Abstract
Expanding the utility of immune-based cancer treatments is a clinical challenge due to tumor-intrinsic factors that suppress the immune response. Here we report the identification of tumoral Ring Finger Protein 2 (RNF2), the core subunit of the Polycomb Repressor Complex 1 (PRC1), as a negative regulator of antitumor immunity in various human cancers, including breast cancer. In syngeneic murine models of triple negative breast cancer, we found that deleting genes encoding PRC1 subunits Rnf2 or BMI1 proto-oncogene, polycomb ring finger (Bmi1), or the downstream effector of Rnf2, Remodeling and Spacing Factor 1 (Rsf1), was sufficient by itself to induce durable tumor rejection and establish immune memory by enhancing infiltration and activation of NK and CD4+ T-cells, but not CD8+ T-cells, into the tumor and enabled their cooperativity. These findings uncover an epigenetic reprogramming of the tumor-immune microenvironment which fosters durable antitumor immunity and memory.
Keywords: RNF2, Polycomb Repressive Complex 1 (PRC1), Cancer Immunity, Immune Evasion, Major Histocompatibility Complex Class II (MHCII)
Tumor growth and progression are normally restrained by cytotoxic innate and adaptive immune cells, but cancer cells have developed mechanisms to evade their immune attack1,2. One of the major means of immune escape is to exclude and/or inactivate cytotoxic immune cells in the tumor, which often results from low tumor immunogenicity1,2. The delicate cellular interaction within the tumor microenvironment (TME), particularly tumor-immune crosstalk, helps to determine tumor response to therapy3-8. Uncovering the factors that regulate this crosstalk may facilitate the understanding of the mechanisms for tumor immune evasion and lead to the development of different immunotherapeutic approaches from current immunotherapies that have faced much resistance9. However, such regulators that shape the tumor-immune landscape and hence tumor immunogenicity remain poorly defined7.
Several epigenetic regulators are reported to shape the TME and modulate the tumor-immune interaction to impact tumor growth and response to therapy8,10-17. The plastic nature of epigenetic changes makes them attractive targets to enhance anti-tumor response. Unfortunately, the outcomes of clinical trials testing inhibitors for histone deacetylase and DNA methyltransferases are not satisfactory18, and the clinical efficacy of other regimens targeting recently identified epigenetic regulators is still unclear.
To identify the tumor-intrinsic factor(s) that may aid immune evasion, we adopted a data driven approach and analyzed published single cell RNA sequencing (RNAseq) datasets from human cancer patients19. This initial data-screen identified multiple epigenetic pathways associated with tumor immune evasion, including Polycomb repressive complex 1 (PRC1), PRC2 and SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 (SMARCB1) (also known as SNF5). Subsequent analysis of a published list including 524 genes encoding epigenetic regulators13 confirmed Ring Finger Protein 2 (RNF2), the enzymatic component of the PRC1 complex, to be one of the top genes significantly correlated to immune cold niches within human tumors. Additional results showed that RNF2 expression was correlated with reduced cytotoxicity of tumor-infiltrating immune cells. These results consistently implicated a general immunosuppressive role of PRC1 complex/RNF2 in human cancers.
Recently, RNF2 is implicated in tumorigenesis20-23 and is a poor prognostic factor in patients with various types of tumors that have high RNF2 expression or RNF2 amplification21,22,24,25. However, the immune-mediated mechanisms by which RNF2 promotes tumorigenesis remain elusive. The identification of PRC1 complex/RNF2 as a suppressor of cancer immunity prompted us to further investigate its role in immune control of tumors. Using syngeneic models of triple negative breast cancer (TNBC) that are inherently refractory to immunotherapy26, we found that deletion of genes encoding PRC1 subunits Rnf2 or Bmi1, or Rsf1 (a downstream executor of Rnf2)27, was sufficient to reject tumors by enhancing the recruitment, activation, and cooperation of NK and CD4+ T-cells, but minimally affected CD8+ T-cells. This coordination between innate (NK cells) and adaptive immunity (CD4+ T-cells) was mediated largely by IFNγ and resulted in a sustained anti-tumor response as well as establishment of immune memory. These effects were not dependent on Rnf2 E3 ligase activity as Rnf2 catalytic-inactive mutant knock-in tumors grew robustly in immunocompetent mice. Mechanistically, ablation of Rnf2 or Rsf1 in in vivo tumors de-repressed multiple tumoral genes related to immune response including those involved in the MHCII-mediated antigen processing/presentation pathway. Subsequent ATACseq analysis of these tumor cells isolated from corresponding tumors implanted in mice demonstrated that immune-related genes, including genes in the MHCII restricted antigen processing/presentation pathway, were significantly more accessible upon deletion of Rnf2/Rsf1, but these genes remained largely inaccessible in Rnf2 catalytic-inactive mutant knock-in tumors. Consistently, these immune-related genes significantly overlapped with Rnf2 bound genes determined by CHIPseq. These findings uncover an epigenetic reprogramming from depleting tumor-expressed RNF2, which reshapes the TME favorably for the immune response and leads to long term immune memory and rejection of tumor.
Results
RNF2 marks the signature of immunologically cold tumors
We first leveraged large human cancer datasets to analyze malignant cell states that facilitate immune evasion. Gene Set Enrichment Analysis (GSEA)28 of a single cell RNAseq dataset interrogating the transcriptome of tumor cells19 revealed significant overlaps of the gene signature of cold tumors with multiple GO terms of PRC1 complex or of its target gene, homeobox A9 (HOXA9) (Extended Data Fig. 1a). We also found that tumoral RNF2 was ranked as the 4th top gene among the total of 248 genes encoding epigenetic writers, erasers and readers associated with the cold tumor gene signature (Extended Data Fig. 1b). This finding was confirmed (RNF2 ranked as the 3rd top gene) by analyzing another list including 524 epigenetic regulators13 (Extended Data Fig. 1c). Additionally, the genes encoding subunits of PRC1 complex, including RNF2, CBX2, CBX4 and CBX8, were among the top-listed epigenetic genes that were inversely correlated with a published tumor-infiltrating immune cell cytotoxicity gene signature29 in multiple human cancers, including breast cancer (BRCA) (Extended Data Fig. 1d-f). Further TCGA analysis revealed that both invasive BRCA, including its deadly subtype, triple negative breast cancer (TNBC), and metastatic BRCA, displayed gene amplification and expressed significantly higher levels of RNF2 and RSF1, which are overexpressed and associated with poor patient prognosis in various other types of cancers30, compared to normal controls at both transcriptional and protein levels (Extended Data Fig. 2a-e). These data are consistent with recent findings of overexpression and amplification of RNF2 in BRCA22,23,31. The overexpression31/amplification of RNF2, CBX2, CBX8 and RSF1 were unfavorable prognostic factors for overall survival in invasive BRCA patients (Extended Data Fig. 2f-i), in addition to the recently reported negative correlation of RNF2 amplification to the survival in patients with the basal subtype of BRCA22.
Deleting Rnf2/Bmi1/Rsf1 induces durable tumor rejection
Based on the above analyses, we hypothesized that ablating tumoral RNF2 may enhance tumor immunogenicity and promote immune activation. To test this hypothesis, we first knocked down the expression of Rnf2 using two sequence-independent short-hairpin RNAs (shRNAs) in the 4T1 cell line, a murine model of TNBC with poor immunogenicity (Extended Data Fig. 3a). Notably, targeting Rnf2 with shRNAs inhibited tumor growth with an almost complete tumor rejection that was correlated with the extent of Rnf2 knockdown (Extended Data Fig. 3b-c). To validate these findings, we next deleted Rnf2 in 4T1 cells using two independent single-guide RNAs (sgRNAs) through CRISPR-Cas9 system (Fig. 1a). Strikingly, tumors with Rnf2 deletion started to shrink at day 5-7 after implantation and were completely cleared within two weeks after injection. Importantly, mice remained tumor free for more than one year (Fig. 1b-g). In contrast, mice injected with the control 4T1 cells had to be euthanized at 40-50 days after injection due to the large tumor burden. We also observed a similar durable tumor rejection when we deleted tumor Rnf2 in another murine TNBC model EMT6 (Fig. 1h-i, Extended Data Fig. 3d). To further validate these results, we established a doxycycline (DOX)-inducible Rnf2 KO 4T1 cell line (Fig. 1j) by introducing a lentiviral-based vector TLCV232, which is an all-in-one dox inducible system and integrates with sgRNA targeting murine Rnf2, and treated mice harboring this inducible 4T1 tumors with DOX 1 day prior to implantation. As shown in Fig. 1k-l, 3 out of 5 DOX-treated mice, exhibited complete rejection of tumor, while 5 out of 5 mice treated with vehicle had robust growth of tumors. Similar anti-tumor activities were observed when DOX was given after the tumors were established to induce Rnf2 KO (Extended Data Fig. 3e-h).
Figure 1. Rnf2/Bmi1 KO induces durable tumor rejection in syngeneic murine models of breast cancer.
a. Immunoblots show the levels of selected proteins in control (Ctrl) and Rnf2 KO (two independent guide RNAs (gRNAs), g1 and g2) 4T1 cells, which represents at least 3 independent experiments.
b-e. The volumes (luminescence intensities) of control or Rnf2 KO 4T1 tumors (two independent gRNAs, g1 (b) and g2 (c)) implanted into the 4th mammary pads of the syngeneic BALB/c mice. d, e. Representative tumor bioluminescence images (BLIs) at indicated days after inoculation. Rnf2 KO tumor phenotypes were repeated at least three times. Mean ± SEM (n = 5 mice [for Ctrl, Rnf2 KO g1 and g2 groups, each mouse harbored one tumor).
f-g. The volumes (f) (luminescence intensities) and representative tumor BLIs (g) at longer time points of panel c. Mean ± SEM. n= 5 mice/group, each mouse harbored one tumor. X, control mice were sacrificed because of the big tumor burdens at the day when images were taken.
h-i. Tumor growth of control and Rnf2 KO EMT6 tumors (two independent gRNAs, g1 (h) and g2 (i)) implanted into the 4th mammary pads of the BALB/c mice. Tumor volumes = length X width2/2. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor).
j-l. j. Immunoblots show the levels of selected proteins in vehicle or doxycycline treated doxycycline inducible Rnf2 KO 4T1 cells, which represents 2 independent experiments. k-l. Tumor growth (k) of doxycycline inducible Rnf2 KO 4T1 tumors in immunocompetent BALB/c mice treated with doxycycline or vehicle (treatment started 1 day before tumor cells were injected). Tumor volumes = length X width2/2. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor). i. Tumor volumes shown in panel k were displayed in individual mice.
m-p. m. Immunoblots show the levels of selected proteins in Ctrl and Bmi1 KO (two independent gRNAs, g1 and g2) 4T1 cells, which represents 2 independent experiments with similar results. n-p. The volumes (luminescence intensities) of control or Bmi1 KO 4T1 tumors (induced by two independent gRNAs, g1 (n) and g2 (o)) implanted into the 4th mammary pads of BALB/c mice. p. Representative tumor BLIs at indicated days after inoculation. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor). Two-way ANOVA with Sidak’s test, in b, c, f, h, i, k, n, o.
PRC1 complex has been biochemically characterized as canonical or non-canonical20,22. Co-immunoprecipitation using anti-Rnf2 antibody coupled with immunoblotting of chromatin bound proteins (chromatin fractionation) isolated from 4T1 cells revealed that Rnf2 interacted with Cbx2, Cbx4, Bmi1, the subunits of canonical PRC1 complex22 (Extended Data Fig. 3i). In contrast, minimal interaction with non-canonical PRC1 complex subunits Pcgf1, Pcgf3, Pcgf522 was observed (Extended Data Fig. 3i). Meanwhile, co-immunoprecipitation using anti-CBX4 antibody pulled-down Rnf2 (Extended Data Fig. 3j). These results suggested that Rnf2 was associated with the canonical PRC1 complex in chromatin. Subsequent targeting of Bmi1 (Fig. 1m) or Rsf127 (Extended Data Fig. 3k) similarly resulted in complete and durable rejection of 4T1 tumors (Fig. 1n-p, Extended Data Fig. 3l-q). These data pointed to the role of the canonical PRC1 complex in mediating tumor growth. Of note, deletion of Rnf2 or Rsf1 did not suppress in vitro cell growth (Extended Data Fig. 3r), and the growth of Rnf2 KO 4T1 tumors in immunodeficient NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice (Extended Data Fig. 3s) was at a significantly greater rate than that of control tumors implanted in immunocompetent BALB/c mice (Extended Data Fig. 3t-u), further supporting the importance of the immune system in the phenotypes we observed.
Rnf2/Rsf1 KO mobilizes both NK and CD4+ T-cells
We next explored the tumor-immune microenvironment in Rnf2 KO and Rsf1 KO 4T1 tumors 7 days post implantation before they were cleared. Compared to control tumors, both Rnf2 KO and Rsf1 KO tumors had significantly higher frequencies of infiltrated NK cells and FoxP3− CD4+ effector T-cells (including CD44+ subsets), but not CD8+ T-cells (Fig. 2a). As Rnf2 KO and Rsf1 KO tumors started to shrink before day 7, these KO tumors were much smaller and had fewer total number of cells compared to control tumors when collected for analysis (day 7) (Extended Data Fig. 3v). Although there were no significant differences for the absolute numbers of NK cells, CD4+ T-cells, and FoxP3−CD4+ effector T-cells (including CD44+ subsets) in Rnf2 KO and Rsf1 KO tumors compared to those of control tumors (Extended Data Fig. 3v), their frequencies relative to tumor-infiltrating CD45+ cells in the KO tumors were significantly higher (Fig. 2a). We also noted that NK cells in KO tumors expressed higher levels of activating receptor NKG2D than NK cells from control tumors (Fig. 2b), indicating their increased activation status in KO tumors. Indeed, both NK and CD4+ T-cells in Rnf2 KO and Rsf1 KO tumors expressed increased levels of interferon γ (IFNγ), tumor necrosis factor α (TNFα) and granzyme B (GZMB) (Fig. 2c-d). The immune profile of Rnf2 KO tumors was also confirmed in another TNBC model EMT6 in immunocompetent mice (Fig. 2e). All these differences were observed only in tumor locally but not the spleens and draining lymph nodes, suggesting that deletion of Rnf2 or Rsf1 induced immune activation mainly in the tumor and had minimal systemic impact. Therefore, it is not surprising that Rnf2 KO tumors did not display “abscopal” like effects33 (Extended Data Fig. 4a-b). These findings suggested that deletion of Rnf2 or Rsf1 in tumor cells preferentially recruited NK and CD4+ T-cell subsets with enhanced effector activity.
Figure 2. Rnf2/Rsf1 KO mobilizes NK and CD4+ T-cells.
a. Frequencies of indicated intratumoral immune cells in Ctrl, Rnf2 KO or Rsf1 KO 4T1 tumors at day 7 post-implantation. n = 10 mice for ctrl and Rsf1 KO for the first 4 panels, n=6 mice for Rsf1 KO in the fifth panel, n = 6 mice for Rnf2 KO. Each mouse harbored one tumor, bars, mean ± SEM. Symbols depict individual mouse.
b. The mean fluorescence intensity (MFI) of NKG2D expression on NK cells in Ctrl, Rnf2 KO or Rsf1 KO 4T1 tumors at day 7 post-implantation. n = 4 mice for Ctrl, n = 5 mice for Rnf2 KO/Rsf1 KO. Each mouse harbored one tumor, bars, mean ± SEM. Symbols depict individual mouse.
c-d. Frequencies of IFNγ+, GzmB+, TNFα+ -NK or -CD4+ T-cells in Ctrl, Rnf2 KO (c) or Rsf1 KO (d) 4T1 tumors at day 7 post-implantation. n = 6/7 mice for Ctrl, n = 7 mice for Rnf2 KO, n = 6/7 mice for Rsf1 KO. Each mouse harbored one tumor, bars, mean ± SEM. Symbols depict individual mouse.
e. Frequencies of indicated intratumoral immune cells in Ctrl, Rnf2 KO (two independent gRNAs, g1 and g2) EMT6 tumors at day 7 post-implantation. n= 5 mice for Ctrl or Rnf2 KO g1, n = 4 mice for Rnf2 KO g2. Each mouse harbored one tumor, bars, mean ± SEM. Symbols depict individual mouse.
f-i. The volumes (f, h) of 4T1 Rnf2 KO tumors (Rnf2 KO g2) (f) or Rsf1 KO tumors (Rsf1 KO g1) (h) in BALB/c mice treated with/without α-asialo GM1 at days 2, 5, 10 post-implantation. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor.). g, i. Representative tumor BLIs at the indicated day.
j-k. The volumes (j) of 4T1 Rnf2 KO tumors (Rnf2 KO g2) in BALB/c mice treated with α-CD4 (GK1.5) or control antibody at days 2, 5 post-implantation. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor.). k. Representative BLIs of tumors at day 45.
l-m. The volumes (l) of control or 4T1 Rnf2 KO tumors (Rnf2 KO g2) in BALB/c mice treated with α-CD8 antibody (Ab) or control Ab at days 2, 5 after tumor implantation. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor.). m. Representative tumor bioluminescence images (BLIs) at indicated day after inoculation.
Unpaired two-tailed Student’s t test in a, b, c, d, e, two-way ANOVA with Sidak’s test in f, h, j, l.
We then performed RNAseq analysis of FACS-sorted NK cells from Rnf2 KO or Rsf1 KO tumors, which revealed their augmented effector activity as evidenced by the increased expression of multiple granzymes and mast cell proteases (Extended Data Fig. 4c-d). To further validate the importance of NK cells in mediating tumor clearance, we depleted NK cells in mice and assessed tumor growth. Although depletion of NK cells did not significantly alter the growth of control tumors (Extended Data Fig. 4e), Rnf2 KO and Rsf1 KO tumors were rescued in mice treated with the NK neutralizing antibody (5 out of 5 mice) (Fig. 2f-i). Importantly, analysis of the human TCGA datasets showed that the high expression of NKG2D that was increased on infiltrated NK cells in Rnf2 KO/Rsf1 KO 4T1 tumors (Fig. 2b) was a significant favorable prognostic factor for invasive BRCA (Extended Data Fig. 4f). Furthermore, RNF2 expression was significantly and negatively correlated with NK cell signature19 in TNBC and multiple other cancer types, including BRCA (Extended Data Fig. 4g-j). Thus, these results established a critical role of tumoral RNF2 in the regulation of NK cell infiltration and activation in the tumor.
Similar to NK cells, we also found increased infiltration of CD4+ T-cells into the Rnf2 KO tumors. Indeed, depletion of CD4+ T-cells using a neutralization antibody rescued the growth of Rnf2 KO tumors (5 out of 5 mice) (Fig. 2j-k). Although depletion of CD4+ T-cells increased the growth rates of control tumors, the extent of this increase did not achieve statistical significance (Extended Data Fig. 4k). CD4 was also a significant predictor of favorable survival for invasive BRCA and was inversely correlated with RNF2 in multiple cancer types, including both BRCA and TNBC (Extended Data Fig. 4l-p).
CD8+ T-cells are considered as the primary cytotoxic immune cells mediating anti-tumor immunity8,34. As expected, depletion of CD8+ T-cells (Extended Data Fig. 4q) in BALB/c mice significantly promoted control tumor growth (Figures 2l-m). However, depletion of CD8+ T-cells failed to rescue the growth of Rnf2 KO tumor in these immunocompetent BALB/c mice (Fig. 2l-m). These results suggested that CD8+ T-cells likely did not contribute significantly to Rnf2 KO induced tumor rejection. Taken together, the above findings demonstrated that NK and CD4+ T-cells but not CD8+ T-cells were essential for the rejection of Rnf2 KO and Rsf1 KO tumors.
NK and CD4+ T-cells in Rnf2 KO tumor activate mutually
The above depletion studies showed that depletion of NK cells in mice harboring Rnf2 KO tumors resulted in significantly reduced infiltration of effector CD4+ T-cells (but not Tregs), diminished CD4+ T-cell activation, and reduced effector activity (IFNγ and TNFα secretion) (Fig. 3a). On the other hand, depletion of CD4+ T-cells attenuated the infiltration, proliferation (Ki67+), activation, and effector function (expressing NKG2D and granzyme B) of NK cells (Fig. 3b). These results suggested that ablating tumoral Rnf2 not only enhanced the effector activity of NK and CD4+ T-cells but also their mutual activation in vivo.
Figure 3. Rnf2 KO/Rsf1 KO induces cooperation between NK and CD4+ T-cells.
a-b. Frequencies of indicated immune cell subsets in 4T1 Rnf2 KO tumors (Rnf2 KO g2) in mice treated with/ without α-asialo GM1 (a) (n = 5 mice/group, each mouse harbored one tumor.) or α-CD4 (GK1.5) (b) (n = 4 mice/group, each mouse harbored one tumor.) at day 7 post-inoculation. Symbols depict individual mouse (bars, mean ± SEM).
c-d. Tumor cells (CD45−) were isolated and enriched from control/Rnf2 KO 4T1 tumors, co-cultured with CD4+ T-cells, or NK cells or both. The anti-NKG2D/control antibody (30 μg/ml) was pre-incubated with NK cells (c) or anti-MHCII/control antibody was added to the co-cultures with CD4+ T-cells (d). Frequencies of IFNγ+CD4+ T-cells or IFNγ+NK cells are shown as mean ± SEM of triplicates. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Data represent one of two independent experiments.
e. Tumor cells (CD45−) were isolated and enriched from control/Rnf2 KO 4T1 tumors, co-cultured with pre-activated CD4+ T-cells, or NK cells or both. The anti-NKG2D/control antibody (30 μg/ml) was pre-incubated with NK cells for 15 min before co-culture. The percent tumor killing is shown as mean ± SEM of triplicates. Each group has 3/4/5/6 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor).
f-g. Tumor cells (CD45−) were isolated and enriched from control or Rnf2 KO 4T1 tumors, and co-cultured with CD4+ T-cells, or NK cells or both. Anti-IFNγ (XMG1.2)/control antibody was added to the co-cultures. Frequencies of IFNγ+CD4+ T-cells (f) or IFNγ+NK cells (g) are shown as mean ± SEM of triplicates. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Data represent one of two independent experiments.
h-k. The volumes of Rnf2 KO tumors (Rnf2 KO g2) (h) or Rsf1 KO tumors (Rsf1 KO g1) (j) in BALB/c mice treated with control/anti-IFNγ antibody (Ab) at day 2, 5 post-implantation. Mean ± SEM (n = 4 mice for control Ab treated Rnf2 KO tumors; n = 5 mice for control Ab treatedRsf1 KO tumors; n = 5 mice for Rnf2 KO or Rsf1 KO tumors treated with anti-IFNγ Ab, each mouse harbored one tumor). i, k, representative tumor BLIs at day 27 after inoculation.
*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (unpaired two-tailed Student’s t test in a, b, c, d; one-way ANOVA with Tukey’s test in e, f, g; two-way ANOVA with Sidak’s test in h, j). The full list of p values can be found in the source data for this figure.
Next, we elucidated the mechanisms by which deletion of tumoral Rnf2 activated CD4+ T-cells or NK cells and enhanced their cooperativity using in vitro co-culture assays. Control or Rnf2 KO 4T1 tumor cells (CD45−) were isolated and enriched from corresponding tumors implanted in mouse mammary pads (at time before KO tumors were cleared), followed by co-culture with NK cells or CD4+ T-cells isolated from spleens of mice bearing control tumors in the presence of IL-2 and soluble CD28 (for CD4+ T-cells). Quantitation of the frequency of NK or CD4+ T-cells expressing IFNγ, a factor indicative of their activation status, revealed that there were more IFNγ+NK or IFNγ+CD4+ T-cells in the co-cultures with Rnf2 KO than control tumor cells (Fig. 3c-d, Left panels, compare group #6 to #2). These increases of IFNγ+NK or IFNγ+CD4+ T-cells were largely abrogated by blockade of either NKG2D or MHCII (Fig. 3c-d, Left panels, compare group #3 to #2 and compare group #7 to #6), which are involved in the direct activation of NK or CD4+ T-cells, respectively’. We also noted that NKG2D ligand (NKG2DL) levels trended higher (p<0.01 for 4T1 Rnf2 KO tumor, p=0.07 for Rsf1 KO 4T1 tumor and Rnf2 KO EMT6 tumors) in Rnf2 KO and Rsf1 KO tumor cells of both 4T1 and EMT6 tumors implanted into the BALB/c mice (Extended Data Fig. 5a). Blockade of NKG2DL in the in vitro co-culture study consistently abolished the activation of NK cells (Extended Data Fig. 5b, Left Panel, compare group #7 to #6). The NKG2D or NKG2DL blocking assays suggested that NKG2D-NKG2DL interaction was involved in NK cell activation.
NKG2D is reported to be expressed on CD4+ T-cells and the engagement of this CD4+ T-cell-expressed NKG2D is capable of stimulating CD4+ T-cells35. However, NKG2D was expressed on infiltrated CD4+ T-cells at similarly low levels among all of groups of control, Rnf2 KO and Rsf1 KO tumors (Extended Data Fig. 5c). Additionally, pre-blocking NKG2D on CD4+ T-cells failed to significantly change the frequencies of IFNγ-expressing CD4+ T-cells or NK cells when co-cultured with either control or Rnf2 KO tumor cells (Extended Data Fig. 5d, Left panel, compare groups #3, 5, 7, 9 to groups #2, 4, 6, 8 respectively, Right panel, compare groups #3, 5 to groups #2, 4 respectively). Therefore, the observed NKG2D and NKG2DL blocking effects in the co-culture system with Rnf2 KO tumors appeared independent of NKG2D expression on CD4+ T-cells.
Notably, the increases in IFNγ+NK or IFNγ+CD4+ T-cells induced by Rnf2 KO tumor cells were significantly enhanced by the presence of the other type of immune cell (CD4+ T-cells or NK cells) in the co-culture, respectively (Fig. 3c-d, Left panels, compare group #8 to groups #4 and #6), suggesting a mutual activation of NK and CD4+ T-cells by Rnf2 KO tumor cells. Interestingly, increased IFNγ+NK cells in the co-culture with CD4+ T-cells and Rnf2 KO tumor cells were largely diminished in the presence of anti-NKG2D neutralization antibody that blocked NK cell activation (Fig. 3c, Left Panel, compare group #9 to #8), or anti-MHCII antibody that blocked CD4+ T-cell activation (Fig. 3d, Right Panel, compare group #5 to #4). Similarly, the proportion of IFNγ+CD4+ T-cells in the co-culture with NK cells and Rnf2 KO tumor cells was decreased by adding anti-MHCII antibody (Fig. 3d, Left Panel, compare group #9 to #8) or anti-NKG2D neutralization antibody (Fig. 3c, Right Panel, compare group #5 to #4). In support of the mutual activation between NK and CD4+ T-cells, blocking NKG2DL also significantly abrogated the activation of CD4+ T-cells (Extended Data Fig. 5b, Right Panel, comparing group #5 to #4), in addition to inhibiting the activation of NK cells in the co-culture of NK, CD4+ T-cells and Rnf2 KO tumor cells (Extended Data Fig. 5b, Left Panel, compare group #9 to #8). The NK/CD4+ T-cell cooperativity in the presence of Rnf2 KO tumor cells was confirmed by using GFP-labeled 4T1 cells (both control and Rnf2 KO) that were enriched by FACS sorting GFP+ populations from corresponding tumors implanted in immunocompetent mice (Extended Data Fig. 5e, both panels, compare group #5 to others). All these results supported a cooperative activation of NK and CD4+ T-cells induced by Rnf2 KO tumor cells.
Finally, we examined whether the cooperation between NK and CD4+ T-cells could directly impact on tumor cells by comparing the in vitro tumoricidal activity of pre-activated NK plus CD4+ T-cells to that of NK or CD4+ T-cells alone. Although CD4+ T-cells displayed tumoricidal activity (Figure 3e, compare column #6 to #1), NK cells appeared to play a dominant role in killing tumor cells (Fig. 3e, compare group #2 to #1, and #7 to #6). This cytolytic function of NK cells was dramatically enhanced by co-cultured Rnf2 KO tumor cells and CD4+ T-cells (Fig. 3e, compare group #9 to groups #2, 4, 7). This enhancement was largely abrogated by NKG2D blockade (Fig. 3e, compare groups #3, 5, 8, 10 to groups #2, 4, 7, 9, respectively), but not by pre-blockade of NKG2D expressed on CD4+ T-cells (Extended Data Fig. 5f, compare groups #3, 7 to groups #4, 8 respectively), suggesting an NKG2D-dependent killing of Rnf2 KO tumor cells by NK cells.
A significant reduction in the expression of MHCI molecules in 4T1 KO tumors was also observed (Extended Data Fig. 5g), which may contribute to increased NK cell activation and cytotoxicity induced by these KO tumors due to “missing” self. This downregulation may also help to explain why CD8+ T-cells were not affected by KO tumors given the critical role of MHCI in activating CD8+ T-cells. Interestingly, the level of MHCI in Rnf2 KO EMT6 tumors was not different from that of control EMT6 tumors (Extended Data Fig. 5h), suggesting that CD8+ T-cells might not also be involved in Rnf2 KO-induced anticancer immunity in the EMT6 model. This discrepancy of MHCI expression could be contextual/tumoral dependent.
IFNγ drives mutual activation of NK-CD4+ T in Rnf2 KO tumor
We next defined potential factor(s) mediating the mutual activation of NK and CD4+ T-cells induced by Rnf2 KO tumors. The increased expression of IFNγ by both NK and CD4+ T-cells in Rnf2 KO or Rsf1 KO tumors (Fig. 2c-d) implicated IFNγ as such a factor, given the positive roles of IFNγ in regulating both NK and CD4+ T-cells36,37. Indeed, neutralization of IFNγ by the addition of anti-IFNγ antibody dramatically reduced IFNγ+CD4+ T-cells or IFNγ+NK cells when NK and CD4+ T-cells were co-cultured with Rnf2 KO tumors (Fig. 3f-g, compare group #9 to #8 in both figures). Pre-blocking IFNγ receptor (IFNγR) on CD4+ T-cells using an anti-IFNγR antibody abrogated both the increases of IFNγ+CD4+ T-cells (Extended Data Fig. 5i, Left panel, compare group #9 to #8) and IFNγ+NK cells (Extended Data Fig. 5i, Right panel, compare group #7 to #6) when these cells were co-cultured with Rnf2 KO tumor cells. Similarly, pre-blocking IFNγR on NK cells abolished the increases of IFNγ+NK cells (Extended Data Fig. 5j, Left panel, compare group #9 to #8) and IFNγ+CD4+ T-cells (Extended Data Fig. 5j, Right panel, compare group #7 to #6) in the co-culture with Rnf2 KO tumor cells.
Because cancer cells also express IFNγR and IFNγ signaling in cancer cells affects cancer immunity4,37, we also examined the potential role of IFNγR on tumor cells. To this end, we deleted IFNγR1 or IFNγR2 or both in Rnf2 KO 4T1 tumor cells to establish double KO (DKO) (IFNγR1 KO-Rnf2 KO or IFNγR2 KO-Rnf2 KO) (Extended Data Fig. 5k-l) or triple KO (TKO) (IFNγR1 KO-IFNγR2 KO-Rnf2 KO) (Extended Data Fig. 5m) 4T1 cells. Interestingly, these DKO and TKO tumors were still efficiently rejected by BALB/c mice (Extended Data Fig. 5n-o), with similar kinetics as Rnf2 KO tumors. TKO tumor cells isolated from these in vivo tumors also activated CD4+ T-cells and NK cells to express IFNγ to similar extents as Rnf2 KO tumors in the co-culture (Extended Data Fig. 5p, compare group #6 to #4 and group #7 to #5 in both panels). These results suggested that Rnf2 KO-induced tumor rejection largely required IFNγ signaling acting in the immune cells.
Given that IFNγ can promote cancer immune surveillance36 and its proposed role in mediating the mutual activation between innate and adaptive immunity38, we reasoned that it might contribute to the enhanced in vivo anti-tumor effects induced by Rnf2 KO or Rsf1 KO. As expected, IFNγ blockade with a neutralization antibody fully rescued growth of both Rnf2 KO and Rsf1 KO 4T1 tumors in immunocompetent mice (5 out of 5 mice in both experiments) (Fig. 3h-k). Although neutralization of IFNγ did promote the growth of control tumors (Extended Data Fig. 5q), the relative ratio of fold change in tumor volumes between anti-IFNγ group and control antibody group (=fold change in mean tumor volumes of group treated with anti-IFNγ antibody divided by fold change in mean tumor volumes of group treated with control antibody) in control tumors is much smaller than that of Rnf2 KO tumors at the end of this experiment (3.73-fold in control tumor vs 46.08-fold in Rnf2 KO tumor) (Extended Data Fig. 5r). Taken together, these results suggested that IFNγ was at least one of the cytokines contributing to tumor rejection of Rnf2 KO tumors via mutual activation of NK and CD4+ T-cells independent of its direct effects on tumor cells.
Activated CD4+ T-cells produce IL-2, which may stimulate NK cells, and IL-2 from NK cells may in turn influence the functional status of CD4+ T-cells39. We also tested whether IL-2 was involved in the mutual activation of NK and CD4+ T-cells induced by Rnf2 KO tumors by including an anti-IL-2 antibody in the in vitro co-culture experiment. However, neutralization of IL-2 did not significantly reduce the increased amount of IFNγ+NK or IFNγ+CD4+ T-cells induced by Rnf2 KO tumor cells (Extended Data Fig. 5s, compare group #5, 7, 9 to group # 4, 6, 8 respectively, in both Left and Right panels). Therefore, IL-2 did not appear to be significantly involved in this crosstalk between NK and CD4+ T-cells in the context of Rnf2 KO.
Rnf2/Rsf1 KO upregulates immune-related genes
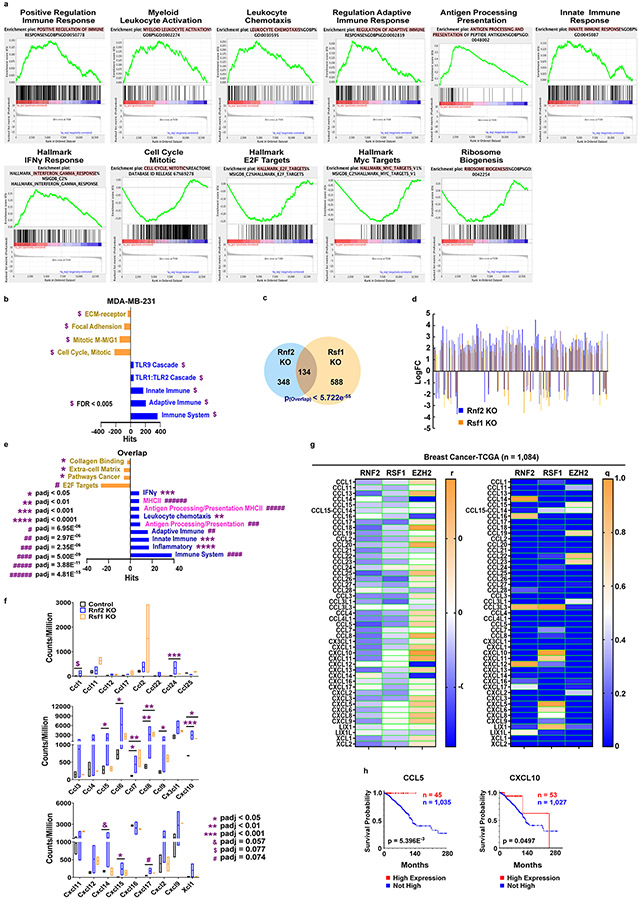
We further explored the molecular mechanism by which deletion of tumoral Rnf2 or Rsf1 elicited both NK and CD4+ T-cell-mediated anticancer immune response. We first profiled the transcriptomes of tumor cells isolated and sorted from Rnf2 KO or Rsf1 KO tumors and compared them to those from control tumors. Notably, multiple immune-related genes were induced in 4T1 tumor cells upon Rnf2 deletion (Extended Data Fig. 6a). Further analysis using NetworkAnalyst40 showed that both Rnf2 KO and Rsf1 KO tumor cells had significantly upregulated genes enriched in both adaptive and innate immune processes, including IFNγ response, antigen processing/presentation, and MHCII (Extended Data Fig. 6b-e). These results suggested a reprogramming of tumor cells upon Rnf2/Rsf1 KO by de-repressing these immune-related genes, leading to increased immune activation. An independent GSEA analysis of the whole transcriptome of tumor cells by clustering the whole gene expression dataset confirmed these findings (Fig. 4a, Extended Data Fig. 6f-g, 7a). Analysis of published transcriptomic data in the human TNBC cell line MDA-MB-23122 also demonstrated significant enrichment in both innate and adaptive immune responses with Rnf2 KO (Extended Data Fig. 7b). Thus, the epigenetic program driven by RNF2 that regulated the immune response appeared to be conserved in human TNBC.
Figure 4. Rnf2/Rsf1 KO tumors upregulate MHCII/CD74.
a. GSEA of DEGs revealed in RNAseq of Rnf2 KO (two groups of cells, Rnf2 KO g1 and Rnf2 KO g2, each group of cells pooled from 25 mice) compared to control 4T1 cells (two groups of cells, each group cells pooled from 25 mice) enriched from tumors in mice (each mouse harbored one tumor) at day-7 post-implantation. The representative enriched gene sets with q values are shown.
b. A floating bars graph (min, max and line at mean) of the counts of MHCII isoforms and Cd74, revealed in RNAseq as in panel a. Rnf2 KO (two groups of cells, each group of cells pooled from 25 mice)/control 4T1 cells (two groups, each group of cells pooled from 25 mice) were enriched from tumors in mice, each mouse harbored one tumor.
c. The MFI of MHCII on CD45− tumor cells and frequencies of CD74+CD45− tumor cells isolated from indicated 4T1 (n = 7/4 mice for Ctrl (7 for Left panel, 4 for Middle panel) , n = 7 mice for Rsf1 KO, n = 6/4 mice for Rnf2 KO (6 for Left panel, 4 for Middle panel)) or EMT6 (n = 5 mice for Ctrl/Rnf2 KO g1, n =4 for Rnf2 KO g2) tumors at day 7 post-implantation. Symbols, individual mouse (bars, mean ± SEM).
d-e. The correlations of RNF2/RSF1/EZH2 to the MHCII isoforms/CD74 (d) or the correlations of MHCII isoforms/CD74 to the survival of invasive breast cancer patients (e), derived from invasive breast cancer TCGA dataset through cBioportal. High expression, mRNA > 2 SD above the mean. n = 1084 (d)/1080 (e) patients total.
f-i. f. Histogram overlays of MHCII expression on Ctrl, Rnf2 KO and Rnf2 KO with additional deletion of MHCII (Rnf2 KO g2-MHCII KO, DKO) 4T1 cells, which represents two independent experiments. g-h. The volumes of indicated tumors in BALB/c mice. g. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor). h. Individual mouse. i. Representative tumor BLIs at day 53.
j-l. Tumor cells (CD45−) were isolated from indicated 4T1 tumors and co-cultured with CD4+ T-cells/NK cells/both (j, k) or pre-activated CD4+ T-cells (l). Frequencies of IFNγ+CD4+ T (j)/IFNγ+NK cells (k)/percent dead tumor cells (CTV+FVD+) (l) are shown as mean ± SEM of triplicates. Each group has 3/4/5 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor).
*p < 0.05 **p < 0.01, ***p < 0.001 and ****p < 0.0001 (padj in b). Two-tailed Wald test in b, unpaired two-tailed Student’s t test in c, l, two-tailed LogRank test in e, two-way ANOVA with Sidak’s test in g, one-way ANOVA with Tukey’s test in j, k. Exact p values for b, j-l are listed in the source data.
We also noted that the differentially expressed genes (DEGs) in Rnf2 KO tumors overlapped significantly with those in Rsf1 KO tumors (Extended Data Fig. 7c), and 98.5% (132 out of 134 DEGs) of these overlapping DEGs exhibited changes of expression in the same direction (Extended Data Fig. 7d). Furthermore, these overlapping DEGs were enriched in immune-activation pathways, including MHCII and antigen processing/presentation (Extended Data Fig. 7e), as well as several genes encoding chemokines (Extended Data Fig. 7f). Consistently, there were inverse associations of expression of RNF2/RSF1, but not EZH2, with the expression of chemokines in human invasive BRCA TCGA dataset (Extended Data Fig. 7g). Among these chemokines, CCL5 and CXCL10, were significant favorable prognostic factors in invasive BRCA (Extended Data Fig. 7h).
Consistent with previous reports41,42, the downregulated DEGs in both Rnf2 or Rsf1 KO tumors were also enriched in cell cycle (Fig. 4a, Extended Data Fig. 6f-g, 7a), Myc and E2F targets, as well as ribosome biogenesis indicative of cold tumors19 (Fig. 4a, Extended Data Fig. 6f-g, 7a), supporting our initial screening of clinical datasets that identified RNF2 as a negative regulator of tumor immunogenicity. In contrast, among those upregulated DEGs in both RNF2 KO and RSF1 KO tumors were multiple gene signatures of the MHCII-mediated antigen processing/presentation pathway (Fig. 4a, Extended Data Fig. 6f). Notably, multiple isoforms of MHCII and Cd74 (also known as HLA class II histocompatibility antigen gamma chain that facilitates MHCII-mediated antigen presentation) that are capable of and essential for activating CD4+ T-cells were significantly upregulated in both Rnf2 KO and Rsf1 KO tumors (Fig. 4b). These increases were confirmed in ex vivo-isolated tumor cells at the protein levels (Fig. 4c). Further analysis of human invasive BRCA TCGA revealed that RNF2 and RSF1 were significantly and negatively correlated with most of the known MHCII isoforms and CD74, both of which were significant favorable prognostic factors for overall survival of invasive BRCA patients (Fig. 4d-e). This is consistent with the previous report that tumoral MHCII is a favorable prognosis factor for TNBC43. Knockout of both H2-Ab1 (encoding the class II antigen A beta 1) and H2-Eb1 (encoding the class II antigen E beta) (Fig. 4f) in Rnf2 KO 4T1 cells at least partially rescued tumor growth in 3 out of 5 mice (Fig. 4g-i). Deletion of H2-Ab1 and H2-Eb1 also failed to increase the frequencies of IFNγ+CD4+ T-cells or IFNγ+ NK cells (Fig. 4j-k, compare group #6 to #4 and group #7 to #5 in both figures), when the double KO tumor cells isolated from in vivo tumors were co-cultured with NK and CD4+ T-cells together. Moreover, these double KO cells displayed significantly reduced capacity to induce CD4+ T-cell cytotoxicity compared to Rnf2 KO and control tumor cells (Fig. 4l). These findings supported the role of tumor-expressed MHCII in promoting anti-tumor responses elicited by tumor-specific deletion of Rnf2.
RNF2 regulates the transcription of multiple immune genes
One of the means by which RNF2 regulates the transcription22,44 is to orchestrate chromatin accessibility22,23, alterations of which in immune related genes are relevant for the sensitivity of cancer cells to immune attack11,15. To investigate whether the chromatin accessibility of immune related genes were altered due to Rnf2 KO, we performed ATACseq analysis of Rnf2 KO (n=2) and control 4T1 tumor cells (n=2) sorted from in vivo tumors by FACS. ATACseq showed that compared to control tumors, Rnf2 KO tumors had thousands of genes with either significantly more or less accessible chromatin sites (Fig. 5a), consistent with the recent reports that PRC1 complex/RNF2 plays a dual role (both opens and closes) in regulating chromatin accessibility and thus transcription22,23,27,45. These more open genes were related to immunity, including antigen presentation via MHCII (Fig. 5b). Other enriched gene signatures were related to development, transcription by RNA polymerase II (RNAPII), and mammary gland development, consistent with the reported functions of PRC1 complex/RNF220,22,23,45. Moreover, the genes with more/less accessible chromatin sites significantly overlapped with up-/down-regulated DEGs in Rnf2 KO tumors revealed by RNAseq (Fig. 5c), including antigen presentation via MHCII (Fig. 5b, 5d).
Figure 5. Rnf2 modulates accessibilities of immune-related genes.
a. The numbers of genes with significantly more/less accessible chromatin sites in Rnf2 KO (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) compared to control 4T1 tumor cells (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor), enriched from the corresponding tumors in mice at day 7 after implantation, were determined by ATACseq.
b. Gene ontology analyses of genes determined in panel a were performed through online tool THE GENE ONTOLOGY RESOURCE79. The representative enriched gene sets with FDR q values are shown (Fisher Test, one-tailed hypergeometric test).
c. The overlap of DEGs (determined by RNAseq in Figure 4a) in Rnf2 KO 4T1 tumors with genes significantly more/less open in Rnf2 KO 4T1 tumors determined in panel a. P values of the overlap are calculated using the web tool Statistical significance of overlap of two groups of genes from Nematode Bioinformatics (SSOTGNB) (http://nemates.org/MA/progs/overlap_stats.html).
d-f. Gene ontology analyses of overlapping genes determined in panel c (d) or in Extended-Figure 8a (e) or in Extended-Figure 8b (f) were performed through online tool THE GENE ONTOLOGY RESOURCE79. The representative enriched gene sets with FDR q values are shown (Fisher Test, one-tailed hypergeometric test).
g. Cut&Run-qPCR analysis of chromatin DNA pulled down by control IgG, or Rnf2 antibody from control or Rnf2 KO 4T1 tumor cell lines. The amplicons are indicated. Data are shown as the percentage of input, n = 3 technical replicates. Fn, Fragment n of the 5’-untranscribed region.
h. The correlations of RNF2 to the levels of its bound immune-related genes that were determined as in Extended-Figure 8f in BRCA patients. The expression levels of these RNF2 bound genes and of RNF2 were extracted from the invasive breast cancer TCGA dataset via cBioportal. The coefficient (yellow) and q value (pink) of each correlation are indicated at the y-axis. The number (n) of RNF2 bound immune-related genes with FDR q values < 0.05, and the percentages of these genes (q < 0.05) among the total RNF2 bound immune-related genes are noted in blue. The blue vertical bars mark the q value at 0.05. Representative immune-related genes with coefficients < 0 or > 0 and q < 0.05 are indicated. Totally 1,070 BRCA patient samples were included.
We next analyzed the profile of Rnf2 target genes in cultured 4T1 cells by CHIPseq. As expected, the Rnf2 target genes significantly overlapped with genes having more/less accessible chromatin sites identified by ATACseq (Extended Data Fig. 8a ) and DEGs determined by RNAseq (Extended Data Fig. 8b) in control and Rnf2 KO tumor cells that were isolated from tumors implanted in immunocompetent mice. Correspondingly, these overlapping genes were also both more accessible/upregulated and less accessible/downregulated upon Rnf2 KO (Fig. 5e-f, Extended Data Fig. 8a-b), consistent with the dual roles (both opens and closes) of Rnf2 in regulating chromatin accessibility and thus transcription22,23,27,45,46. Importantly, the overlapping genes with more open chromatin sites/upregulated were significantly and highly enriched with gene signatures of immunity (Fig. 5e-f). Other enriched gene signatures were development, transcription by RNAPII, and mammary gland development, which are related to the well-established functions20 and the recently reported activity of PRC1 complex/RNF2 in estradiol signaling22,23.
The screenshots of RNAseq and ATACseq of MHCII genes H2-Ab1 and H2-Eb1 showed that they were significantly open and upregulated upon Rnf2 KO (Extended Data Fig. 8c). The occupancy by Rnf2 was determined by Cut&RUN-qPCR (Figure 5g). Further evidence obtained from reanalyzing published datasets of Rnf2/RING1B CHIPseq in mouse embryonic stem cells (mES)45 (Extended Data Fig. 8d) and human triple negative breast cancer cell MDA-MB-23122 (Extended Data Fig. 8g, shown below) all demonstrated the occupancy of Rnf2/RING1B on these MHCII genes. Furthermore, upon Rnf2 KO, H2-Ab1 expression was also increased in mES cells demonstrated by analyzing published RNA microarray dataset45,47 (Extended Data Fig. 8e).
We next examine the potential correlation of RNF2 to immune-related genes in human cancers by analyzing published RNF2 CHIPseq dataset for the human TNBC cell line MDA-MB-23122. RNF2 was shown to bind to multiple genes involved in immune processes (11.48%, 164 genes out of total 1428 RNF2 bound genes), including MHCII genes-HLA-DPA1, and HLA-DPB1 (Extended Data Fig. 8f-g). GSEA analysis confirmed the association of these genes with immunity (Extended Data Fig. 8h). We then analyzed the correlation of the expression of the above RNF2 bound genes identified in MDA-MB-231 cells22 (Extended Data Fig. 8i) to RNF2 expression using the RNAseq dataset of TCGA human invasive BRCA patients. This analysis showed that 34.9% of RNF2 bound immune-related genes (58 out of 166) (Fig. 5h) and 17.2% of all RNF2 bound genes (246 out of 1428) (Extended Data Fig. 8h) were significantly and negatively correlated with RNF2 expression levels, consistent with the putative role of PRC1/RNF2 in repressing transcription of target genes20,48. Notably, these genes included those related to immune activation and high tumor immunogenicity, such as HLA-DPB1, CCL20 and SLAMF7 (Fig. 5h). Consistent with the possible role of PRC1/RNF2 in transcriptional activation22,23,27,45,46, a subset of RNF2 bound genes, including immune-suppressive genes (e.g., FLRT349, IGFR150, FOXP1 and SOCS551), were positively associated with RNF2 (Fig. 5h), supporting the notion that the PRC1 complex/RNF2 mediated immune suppression. Taken together, RNF2 repressed immunogenicity by controlling the accessibility of the immune genes, and knockout of Rnf2 resulted in de-repression of genes that mediated immune activation.
We also performed ATACseq analysis of Rsf1 KO tumor cells (n=2), which revealed that the more/less accessible chromatin genes (Fig. 6a) were significantly overlapped with those of Rnf2 KO tumors (Fig. 6b). Interestingly, these overlapped genes that were more accessible were also enriched in the signatures related to immunity, including antigen presentation through MHCII (Fig. 6c). Moreover, up-/down-regulated DEGs shared by in vivo Rnf2 KO and Rsf1 KO tumors revealed by RNAseq (Extended Data Fig. 7c-e) largely overlapped with shared genes with more/less accessible chromatin sites between Rnf2 KO and Rsf1 KO tumors, respectively (Fig. 6d). These overlapping genes with more accessible chromatin sites and upregulated in Rnf2 KO/Rsf1 KO tumors were also significantly enriched in gene signatures of immunity, including MHCII restricted antigen presentation (Fig. 6e). Taken together, Rsf1 KO and Rnf2 KO tumors displayed enhanced immuno-eliciting characteristics due to the direct changes of gene transcription by Rnf2/Rsf1.
Figure 6. Rsf1 modulates accessibilities of an overlapping group of immune-related genes with Rnf2.
a. The numbers of genes with significantly more/less accessible chromatin sites in Rsf1 KO (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) compared to control 4T1 tumor cells (n = 2, two groups of cells, each group cells pooled from 25 mice, each mouse harboring one tumor), enriched from the corresponding tumors in mice at day 7 after implantation, were determined by ATACseq similar to that described in Figure 5a.
b. Overlap of genes with significantly more/less accessible chromatin sites in Rnf2 KO and Rsf1 KO 4T1 tumors measured by ATACseq determined in panel a and Figure 5a, respectively. P values of the overlap were calculated using the web tool SSOTGNB (http://nemates.org/MA/progs/overlap_stats.html) similar to that described in Figure 5c.
c. Gene ontology analyses of overlapping genes determined in panel b were performed through online tool THE GENE ONTOLOGY RESOURCE, similar to that described in Figure 5b. The representative enriched gene sets with FDR q values are shown (Fisher Test, one-tailed hypergeometric test).
d-e. d. The overlap of shared significantly upregulated (Left)/downregulated (Right) DEGs of Rnf2 KO and Rsf1 KO 4T1 tumors (determined by comparing to control 4T1 tumors shown in Extended-Figure 7c) with genes showing significantly more (Left)/less (Right) accessible chromatin sites in Rnf2 KO and Rsf1 KO 4T1 tumors (determined by ATACseq by comparing to control 4T1 tumors shown in panel a and Figure 5a, respectively). e. Gene ontology analyses of overlapping significantly upregulated and more accessible genes determined in panel d were performed through online tool THE GENE ONTOLOGY RESOURCE. The representative enriched gene sets with FDR q values are shown (Fisher Test, one-tailed hypergeometric test), similar to that in Figure 5b.
Catalytic dead mutant Rnf2 does not elicit immune response
We investigated the role of the E3 ubiquitin ligase activity of Rnf2 in its transcriptional regulation and suppression of tumor immunity. We used CRISPR/Cas9 technology to generate a catalytic dead mutant of Rnf2 (Rnf2I53A/I53A)21,23 knock-in 4T1 cell line (Fig. 7a). This amino acid change (I to A) disrupts the interaction of Rnf2 with the E2 UBCH5C and ablates the ability of Rnf2 to act as an E3 ligase23, without affecting the incorporation of Rnf2 into canonical and variant PRC1 complexes. Although the level of endogenous H2aK119ub1 that is catalyzed by the Rnf2 E3 ligase was largely reduced (Fig. 7b), this tumor cell line was able to grow robustly in syngeneic BALB/c mice at similar rates as control tumors (Fig. 7c-d). Similar immune profiles within the microenvironment of Rnf2I53A/I53A tumor and the tumoral MHCII level were also observed to those of control 4T1 tumors (Fig. 7e). All these data suggested that the E3 ligase activity of Rnf2 may be dispensable for its function in suppressing antitumor immunity that we demonstrated with Rnf2 KO.
Figure 7. The E3 ligase activity of Rnf2 is dispensable for its regulation of anti-tumor immunity.
a. The sequencing result of the I53A knockin mutant. The mutated genomic sequence encoding Alanine is highlighted.
b. Immunoblots show the protein levels of H2AK119ub1 in control and Rnf2I53A/I53A knockin 4T1 tumor cells. The reduction of H2aK119ub1 level has been independently confirmed twice.
c-d. The volumes (c) (luminescence intensities) of control 4T1 tumors or Rnf2I53A/I53A 4T1 tumors implanted into the 4th mammary pads of the syngeneic BALB/c mice and representative tumor bioluminescence images (BLIs) (d) at indicated days after inoculation. Mean ± SEM (n = 5 mice/group, each mouse harbored one tumor, two-way ANOVA with Tukey’s test). The in vivo growing phenotype of Rnf2I53A/I53A 4T1 tumor has been independently confirmed.
e. Frequencies of indicated immune cell subsets in 4T1 control and Rnf2I53A/I53A tumors injected into the 4th mammary fat pads of syngeneic BALB/c mice at day 7 after tumor inoculation. Symbols, individual mouse (bars, mean ± SEM, n = 4 mice for Ctrl, n = 5 mice for Rnf2I53A/I53A, each mouse harboring one tumor, unpaired two-tailed Student’s t test).
f. The numbers of genes with significantly more/less accessible chromatin sites in 4T1 Rnf2I53A/I53A tumor cells (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) compared to 4T1 control tumor cells (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) were determined by ATACseq, similar to that described in Figure 5a. These tumor cells were enriched by FACS from the corresponding tumors implanted into the 4th mammary fat pads of syngeneic BALB/c mice. The tumors were removed at day 7 after implantation.
g. Overlap of genes with more/less accessible chromatin sites in Rnf2 KO (determined in Figure 5a) and Rnf2I53A/I53A (panel f) 4T1 tumors, which were measured by ATACseq and determined by compared to control 4T1 tumors. These tumor cells were enriched by FACS from corresponding tumors implanted in BALB/c mice. Both Rnf2 KO and Rnf2I53A/I53A had two group of tumor cells with each group of cells pooled from 25 mice (each mouse harboring one tumor). P values of the overlap were calculated using the web tool SSOTGNB (http://nemates.org/MA/progs/overlap_stats.html), similar to that described in Figure 5c.
h-j. Gene ontology analyses of overlap (h) and not overlap genes (I, j) determined in panel g were performed through online tool THE GENE ONTOLOGY RESOURCE. The representative enriched gene sets with FDR q values are shown (Fisher Test, one-tailed hypergeometric test), similar to that described in Figure 5b.
We also performed ATACseq analysis of Rnf2I53A/I53A 4T1 tumor cells (n=2) isolated and enriched from tumors implanted into syngeneic BALB/c mice (Fig. 7f). We observed significant overlaps of both more and less accessible genes between Rnf2 KO and Rnf2I53A/I53A 4T1 tumors (Fig. 7g). These overlap genes were enriched in the signatures of development and transcription, consistent with the well-established functions of PRC1 Complex/Rnf2 (Fig. 7h), and in the signature of response to estradiol consistent with the recently reported role of RNF2 in estrogen signaling that appeared to involve the E3 ligase activity of RNF222,23. Although there were overlap genes related to inflammatory responses, cytokine production, chemotaxis, and myeloid leukocyte differentiation (Fig. 7h), the more accessible genes in Rnf2I53A/I53A tumors did not include those of MHCII and MHCII restricted antigen presentation which were enriched in Rnf2 KO tumors (Fig. 7i). Moreover, genes that were more/less accessible in Rnf2I53A/I53A but not in Rnf2 KO tumors were not directly related to immune process (Fig. 7j). These results suggested that transcriptional regulation of MHCII and MHCII restricted antigen presentation by Rnf2 appeared not to require the E3 ligase activity.
Rnf2/Rsf1 KO induces durable anti-tumor memory
The findings that mice implanted with Rnf2 KO or Rsf1 KO tumors remained long-term tumor free post-rejection (Fig. 1f-g, Extended Data Fig. 3p-q) suggested that these mice potentially developed anti-tumor immune memory. To test this hypothesis, we re-challenged mice that initially cleared Rnf2 KO or Rsf1 KO tumors by injecting wildtype (WT) parental 4T1 tumor cells into the contralateral 4th mammary fat pads. Notably, these mice rejected WT tumors completely and efficiently in a short period and remained tumor free for a long period of time (> 300 days) (Fig. 8a, Extended Data Fig. 9a). The similar eradication of re-challenged WT EMT6 tumor was observed in immunocompetent mice that were previously exposed to and cleared Rnf2 KO EMT6 tumors (Fig. 8b-d). In contrast, re-challenging mice that were previously exposed to Rnf2 KO 4T1 tumors with TSA, another murine TNBC cell line that does not share antigens with 4T152, failed to induce rejection (Extended Data Fig. 9b). These results suggested that deletion of Rnf2 or Rsf1 programmed a specific anti-tumor recall response characteristic of immunological memory. Profiling immune cells of these WT 4T1 tumors prior to being cleared in mice that previously rejected Rnf2 KO or Rsf1 KO primary tumors consistently showed significantly more CD4+ T-cells, NK cells and KLRG1+ effector subsets among these cells than those of WT tumors in naïve mice (Fig. 8e, Extended Data Fig. 9c). Moreover, NK cells, but not CD4+ or CD8+ T-cells, expressed higher levels of granzyme B (Fig. 8e, Extended Data Fig. 9c).
Figure 8. Ablation of Rnf2/Rsf1 induces anti-tumor memory response that is dependent of CD4+ T-cells.
a. BALB/c mice were inoculated with Rnf2 KO- (Rnf2 KO g2) (Left) or Rsf1 KO-4T1 tumors (Rsf1 KO g1) (middle) in the left 4th mammary pads. At day 45 after the primary tumors were rejected, wildtype 4T1 tumors were implanted into the right 4th mammary pads of these mice or of naïve mice. n = 5 mice/group, each mouse harboring one tumor. Left/middle, Volumes (luminescence intensities) (mean ± SEM). Right, Representative BLIs of wildtype 4T1 tumors at day 42 after the 2nd challenge.
b-d. BALB/c mice were inoculated with Rnf2 KO EMT6 tumors (Rnf2 KO g1 and g2) in the left 4th mammary pads. At day 45 after the primary tumors were rejected, wildtype EMT6 tumors were implanted into the right 4th mammary pads of these mice or of naïve mice. Tumor volumes (b) were calculated by length X width2/2. Mean ± SEM (n = 5 mice in naïve mice group, n = 4 mice for Rnf2 KO g1 or g2 group, each mouse harboring one tumor). The weights (c) and image (d) of the tumors at the end of the study.
e. Frequencies of indicated immune cells in wildtype 4T1 tumors at day 7 after the 2nd challenge, as in panel a. Symbols, individual mouse (bars, mean ± SEM). n = 3 mice for naïve and Rsf1 KO, n = 4 mice for Rnf2 KO, each mouse harboring one tumor.
f-g. Mice were inoculated with Rnf2 KO 4T1 tumors and re-challenged with wildtype 4T1 tumors on the contralateral, as in panel a. Anti-CD4 (GK1.5) or control antibody was injected at days −1 and +1 relative to tumor implantation. f. Volumes (luminescence intensities) of wildtype 4T1 tumors. Mean ± SEM (n = 4 mice for control antibody, n = 5 mice for GK1.5, each mouse harboring one tumor). g. Representative tumor BLIs at day 48 after the 2nd challenge.
h-i. Working models.
Two-way ANOVA with Sidak’s test in a, b, f; one-way ANOVA with Tukey’s test in c; unpaired two-tailed Student’s t test in e.
We also determined which of these cells could potentially mediate the anti-tumor memory response by depleting each cell type using various neutralizing antibodies. Depletion of NK cells in mice that were previously exposed to Rnf2 KO tumors rescued the tumor growth at the early time points (day 12). However, 4 out of 5 mice eventually cleared the tumors after day 48 (Extended Data Fig. 9d-e). Depletion of CD8+ T-cells partially restored the tumor growth in 2 out of 5 mice (Extended Data Fig. 9f-h). In contrast, depleting CD4+ T-cells fully rescued tumor growth in 5 out of 5 mice (Fig. 8f-g, Extended Data Fig. 9i), suggesting the central role of CD4+ T-cells in the regulation of anti-tumor memory responses induced by ablation of Rnf2.
Discussion
The nature and effector status of tumor-infiltrating immune cells are predictors of clinical outcomes for many solid tumors, while factors expressed by the tumor itself influence immune profiles within the TME. It is critical to define the tumor-immune interactions for therapeutic strategies aiming at inducing optimal anti-tumor immunity3,4. Here we report an epigenetic reprogramming of tumor cells by depleting tumor-expressed Rnf2/Bmi1/Rsf1, which de-repressed tumor genes related to immune responses, including those involved in the MHCII-mediated antigen processing and presentation pathway. Mechanistically, the immune genes regulated by Rnf2/Rsf1 were rendered more accessible upon ablation of Rnf2 or Rsf1 in in vivo tumors. The subsequent remodeling of the TME not only promoted the recruitment of NK and CD4+ T-cells into the tumor, but also reinvigorated their anti-tumor activity. The resultant enhanced co-operative anti-tumor responses by these innate and adaptive immune cells led to immune memory and durable rejection of the tumor (please see the proposed model in Fig. 8h-i).
Our results support that the anticancer activity induced by Rnf2 KO required an intact immune system, consistent with a recent report that human breast cancer cells T47D and MDA-MB-231 with RNF2 knockdown do not exhibit apparent growth defects in immunocompromised mice22. CD8+ T-cells are traditionally thought as the major immune cell type against cancer36. Unexpectedly, knocking out tumor Rnf2/Rsf1 induced a preferential and concomitant activation of NK and CD4+ T-cells, but not CD8+ T-cells. Depletion of NK or CD4+ T-cells but not CD8+ T-cells rescued growth of Rnf2/Rsf1 KO tumors in immunocompetent mice, supporting their anti-tumor activity in our system. Although the anticancer capacity of invariant T-cells are reported recently, the requirement of MHCII for anti-tumor responses upon Rnf2 KO and the rescue of Rnf2 KO tumor growth after the depletion of CD4+ T-cells argue against the possible involvement of invariant T-cells in our models.
NK cells are one of the components of innate immunity mediating tumor killing, and their killing capacity does not require a prior sensitization, which may explain that depletion of NK cells in the immunocompetent host resulted in a quicker rescue of Rnf2 KO tumors than that of deletion of CD4+ T-cells. Most solid tumors present a barrier to NK cell infiltration, and intratumoral NK cells are often dysfunctional and exhausted. The influx and activation of NK cells induced by Rnf2/Rsf1 KO may result from increased immune responses in these tumors that upregulate NKG2D on NK cells, and are further enhanced by CD4+ T-cells.
Deficiencies in the activities and infiltration of CD4+ T-cells within the TME can also promote tumorigenesis36, emphasizing the critical role of these cells in the regulation of anti-cancer immunity. Provided with the CD28-mediated co-stimulatory signals, Rnf2 KO tumor cells could directly activate CD4+ T-cells in vitro that was abrogated by MHCII blockade/deletion, suggesting that deletion of tumor Rnf2 reprogrammed tumor cells to acquire the potential capability of antigen presentation and activate CD4+ T-cells in the absence of professional antigen-presenting cells (APCs). This CD4+ T-cell activation appeared not to be a bystander effect considering the decreased activation of CD4+ T cells and the rescue of in vivo growth of Rnf2 KO tumor induced by additional KO of MHCII.
Although our system could not fully exclude the possible involvement of other cell types within the CD45− population in the activation of CD4+ T-cells and NK/CD4+ T-cells from mice bearing control tumors displayed basal activation in the coculture studies, our additional analysis of NK/CD4+ T-cell activation using sorted GFP+ tumor cells from in vivo tumors further support the capacity of Rnf2 KO tumor cells to activate CD4+ T-cells directly along with the CD28-CD80/CD86 co-stimulation. At least some of these activated CD4+ T-cell subsets are reactive to tumor associated antigens as judged by the presence of CD4-dependent anti-tumor specific memory response, although the identity of antigens presented by MHCII in Rnf2 KO tumors requires further investigation. Given the recently emerging appreciation of MHCII-restricted CD4+ T-cell responses in anti-tumor immunity53, our findings that the additional ablation of MHCII in Rnf2 KO tumor rescued tumor growth in vivo have revealed a previously unrecognized epigenetic mechanism controlling MHCII expression on tumor cells, thus leading to increased CD4+ T-cell activation and the subsequent enhanced anti-tumor immunity.
The importance of CD4+ T-cells in Rnf2/Rsf1 KO-induced anti-tumor effects is further emphasized by the observation that deletion of tumoral Rnf2/Rsf1 facilitated the generation of anti-tumor specific immunological memory that was abrogated by depletion of CD4+ T-cells. This immune memory and prolonged tumor clearance could be leveraged to design more effective immunotherapy, considering the poor induction of sustained anti-tumor responses by current immunotherapies that focus on CD8+ T-cells54. The observed activities of CD4+ T-cells in enhancing NK cell function, in killing tumor cells55 (albeit less potent than NK cells in our in vitro cytotoxicity assays), and in anticancer memory may explain why the extent of CD4+ T-cells correlating to BRCA patient survival appeared greater than that of NK cells.
Perhaps one of the most intriguing findings in our study is the cooperative activation of innate NK and adaptive CD4+ T-cells as a result of deletion of Rnf2/Rsf1 in tumors. Although the interplay of NK and CD4+ T-cells has been implicated in diverse settings56,57, the activation of these two types of immune cells and their mutual dependence as a result of specific targeting of a single tumoral epigenetic regulator constitutes an unconventional mechanism for anti-tumor immunity. Despite that CD4+ T-cells and NK cells may function with different tempos reflected by the different growth rates of the rescued Rnf2 KO tumors upon depletion of NK and CD4+ T-cells in immunocompetent mice, our data support a mutual activation of NK and CD4+ T-cells in the context of Rnf2 KO.
Although the mechanisms by which NK and CD4+ T-cells cooperate against tumors require further analysis, our study has revealed that IFNγ may serve as one of the critical factors mediating their interplay to create an inflamed TME in Rnf2/Rsf1 KO tumors. Further investigation is also required to precisely define how NK and CD4+ T-cells communicate in real time during tumor progression. However, our results may suggest a sequential model in which NK cells as the first line of defense kill Rnf2 KO tumors in a NKG2D-NKG2DL dependent pathway, followed by NK cell activation in the recognition of dead tumor cell-associated danger signals. Activated NK cells produce proinflammatory cytokines, like IFNγ, that function as the signal 3 for CD4+ T-cell activation. The increased expression of IFNγ along with TNFα may also render CD4+ T-cells to acquire the T-helper-1 (Th1) phenotype, in turn further enhancing NK cell function36, including tumor killing activity. The resultant cooperative interactions by these two types of cells efficiently cleared Rnf2/Rsf1 KO tumors and likely prevented them from constant exposure to the tumor cells/antigens and becoming exhausted. Rnf2/Rsf1 KO also induced the generation of memory CD4+ T-cells, which explains the long-term rejection of primary Rnf2/Rsf1 KO tumors. Future characterization of memory CD4+ T-cell subsets and the mechanisms by which IFNγ induces the cooperation between NK and CD4+ T-cells leading to memory cells are expected to inform different strategies from current immunotherapies that have faced much resistance in cancer patients.
Given its plastic and druggable nature, the importance of epigenetic regulation in cancer immune evasion has been appreciated. Epigenetic regulators are reported to mediate silencing of endogenous retroviruses, chemokines, cancer testis antigens, IFN-responsive genes and MHCI to primarily dampen CD8+ T-cell-mediated anti-tumor responses10,11,13-17. It is known that the PRC1 complex and the PRC2 complex regulate transcription cooperatively or independently20. Our results suggest that the mechanistic action of RNF2 in the regulation of tumor immunogenicity beyond its conventional roles in embryonic development and tumorigenesis appears distinct from and independent of the reported immune modulating functions of other epigenetic regulators, including EZH2. For example, analyzing the human invasive BRCA TCGA dataset revealed that the significant and negative correlations of Rnf2/Rsf1 to most of the MHCII genes and chemokines were not observed for EZH2. The observed positive correlations of EZH2 to chemokines in BRCA differ from a published report showing that EZH2 represses tumor production of CXCL9 and CXCL10 in ovarian cancer14. Although EZH2 is conventionally viewed as a transcription silencer, it has been recently reported to act as a contextual transcription activator58. The transition from transcription silencer to activator is thought to be driven by the changes in the posttranslational modifications, such as phosphorylation of the EZH2 protein. It is possible that in breast cancer these modifications may allow EZH2 to act as a transcription activator to express certain chemokines, which does not occur in ovarian cancer.
It remains controversial regarding the requirement of E3 ligase activity of RNF2 for its function21,23,59,60. Here, by employing an Rnf2 catalytic dead mutant knockin 4T1 cell line Rnf2I53A/I53A, we demonstrated that its E3 ligase activity was dispensable for repressing immune gene transcription. RNF2 has been recently shown to directly compact chromatin independent of its E3 ligase activity59. This process is likely mediated by the canonical RNF2/PRC1 complex independent of histone H3K27me3 that is the substrate of PRC2 complex/EZH2/EZH159. Further studies are warranted to explore whether the repression of immune related genes by RNF2 is related to its function to compact chromatin.
We previously identified Rsf1 as a ubiquitinated histone H2A binding protein from the ubiquitinated H2A containing nucleosome. Thus, it is expected to observe in ex vivo-isolated Rnf2 KO and Rsf1 KO tumor cells the overlaps of DEGs revealed by RNAseq and of a large number of genes with more/less accessible chromatin sites identified by ATACseq. It is also anticipated that Rsf1 KO displayed similar phenotype as Rnf2 KO to induce tumor rejection. Interestingly, Rnf2I53A/I53A tumor grew robustly in immunocompetent mice. H2A ubiquitination is catalyzed by PRC1 complex/Rnf2. It is possible that, in addition to ubiquitinated H2A, subunits of PRC1 complex or their interacting proteins recruit Rsf1 to certain immune related genes that are rendered open in Rnf2 KO/Rsf1 KO but not Rnf2I53A/I53A tumors to regulate their transcription. These immune genes may subsequently contribute to the different phenotypes of Rnf2 KO/Rsf1 KO tumors versus Rnf2I53A/I53A tumors. The findings that additional KO of MHCII in Rnf2 KO tumor abrogated the activation of NK, CD4+ T-cells and their cooperativity in vitro as well as rescued Rnf2 KO tumor in vivo may support this notion. Further dissection of the detailed molecular mechanisms by which Rsf1 is recruited to modulate the expression of these immune genes, including MHCII, is warranted.
In summary, we have identified a previously unrecognized epigenetic reprogramming of tumor cells by abrogating tumoral Rnf2/Rsf1. The resultant mobilization and cooperation of NK and CD4+ T-cells, but not CD8+ T-cells, constitutes unappreciated mechanisms regulating the tumor-immune interaction within the TME. These findings may suggest supplemental therapeutic approaches capable of complementing current immunotherapies that focus on boosting CD8+ T-cell responses. Moreover, specific deletion of tumoral Rnf2/Rsf1 induced durable tumor elimination without affecting systemic response may suggest additional advantages of targeting RNF2/RSF1 over other epigenetic regulators. Importantly, the inverse correlation of RNF2/RSF1 to immunogenicity and patient outcomes was observed in multiple human cancer types, indicating the potential general applicability for targeting these molecules in human cancers.
Methods
The research presented in this report complies with all relevant ethical regulations. All animal procedures were performed in compliance with federal laws and institutional guidelines as approved by the UAB’s Animal Care and Use Committee. All reagents or resources are listed in Supplementary Table 1 and 2, if not specified in the text. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Cell Lines
Murine triple negative breast cancer cell lines TSA, luciferase-expressing 4T161 and EMT6 were kindly provided by Drs. Lizhong Wang and Runhua Liu, and Dr. Lalita Shevde-Samant, and Dr. Narendra Wajapeyee (all at University of Alabama at Birmingham), respectively. 4T1 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/Hams F-12 50/50 Mix (Corning, Cat# 10-090-CV) supplemented with 10% Fetal Bovine Serum (FBS) (MilliporeSigma, Cat# 12306C), and 10 units of Penicillin and 10 μg/mL Streptomycin (GE, Cat# SV30010), and 50 μg/mL Geneticin (ThermoFisher Scientific, Cat# 10131035). TSA and EMT6 cells were cultured in DMEM (MilliporeSigma, Cat# D6429) supplemented with 10% FBS, and 10 units of Penicillin and 10 μg/mL Streptomycin. All the cell lines used were confirmed pathogens free, including Mycoplasma, by Charles River Research Animal Diagnostic Services.
Animal Studies
6-8 week old female BALB/c mice and immunodeficient NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice (Charles River Laboratories) were allowed to acclimatize for 1 week prior to experiments, except that age-matched 12-14 week old mice were used for the immune memory studies. In general, mice were randomly assigned to groups for each experiment in an unblinded fashion. The procedure to establish orthotopic 4T1 model was conducted as described previously61. Briefly, 4 x 105 control, or Rnf2 KO, Rsf1 KO 4T1 or doxycycline inducible Rnf2 KO cells suspended in 100 μL HBSS (ThermoFisher Scientific, Cat# 14170112) were injected into the left 4th mammary fat pads of BALB/c or NCG mice using 28G 0.5 mL Insulin Syringes (BD Biosciences, Cat# 329461), and tumors were monitored weekly using the bioluminescence imaging system starting at day 0 after injection. For immunological memory studies, 4 x 105 Rnf2 KO or Rsf1 KO 4T1/EMT6 cells were injected into BALB/c mice as described above. At day 45 when Rnf2 KO or Rsf1 KO 4T1/EMT6 cells were cleared, 4 x 105 wildtype 4T1/EMT6 or TSA cells were injected into the right 4th mammary fat pads. A group of age-matched naïve mice was included as controls. 4T1 tumors were monitored weekly using the bioluminescence imaging system starting at day 0 after injection. EMT6 or TSA tumors were measured using a caliper every 3 days starting at day 4 after injection.
For luciferase bioluminescence assay, animals were intraperitoneally administrated with 2.5 mg/100ul solution of XenoLight D-luciferin (PerkinElmer). The tumor images were captured using an IVIS 100 imaging system (PerkinElmer). In some experiments, antibodies to IFNγ, NK, CD4+ or CD8+ T cells (BioXcell) were intraperitoneally injected into mice at indicated time points, as described in the legends. To induce Rnf2 KO in doxycycline inducible 4T1 cells in BALB/c mice, doxycycline (Dox) (MilliporeSigma, Cat# D9891) were injected intraperitoneally at 50 mg/kg suspended in saline with a concentration as 5 μg/μL as described in a published protocol62. Dox administration either started one day before tumor cells were injected or after the tumors were palpable. No obvious toxicity was noticed. For “abscopal” tumor models, 4 x 105 control and Rnf2 KO 4T1 cells suspended in 100 μL HBSS were simultaneously injected into left and right 4th mammary pads of the BALB/c mice, respectively. These tumors were monitored weekly using the bioluminescence imaging system starting at day 0 after injection. The maximal tumor volume is permitted by UAB’s Animal Care and Use Committee Tumor burden is 10% of the normal body weight which was not exceeded in the animal studies. Once the tumor reaches this limit, the mouse bearing the tumor is sacrificed.
Generation of Rnf2/Rsf1/Bmi1 knockdown/knockout and double/triple knockout cells
The establishment of stable 4T1 cells with shRNAs targeting Rnf2 (MilliporeSigma) was performed as described previously27. Briefly, lentiviral particles were packaged in 293T cells (ATCC) and transduced into 4T1 cells in the presence of Hexadimethrine bromide (8 μg/mL) (MilliporeSigma). The positive transduced cells were selected by puromycin and the knockdown efficiency was verified by immunoblots. The process of knocking out (KO) Rnf2, Rsf1 or Bmi1 in 4T1/EMT6 cells was conducted as described previously27. Oligonucleotides encoding guide RNAs were synthesized from the Integrated DNA Technologies (IDT) and cloned into lentiviral vector lentiCRISPRv2 (Addgene) using BsmBI restriction sites. To knock out MHCII I-A and I-E, two separate oligonucleotides encoding guide RNAs targeting I-A and I-E, synthesized from IDT, were simultaneously cloned into pKLV2.2-h7SKgRNA5(SapI)-hU6gRNA5(BbsI)-PGKpuroBFP-W (Addgene) using SapI and BbsI restriction sites, respectively. Similar strategy was adopted for knocking out IFNγR1 and IFNγR2 in 4T1 Rnf2 KO cells to establish double KO and triple KO cells. After verification of the plasmids by sequencing, Rnf2 KO 4T1 cells were transduced and sorted into single cells that were BFP positive into 96-well plate with one cell in each well by FACS (UAB Flowcytometry Core Facility). The screening of positive clones were performed by Flowcytometry to analyze the MHCII or IFNγR1/IFNγR2 stained single cell populations. To establish doxycycline inducible Rnf2 KO 4T1 cells, oligonucleotides encoding guide RNAs targeting Rnf2 described above were cloned into lentiviral vector TLCV232 (Addgene) that is an all-in-one dox inducible system using BsmBI restriction sites. After verification of the plasmids by sequencing, 4T1 cells were transduced, selected by puromycin and maintained in Tet System Approved FBS (TaKaRa, Cat#631106). The screening of dox-inducible clones were performed in vitro as we described.
Establishment of Rnf2I53A/I53A knockin 4T1cell by CRISPR/Cas9
To establish Rnf2I53A/I53A knockin 4T1 cells, our published protocol has been adopted63. Briefly, 106 cells were washed with Opti-MEM medium (ThermoFisher Scientific) and resuspended in 100 μl Nucleofector Solution (Lonza). 400 pmol of HiFi Cas9 (IDT) was mixed with 500 pmol sgRNA thoroughly. The mixture was incubated in room temperature for 10 minutes. 500 pmol ssODN was then mixed with the Cas9 RNP. The final mixture was added into Nucleofector Solution containing the cells and gently mixed before electroporation (program T-024, Lonza). Half of Electroporated cells were cultured for four days and then the genomic DNA was extracted. Gene targeting efficiency was examined by droplet digital PCR (ddPCR) as following composition: 11 μl 2X ddPCR Supermix for Probes (Bio-Rad), 1 μl mRnf2-f2 (5’-GTGGTTTCACCTAGAAGTCTACACA-3’, 20 μM), 1μl mRnf2-r2 (5’-CCACTTCTAAGGGCTGTGATAA-3’, 20 μM), 1μl mRnf2-I53A-FAM (5’-ATGTGCCCAGCCTGTTTGGATATG-3’, 5 μM), 1 μl R381 (5’-GGCCACCTTGAGCTTCTCA-3’, 20 μM), 1μl R382 (5’-CAAGATGATGACTTGATATAGGCA-3’, 20 μM), 1μl R379-HEX (5’-CACAGGGCAGTAAGGGCAGC-3’, 5 μM), 50 ng genomic DNA, add ddH2O to 22 μl of total volume). 20 μl of the PCR mix was transferred to DG8 Catridges (Bio-rad) and generated droplet with a QX200 Droplet Generator (Bio-rad). The droplets were transferred into a 96-well twin.tec PCR plate (Eppendorf), sealed with PX1 PCR Plate Sealer (Bio-rad) and perform PCR on a C1000 Touch Thermal Cycler (Bio-rad). The thermal cycling program conducted was: Step 1: 95°C 10 min; Step 2: 95°C 30s; Step 3: 56°C 1min; repeat steps 2-3 for 39 times; Step 4: 98°C 10min; Step 5: 8°C hold. After the PCR, the droplets were analyzed using a QX200 Droplet Reader (Bio-rad) with the “absolute quantification” option. Once I53A gene targeting was verified, the other half of the electroporated 4T1 cells were used for isolating single cell colonies with sib-selection as described63.
Cell proliferation
Cell proliferation assays were performed as described previously. Briefly, cells were seeded in 24-well plates and harvested at the indicated times. Cells were washed with PBS, trypsinized, and diluted 1:20 in isotonic saline solution (RICCA Chemical) followed by counting on a Beckman Z1 Coulter particle counter.
Chromatin bound and unbound fractionation
Fractionation of chromatin bound and unbound cell lysates were performed following a modified published protocol64. Briefly, cells cultured in 10 cm dishes were fixed in culturing medium supplemented with 1% formaldehyde (ThermoFisher Scientific, Cat# 28908) for 15 min at room temperature followed by quench in 125 mM Glycine (MilliporeSigma) for 5 min at room temperature. After thoroughly washed in PBS for 3 times, cell were scraped in PBS and centrifuged at 2,000 rpm for 4 min at 4°C. Cell pellets were lysed by Dounce homogenizer on ice (50 times) after being incubated with lysis buffer A (5mM HEPES pH7.9, 0.75mM MgCl2, 5mM KCl, 0.25mM dithiothreitol [DTT]) on ice for 30 min. After centrifuging for 15 min at 3,300g at 4 °C, supernatant (S1) was collected, and the pellet was resuspended in buffer B (buffer A supplemented with 0.34M sucrose, 10% glycerol, 10mM NaF and 1mM Na3VO4) plus 0.5% NP40. Next, samples were incubated on ice for another 30 min and centrifuged for 10 min at 3,300g at 4 °C. Supernatant (S2) was added to (S1) to make the “unbound” fraction. The pellet was the “bound” fraction that was used for subsequent immunoprecipitation.
Immunoprecipitation in chromatin bound proteins
The isolated chromatin bound protein pellets were dissolved in RIPA buffer (50 mM Tris.HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS) by sonication multiple times in Sonic 60 Dismembrator on ice. The supernatants (after centrifuge at 13,300 rpm at 4 °C for 15 min) were diluted 1:1 with RIPA buffer without Sodium Deoxycholate, SDS and precleared with 1 μg IgG (Cell Signaling Technology, Cat# 2729) and 5 μL Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Cat# sc-2003), followed by incubation with agarose-crosslinked Rnf2 antibody (Cell Signaling Technology, Cat# 5694) or control rabbit IgG (Cell Signaling Technology, Cat# 2729) overnight at 4 °C. After extensive wash, twice with wash buffer (provided by Pierce™ Crosslink IP Kit (ThermoFisher Scientific, Cat# 26147)), once with high salt buffer (20 mM Tris.HCl, pH 7.9, 10% Glycerol, 0.5 M KCl, 0.2 mM EDTA), twice with wash buffer, the immunoprecipitation were eluted from agarose with Elution Buffer provided by Pierce™ Crosslink IP Kit. To minimalize the backgrounds, during each washing cycle, the beads were agitated on a vortex mixer. The elutes were immediately neutralized with Tris.HCl, pH 9.4 and boiled before being loaded on SDS-PAGE.
Phenotyping of blood cells
Blood from mouse tail vein was collected into 100 μl PBS/1% heparin solution and mixed immediately. After stained with the fixable viability dye (Biolegend), blood was incubated with an antibody cocktail for 30 min at 4°C followed by red blood cell lysis, wash and flow cytometry analysis.
Tumor cell isolation from mice
To isolate mouse breast cancer cells, each tumor was cut into small pieces (<3mm) and incubated in 3ml dissociation solution (PBS supplemented with 2% FBS, 1mg/ml collagenase/Dispase and 0.5 mg/ml DNase I) for 1 hour at 37°C with gentle shaking. Cell suspension was then passed through a 70 μm cell strainer and washed with DMEM/2% FBS followed by flow cytometry analysis.
Flow Cytometry and Sorting
Single cell suspension was first stained with the fixable viability dye (Biolegend) at 1:1000 in PBS for 10 min. After washed with FACS buffer (PBS/2% FBS), cells were then incubated with Fc block (anti-mouse CD16/32 antibody) at 1:100 for 10 min, followed by incubation with indicated antibody mixtures for 30 min before washing and flow cytometry analysis. For intracellular staining, cells were fixed and permeabilized using the FoxP3 staining buffer set (ThermoFisher Scientific) according to the manufacturer’s protocol, followed by incubation with Fc block and intracellular antibodies for 30 min prior to wash and flow cytometry analysis. All of steps were performed at 4°C. Cells were acquired on a BD LSRII using FACSDiva software (BD Biosciences) and analyzed with FlowJo v10 software (Treestar). For cell sorting, single-cell suspension of tumor cells was labeled with the fixable viability dye and antibodies to the surface antigens (CD45, CD3, NKp46, NK1.1), as described above, followed by sorting on a FACSAria II using FACSDiva software (BD Biosciences). CD45− live cells were collected as tumor cells and CD45+CD3−NK1.1+NKp46+ live cells were NK cells.
Microbead-based cell enrichment
Single-cell suspension isolated from mouse spleens or tumors was enriched for CD4+ T cells, NK cells, or tumor cells using CD4, NK or CD45 microbeads (Miltenyi Biotec), respectively. CD45− cells in the effluent fluid after enrichment were collected as tumor cells.
In vitro tumor killing assay by activated CD4+ T cells or NK cells
Enriched CD4+ T cells were stimulated with plate-coated anti-CD3 antibody (5 μg/ml) and soluble anti-CD28 antibody (2 μg/ml) in the presence of recombinant human IL-2 (hIL-2) (50 U/ml) for 72 h. Enriched NK cells were incubated with hIL-2 (500 U/ml) for 72 h. Enriched tumor cells (Control or Rnf2 KO) were labeled with CellTrace Violet (CTV) dye (5 μM) according to the manufacturer’s protocol (ThermoFisher Scientific) followed by co-culture in triplicates without or with activated CD4+ T cells, NK cells or CD4+ T cells plus NK cells (the ratio of tumor: immune effector cells = 1:5) in the presence of hIL-2 (50 U/ml) for 16 h prior to staining with the fixable viability dye and flow cytometry analysis. In some cases, 30 μg/ml anti-NKG2D or rat IgG1 isotype antibody (Biolegend) was pre-incubated with NK cells or CD4+ T cells for 15 min before addition to the co-culture. Cells positive for both CTV and fixable viability dye are defined as dead cells. The percent tumor killing is calculated using the following equation: [%dead cells (experimental group)-% dead cells (tumor alone)]/% dead cells (tumor alone).
In vitro activation of CD4+ T cells or NK cells by tumor cells
NK and CD4+ T cells were isolated from spleens of mice harboring control tumors and enriched as described above. Tumor cells (Control or Rnf2 KO) were enriched as CD45− cells as described above, followed by co-culture in triplicates with CD4+ T cells, NK cells or CD4+ T cells plus NK cells (tumor: immune = 1:5) for 16 h in the presence of hIL-2 (50 U/ml) and soluble anti-CD28 (2 μg/ml, for groups with CD4+ T cells). Cells were then incubated with the Golgi Plug (BD Biosciences) plus Golgi Stop (BD Biosciences) for 5 h prior to flow cytometry analysis of IFNγ production by CD4+ T cells or NK cells. In some groups, 30 μg/ml anti-NKG2D or rat IgG1 isotype antibody (Biolegend) was pre-incubated with NK cells or CD4+ T cells for 15 min before addition to the co-culture, and 10 μg/ml anti-IFNγR1 or rat IgG2a isotype antibody (Fisher) was pre-incubated with NK cells or CD4+ T cells for 30 min before addition to the co-culture. 10 μg/mL anti-MHCII or rat IgG2b isotype antibody (BioXcell), or 10 μg/mL antibodies to IFNγ, IL-2 or recombinant mouse NKG2D Fc (blocking NKG2DL) were added into some groups. Groups with CD4+ T cells or NK cell alone were also included for analysis.
RNA isolation and sequencing
Total RNA from sorted tumor cells and NK cells was extracted using a QIAshredder kit (QIAGEN) and an RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s protocol. RNA sequencing was performed at Genewiz (South Plainfield, NJ) using Ultra-Low Input RNA-Seq Service. Briefly, mRNA was specifically enriched from total RNA by removing rRNA and hydrolyzed into small pieces. The fragments were then reverse-transcribed into first-strand cDNA using random hexamer primers, followed by second strand synthesis. The short cDNA strands were ligated with 3′- and 5′-adapters for amplification and sequencing using the Illumina® platforms (HiSeq 2 x 150 bp, single index, per lane). The RNA-seq data have been deposited in the NCBI GEO under accession number GSE143352.
RNA-seq data analysis
Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Mus musculus GRCm38 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b65. The STAR aligner is a splice aligner that detects splice junctions and incorporates them to help align the entire read sequences. BAM files were generated as a result of this step. Unique gene hit counts were calculated using feature Counts from the Subread package v.1.5.2. Only unique reads that fell within exon regions were counted. Since a strand-specific library preparation was performed, the reads were strand-specifically counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq266, a comparison of gene expression between the control groups and KO groups of samples was performed. The Wald test was used to generate p-values and log2 fold changes. Genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 were called as differentially expressed genes for each comparison. These differential gene expression data were uploaded into the published web tool NetworkAnalyst to generate the heatmap and the enriched gene sets40.
Independently, Gene Set Enrichment Analysis (GSEA) was performed according to the previous protocols67. Gene set database “Mouse_GOBP_AllPathways_no_GO_iea_March_01_2019_symbol.gmt” was downloaded from http://baderlab.org/GeneSets. It contains only gene sets from GO biological process excluding annotations that have evidence code IEA (inferred from electronic annotation), ND (no biological data available), and RCA (inferred from reviewed computational analysis) and all pathway resources. Genes were ranked by fold change and subject to the analysis under the “GSEAPreranked” module. The method “classic” was used as the enrichment statistic method and pathways with the size 15 to 500 were used for analysis. The enriched pathways were defined as FDR value < 0.05.
ATACseq
Control, Rnf2 KO, Rsf1 KO, or Rnf2I53A/I53A tumor cells were isolated and enriched from in vivo tumors implanted into the 4th mammary fat pads at day 7 after tumor cell injection by FACS described above. Before being subjected to FACS, the cells were treated with 0.5 mg/ml DNase I for 1 hour at 37°C with gentle shaking to get rid of DNA released from dead/damaged cells. The isolated cells were snap frozen in 70% DMEM, 20%FBS and 10%DMSO and delivered on dry ice to Active Motif Epigenetic Services who performed subsequent ATACseq using their standard procedure. Upon received, the cells were then thawed in a 37°C water bath, pelleted, washed with cold PBS, and tagmented as previously described, with some modifications based on a published protocol68 to increase the signal-to-background ratio and remove the contamination of mitochondria genome. Briefly, cell pellets were resuspended in lysis buffer, pelleted, and tagmented using the enzyme and buffer provided in the Nextera Library Prep Kit (Illumina). Tagmented DNA was then purified using the MinElute PCR purification kit (Qiagen), amplified with 10 cycles of PCR, and purified using Agencourt AMPure SPRI beads (Beckman Coulter). Resulting material was quantified using the KAPA Library Quantification Kit for Illumina platforms (KAPA Biosystems), and sequenced with PE42 sequencing on the NextSeq 500 sequencer (Illumina).
ATACseq data analysis
Raw sequencing reads were aligned to the mouse reference genome (mm10) with BWA69. Only reads mapped uniquely to the nuclear genome (very few reads were mapped to mitochondria genome) and aligned with less than 3 mismatches were used for peak calling. Duplicate reads arose from PCR were also removed. Normalization was performed by randomly sampling reads of each sample to the number of reads present in the smallest sample. Peaks were identified by MACS270 with options “--nomodel -g 1.87e9 --bw 200”. BigWig files were generated with BamCoverage from deepTools71 and were used to visualize peaks in the genome browser. To compare peak metrics between samples, overlapping peak intervals were grouped into merged regions. Different accessible sites were identified by DESeq266 with input of read counts for all merged regions.
TCGA dataset analysis
For breast cancer samples, TCGA biolinks was implemented for data analysis72. RNA-seq data for each tumor sample was downloaded from GDC Data Portal (https://portal.gdc.cancer.gov/). Differential gene expression analysis was performed for the cancer samples compared to the normal samples. The program edgeR was employed to normalize gene expressions and perform differential expression analysis73. Quantile filtering was implemented to filtering low expressed genes. Differentially expressed genes (DEGs) were identified with at least two-fold change and FDR<=0.01. For pan-cancer analysis, the standardized, normalized, batch corrected and platform-corrected datasets were downloaded from the TCGA consortium (https://gdc.cancer.gov/about-data/publications/pancanatlas). Copy number variation was downloaded from Broad GDAC firehose (http://gdac.broadinstitute.org/runs/analyses__latest/data/). To analyze the correlation between RNF2 and other genes, linear regression model was used to calculate correlation coefficients and p value. To analyze immune cell cytolytic activity, immune cytolytic score was calculated by averaging log transformed transcript levels of two key cytolytic effectors, granzyme A (GZMA) and perforin (PRF1)29. To analyze ICPi responsive gene signature, the gene expression profile score was calculated by averaging log transformed transcript levels of 18 genes.
For survival analysis, clinical information of each patient was downloaded by TCGA biolinks. The survival time was calculated based on their live status. Survival time was chosen as “days to death” or “days to last follow up” based on patient alive status. “Surv” function in “Survival” package of R was used to build the standard survival object. The variable time records survival time, and the status indicates whether the patient’s death was observed (status = 1) or that survival time was censored (status = 0). A survival curve was computed for the censored data using the Kaplan-Meier method. To investigate whether the gene expression level can influence a patient’s survival, the median gene expression was calculated, and patient samples were grouped into high expression or low expression group based on their expression levels. Then survival curves were computed for the patient with each group. The log rank test was used to define whether patients with different gene expressions have significantly different survival time (p < 0.05).
ChIPSeq and ChIPSeq analysis
Control 4T1 cells were snap frozen and delivered on dry ice to Active Motif Epigenetic Services who performed subsequent CHIPseq using their standard procedure. ChIP-Seq data were aligned to the mouse reference genome (mm10) using BWA69. Samples were normalized by randomly sampling reads to the same number. Peaks were identified with MACS270 with input as control sample and “-g 1.87e9 -f BAM --nomodel -p 1.00e-07 --extsize 200”. Peaks were annotated by comparing to the Ensembl gene annotation (release 99 for GRCm38) with custom Python script. Read depth was computed by DeepTools bamCoverage71 at bin width of 50bp. The bigWig files were displayed in the UCSC genome browser.
ChIP-seq analysis of Published Data
The samples, GSM2862178 (control) and GSM2862179 (RNF2), from the published ChIP-seq data (GSE107176)22 of MDA-MB-231 cell line were downloaded from the NCBI Gene Expression Omnibus (GEO) database. Trimmomatic (v0.36) were implemented to remove low-quality reads. Then the raw reads were aligned to the human genome hg19 using Bowtie2 (v2.3.5.1)74. Duplicate reads were removed using Samblaster (v0.1.24)75.
Peaks were called using MACS2.1 (v2.1.2) with default parameters with 0.05 q value as a cutoff 70. Peaks with fold change > 4 were used for downstream analysis. The resulted bedgraph files were visualized by IGV (v2.6)76. The RNF2 bound genes were uploaded to the published web tool GSEA28 to generate the enriched gene sets by overlapping with Reactome gene sets77.
Cut&Run-qPCR
Cut&Run assay was performed using kit purchased from Cell Signaling Technology (Cat# #86652) according its standard protocol. The pulled-down chromatin DNA was amplified as described previously27.
Public IHC Data
The images of IHC of RNF2 and RSF1 were downloaded from Human Protein Atlas (http://www.proteinatlas.org)78.
Statistical analysis and Reproducibility
Statistical analyses were performed using two-tailed, unpaired Student’s t test, one-way or two-way ANNOVA with GraphPad Prism V8 software, as indicated. A p value of < 0.05 was considered to be statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). No exclusion of data points was used. Sample size was not specifically predetermined, but the number of mice used was consistent with previous experience with similar experiments. The experiments were not randomized and the Investigators were not blinded to allocation during experiments and outcome assessment. Most studies were either exactly independently repeated or confirmed independently with different models/systems.
Data availability
ChIP-seq, RNA–seq and ATAC-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE143352. Previously published ChIP-seq data that were re-analyzed here are available under accession code GSE1057347, GSE3452045 and GSE10717622.
The human pan-cancer and breast cancer data were derived from the TCGA Research Network: https://portal.gdc.cancer.gov/. For pan-cancer analysis, the standardized, normalized, batch corrected and platform-corrected datasets were downloaded from the TCGA consortium (https://gdc.cancer.gov/about-data/publications/pancanatlas). Copy number variation was downloaded from Broad GDAC firehose (http://gdac.broadinstitute.org/runs/analyses__latest/data/). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The software and methods to generate the results have been described in detail in the Method part. No additional code needs to be deposited.
Extended Data
Extended Data Fig. 1. RNF2 marks the signature of immunologically cold tumors and is negatively associated with cytotoxicity of immune cells.
a. GSEA analysis of the gene signature of cold tumor (noted as T cell exclusion) revealed in single cell RNAseq of tumor cells from melanoma patients using oncogenic gene sets (C6). Gene sets related to epigenetic pathways are shown. n = 7,186 single tumor cells from 33 human melanoma tumors (from 31 patients).
b-c. The ranking of 248 (b) or 524 (c) epigenetic regulators based on their median expression levels in relation to the high and low expression of “cold” gene signature, extracted from the dataset in panel a via the Single Cell Portal. Each value at the y-axis for each gene is defined as its median expression level in the single tumor cells with high expression of cold gene signature minus that in the tumor cells with low expression of cold gene signature. These values were used to sort and plot corresponding genes on a two-dimensional plane. The x-axis denotes the rank of these genes. RNF2 is ranked as the top 4th/3rd gene. The number (n) of single tumor cells included in this single cell RNAseq is noted.
d-f. Analysis of genes extracted from TCGA datasets. Heatmaps (Left) with the top parts zoomed in (Right) show correlations between expression levels of 248 epigenetic genes with the immune cell cytotoxicity gene signature, GZMA and PRF1, in diverse human cancers. The expression levels of these genes are extracted from TCGA. The coefficients (r) (d), and p values of these correlations (e) are shown. The zoomed in images of the top parts of the graph are shown in the right. f. The volcano plot shows the correlation of RNF2 to GZMA and PRF1 in human cancers. The y-axis and x-axis denote Log10(FDR) and coefficients (r) of correlations, respectively. The case numbers (n) are noted. n = 11,160 patients.
Extended Data Fig. 2. RNF2/CBX2/CBX8/RSF1 is amplified/overexpressed in human breast cancer patients, and associated with shorter survival time.
a. The box plot shows the expression levels of RNF2 in human normal breast tissue, invasive breast cancer tissues (Left) and triple negative breast cancer (TNBC) tissues (Right). Log2 fold change (Log2Fold) of cancer tissues relative to normal tissues and significance (p) of the correlations are displayed. The lower and upper bound of box plot represent the first quartile (Q1, 25% of data) and third quartile (Q3, 75% of data) of the data. Center line within the box represents the median value (also the second quartile). The whisker marks 1.5*IQR (Inter Quartile Range, the distance between Q1 and Q3) at both side of the box. Dots are the outliers, which are the values outside the whiskers (> Q3 + 1.5*IQR or < Q1 - 1.5*IQR). Normal controls for breast cancer patients: n = 125; breast cancer patients: n = 1,097; normal controls for TNBC patients: n = 112; TNBC patients: n = 123.
b. The percentages of primary invasive breast cancer (BRCA), primary TNBC and metastatic breast cancer patients with RNF2 amplification. The numbers of patients (n) are indicated.
c. Images with patient IDs from The Human Protein Atlas (http://www.proteinatlas.org) display the expression of RNF2 protein in normal breast tissues and human breast cancer tissues by immunohistochemistry staining. Scale bars: 100 (Upper)/50 (Bottom) μm. The website link for each image is provided in the Resources Table.
d. The percentages of invasive breast cancer patients (n = 1,981) with the gain and amplification of RSF1, obtained from the METABRIC dataset via cBioPortal.
e. Images with patient IDs from The Human Protein Atlas (http://www.proteinatlas.org) display the expression of RSF1 protein in normal breast tissues and human breast cancer tissues by immunohistochemistry staining. Scale bars: 100 (Upper)/50 (Bottom) μm. The website link for each image is provided in the Resources Table.
f. The percentages of breast cancer patients with the amplification of CBX2 and CBX8 obtained from METABRIC dataset via cBioPortal. The numbers of patients (n) are indicated.
g-i. The correlations of amplification of RNF2 (g) or Rsf1 (h) or CBX2/CBX8 (i) to the survival of invasive breast cancer patients (PanCancer Atlas) (g), or breast cancer patients (METABRIC dataset) (h, i) obtained from cBioPortal. P values are generated using two-tailed LogRank Test. The numbers of BRCA patients (n) with (red color) or without (blue color) amplification/amplification + gain of indicated gene are shown.
Extended Data Fig. 3. Targeting Rnf2/Rsf1 results in tumor rejection in syngeneic murine breast cancer models.
a-c. a. Immunoblots of indicated proteins in 4T1 cells transduced with the scrambled shRNA or two independent shRNAs targeting Rnf2 gene, which represents two independent experiments with similar results. The knockdown efficiency has been independently confirmed. b-c. The volumes (b) and representative BLIs at day 28 after inoculation (c) of tumors transduced with shRNAs as described in panel a (mean ± SEM, n = 5 mice/group, each mouse harboring one tumor) in mice.
d. Immunoblots of indicated proteins in Ctrl and Rnf2 KO (two independent gRNAs, g1 and g2) EMT6 cells. The knockout efficiency was independently confirmed at least twice.
e-h. e. Treating regimen. f. Tumor volumes of doxycycline inducible Rnf2 KO tumors at the indicated days after implantation in BALB/c mice. Tumor volumes were calculated by length X width2/2. g, h. The weights (g) and images (h) of the tumors at the end of the study. n = 4 mice for - Dox, n = 5 mice for + Dox, each mouse harboring one tumor.
i-j. Immunoblots show the selective subunits of PRC1 complex interacting with Rnf2 that was immunoprecipitated from chromatin of 4T1 cells (i) or Rnf2 interacting with Cbx4 that was immunoprecipitated from whole cell lysates of 4T1 cells (j). Ft, Flowthrough, E, Elute, which represents two independent experiments with similar results.
k. Immunoblots of Rsf1/β-actin in Ctrl and Rsf1 KO (two independent gRNAs, g1 and g2) 4T1 cells. The knockout efficiency was independently confirmed at least twice.
l-q. l, m. The tumor volumes (mean ± SEM) in mice implanted with control or Rsf1 KO 4T1 tumor cells (guide 1 (n = 4 mice/group) (l) or guide 2 (n = 5 mice in control group, n = 4 mice in Rsf1 KO group) (each mouse harboring one tumor). (m). n-q. Representative tumor BLIs of tumors at day 22/47 after inoculation (n, o) or at longer time points (p, q). X, mice were sacrificed because of the big tumor burdens at the day when images were taken.
r. The proliferation (mean of quadruplicates, n = 4 for technical replicates), (fold changes in cell numbers), of control or Rnf2 KO (g1 and g2) or Rsf1 KO (g1 and g2) 4T1 cells in vitro. Cell numbers at day 0 are set as 1.
s-u. s. The volumes of control or Rnf2 KO 4T1 tumors (g1 and g2) implanted into the 4th mammary pads of the immuno-compromised NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor). t-u. The volumes of control tumors or Rnf2 KO 4T1 tumors ((g1) (t) or (g2) (u)) implanted into the immuno-compromised NCG mice (as in panel s) or immune competent BALB/c mice (as in Fig. 1b-c). Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor).
v. Numbers of total cells and of intratumoral immune cells of 4T1 tumors (Ctrl, Rnf2 KO and Rsf1 KO) displayed in Fig. 2a-b. n = 6 mice for Ctrl /Rnf2 KO; n = 5 mice for Rsf1 KO, each mouse harboring one tumor. Symbols, individual mouse (bars, mean ± SEM).
Two-way ANOVA with Tukey’s test in b, s, t, u; unpaired two tailed Student’s t test in f, g, r; two-way ANOVA with Sidak’s test in l, m.
Extended Data Fig. 4. Rnf2/Rsf1 is negatively correlated to infiltration/activation of NK and CD4+ T-cells.
a-b. BALB/c mice were inoculated with control 4T1 tumor into the left 4th mammary fat pads or simultaneously with control tumor into the left 4th mammary fat pads and Rnf2 KO 4T1 tumors (Rnf2 KO g2) into the right 4th mammary pads. a. Tumor volumes (mean ± SEM, n = 5 mice in the group of 4T1 tumor injected into the left 4th mammary fat pads (Control Tumor), each mouse harboring one tumor; n = 4 mice in the group of simultaneously injected with control tumor into the left 4th mammary fat pads (Control Tumor-Abscopal) and Rnf2 KO 4T1 tumors into the right 4th mammary pads (Rnf2 KO-Abscopal)), each mouse harboring two tumors: one is control tumor injected in the left mammary fat pad, the other one is Rnf2 KO tumor injected into the right mammary fat pad. p < 0.0001 (control tumors injected alone (Control Tumor) vs. Rnf2 KO tumors injected into the right mammary pads of the mice in which control tumors were injected simultaneously into the left mammary pads (Rnf2 KO-Abscopal)); p<0.0001 (control tumor injected into the left mammary pads of the mice in which Rnf2 KO tumors were injected simultaneously into the right mammary pads (Control Tumor-Abscopal) vs. Rnf2 KO tumors injected into the right mammary pads of the mice in which control tumors were injected simultaneously into the left mammary pads (Rnf2 KO-Abscopal)); not statistically significant (control tumors injected alone (Control Tumor) vs. Control tumor injected into the left mammary pads of the mice in which Rnf2 KO tumors were injected simultaneously into the right mammary pads (Control Tumor-Abscopal)) (two-way ANOVA with Tukey’s test). b. Representative BLIs of tumors at indicated days.
c-d. The counts of genes encoding granzymes (GZMB, GZMC, GZMD, GZME, GZMF) and mast cell proteases (MCPT1, MCP2, MCPT8), revealed in RNAseq of FACS-sorted NK cells from control 4T1 tumors compared to those in Rnf2 KO tumors (Upper) or Rsf1 KO tumors (Bottom), at day 7 after implantation. The RNAseq was performed in control tumors (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor), Rnf2 KO tumors (n = 2, two groups of cells, one group of cells pooled from 25 mice, each mouse harboring one tumor of Rnf2 KO g1, the other group of cells pooled from another 25 mice, each mouse harboring one tumor of Rnf2 KO g2), Rsf1 KO tumors (n = 2, two groups of cells, one group of cells pooled from 25 mice, each mouse harboring one tumor of Rsf1 KO g1, the other group of cells pooled from another 25 mice, each mouse harboring one tumor of Rsf1 KOg2). The full list of p values can be found in the source data for this figure (two-tailed Wald test).
e. The volumes of 4T1 control tumors in BALB/c mice treated with/without α-asialo GM1 at days 2, 5, 10 post-implantation. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor. Two-way ANOVA with Sidak’s test.
f. The correlation of expression of NKG2D with the overall survival of invasive breast cancer patients (TCGA dataset) via cBioPortal. High, expression level > 1.5 SD above the mean. P value is generated using two-tailed LogRank test. Numbers of BRCA patients (n) with (red color)/without (blue color) high expression of NKG2D are indicated.
g. The correlation of RNF2 expression to the published NK cell signature in TNBC was analyzed by Pearson correlation (two-tailed, no adjustment for multiple comparisons because of one correlation test for a gene pair). The expression levels of these genes are extracted from the TCGA dataset. The values of the coefficients (r) and significance (p) are indicated. Shaded area, 95% confidential interval. n = TNBC patient numbers.
h-j. The expression levels of RNF2 and indicated genes from the published NK cell gene signature are extracted from TCGA RNAseq datasets of various cancer types (n = 11,160 patients). The correlations of RNF2 expression to the levels of these genes as a whole (h), or individually (r in i; p values in j) are shown.
k. The volumes of control 4T1 tumors in BALB/c mice treated with α-CD4 (GK1.5) or its isotype control antibody at days 2, 5 post-implantation. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor.). Two-way ANOVA with Sidak’s test.
l. The correlation of RNF2 to CD4 expression level in TNBC was analyzed by Pearson correlation (two-tailed, no adjustment for multiple comparisons because of one correlation test for a gene pair). The expression levels of these genes are extracted from the TCGA dataset. The values of the coefficients (r) and significance (p) of the correlations are displayed. Shaded area, 95% confidential interval. n = TNBC patient numbers.
m.The correlation of CD4 expression with the overall survival of invasive breast cancer patients (TCGA dataset) via cBioPortal. P value is generated using LogRank test. High, the expression level > 2 SD above the mean. P value is generated using two-tailed LogRank test. Numbers of BRCA patients (n) with (red color)/without (blue color) high expression of CD4 are indicated.
n-p. The expression levels of RNF2 and genes encoding T cell markers and MHCII are extracted from TCGA RNAseq datasets of various cancer types (n = 11,160 patients). The correlations of RNF2 expression to the levels of these genes as a whole (n) or individually (r in o; p values in p) are shown.
q. Frequencies of peripheral blood CD8+ T-cells in mice bearing control or Rnf2 KO tumors at day 10 after injection of anti-CD8 (2.43) or control antibody. Symbols depict individual mouse (bars, mean ± SEM). n = 5 mice/group, each mouse harboring one tumor. One-way ANOVA with Tukey’s test.
Extended Data Fig. 5. Tumoral NKG2DL and NK/CD4+ T cell-expressed IFNγR are required for the activation of these immune cells by Rnf2 KO tumors.
a. The frequencies of NKG2DL+CD45− tumor cells isolated from Ctrl/Rnf2 KO/Rsf1 KO 4T1 tumors (Left, n = 5 mice for Ctrl, n = 4 mice for Rnf2 KO and Rsf1 KO, each mouse harboring one tumor) or EMT6 tumors (Right, n = 5 mice for Ctrl and Rnf2 KO g1, n = 4 mice for Rnf2 KO g2, each mouse harboring one tumor) at day 7 post-implantation. Symbols, individual mouse (bars, mean ± SEM).
b. Tumor cells (CD45−) were isolated and enriched from indicated tumors, co-cultured with NK/CD4+ T-cells/both. The coculture setup is similar to that described in Fig. 3c. Anti-NKG2DL/control antibody was added into the co-culture. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+NK cells (Left) or IFNγ+CD4+ T-cells (Right) are shown as mean ± SEM of triplicates.
c. Frequencies of intratumoral NKG2D+ cells of CD4+ T effector cells of control, Rnf2 KO and Rsf1 KO 4T1 tumors at day 7 post-implantation. Symbols, individual mouse (bars, mean ± SEM). n = 5 mice/group, each mouse harboring one tumor.
d. In vitro co-culture experiment was set up as that shown in panel b, except the addition of anti-NKG2DL antibody. Instead, CD4+ T-cells were pre-incubated with anti-NKG2D antibody for 30 min before being added into the co-culture. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+CD4+ T-cells (Left) or IFNγ+NK cells (Right) are shown as mean ± SEM of triplicates.
e. Tumor cells (GFP+) were isolated and enriched by FACS for GFP positive populations from indicated 4T1 tumors in mice. They were co-cultured with NK/CD4+ T cells/both. The coculture setup is similar to that described in Fig. 3c. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+NK cells (Left) or IFNγ+CD4+ T cells (Right) are shown as mean ± SEM of triplicates.
f. Tumor cells (CD45−) were enriched from indicated 4T1 tumors in mice and co-cultured with pre-activated NK/CD4+ T-cells/both. CD4+ T-cells were pre-incubated with anti-NKG2D antibody for 30 min before being added into the co-culture. The coculture setup is similar to that described in Fig. 3e. Each group has 3/4/5/6 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). The percent tumor killing is shown as mean ± SEM of triplicates.
g-h. The frequencies of MHCI+CD45− tumor cells and/or the MFI of MHCI expression on CD45− tumor cells isolated from control (Ctrl)/Rnf2 KO cells of 4T1 tumors (g, n = 4 mice for Ctrl, n = 5 mice for Rnf2 KO, each mouse harboring one tumor) or EMT6 tumors (h, n = 5/group, each mouse harboring one tumor) at day 7 post-implantation. Symbols depict individual mouse (bars, mean ± SEM).
i-j. Tumor cells (CD45−) were enriched from indicated 4T1 tumors in mice, co-cultured with NK/CD4+ T-cells/both. The coculture setup is similar to that described in Fig. 3c. CD4+ T-cells (i) or NK cells (j) were pre-incubated with anti-IFNγ receptor (IFNγR) antibody (GR-20) for 30 min before being added into the co-culture. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+CD4+ T-cells or IFNγ+NK cells are shown as mean ± SEM of triplicates.
k-m. Expression levels of IFNγR1 and IFNγR2 in Rnf2 KO plus IFNγR1 KO (DKO1) (k), Rnf2 KO plus IFNγR2 KO (DKO2) (l), and Rnf2 KO plus IFNγR1 KO and IFNγR2 KO (TKO) (m) 4T1 cells, measured by Flowcytometry (Histogram), which represents two independent experiments.
n-o. n. The volumes (n) and representative BLIs at day 49 (o) of indicated 4T1 tumors in BALB/c mice. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor). DKO 1: Rnf2 KO + IFNγR1 KO; DKO 2: Rnf2 KO + IFNγR2 KO. TKO: Rnf2 KO + IFNγR1 KO + IFNγR2 KO. ****, control tumor vs. Rnf2 KO + IFNγR1 KO tumor; ****, control tumor vs. Rnf2 KO + IFNγR2 KO tumor; ****, control tumor vs. Rnf2 KO + IFNγR1 KO + IFNγR2 KO tumor; ****, control tumor vs. Rnf2 KO tumor.
p. Tumor cells (CD45−) were enriched from indicated tumors in mice, co-cultured with NK/CD4+ T-cells/both. The coculture setup is similar to that described in Fig. 3c. Each group has 3 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+CD4+ T-cells (Left) or IFNγ+NK cells (Right) are shown as mean ± SEM of triplicates.
q-r. q. The volumes of control 4T1 tumors in BALB/c mice treated with control/anti-IFNγ antibody at day 2, 5 post-implantation. Mean ± SEM (n = 5 mice/group, each mouse harboring one tumor).
r. The relative ratio of the fold changes in the tumor volumes (=Average of Fold Changes in tumor volumes of the group of anti-IFNγ divided by Average of Fold Changes in tumor volumes of the group of Control antibody) in control 4T1 tumors (panel q) or Rnf2 KO tumors (shown in Fig. 3h) at the end of the study. n = 5 mice for Ctrl Tumor treated with Ctrl Antibody; n = 4 mice for Ctrl Tumor treated with IFNγ Antibody; n = 4 mice for Rnf2 KO treated with Ctrl Antibody; n = 5 mice for Rnf2 KO treated with IFNγ Antibody. Each mouse harbored one tumor.
s. Tumor cells (CD45−) were enriched from indicated 4T1 tumors in mice, co-cultured with NK/CD4+ T-cells/both. The coculture setup is similar to that described in Fig. 3c. Control/anti-IL-2 antibody (JES6-1A12) was supplemented in the co-culture. Each group has 3/4 replicates of co-culture. Tumor cells (Ctrl/Rnf2 KO) in each replicate of co-culture were pooled from 2-3 tumors from 2-3 mice (each mouse harboring one tumor). Frequencies of IFNγ+CD4+ T-cells (Left) or IFNγ+NK cells (Right) are shown as mean ± SEM of triplicates.
*p<0.05, **p<0.01, ***p<0.001,****p<0.0001, n.s., not statistically significant (unpaired two-tailed Student’s t test in a, b, c, f, g, h, p; one-way ANOVA with Tukey’s test in d, e, i, j, s; two-way ANOVA with Tukey’s test in n; two-way ANOVA with Sidak’s test in q). The full list of p values can be found in the source data for this figure.
Extended Data Fig. 6. Rnf2/Rsf1 KO tumors upregulate genes related to immunity.
a. Log2FC (fold change) (Left) and adjusted p values (padj) (Right) of the upregulated immune cell markers expressed in Rnf2 KO 4T1 tumor cells (n = 2, two groups of tumor cells, one group of cells was from Rnf2 KO g1, the other group was from Rnf2 KO g2, each group of cells were pooled from 25 mice, each mouse harboring one tumor) compared to those in control tumor cells (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) isolated and sorted from in vivo tumors at day 7 post-implantation followed by RNAseq analysis, as in Fig. 4a.
b-e. Analysis of differentially expressed genes revealed in RNAseq of FACS-sorted Rnf2 KO 4T1 tumor cells (as described in panel a) (n = 2, two groups of cells, one group of cells was from Rnf2 KO g1, the other group was from Rnf2 KO g2, each group of cells pooled from 25 mice) (b), or Rsf1 KO 4T1 tumor cells (similar to that described in panel a) (n = 2, two groups of cells, one group of cells was from Rsf1 KO g1, the other group was from Rsf1 KO g2, each group of cells pooled from 25 mice) (c) compared to control 4T1 tumors cells (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) isolated from in vivo tumors at day 7 post-tumor implantation, using Network Analyst. The representative enriched gene sets with FDR q values are shown (b, c). Heatmaps display these DEGs and representative genes related to immunity are noted (d, e).
f. GSEA analysis of differentially expressed genes (DEGs) revealed in RNAseq of FACS-sorted Rsf1 KO 4T1 tumor cells (as described in panel c) (n = 2, Rsf1 KO g1 and g2, two groups of cells, each group of cells pooled from 25 mice) compared to control 4T1 tumors cells (n = 2, two groups of cells, each group of cells pooled from 25 mice)isolated from corresponding tumors implanted in mice at day 7 post-tumor implantation. The representative enriched gene sets with FDR q values are shown.
g. Representative GSEA Enrichment plots (score curves) of DEGs determined by RNAseq in Rnf2 KO compared to control tumors. GSEA analysis of DEGs revealed in RNAseq of 4T1 tumors is performed as in Fig. 4a. The representative Enrichment plots of positively or negatively regulated genes in Rnf2 KO tumor cells compared to control tumor cells are shown.
Extended Data Fig. 7. Rsf1 regulates expression of an overlapping group of immune-related genes with Rnf2.
a. Representative GSEA Enrichment plots (score curves) of DEGs in Rsf1 KO compared to control tumors. GSEA analysis of DEGs revealed in RNAseq of 4T1 tumors is performed as in Extended-Fig. 6f. The representative Enrichment plots are shown.
b. Analysis of DEGs revealed in a published RNAseq of control siRNA- compared to RNF2 siRNA-treated human TNBC cell line MDA-MB-231 (log2FC≥ 1 and log2FC≤ −1) using Network Analyst. The representative enriched gene sets with FDR q values are shown.
c. The overlap of DEGs in Rnf2 KO and Rsf1 KO 4T1 tumor cells compared to control 4T1 tumors cells, which were isolated by FACS from in vivo tumors. DEGs are determined and described in Fig. 4a and Extended-Fig. 6f. P value of overlap is calculated using web tool SSOTGNB (http://nemates.org/MA/progs/overlap_stats.html), similar to that described in Fig. 5c.
d. The log2FC (Fold Change) of overlapping DEGs in Rnf2 KO tumors and Rsf1 KO tumors. DEGs are determined and described in Fig. 4a and Extended-Fig. 6f.
e. g:Profiler analysis of the 134 overlapped DEGs in panel c. The adjust p value (padj) is indicated (one-tailed hypergeometric test with adjustment for multiple comparisons).
f. A floating bars graph (min, max and line at mean) depicts the counts of genes encoding chemokines revealed in RNAseq of FACS-sorted tumors cells, as in Fig. 4a and Extended Fig. 6f. These Rnf2 KO (n = 2, two groups of cells, one group was from Rnf2 KO g1, the other group was from Rnf2 KO g2, each group of cells pooled from 25 mice, each mouse harboring one tumor), Rsf1 KO (n = 2, two groups of cells, one group was from Rsf1 KO g1, the other group was from Rsf1 KO g2, each group of cells pooled from 25 mice, each mouse harboring one tumor) and control (n = 2, two groups of cells, each group of cells pooled from 25 mice, each mouse harboring one tumor) 4T1 tumors cells were isolated by FACS from corresponding in vivo tumors. The adjust p values (padj) are determined by the two-tailed Wald test and can be found in the source data for this figure.
g. The correlations of expressions of RNF2, RSF1 or EZH2 to the levels of human chemokine genes were analyzed using the RNAseq data extracted from invasive breast cancer TCGA dataset. Heatmaps show the correlation coefficients (Left) and q values of each correlation (Right). n = 1,084 BRCA patients.
h. The correlations of the chemokine gene CCL5 or CXCL10 to the overall survival of invasive breast cancer patients, extracted from the TCGA dataset via cBioPortal. High expression is defined as the expression levels greater than 2 SD above the mean. P values are generated using two-tailed LogRank test. Numbers (n) of BRCA patients with (red color)/without (blue color) high expression of CCL5 (left)/CXCL10 (right) are indicated.
Extended Data Fig. 8. Rnf2 binds to immune related genes in mouse and human breast cancer cells.
a-b. The overlap of Rnf2 target genes in cultured 4T1 cells determined by Rnf2 CHIPseq with genes displaying significantly more/less accessible chromatin sites in Rnf2 KO 4T1 tumors determined by ATACseq as in Fig. 5a (a) or with DEGs (determined by RNAseq shown in Fig. 4a) in Rnf2 KO 4T1 tumors (compared to control 4T1 tumors) (b). P values of the overlap are calculated using web tool SSOTGNB (http://nemates.org/MA/progs/overlap_stats.html), similar to that described in Fig. 5c.
c-d. Screenshots of genes H2-Ab1 and H2-Eb1 in both control and Rnf2 KO 4T1 tumor cells obtained from RNAseq (determined and described in Fig. 4a) and ATACseq (determined and described in Fig. 5a) (c) or of gene H2-Ab1 in mouse embryonic stem cells (mES) obtained by reanalyzing published datasets of Rnf2 CHIPseq (GSE34520) (d).
e. Expression level of gene H2-Ab1 in control and Rnf2 KO mES cells obtained by reanalyzing published dataset of RNA microarray (GSE10573).
f. Genes occupied by RNF2 were identified by CHIPseq analysis of human TNBC cell line MDA-MB-231 using anti-RNF2 antibody compared to Input (GSE107176).
g. Integrative genomics viewer (IGV) screenshots of Input or RNF2 ChIPseq (as in panel f) tracks (scale bar, 40) of HLA-DPA1, HLA-DPB1, CCL20 and CXCL8.
h. GSEA analysis (REACTOME) of genes occupied by RNF2 revealed in published CHIPseq of human TNBC cell line MDA-MB-231 using anti-RNF2 antibody compared to Input (as in panel f). The representative enriched pathways with FDR q values are shown. The full list of q values is provided in the Source Data File associated with this panel.
i. The correlations of RNF2 expression in BRCA patients to the levels of its bound genes that were determined as in panel f. The correlation analyses were performed similar to that described in Fig. 5h. The expression levels of these RNF2 bound genes and of RNF2 were extracted from the invasive breast cancer TCGA dataset. The coefficient (yellow) and q value (purple) of each correlation are indicated at the y-axis (two-tailed Spearman correlation analysis). The number (n) of RNF2 bound genes with FDR q values (indicative of the significance of the correlations of these genes to RNF2 expression) < 0.05, and the percentages of these genes (q < 0.05) among the total RNF2 bound genes are noted in blue. The blue vertical bars mark the q value at 0.05. Totally 1,070 BRCA patient samples were included.
Extended Data Fig. 9. Ablation of Rnf2/Rsf1 induces anti-tumor memory response.
a. BALB/c mice were inoculated with Rnf2 KO 4T1 tumors (Rnf2 KO g2) or Rsf1 KO 4T1 tumors (Rsf1 KO g1) in the left 4th mammary pads. At day 45 after the primary tumors were rejected, wildtype 4T1 tumors were implanted into the right 4th mammary pads or naïve mice. n = 5 mice /group, each mouse harboring one tumor. Representative BLIs of wildtype 4T1 tumors at days 97, 131, 182 and 307 after the 2nd challenge are shown. X, mice were sacrificed because of the big tumor burdens at the day when images were taken.
b. BALB/c mice were inoculated with Rnf2 KO 4T1 tumors (Rnf2 KO g2) in the left 4th mammary pads. At day 45 after the Rnf2 KO tumors were rejected, TSA tumors were implanted into the right 4th mammary pads or naïve mice. The tumor volumes were measured by caliper (mean ± SEM, n = 5 mice for naïve, n=4 mice for Rnf2 KO, each mouse harboring one tumor).
c. Frequencies of intratumoral indicated immune cells in wildtype 4T1 tumors at day 7 after the 2nd challenge, measured similar to that described in Fig. 8e. Symbols, individual mouse (bars, mean ± SEM). n = 3 mice for naïve and Rsf1 KO, n = 4 mice for Rnf2 KO, each mouse harboring one tumor.
d-e. Mice were inoculated with Rnf2 KO 4T1 tumors and re-challenged with wildtype 4T1 tumors on the contralateral, as in panel a and Fig. 8a. α-asialo GM1 antibody was injected at days −1, +1, +4 relative to the 2nd challenge. d. Volumes (Left, luminescence intensities, mean ± SEM, n = 5 mice/group, each mouse harboring one tumor; Right, individual mouse) of wildtype 4T1 tumors. e. Representative BLIs of wildtype 4T1 tumors at indicated days after the 2nd challenge.
f. Frequencies of peripheral blood CD8+ T-cells in Rnf2 KO mice at day 10 after injection of anti-CD8 (2.43) or control antibody. Symbols, individual mouse (bars, mean ± SEM). n = 5 mice/group, each mouse harboring one tumor.
g-h. g. Mice were inoculated with Rnf2 KO 4T1 tumors (Rnf2 KO g2) and re-challenged with wildtype 4T1 tumors on the contralateral, as in panel a and Fig. 8a. Anti-CD8 or control antibody was injected at days −1 and +1 relative to the 2nd challenge. g. Volumes (Left, luminescence intensities, mean ± SEM, n = 5 mice/group, each mouse harboring one tumor; Right, individual mouse) of wildtype 4T1 tumors. h. Representative BLIs of wildtype 4T1 tumors at day 26 or day 96 after the 2nd challenge.
i. Frequencies of peripheral blood CD4+ T-cells in Rnf2 KO mice at day 10 after injection of anti-CD4 (GK1.5) or control antibody. Symbols, anti-tumor memory response. individual mouse (bars, mean ± SEM). n = 5 mice/group, each mouse harboring one tumor.
Unpaired two-tailed Student’s t test in c, f, i; two-way ANOVA with Sidak’s test in b, d, g.
Extended Data Fig. 10. Gating Strategy used for flow cytometry.
a. Gating Strategy used for analysis of immune cells from tumors isolated from tumor-bearing mice presented on Fig. 2a-e, Fig. 3a-b, Fig. 4c, Fig. 7e, Fig. 8e, Extended Fig. 3v, Extended Fig. 5a,c,g-h, Extended Fig. 9c.
b. Gating Strategy used for analysis of each subset of CD4+ T-cells in a (5-2).
c. Gating Strategy used for analysis of each subset or marker of NK cells in a (5-3).
d. Gating Strategy used for analysis of each subset or marker of CD45–tumor cells in a.
e. Gating Strategy used for analysis of CD4+ T-cells or NK cells from the in vitro co-culture assays presented on Fig. 3c-d, f-g, Fig. 4j-k, Extended Fig. 5b, d-e, Extended Fig. 5i-j, p, s.
f. Gating Strategy used for analysis of dead tumors in the in vitro killing assays presented on Fig. 3e, Fig 4l, Extended Fig. 5f.
g. Gating Strategy used for sorting of tumor cells (CD45–) and NK cells (CD45+NKp46+CD3−) for RNA-Seq and ATAC-Seq presented on Fig. 4a-b, Fig. 5a-f, Fig. 6, Fig. 7f-j, Extended Fig. 4c-d, Extended Fig. 6, Extended Fig. 7a, c-f, Extended Fig. 8a-c.
Supplementary Material
Supplementary Table 1-Reagents (chemicals, antibodies and others).
Supplementary Table2- Reagentsoligonucleotides.
Acknowledgments
We thank Drs. Lewis Zhichang Shi, and Hengbin Wang for helpful discussion on this project. We are grateful to Drs. Hongxing Shen and Suresh Bugide for the technical assistance. We also thank Vidya Sagar Hanumanthu and the UAB Comprehensive Flow Cytometry Core for their assistance with FACS analysis and cell sorting. Eddy S Yang is a ROAR Southeast Cancer Foundation Endowed Chair. This work was supported by grants from Autotech LLC (to E.S.Y.), from Breast Cancer Research Foundation of Alabama (to E.S.Y.), from American Association for Cancer Research/Triple Negative Breast Cancer Foundation (15-20-43-YANG) (to E.S.Y.), from the Start-up funds from the University of Alabama at Birmingham (to J.W.L.). J.W.L. is also supported by the DoD PRCRP Career Award (W81XWH-18-1-0315) and NIH R01AI148711. N.W. is supported by the DoD grant W81XWH-19-1-0084.
Footnotes
Competing Interests Statement
Eddy S Yang is the consultant for AstraZeneca Plc, Eli Lilly and Company and is on the Advisory Board for Bayer Pharmaceuticals, AstraZeneca Plc, Clovis Oncology, and Strata Oncology. The research in Eddy S Yang’s laboratory is also supported by the funding from Novartis International AG, Eli Lilly and Company, Clovis Oncology and American Society of Clinical Oncology. The remaining authors declare no competing interests.
References
- 1.Leach DR, Krummel MF & Allison JP Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736, doi: 10.1126/science.271.5256.1734 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Patel SA & Minn AJ Combination Cancer Therapy with Immune Checkpoint Blockade: Mechanisms and Strategies. Immunity 48, 417–433, doi: 10.1016/j.immuni.2018.03.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankel T, Lanfranca MP & Zou W The Role of Tumor Microenvironment in Cancer Immunotherapy. Adv Exp Med Biol 1036, 51–64, doi: 10.1007/978-3-319-67577-0_4 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Benci JL et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 178, 933–948 e914, doi: 10.1016/j.cell.2019.07.019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegde PS & Chen DS Top 10 Challenges in Cancer Immunotherapy. Immunity 52, 17–35, doi: 10.1016/j.immuni.2019.12.011 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Yu J et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med, doi: 10.1038/s41591-020-1131-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalbasi A & Ribas A Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 20, 25–39, doi: 10.1038/s41577-019-0218-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bian Y et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282, doi: 10.1038/s41586-020-2682-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanmamed MF & Chen L A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 175, 313–326, doi: 10.1016/j.cell.2018.09.035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng W et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 174, 549–563 e519, doi: 10.1016/j.cell.2018.05.052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan D et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775, doi: 10.1126/science.aao1710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adeegbe DO et al. Synergistic Immunostimulatory Effects and Therapeutic Benefit of Combined Histone Deacetylase and Bromodomain Inhibition in Non-Small Cell Lung Cancer. Cancer Discov 7, 852–867, doi: 10.1158/2159-8290.CD-16-1020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov 10, 270–287, doi: 10.1158/2159-8290.CD-19-0780 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng D et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253, doi: 10.1038/nature15520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest 130, 2712–2726, doi: 10.1172/JCI134402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burr ML et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 36, 385–401 e388, doi: 10.1016/j.ccell.2019.08.008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su W et al. The Polycomb Repressor Complex 1 Drives Double-Negative Prostate Cancer Metastasis by Coordinating Stemness and Immune Suppression. Cancer Cell 36, 139–155 e110, doi: 10.1016/j.ccell.2019.06.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly RM et al. Combination Epigenetic Therapy in Advanced Breast Cancer with 5-Azacitidine and Entinostat: A Phase II National Cancer Institute/Stand Up to Cancer Study. Clin Cancer Res 23, 2691–2701, doi: 10.1158/1078-0432.CCR-16-1729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerby-Arnon L et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 175, 984–997 e924, doi: 10.1016/j.cell.2018.09.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piunti A & Shilatifard A Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352, aad9780, doi: 10.1126/science.aad9780 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Rai K et al. Dual Roles of RNF2 in Melanoma Progression. Cancer Discov 5, 1314–1327, doi: 10.1158/2159-8290.CD-15-0493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan HL et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat Commun 9, 3377, doi: 10.1038/s41467-018-05728-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y et al. Estrogen induces dynamic ERalpha and RING1B recruitment to control gene and enhancer activities in luminal breast cancer. Sci Adv 6, eaaz7249, doi: 10.1126/sciadv.aaz7249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laugesen A & Helin K Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14, 735–751, doi: 10.1016/j.stem.2014.05.006 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Piunti A & Shilatifard A The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol 22, 326–345, doi: 10.1038/s41580-021-00341-1 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Adams S et al. Current Landscape of Immunotherapy in Breast Cancer: A Review. JAMA Oncol, doi: 10.1001/jamaoncol.2018.7147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z et al. Role of remodeling and spacing factor 1 in histone H2A ubiquitination-mediated gene silencing. Proc Natl Acad Sci U S A 114, E7949–E7958, doi: 10.1073/pnas.1711158114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61, doi: 10.1016/j.cell.2014.12.033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai G, Yang Q & Sun W RSF1 in cancer: interactions and functions. Cancer Cell Int 21, 315, doi: 10.1186/s12935-021-02012-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J et al. The oncogenic impact of RNF2 on cell proliferation, invasion and migration through EMT on mammary carcinoma. Pathol Res Pract 215, 152523, doi: 10.1016/j.prp.2019.152523 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Barger CJ, Branick C, Chee L & Karpf AR Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers (Basel) 11, doi: 10.3390/cancers11020251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shabason JE & Minn AJ Radiation and Immune Checkpoint Blockade: From Bench to Clinic. Semin Radiat Oncol 27, 289–298, doi: 10.1016/j.semradonc.2017.03.002 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Wang W et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274, doi: 10.1038/s41586-019-1170-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babic M et al. NK cell receptor NKG2D enforces proinflammatory features and pathogenicity of Th1 and Th17 cells. J Exp Med 217, doi: 10.1084/jem.20190133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borst J, Ahrends T, Babala N, Melief CJM & Kastenmuller W CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 18, 635–647, doi: 10.1038/s41577-018-0044-0 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Minn AJ Interferons and the Immunogenic Effects of Cancer Therapy. Trends Immunol 36, 725–737, doi: 10.1016/j.it.2015.09.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng M et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer 19, 568–586, doi: 10.1038/s41568-019-0183-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross SH & Cantrell DA Signaling and Function of Interleukin-2 in T Lymphocytes. Annu Rev Immunol 36, 411–433, doi: 10.1146/annurev-immunol-042617-053352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia J, Gill EE & Hancock RE NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nature protocols 10, 823–844, doi: 10.1038/nprot.2015.052 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Klusmann I et al. Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc Natl Acad Sci U S A 115, E11311–E11320, doi: 10.1073/pnas.1809592115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HS et al. The chromatin remodeler RSF1 controls centromeric histone modifications to coordinate chromosome segregation. Nat Commun 9, 3848, doi: 10.1038/s41467-018-06377-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forero A et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol Res 4, 390–399, doi: 10.1158/2326-6066.CIR-15-0243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morey L & Helin K Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35, 323–332, doi: 10.1016/j.tibs.2010.02.009 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Brookes E et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10, 157–170, doi: 10.1016/j.stem.2011.12.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen I et al. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev 33, 55–60, doi: 10.1101/gad.319939.118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endoh M et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524, doi: 10.1242/dev.014340 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878, doi: 10.1038/nature02985 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Yasinska IM et al. The Tim-3-Galectin-9 Pathway and Its Regulatory Mechanisms in Human Breast Cancer. Front Immunol 10, 1594, doi: 10.3389/fimmu.2019.01594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. J Immunother Cancer 7, 88, doi: 10.1186/s40425-019-0563-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toniolo PA et al. Deregulation of SOCS5 suppresses dendritic cell function in chronic lymphocytic leukemia. Oncotarget 7, 46301–46314, doi: 10.18632/oncotarget.10093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q et al. Adoptive transfer of tumor reactive B cells confers host T-cell immunity and tumor regression. Clin Cancer Res 17, 4987–4995, doi: 10.1158/1078-0432.CCR-11-0207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alspach E et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701, doi: 10.1038/s41586-019-1671-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribas A et al. Association of Pembrolizumab With Tumor Response and Survival Among Patients With Advanced Melanoma. JAMA 315, 1600–1609, doi: 10.1001/jama.2016.4059 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Hirschhorn-Cymerman D et al. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med 209, 2113–2126, doi: 10.1084/jem.20120532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kerdiles Y, Ugolini S & Vivier E T cell regulation of natural killer cells. J Exp Med 210, 1065–1068, doi: 10.1084/jem.20130960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waggoner SN, Cornberg M, Selin LK & Welsh RM Natural killer cells act as rheostats modulating antiviral T cells. Nature 481, 394–398, doi: 10.1038/nature10624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao L et al. A partially disordered region connects gene repression and activation functions of EZH2. Proc Natl Acad Sci U S A 117, 16992–17002, doi: 10.1073/pnas.1914866117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle S et al. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev 34, 931–949, doi: 10.1101/gad.336487.120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Illingworth RS et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 29, 1897–1902, doi: 10.1101/gad.268151.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 61.Hanna A et al. Inhibition of Hedgehog signaling reprograms the dysfunctional immune microenvironment in breast cancer. Oncoimmunology 8, 1548241, doi: 10.1080/2162402X.2018.1548241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zabala M et al. Optimization of the Tet-on system to regulate interleukin 12 expression in the liver for the treatment of hepatic tumors. Cancer Res 64, 2799–2804, doi: 10.1158/0008-5472.can-03-3061 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Li C et al. Novel HDAd/EBV Reprogramming Vector and Highly Efficient Ad/CRISPR-Cas Sickle Cell Disease Gene Correction. Sci Rep 6, 30422, doi: 10.1038/srep30422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev 27, 1581–1595, doi: 10.1101/gad.211037.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reimand J et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 14, 482–517, doi: 10.1038/s41596-018-0103-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962, doi: 10.1038/nmeth.4396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. , 1303.3997 (2013). [Google Scholar]
- 70.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colaprico A et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44, e71, doi: 10.1093/nar/gkv1507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faust GG & Hall IM SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505, doi: 10.1093/bioinformatics/btu314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fabregat A et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, D649–D655, doi: 10.1093/nar/gkx1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419, doi: 10.1126/science.1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1-Reagents (chemicals, antibodies and others).
Supplementary Table2- Reagentsoligonucleotides.
Data Availability Statement
ChIP-seq, RNA–seq and ATAC-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE143352. Previously published ChIP-seq data that were re-analyzed here are available under accession code GSE1057347, GSE3452045 and GSE10717622.
The human pan-cancer and breast cancer data were derived from the TCGA Research Network: https://portal.gdc.cancer.gov/. For pan-cancer analysis, the standardized, normalized, batch corrected and platform-corrected datasets were downloaded from the TCGA consortium (https://gdc.cancer.gov/about-data/publications/pancanatlas). Copy number variation was downloaded from Broad GDAC firehose (http://gdac.broadinstitute.org/runs/analyses__latest/data/). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.