 113 | 15 | 1 | 51 | 37 | 6 | 11 | 2 | 4 | 11 | ||||||
| Deltaproteobacteria c | 71 | 12 | 217 | 19 | 84 | 11 | 526 | 102 | 14 | 5 | 116 | 25 | 115 | 37 | 20 | 74 | 9 | 285 | 20 | |||
| Euryarchaeota | 1 | 20 | 3 | 10 | 4 | 1 | 8 | 149 | 31 | |||||||||||||
| Gammaproteobacteria d | 28 | 45 | 105 | 63 | 86 | 142 | 669 | 432 | 86 | 87 | 448 | 211 | 158 | 49 | 119 | 106 | 71 | 37 | 8 | |||
| Nitrospirae | 9 | 300 | 3 | 9 | 38 | 10 | 1 | 157 | 65 | 1 | 30 | 1 | 13 | 1 | ||||||||
| Planctomycetes | 1 | 6 | 2 | 9 | 22 | 16 | 13 | 5 | 11 | 6 | 8 | 3 | ||||||||||
| Verrucomicrobia | 27 | 6 | 10 | 26 | 39 | 10 | 45 | 11 | 5 | 14 | 32 | 1 | 7 | 17 | 5 | 6 | ||||||
The numbers are the sum of gpm per gene per lake and phyla. The most abundant phyla are highlighted in boldface for Lake Baldegg and underlining for Lake Sarnen. The most abundant classes of the Proteobacteria are listed in the footnotes below. The full list of Proteobacteria orders per gene can be found in Table S5. Note that Gammaproteobacteria include Betaproteobacteria.
The most assigned orders of Alphaproteobacteria were Caulobacterales (most often assigned in Lake Sarnen), Rhizobiales, Rhodobacterales (most often assigned in Lake Sarnen), and Rhodosprillales.
The most abundant orders of Deltaproteobacteria were Desulfobacterales, Desulfuromonoadales, and Syntrophobacterales.
The most often assigned Gammaproteobacteria (including Betaproteobacteria) orders were Acidiferrobacterales (most often assigned in Lake Sarnen), Aeromonadales (most often assigned in Lake Sarnen), Chromatiales, Enterobacterales (most abundant in Lake Baldegg), Methylococcales, Tiotrichales (most abundant in Lake Baldegg), Burkolderiales, Nitrosmonadales, and Rhodocyclales.
Overview of the mapping rates of the N removal genes to Kaiju database of Proteobacteria (top 12 phyla) for the significantly different gene families, plus nirS. The numbers are the total sum of gpm per genes per lake and phylum. The most abundant are highlighted in green for Lake Baldegg and yellow for Lake Sarnen. We listed Betaproteobacteria here separately, as Kaiju assign them separately and does not have the SILVA132 database assignment. In the paper, Beta- and Gammaproteobacteria are summed (92, 93). Download Table S5, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Distinct microbial communities and pore water chemistry.
The pore water chemistry in the eutrophic and oligotrophic lake sediments studied was clearly a driver of microbial community composition. However, we observed that while the chemical sediment pore water signature was seasonally driven to a considerable extent (20), the microbial community composition was mainly affected by location (Fig. 4; see also Fig. S2 in the supplemental material). The sediment pore water profiles of eutrophic Lake Baldegg indicated a high availability of inorganic N (Fig. 2). The rapid depletion of O2, NO3−, NO2−, and SO42− and the increasing concentration of NH4+ with sediment depth indicated a high aerobic and anaerobic respiration activity, in agreement with previous reports from Lake Baldegg (20, 37, 38). Lake Sarnen, in contrast, had sediment pore water profiles that were similar to those of other oligotrophic lakes, e.g., Lakes Lucerne (37), Brienz, and Thun (38), and lower concentrations of inorganic N than in Lake Baldegg. The O2 concentrations and penetration depth, as well as the SO42− concentrations, were higher in Lake Sarnen, indicating lower respiratory activity (Fig. 2). Overall, the pore water data confirmed that the lake sediments chosen for this study were suitable endmembers of the oligotrophic to eutrophic spectrum of lakes with aerobic water columns in Switzerland.
The two lakes share many microbial taxa. The most abundant phyla occurring in both lakes (Fig. 3) have previously been reported to also dominate in other lake sediments (25, 39–44). Nonetheless, the alpha and beta diversity analysis indicated a very distinct community composition (Fig. 4 and Fig. S1 and S2). We hypothesize that the significantly higher species richness in Lake Sarnen is explained by a greater number of fastidious oligotrophic microbes due to the low nutrient content. In contrast, the high nutrient abundance in Lake Baldegg appears to favor a smaller number of competitive, copiotrophic microbes. Similar observations have been made for the planktonic diversity in oligotrophic mountain lakes (45–48). Further studies are required to test this hypothesis.
The microbial community composition changes over sediment depth are directly linked to the sediment's redox cascade (Fig. S2A and B). For example, the high abundance of Nitrospirae in the surface sediment, especially in Lake Sarnen (Fig. S4), can be linked to low NH4+ and high O2 concentrations. Nitrospirae is a known aerobic NOB occurring at low NH4+ concentrations, as in Lake Sarnen (49). The same habitat preferences were reported for Nitrososphaeria, ammonia-oxidizing archaea (AOA) (50), which were only found in Lake Sarnen sediments. The absence of NOB and AOA in Lake Baldegg sediments can be explained by the rapid O2 depletion and high NH4+ concentrations in the first few millimeters. In contrast, the Deltaproteobacteria, among which many anaerobic sulfate-reducing bacteria are found (51), and Methanomicrobia (anaerobic Euryarchaeota) became more dominant with increasing sediment depth in Lake Baldegg (Fig. S3). Similar trends in Lake Baldegg were reported in 2016 by Han et al. (39).
Spatial (columns), sediment depth (rows), and seasonal (x axis) distribution of the top 500 ASVs (relative abundance) of Lake Baldegg. Classes that belong to the same phylum are grouped together, and the phylum is written above each class group. The data show the sediment depth and temporal average for each station per lake separately. The ASVs were retrieved with DADA2 and assigned using the SILVA132 database. Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Spatial (columns), sediment depth (rows), and seasonal (x axis) distribution of the top 500 ASVs (relative abundance) of Lake Sarnen. Classes that belong to the same phylum are grouped together, and the phylum is written above each class group. The data show the sediment depth and temporal average for each station per lake separately. The ASVs were retrieved with DADA2 and assigned using the SILVA132 database. Download FIG S4, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Within lakes, locations differed considerably. We found the most distinctive microbial community composition at the shallow inflow location of Lake Baldegg, which could be explained by high nutrient and organic matter input from the tributary. The deeper and intermediate stations also contained distinct communities. One explanation for spatial differences can be sediment focusing. Both lakes have rather steep shorelines (Fig. 1), which can lead to sediment movements toward the deeper parts of the lake in a process called sediment focusing (52). The overall greater community similarity among samples from different locations in Lake Sarnen could be explained by the influence of its alpine river tributaries, which can have very high flow rates and transport large amounts of terrestrial material (53). During storm events, we measured an increase in sedimentation rates from ∼8 g m−2 day−1 to 556 g m−2 day−1 in June 2017 and 112 g m−2 day−1 in January 2018 at the deepest station ∼2 km from the inflow (20). Therefore, we assume that the larger input of terrestrial OM and river sediments and its wide distribution in the lake contribute to lower local niche diversity in Lake Sarnen compared to Lake Baldegg.
Seasonal changes in microbial community composition were only apparent among surface sediment samples, in which Cyanobacteria became more abundant during August and September, mainly in Lake Baldegg (Fig. S3) and less strongly in Lake Sarnen (Fig. S4). This finding likely reflects the seasonal sedimentation of biomass from cyanobacterial blooms in the surface water in late spring and summer, which has also been observed in sediment trap samples (20).
Microbial nitrogen transformation potential.
Our metagenomic analysis revealed that both lakes harbor microbial communities with the potential to perform almost all known N transformation processes (Fig. 8). This is in agreement with metagenomic studies of various ecosystems recovering the genetic potential for most, if not all, N transformation pathways (26, 54–57). Despite the overall similarities in the N cycling potential, ecosystems harbor unique N transforming microbial communities forming complex networks (1). Our work showed that this is also true for different freshwater lake systems and even for different sediment patches. The N gene composition and taxonomic data indicated spatial differences in the microbial community composition and N transformation potential between and within both lakes. This is in agreement with previous observations of a local difference in denitrification rates in lake sediments (25). This finding emphasizes the importance of including local differences of microbial community composition and function in lake sediment studies.
FIG 8.
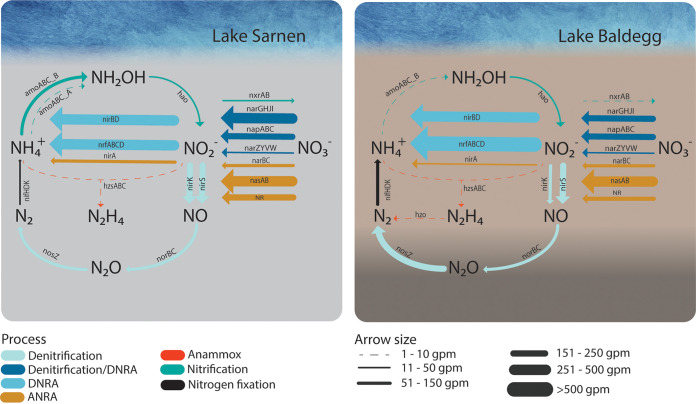
Conceptual overview of the N transformation processes by gene families in Lake Sarnen (left) and Lake Baldegg (right). The arrow thickness indicates the average gpm per lake for the different transformation processes. The gene families are colored in the different transformation processes they belong to nitrification (turquoise), ANRA (brown), denitrification_DNRA nitrate reductase genes (dark blue), DNRA (bright blue), denitrification (bright turquoise), anammox (red), and nitrogen (N2) fixation (N_fixation in black).
We saw little evidence for seasonal variation in the sediment microbial communities or their N cycling potential. This indicates that the assembly of the largely sedentary microbial community in the sediment, together with its N transformation potential, plays out over longer time scales, and these communities do not adapt rapidly to minor seasonal fluctuations in environmental conditions. This does not exclude short-term responses of activity, which can only be clarified by N transformation rate measurements and by applying methods that characterize the active microbial community, e.g., metatranscriptomics or proteomics.
Nitrogen inputs: organic N transformation and N2 fixation.
Organic N transformation (N_degradation) was highly abundant in both lakes, indicating that microbes can degrade organic N compounds in the OM to produce reactive N (Fig. 5). The ecology of organic N mineralization is not well characterized. Our results suggest that this function is more widespread among sediment microbial communities than the more specialized respiratory N transformations. Another way for the ecosystem to get reactive N is N2 fixation. In the water column of lakes, N2 fixation is a well-known process (3, 22, 36, 58). Nitrogenase genes were found in all samples in this study (Fig. 5). However, despite our observation of seasonal deposition of cyanobacteria, cyanobacterial nitrogenase genes were not identified in the sediment of Lake Baldegg. Instead, the abundant N2 fixation genes in Lake Baldegg were mainly assigned to methanogenic Euryarchaeota (Methanomicrobiales) and Proteobacteria (Table 1). Methanogenic Euryarchaeota has been reported from wetland soils, where they actively transcribed nifH genes (59). This leads to the assumption that Lake Baldegg's sediment microbial community could respond to N scarcity. However, given the high NH4+ concentration in the sediments, this is unlikely to represent an important way to obtain reactive N under the current conditions. Further research would be required to verify this assumption.
Nitrification.
Lake Sarnen's higher nitrification potential compared to Lake Baldegg is expected due to its higher O2 concentration and penetration depth, since nitrification is an aerobic process. Similar results have been reported for river freshwater sediments (60). The shallower O2 penetration depth in Lake Baldegg sediments likely constrains the development of an abundant nitrification community, which is supported by our rate measurements (Callbeck et al., unpublished). Here, instead, nitrification dominates in the overlaying water where O2 is more readily available (Callbeck et al., unpublished). Lake Baldegg showed the highest abundance for Nitrosomonadales among identified AOB. This taxon mediates partial nitrification by oxidizing NH4+ to NO2− (encoded by amoABC) and can thrive under low O2 concentrations (1, 23). Nitrospirales, well-known NOBs (1, 36, 50, 60), were the main taxa assigned to nxrB in Lake Sarnen (Table 1). Interestingly, amoABC_B genes were also assigned to Nitrospirales in Lake Sarnen, indicating the potential for comammox (30, 31, 61). Comammox Nitrospira organisms have been reported from wetlands, agricultural and forest soil, wastewater treatment plants, drinking water systems (62), and sediments from Lake Superior (25). To test the hypothesis that comammox Nitrospira are abundant in Lake Sarnen, we mapped the bacterial amoABC genes against four comammox Nitrospira and two noncomammox Nitrospira genomes (30, 61, 63, 64). We found amoABC and nxrB genes from Lake Sarnen mapping to the comammox Nitrospira genomes but not those from Lake Baldegg, which supports our hypothesis (Table S4). Unfortunately, due to insufficient sequencing depth, we were unable to retrieve enough high-quality metagenome-assembled genomes (MAGs) to confirm the presence of comammox Nitrospira. Hence, at this time, we cannot provide further evidence that comammox contributes to NH4+ oxidation in Lake Sarnen sediments.
Mapping results (number and percentage of reads) of the amoABC and nxrB genes against 4 comammox Nitrospira genomes, Ca.N.nitrosa (SAMEA3638995), and Ca.N.nitrificans (SAMEA3638996) (2), Ca.N.inopinata (SAMEA3560316) (3), and Nitrospira sp. strain GA0074138 (SAMN04254488) (4), and 2 non-comammox Nitrospira genomes, N. defluvii (SAMEA2272290), and N. moscoviensis (SAMN03702441) (5). The genes were mapped against the genomes using bbmap (minid=default (0.76), 0.85, 0.9) (version 38.49) (6). Download Table S4, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NO3− and NO2− reduction processes.
Interestingly, we measured lower rates for DNRA than for denitrification in both lakes (Callbeck et al., unpublished), even though DNRA genes were more abundant than denitrification genes (Table S4). It seems that many microbes in these lakes have the capacity to perform DNRA if conditions are favorable for this process but generally rely more on denitrification. This is probably because denitrification has a higher energy yield than DNRA (36). Many environmental variables play a role in the competition between denitrification and DNRA. For example, higher input of labile organic carbon (C), an increasing C-N ratio, the ratio of electron donors to electron acceptors, and sulfide concentrations are all known to affect the balance between these processes (18, 27, 36, 65–67). The more balanced ratio of denitrification to DNRA activity in Lake Sarnen measured by Callbeck et al. (unpublished) may be a consequence of the higher C:N ratios or the presumably higher sulfide levels from sulfate reduction, both of which can promote DNRA (18, 36). On the other hand, inhibition of DNRA due to high NO3− concentrations has been observed (65), which could explain the low DNRA rates in Lake Baldegg. However, it is not yet fully understood what conditions favor a switch from denitrification to DNRA in lake sediments (36).
There are other processes potentially competing for NO2−, such as anammox. Callbeck et al. (unpublished) measured low anammox rates relative to denitrification rates in the deepest station of Lake Baldegg. In contrast, in Lake Sarnen, the anammox rates in the surface sediment were as high as the denitrification rates. Interestingly, we could find some but not the complete set of anammox genes in Lake Sarnen. This suggests a low abundance of anammox bacteria, confirmed by the low abundance of Planctomycetes (the phylum containing all known anammox bacteria) in both lakes (Lake Baldegg, 0.01%; Lake Sarnen, 0.1% from all 16S rRNA sequences). However, a low abundance of anammox bacteria does not mean that anammox is not an active process. There have been reports of other aquatic ecosystems with high anammox rates and low abundance of Planctomycetes (11, 15, 68, 69). Furthermore, Crowe et al. (28) stated that anammox might be an important N loss process (up to 50% of the N loss in Lake Superior) for freshwater environments with low productivity, such as high-latitude lakes. However, more research is needed to resolve the role and activity of anammox in the oligotrophic Lake Sarnen.
Denitrification.
The higher abundance of nosZ in Lake Baldegg compared to Lake Sarnen, especially at the deep station (Fig. 6), agrees with the reported higher denitrification rates and N removal rates in Lake Baldegg (20 and Callbeck et al., unpublished). Graf et al. (29) found that nosZ genes have a higher cooccurrence with nirS than nirK, suggesting that nirS denitrifiers are more likely to be capable of complete denitrification. Our observation of a higher nirS:nirK ratio in Lake Baldegg compared to Lake Sarnen suggests that the capability for complete denitrification is more important in this lake. nirK denitrifiers are reported to respond differently to environmental gradients than nirS denitrifiers and occupy a different ecological niche (29). Our results allow the formation of a hypothesis according to which lakes with higher trophic status harbor more denitrifiers with the nirS gene and the capacity for complete denitrification, while oligotrophic lakes provide an ecological niche for nirK denitrifiers. Thus, further studies should determine whether the nirS:nirK ratio differs in lakes with different trophic states.
Nitrogen removal in a eutrophic and oligotrophic lake from the microbe's perspective.
Our data show that the trophic state did not impact the two lakes' overall microbial N transformation potential. However, internal N recycling through nitrification (including comammox), DNRA, and anammox appeared to be more important in the oligotrophic Lake Sarnen than in the eutrophic Lake Baldegg. In addition, the key players involved in N cycling clearly differ. Our findings are reflected in different N transformation rates and N removal efficiencies in the lakes studied. Müller et al. (20) calculated a much higher N removal rate (NRR) (20 ± 6.6 g N m−2 year−1) and N removal efficiency (NRE; 66%) for Lake Baldegg compared to Lake Sarnen, with an NRR of 3.2 ± 4.2 g N m−2 year−1 and an NRE of 33%. In agreement with that study, Callbeck et al. (unpublished data) measured more than 15 times higher denitrification rates in Lake Baldegg sediments (7.7 g N m−2 year−1) than in Lake Sarnen (0.5 g N m−2 year−1). Low denitrification activity in oligotrophic lakes is a known phenomenon reported from the Laurentian Great Lakes (25). This is because oligotrophic lakes might have a microbial N activity threshold and cannot achieve higher denitrification rates even under replete NO3− concentrations (25). The higher denitrification rates in Lake Baldegg were linked to higher nirS:nirK ratios. Furthermore, the higher total organic carbon (TOC) input in Lake Baldegg stimulates the microbial N cycling as TOC acts as an electron donor for various N reduction pathways in organotrophic N reducing reactions, such as denitrification (1, 18, 25, 36). Additionally, we found that higher NO3− concentrations in Lake Baldegg enhanced denitrification while inhibiting DNRA (70). Both mechanisms could explain the lower denitrification activity in Lake Sarnen. The considerable diversity and abundance of N transformation genes in the sediment surface layer show the tight connection of the various processes as reported in other studies focusing on the coupling of nitrification and denitrification in sediments (19). The combination of high TOC deposition, high NO3− concentrations, and the artificial aeration of Lake Baldegg appears to create an optimal ecological niche for denitrifying bacteria. Lake Sarnen, in contrast, is less efficient in terms of N removal, but as an oligotrophic lake, it nevertheless exports little N with the outflow. Although less efficient at removing N, the oligotrophic Lake Sarnen harbors a high microbial diversity with important ecosystem functions.
Conclusions.
Lake sediments were shown to harbor the genetic potential for all considered nitrogen cycle processes but in variable abundance, diversity, and performed by distinct microbial networks. Judging from the contrasting results obtained from the lakes studied here, the microbial potential for N transformation in lake sediments can differ considerably in individual freshwater lakes and varies with location within each lake. We have demonstrated that a detailed quantitative description of the N functional genes and potential microbial key players in lake sediments can be obtained through assembly-based metagenomic analysis. This microbial N gene potential forms the foundation of N removal on the ecosystem scale (20) and the specific N transformation rates of sediments (Callbeck et al., unpublished). N transformation potential in lake sediments clearly reflects the environmental conditions (e.g., availability of electron acceptors, OM input, etc., most of which are at least partly controlled by the lakes’ trophic state). This results in distinct microbial communities with a different representation of the processes linked to N removal. The response of N removal processes to changing environmental conditions may, at least in the short term, depend on the properties of the established microbial communities. Therefore, these responses are expected to differ between lake sediments with contrasting N cycling microbial networks.
We suggest two hypotheses for further study: that the nirS:nirK ratio should be considered a possible indicator of N removal efficiency and that oligotrophic lake sediments harbor higher microbial diversity. Finally, we report the first evidence of possible comammox Nitrospira in oligotrophic Lake Sediments in Switzerland.
This study not only provides a comprehensive view on the biology and ecology that underpins N removal in freshwater lakes, which is an important ecosystem service, but also shows the value of including metagenomic analysis to better understand the microbially mediated N cycle in natural systems.
MATERIALS AND METHODS
Study sites.
Lakes Baldegg and Sarnen are situated on the Swiss plateau and in the peri-alpine region, respectively. Both are small lakes, with a surface of 7.15 km2 and a maximum depth of 49 m for Lake Sarnen and 5.22 km2 and 66 m for Lake Baldegg (20). Lake Sarnen is surrounded by mountains, forests, and extensive agriculture and therefore remained oligotrophic throughout the 20th century, maintaining a total phosphorus (TP) concentration of ∼5 mg P m−3. Lake Baldegg, in comparison, receives high P loads due to pig and cattle farming in the catchment. In the second half of the 20th century, this lake experienced pronounced eutrophication that peaked in the 1970s. To prevent bottom water anoxia, the lake has been artificially aerated with O2 during the stratified season since 1982 (71). Today, the lake is still eutrophic with TP concentrations of ∼24 mg P m−3 (37, 72). Sediment trap data from our parallel study in 2017 and 2018 found a higher sedimentation rate of N-enriched OM in Lake Baldegg (56.8 g C m−2 year−1 and 11.2 gN m−2 year−1, C:N 5.9) compared to Lake Sarnen (28.6 g C m−2 year−1 and 4.1 gN m−2 year−1, C:N 8.5) (20). Both lakes are dimictic with an oxic water column throughout the year; thus, the oxic-anoxic transition zone, where most N transformation processes occur, extends over the first few millimeters of the sediments. To study spatial variability, we selected five sampling locations representing different depths between the inflow and outflow of the lakes (Fig. 1). The seasonal dynamics were investigated by sampling each station in March, May, August, and September 2018 (for details, see Müller et al. [20]). The inflow location of Lake Baldegg was additionally sampled in August and September. Sediment depth was resolved by analyzing pore water through sampling ports or cutting layers from the core, as described below.
Sediment sampling and chemical and statistical analyses.
The sediment sampling, pore water analyses, and oxygen measurements are described in Müller et al. (20). Briefly, we used a gravity corer for the sediment sampling with PVC tubes (5.9-cm diameter and 60-cm length; Uwitec, Austria). The PVC tubes had predrilled ports to extract pore water samples (∼200 μl) in a 0.25-cm depth resolution using MicroRhizon filter tubes (0.2 μm pore size; 0.8 mm diameter; Rhizosphere Research Products, Wageningen, Netherlands). The pore water was sampled onshore immediately after core retrieval, transferred to 1.5-ml tubes, stored on ice in the dark, and analyzed within 24 h. NO3−, NO2−, SO42−, and NH4+ were analyzed using two ion chromatography devices (cations, 882 Compact IC plus; anions, 881 Compact IC pro; Metrohm, Switzerland). In addition, vertical O2 concentration profiles were recorded with an O2 micro-optode mounted to an automated micromanipulator (Presens, Germany) immediately after core retrieval. The O2 and pore water profiles were visualized using ggplot2, and the spatial and temporal differences were calculated using analysis of variance (ANOVA) from the base statistical packages in R (version 3.6.1) (73, 74).
DNA sampling and extraction.
During each sampling event, a sediment core was retrieved for DNA analysis. The core was cut with a metal slicing device into the following sections: 0 to 0.5 cm, 0.5 to 1 cm, 1 to 1.5 cm, 1.5 to 2 cm, 2 to 3 cm, and 3 to 5 cm. The slicing device was washed with 80% ethanol and molecular grade water (DNase/RNase free; SigmaAldrich) before each core sampling and with molecular-grade water during the slicing of cores. Three subsamples (∼2 ml) were taken from each section with a 3-ml syringe and immediately fixed in 2 ml of RNAlater (Sigma Life Science) in a 5-ml tube (DNase/RNase free; Qiagen). The sediment samples were kept at 4°C until the next day and then stored at −80°C. Prior to nucleic acid extraction, samples were thawed and washed three times with 2.5 ml 1× Tris-EDTA buffer to remove excess salt from RNAlater. Nucleic acids were then extracted using the RNeasy PowerSoil total RNA kit in combination with the RNAeasy PowerSoil DNA elution kit (Qiagen) according to the manufacturer's instructions. The DNA quality and quantity were measured with a NanoDrop spectrophotometer (NanoDrop One), and extracts were stored at −80°C until sequencing. Extraction blanks were prepared to check for contamination and used as negative controls in sequencing.
Amplicon sequencing, pipeline, and statistical analysis.
All amplicon sequencing was performed by Novogene (Hong Kong) by following standard sequencing and quality control protocols. Extracted DNA from all stations and all seasons (120 samples from Lake Baldegg and 96 samples from Lake Sarnen) was amplified with universal 16S rRNA gene primers 341F (5′-CCT AYG GGR BGC ASC AG-3′) and 806R (5′-GGA CTA CNN GGG TAT CTA AT-3′), targeting the V3-V4 region (fragment length, ∼466 bp), and sequenced with Illumina HiSeq technology to generate paired-end reads. This resulted in 119 samples with an average of 112,857 raw paired-end reads in Lake Baldegg and 90 samples with an average of 139,822 raw paired-end reads in Lake Sarnen. The sequences were analyzed separately for each lake using the DADA2 pipeline in R (version 3.5.1) (74, 75). Briefly, after a quality check, the raw sequence adapters were trimmed and dereplicated, and error rates were calculated before applying the DADA2 core sample inference algorithm. Next, the forward and reverse reads were merged to obtain the amplicon sequence variants (ASVs). Following chimera removal with the removeBimeraDenovo algorithm, the taxonomic assignment of the ASVs was performed using a Naïve Bayesian classifier method based on the SILVA database 132 for the V3-V4 region (76). (Note that the SILVA database release 132 implemented numerous changes to the taxonomy and, e.g., assigned the former Betaproteobacteria to the class of Gammaproteobacteria). Unclassified and rare ASVs (less than five instances in at least 10% of all samples) were filtered out. The ASV abundance tables and the corresponding environmental parameters were further analyzed by calculating the alpha and beta diversity (redundancy analysis [RDA]), testing for significantly different microbial community composition between the lakes, locations, sampling months, and sediment depth using adonis and visualized with the phyloseq, ggplot2, vegan, and microbiomeSeq packages using R statistical software (version 3.6.3) (73, 74, 77–79). Details of the 16S rRNA gene amplicon library can be found in the supplemental material (Table S1), and the abundance table was added to the Eawag data package (see “Data availability,” below).
Sequencing overview for the 16S amplicon sequencing data (PE, paired end). Download Table S1, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metagenomic sequencing, pipeline, and statistical analysis.
For metagenomic sequencing, DNA extracts from all samples of one core were pooled using 1 μg of DNA from each sampling depth (0 to 0.5 cm, 0.5 to 1 cm, 1 to 1.5 cm, 1.5 to 2 cm, and 2 to 3 cm) after measuring the DNA concentration with a Qubit (Qubit 2.0 fluorometer; Invitrogen). We obtained eight metagenomes from Lake Sarnen (deepest and shallow_inflow station for March, May, August, and September) and 10 metagenomes from Lake Baldegg (deepest and shallow_outflow locations for March, May, August, and September and shallow_inflow for August and September). The metagenomes were sequenced using the Illumina NextSeq platform to generate 150-bp paired-end reads (averaging ∼350 bp in length) by Novogene (Hong Kong). The quality of each metagenome was evaluated using FastQC (80). Prinseq was used to trim metagenomic reads (minimum quality mean, 20) (81). Each sample was assembled using Megahit with the options meta-sensitive and min_contig length 500 (82), and reads were mapped back using BBmap (minid = 0.95) (83). The sam files were converted to bam files using sambamba (84) and SAMtools (85), and the reads were subsequently counted using featureCounts from subread 1.6.4 (86). For functional annotation, we used prokka v1.13 (87). The N transformation genes (Table 2) were obtained by mapping the protein output from prokka against the N cycle database (NCycDB; accessed July 2019), applying a 100% identity criterion (32). The genes and representative gene families were assigned to N cycle processes based on information provided by NCycDB (see Table 1 in Tue et al. [32]); if a gene was assigned to more than one process in this table, we made a new category naming all processes (e.g., Denitrification_DNRA) (Table 2). Further analysis and visualization were done in R (version 3.6.3) (74). The quantified genes were normalized for gene lengths and sequencing depth by calculating genes per million (gpm) for the visualization. We calculated the nirS:nirK ratio from the gpm data to indicate the ratio of complete denitrifiers versus incomplete denitrifiers (29). A principal component analysis was used to test for significant differences in the abundance of N transformation genes between lakes (alpha = 0.001) using DESeq2 (88). We used the nonnormalized feature count files because DESeq2 applies its own normalization step (by following the workflow from Love et al., accessed 1 June 2019 [89]). We used the Wald test, with the condition Lake Baldegg versus Lake Sarnen, to extract the significantly different N transformation genes and the Benjamini-Hochberg method from DESeq2 to correct P values for multiple testing (88, 89). The results were visualized with ggplot2 and ComplexHeatmap (90) after calculating the z score [z = (x − mean(x))/SD(x), where x is N gene counts and SD is standard deviation] for all significantly different N transformation genes to emphasize the differences. Finally, candidate key players containing the significantly different genes and nirS were resolved by taxonomic assignment of the prokka output using Kaiju (91). Kaiju is a taxonomic assignment tool developed for short reads and might be biased when used for long reads such as assembled contigs. Kaiju still contains the former class Betaproteobacteria; we assigned it manually to Gammaproteobacteria, considering the taxonomical assignment of the SILVA database 132.
TABLE 2.
Overview of the nitrogen transformation pathways, processes, and corresponding gene families analyzed using NCycDBa
| Pathway | Enzyme | Gene family |
|---|---|---|
| Nitrification | Ammonia monooxygenase | amoABC |
| Hydroxylamine dehydrogenase | hao | |
| Nitrite oxidoreductase | nxrAB | |
| Denitrification or DNRA | Nitrate reductase | napABC, narGHJI, narZYVW |
| DNRA | Nitrite reductase | nirBD, nrfABCD |
| Denitrification | Nitrite reductase | nirK (Cu), nirS (cytochrome cd1) |
| Nitric oxide reductase | norBC | |
| Nitrous oxide reductase | nosZ | |
| Anammox | Hydrazine oxidoreductase | hzo |
| Hydrazine synthase | hzsABC | |
| Hydrazine dehydrogenase | hdh | |
| ANRA | Nitrate reductase | nasAB, NR, narBC |
| Nitrite reductase | nirA | |
| Nitrogen fixation | Nitrogenase | nifDHKW, anfG |
| Organic nitrogen degradation | Urease | ureABC |
| Nitroalkane oxidase | nao | |
| Nitronate monooxygenase | nmo | |
| Glutamate dehydrogenase | gdh | |
| Glutamate synthase | gs | |
| Glutaminase | glsA | |
| Glutamine synthetase | glnA | |
| Asparagine synthase | asnB | |
| Glutamin-(aspargin-)ase | ansB | |
| Others | Hydroxylamine reductase | hcp |
| Particulate methane monooxygenase | pmoABC |
Data are adapted from Tu et al. (32). Categories of pathways are exclusive if not stated otherwise.
The sequencing depth, quality, filtered reads, mean contig length, and mapped reads for each sample are summarized in the supplemental material (Table S2).
Data availability.
All sequences can be found under the NCBI BioProject PRJNA726540. The 119 16S rRNA gene sequences and the metadata from Lake Baldegg have the accession numbers SAMN18962412–SAMN18962530. The 90 16S rRNA gene sequences and the metadata of Lake Sarnen are deposited under the accession numbers SAMN18978354–SAMN18978443. The 10 metagenomes and the metadata can be found under the accession numbers SAMN19067306–SAMN19067315. The eight metagenomes and the metadata from Lake Sarnen can be found under the accession numbers SAMN19030157–SAMN19030164. All other data, such as metadata and the abundance tables, are available at the institutional data repository of Eawag (ERIC) (https://opendata.eawag.ch/) in accordance with FAIR data sharing principles (https://doi.org/10.25678/0005HX).
ACKNOWLEDGMENTS
We thank the technical staff of Eawag, namely, Patrick Kathriner, Karin Beck, Alois Zwyssig, Serge Robert, and Nina Studhalter, for their help during fieldwork and in the analytical, biogeochemical, and microbial laboratory. Thanks goes to Xinguo Han for his support in troubleshooting DNA/RNA extraction from the two lakes. We are grateful to the cantonal authorities Obwalden (Lake Sarnen) and ProNatura and Canton Lucerne (Lake Baldegg) for supporting our study by granting access to the lakes for sampling and permission to deploy sediment traps. We thank the genomic diversity center (GDC) and Euler Cluster at ETH for the bioinformatic support and access to their computational resources for the metagenomic analysis. We also thank Martin Schmid and Bernhard Wehrli for the fruitful discussions of the data.
This work was financially supported by the Swiss National Science Foundation grant Regulation of Nitrogen Turnover in Lakes (205321_169142).
Contributor Information
Helmut Bürgmann, Email: helmut.buergmann@eawag.ch.
Katherine McMahon, University of Wisconsin-Madison.
REFERENCES
- 1.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 2.Isobe K, Ohte N. 2014. Ecological perspectives on microbes involved in N-cycling. Microbes Environ 29:4–16. doi: 10.1264/jsme2.me13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zehr JP, Kudela RM. 2011. Nitrogen cycle of the open ocean: from genes to ecosystems. Annu Rev Mar Sci 3:197–225. doi: 10.1146/annurev-marine-120709-142819. [DOI] [PubMed] [Google Scholar]
- 4.Gruber N, Galloway JN. 2008. An earth-system perspective of the global nitrogen cycle. Nature 451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 5.Seitzinger S, Harrison JA, Böhlke JK, Bouwman AF, Lowrance R, Peterson B, Tobias C, Van Drecht G. 2006. Denitrification across landscapes and waterscapes: a synthesis. Ecol Appl 16:2064–2090. doi: 10.1890/1051-0761(2006)016[2064:DALAWA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 6.Heldstab J, Reutimann J, Biedermann R, Leu D. 2010. Stickstoffflüsse in der Schweiz. Stoffflussanalyse für das Jahr 2005. Umwelt-Wissen nr. 1018. Bundesamt für Umwelt, Bern, Switzerland. https://www.bafu.admin.ch/dam/bafu/de/dokumente/chemikalien/uw-umwelt-wissen/stickstofffluesseinderschweiz.pdf.download.pdf/stickstofffluesseinderschweiz.pdf. [Google Scholar]
- 7.Holland EA, Braswell BH, Sulzman J, Lamarque J-F. 2005. Nitrogen deposition onto the United States and western Europe: synthesis of observations and models. Ecol Appl 15:38–57. doi: 10.1890/03-5162. [DOI] [Google Scholar]
- 8.Eawag. 2018. National long-term surveillance of Swiss rivers. https://opendata.eawag.ch/group/naduf-national-long-term-surveillance-of-swiss-rivers. Accessed 10 April 2018.
- 9.Billen G, Silvestre M, Grizzetti B, Leip A, Garnier J, Voss M, Howarth R, Bouraoui F, Lepistö A, Kortelainen P, Johnes P, Curtis C, Humborg C, Smedberg E, Kaste Ø, Ganeshram R, Beusen A, Lancelot C. 2011. Nitrogen flows from European regional watersheds to coastal marine waters, p 271–297. In The European Nitrogen Assessment. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 10.Bernhardt ES. 2013. Ecology. Cleaner lakes are dirtier lakes. Science 342:205–206. doi: 10.1126/science.1245279. [DOI] [PubMed] [Google Scholar]
- 11.Schubert CJ, Durisch-Kaiser E, Wehrli B, Thamdrup B, Lam P, Kuypers MMM. 2006. Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ Microbiol 8:1857–1863. doi: 10.1111/j.1462-2920.2006.01074.x. [DOI] [PubMed] [Google Scholar]
- 12.Finlay JC, Small GE, Sterner RW. 2013. Human influences on nitrogen removal in lakes. Science 342:247–250. doi: 10.1126/science.1242575. [DOI] [PubMed] [Google Scholar]
- 13.Thamdrup B. 2012. New pathways and processes in the global nitrogen cycle. Annu Rev Ecol Evol Syst 43:407–428. doi: 10.1146/annurev-ecolsys-102710-145048. [DOI] [Google Scholar]
- 14.Penton CR, Devol AH, Tiedje JM. 2006. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl Environ Microbiol 72:6829–6832. doi: 10.1128/AEM.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jørgensen BB, Kuenen JG, Sinninghe Damsté JS, Strous M, Jetten MSM. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608–611. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- 16.Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuña-González J. 2003. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422:606–608. doi: 10.1038/nature01526. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch MD, Long ZT, Song B. 2011. Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoa/hzob). Microb Ecol 61:264–276. doi: 10.1007/s00248-010-9743-1. [DOI] [PubMed] [Google Scholar]
- 18.Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, Geelhoed JS, Strous M. 2014. Nitrogen cycling. The environmental controls that govern the end product of bacterial nitrate respiration. Science 345:676–679. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- 19.Marchant HK, Holtappels M, Lavik G, Ahmerkamp S, Winter C, Kuypers MMM. 2016. Coupled nitrification–denitrification leads to extensive n loss in subtidal permeable sediments. Limnol Oceanogr 61:1033–1048. doi: 10.1002/lno.10271. [DOI] [Google Scholar]
- 20.Müller B, Thoma R, Baumann KBL, Callbeck CM, Schubert CJ. 2021. Nitrogen removal processes in lakes of different trophic states from on-site measurements and historic data. Aquat Sci 83:37. doi: 10.1007/s00027-021-00795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalvelage T, Lavik G, Lam P, Contreras S, Arteaga L, Löscher CR, Oschlies A, Paulmier A, Stramma L, Kuypers MMM. 2013. Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone. Nat Geosci 6:228–234. doi: 10.1038/ngeo1739. [DOI] [Google Scholar]
- 22.Callbeck CM, Ehrenfels B, Baumann KBL, Wehrli B, Schubert CJ. 2021. Anoxic chlorophyll maximum enhances local organic matter remineralization and nitrogen loss in Lake Tanganyika. Nat Commun 12:830. doi: 10.1038/s41467-021-21115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bristow LA, Dalsgaard T, Tiano L, Mills DB, Bertagnolli AD, Wright JJ, Hallam SJ, Ulloa O, Canfield DE, Revsbech NP, Thamdrup B. 2016. Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc Natl Acad Sci USA 113:10601–10606. doi: 10.1073/pnas.1600359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rissanen AJ, Tiirola M, Hietanen S, Ojala A. 2013. Interlake variation and environmental controls of denitrification across different geographical scales. Aquat Microb Ecol 69:1–16. doi: 10.3354/ame01619. [DOI] [Google Scholar]
- 25.Small GE, Finlay JC, McKay RML, Rozmarynowycz MJ, Brovold S, Bullerjahn GS, Spokas K, Sterner RW. 2016. Large differences in potential denitrification and sediment microbial communities across the Laurentian great lakes. Biogeochemistry 128:353–368. doi: 10.1007/s10533-016-0212-x. [DOI] [Google Scholar]
- 26.Tu Q, He Z, Wu L, Xue K, Xie G, Chain P, Reich PB, Hobbie SE, Zhou J. 2017. Metagenomic reconstruction of nitrogen cycling pathways in a CO2-enriched grassland ecosystem. Soil Biol Biochem 106:99–108. doi: 10.1016/j.soilbio.2016.12.017. [DOI] [Google Scholar]
- 27.Giblin A, Tobias C, Song B, Weston N, Banta G, Rivera-Monroy V. 2013. The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanography 26:124–131. doi: 10.5670/oceanog.2013.54. [DOI] [Google Scholar]
- 28.Crowe SA, Treusch AH, Forth M, Li J, Magen C, Canfield DE, Thamdrup B, Katsev S. 2017. Novel anammox bacteria and nitrogen loss from Lake Superior. Sci Rep 7:13757. doi: 10.1038/s41598-017-12270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graf DRH, Jones CM, Hallin S. 2014. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9:e114118. doi: 10.1371/journal.pone.0114118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch H, van Kessel MAHJ, Lücker S. 2019. Complete nitrification: insights into the ecophysiology of comammox nitrospira. Appl Microbiol Biotechnol 103:177–189. doi: 10.1007/s00253-018-9486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu Q, Lin L, Cheng L, Deng Y, He Z. 2019. NcycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics 35:1040–1048. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- 33.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson MB, Martiny AC, Martiny JBH. 2016. Global biogeography of microbial nitrogen-cycling traits in soil. Proc Natl Acad Sci USA 113:8033–8040. doi: 10.1073/pnas.1601070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT. 2014. Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e01193-14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damashek J, Francis CA. 2018. Microbial nitrogen cycling in estuaries: from genes to ecosystem processes. Estuaries Coast 41:626–660. doi: 10.1007/s12237-017-0306-2. [DOI] [Google Scholar]
- 37.Fiskal A, Deng L, Michel A, Eickenbusch P, Han X, Lagostina L, Zhu R, Sander M, Schroth MH, Bernasconi SM, Dubois N, Lever MA. 2019. Effects of eutrophication on sedimentary organic carbon cycling in five temperate lakes. Biogeosci Discuss 16:1–35. [Google Scholar]
- 38.Steinsberger T, Schwefel R, Wüest A, Müller B. 2020. Hypolimnetic oxygen depletion rates in deep lakes: effects of trophic state and organic matter accumulation. Limnol Oceanogr 65:3128–3138. doi: 10.1002/lno.11578. [DOI] [Google Scholar]
- 39.Han X, Schubert CJ, Fiskal A, Dubois N, Lever MA. 2020. Eutrophication as a driver of microbial community structure in lake sediments. Environ Microbiol 22:3446–3462. doi: 10.1111/1462-2920.15115. [DOI] [PubMed] [Google Scholar]
- 40.Wang NF, Zhang T, Yang X, Wang S, Yu Y, Dong LL, Guo YD, Ma YX, Zang JY. 2016. Diversity and composition of bacterial community in soils and lake sediments from an arctic lake area. Front Microbiol 7:1170. doi: 10.3389/fmicb.2016.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao B, Yan X, Zhang J, Chen M, Li Y, Huang J, Lei M, He H, Wang J. 2019. Microbial community composition in alpine lake sediments from the Hengduan mountains. Microbiologyopen 8:e00832. doi: 10.1002/mbo3.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rissanen AJ, Peura S, Mpamah PA, Taipale S, Tiirola M, Biasi C, Mäki A, Nykänen H. 2019. Vertical stratification of bacteria and archaea in sediments of a small boreal humic lake. FEMS Microbiol Lett 366:fnz044. doi: 10.1093/femsle/fnz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Chen X, Jiang X, Zheng B. 2017. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. MicrobiologyOpen 6:e00503. doi: 10.1002/mbo3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Zhao T, Wang Q, Li L, Shen T, Gao G. 2019. Bacterial community composition in aquatic and sediment samples with spatiotemporal dynamics in large, shallow, eutrophic lake chaohu, china. J Freshw Ecol 34:575–589. doi: 10.1080/02705060.2019.1635536. [DOI] [Google Scholar]
- 45.Moss B. 1973. Diversity in fresh-water phytoplankton. Am Midl Nat 90:341–355. doi: 10.2307/2424458. [DOI] [Google Scholar]
- 46.Ortiz-Álvarez R, Triadó-Margarit X, Camarero L, Casamayor EO, Catalan J. 2018. High planktonic diversity in mountain lakes contains similar contributions of autotrophic, heterotrophic and parasitic eukaryotic life forms. Sci Rep 8:4457. doi: 10.1038/s41598-018-22835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maron P-A, Sarr A, Kaisermann A, Lévêque J, Mathieu O, Guigue J, Karimi B, Bernard L, Dequiedt S, Terrat S, Chabbi A, Ranjard L. 2018. High microbial diversity promotes soil ecosystem functioning. Appl Environ Microbiol 84:e02738-17. doi: 10.1128/AEM.02738-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron P-A. 2013. Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7:1609–1619. doi: 10.1038/ismej.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herber J, Klotz F, Frommeyer B, Weis S, Straile D, Kolar A, Sikorski J, Egert M, Dannenmann M, Pester M. 2020. A single thaumarchaeon drives nitrification in deep oligotrophic Lake Constance. Environ Microbiol 22:212–228. doi: 10.1111/1462-2920.14840. [DOI] [PubMed] [Google Scholar]
- 50.Pester M, Schleper C, Wagner M. 2011. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wörner S, Pester M. 2019. The active sulfate-reducing microbial community in littoral sediment of oligotrophic Lake Constance. Front Microbiol 10:247. doi: 10.3389/fmicb.2019.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blais JM, Kalff J. 1995. The influence of lake morphometry on sediment focusing. Limnol Oceanogr 40:582–588. doi: 10.4319/lo.1995.40.3.0582. [DOI] [Google Scholar]
- 53.Monecke K, Anselmetti FS, Becker A, Sturm M, Giardini D. 2004. The record of historic earthquakes in lake sediments of central Switzerland. Tectonophysics 394:21–40. doi: 10.1016/j.tecto.2004.07.053. [DOI] [Google Scholar]
- 54.Lüke C, Speth DR, Kox MAR, Villanueva L, Jetten MSM. 2016. Metagenomic analysis of nitrogen and methane cycling in the Arabian sea oxygen minimum zone. PeerJ 4:e1924. doi: 10.7717/peerj.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran PQ, Bachand SC, McIntyre PB, Kraemer BM, Vadeboncoeur Y, Kimirei IA, Tamatamah R, McMahon KD, Anantharaman K. 2021. Depth-discrete metagenomics reveals the roles of microbes in biogeochemical cycling in the tropical freshwater Lake Tanganyika. ISME J 15:1971–1986. doi: 10.1038/s41396-021-00898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao R, Summers ZM, Christman GD, Yoshimura KM, Biddle JF. 2020. Metagenomic views of microbial dynamics influenced by hydrocarbon seepage in sediments of the Gulf of Mexico. Sci Rep 10:5772. doi: 10.1038/s41598-020-62840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orellana LH, Chee-Sanford JC, Sanford RA, Löffler FE, Konstantinidis KT. 2018. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl Environ Microbiol 84:e01646-17. doi: 10.1128/AEM.01646-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beversdorf LJ, Miller TR, McMahon KD. 2013. The role of nitrogen fixation in cyanobacterial bloom toxicity in a temperate, eutrophic lake. PLoS One 8:e56103. doi: 10.1371/journal.pone.0056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bae H-S, Morrison E, Chanton JP, Ogram A. 2018. Methanogens are major contributors to nitrogen fixation in soils of the Florida Everglades. Appl Environ Microbiol 84:e02222-17. doi: 10.1128/AEM.02222-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altmann D, Stief P, Amann R, De Beer D, Schramm A. 2003. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ Microbiol 5:798–803. doi: 10.1046/j.1469-2920.2003.00469.x. [DOI] [PubMed] [Google Scholar]
- 61.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun D, Tang X, Zhao M, Zhang Z, Hou L, Liu M, Wang B, Klümper U, Han P. 2020. Distribution and diversity of comammox nitrospira in coastal wetlands of China. Front Microbiol 11:589268. doi: 10.3389/fmicb.2020.589268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinto AJ, Marcus DN, Ijaz UZ, Bautista-de Lose Santos QM, Dick GJ, Raskin L. 2016. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere 1:e00054-15. doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakoula D, Nowka B, Spieck E, Daims H, Lücker S. 2018. The draft genome sequence of “Nitrospira lenta” strain bs10, a nitrite oxidizing bacterium isolated from activated sludge. Stand Genomic Sci 13:32. doi: 10.1186/s40793-018-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heo H, Kwon M, Song B, Yoon S. 2020. Involvement of NO3− in ecophysiological regulation of dissimilatory nitrate/nitrite reduction to ammonium (DNRA) is implied by physiological characterization of soil DNRA bacteria isolated via a colorimetric screening method. Appl Environ Microbiol 86:e01054-20. doi: 10.1128/AEM.01054-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bu C, Wang Y, Ge C, Ahmad HA, Gao B, Ni S-Q. 2017. Dissimilatory nitrate reduction to ammonium in the yellow river estuary: rates, abundance, and community diversity. Sci Rep 7:6830. doi: 10.1038/s41598-017-06404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgin AJ, Hamilton SK. 2007. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ 5:89–96.2.0.CO;2]. doi: 10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2. [DOI] [Google Scholar]
- 68.Pollet T, Humbert J-F, Tadonléké RD. 2014. Planctomycetes in lakes: poor or strong competitors for phosphorus? Appl Environ Microbiol 80:819–828. doi: 10.1128/AEM.02824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tadonléké RD. 2007. Strong coupling between natural planctomycetes and changes in the quality of dissolved organic matter in freshwater samples. FEMS Microbiol Ecol 59:543–555. doi: 10.1111/j.1574-6941.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- 70.Hardison AK, Algar CK, Giblin AE, Rich JJ. 2015. Influence of organic carbon and nitrate loading on partitioning between dissimilatory nitrate reduction to ammonium (DNRA) and N2 production. Geochim Cosmochim Acta 164:146–160. doi: 10.1016/j.gca.2015.04.049. [DOI] [Google Scholar]
- 71.Gächter R, Wehrli B. 1998. Ten years of artificial mixing and oxygenation: no effect on the internal phosphorus loading of two eutrophic lakes. Environ Sci Technol 32:3659–3665. doi: 10.1021/es980418l. [DOI] [Google Scholar]
- 72.Steinsberger T, Müller B, Gerber C, Shafei B, Schmid M. 2019. Modeling sediment oxygen demand in a highly productive lake under various trophic scenarios. PLoS One 14:e0222318. doi: 10.1371/journal.pone.0222318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham H. 2011. ggplot2. WIREs Comp Stat 3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 74.R Core Team. 2017. R: a language and environment for statistical computing. https://www.r-project.org/. Accessed 9 January 2017.
- 75.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. Dada2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callahan B. Dada2 pipeline tutorial. https://benjjneb.github.io/dada2/tutorial.html Accessed 3 January 2020.
- 77.Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ, Suggests M. 2007. The vegan package. Comm Ecol Package 10:719. [Google Scholar]
- 78.McMurdie PJ, Holmes S. 2013. Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ssekagiri A, Sloan WT, Ijaz UZ. 2018. Microbiomeseq: an R package for microbial community analysis in an environmental context. https://github.com/umerijaz/microbiomeSeq.
- 80.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 81.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. Megahit: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 83.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Laboratories, Berkeley, CA. [Google Scholar]
- 84.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and samtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 87.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 88.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love MI, Anders S, Huber W. 2019. Analyzing RNA-seq data with deseq2. http://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html. Accessed 6 January 2019.
- 90.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 91.Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Broadbent AAD, Snell HSK, Michas A, Pritchard WJ, Newbold L, Cordero I, Goodall T, Schallhart N, Kaufmann R, Griffiths RI, Schloter M, Bahn M, Bardgett RD. 2021. Climate change alters temporal dynamics of alpine soil microbial functioning and biogeochemical cycling via earlier snowmelt. ISME J 15:2264–2275. doi: 10.1038/s41396-021-00922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi D, Feng F, Fu Y, Ji X, Liu X. 2020. Effects of soil microbes on forest recovery to climax community through the regulation of nitrogen cycling. Forests 11:1027. doi: 10.3390/f11101027. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the alpha diversity measures (A and B) for the total microbial community composition based on the 16S rRNA of Lake Baldegg (green, 119 samples) and Lake Sarnen (yellow, 90 samples) and the significant differences between the alpha diversity of the two lakes. (C) Overview of the beta diversity measures for the whole microbial community composition based on the 16S rRNA of Lake Baldegg (green, 119 samples) and Lake Sarnen (yellow, 90 samples). All analyses were done in R with the vegan package by following the tutorial from the Küsel lab. Download FIG S1, EPS file, 1.4 MB (1.5MB, eps) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Constrained ordination biplots (redundancy analysis [RDA] on weighted UniFrac distance) of microbial community composition and the different environmental drivers (O2, NO3−, NO2−, NH4+, and SO42−) for Lake Baldegg (A and C) and Lake Sarnen (B and D). The samples are colored by sediment depth (A and B) and month (C and D). All analyses were done in R with the vegan package by following the tutorial from the Küsel lab. Download FIG S2, EPS file, 0.1 MB (68.1KB, eps) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing overview for the metagenomic data, with the number of raw reads, raw bases (G, giga), GC content, number of contigs (assembled sequences), number of contigs aligned to NCycDB, and number of total N genes (count N genes from NCycDB pipline) per sample (1). Download Table S2, XLSX file, 0.01 MB (12.2KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of all nitrogen cycle genes found in Lake Baldegg and Lake Sarnen from the different locations and months. The numbers are the sum of gpm (genes per million) per sample. The genes are clustered in the N transformation process they belong to. Download Table S3, XLSX file, 0.01 MB (37.4KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the mapping rates of the N removal genes to Kaiju database of Proteobacteria (top 12 phyla) for the significantly different gene families, plus nirS. The numbers are the total sum of gpm per genes per lake and phylum. The most abundant are highlighted in green for Lake Baldegg and yellow for Lake Sarnen. We listed Betaproteobacteria here separately, as Kaiju assign them separately and does not have the SILVA132 database assignment. In the paper, Beta- and Gammaproteobacteria are summed (92, 93). Download Table S5, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Spatial (columns), sediment depth (rows), and seasonal (x axis) distribution of the top 500 ASVs (relative abundance) of Lake Baldegg. Classes that belong to the same phylum are grouped together, and the phylum is written above each class group. The data show the sediment depth and temporal average for each station per lake separately. The ASVs were retrieved with DADA2 and assigned using the SILVA132 database. Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Spatial (columns), sediment depth (rows), and seasonal (x axis) distribution of the top 500 ASVs (relative abundance) of Lake Sarnen. Classes that belong to the same phylum are grouped together, and the phylum is written above each class group. The data show the sediment depth and temporal average for each station per lake separately. The ASVs were retrieved with DADA2 and assigned using the SILVA132 database. Download FIG S4, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mapping results (number and percentage of reads) of the amoABC and nxrB genes against 4 comammox Nitrospira genomes, Ca.N.nitrosa (SAMEA3638995), and Ca.N.nitrificans (SAMEA3638996) (2), Ca.N.inopinata (SAMEA3560316) (3), and Nitrospira sp. strain GA0074138 (SAMN04254488) (4), and 2 non-comammox Nitrospira genomes, N. defluvii (SAMEA2272290), and N. moscoviensis (SAMN03702441) (5). The genes were mapped against the genomes using bbmap (minid=default (0.76), 0.85, 0.9) (version 38.49) (6). Download Table S4, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing overview for the 16S amplicon sequencing data (PE, paired end). Download Table S1, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2022 Baumann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All sequences can be found under the NCBI BioProject PRJNA726540. The 119 16S rRNA gene sequences and the metadata from Lake Baldegg have the accession numbers SAMN18962412–SAMN18962530. The 90 16S rRNA gene sequences and the metadata of Lake Sarnen are deposited under the accession numbers SAMN18978354–SAMN18978443. The 10 metagenomes and the metadata can be found under the accession numbers SAMN19067306–SAMN19067315. The eight metagenomes and the metadata from Lake Sarnen can be found under the accession numbers SAMN19030157–SAMN19030164. All other data, such as metadata and the abundance tables, are available at the institutional data repository of Eawag (ERIC) (https://opendata.eawag.ch/) in accordance with FAIR data sharing principles (https://doi.org/10.25678/0005HX).