Abstract
PCR amplification-restriction analysis (PRA) of rpoB DNA (342 bp), which comprises the Rifr region, was used for the differential identification of 49 mycobacteria. The DNA had been used previously for the identification of mycobacterial species by comparative sequence analysis (B. J. Kim et al., J. Clin. Microbiol. 37:1714–1720, 1999). Digestion with four restriction enzymes (HaeIII, HindII, MvaI, and AccII), which were selected on the basis of rpoB DNA sequences, generated distinctive PRA patterns that allowed not only the reference strains but also the clinical isolates of mycobacteria to be distinguished. Both rapidly and slowly growing mycobacteria were distinctly differentiated by HaeIII digestion of the amplified rpoB DNA. By HindII digestion the Mycobacterium tuberculosis complex was distinguished from the other mycobacteria. Furthermore, six subspecies of Mycobacterium kansasii (subspecies I to VI) as well as the closely related Mycobacterium gastri, and other closely related species, were distinguished by simultaneous digestion of MvaI and AccII. According to the rpoB PRA scheme, 240 strains of clinical isolates could be identified. It was also possible to detect and identify M. tuberculosis directly from sputa and bronchoalveolar lavage specimens. These results suggest that PRA of rpoB DNA is a simple and feasible method not only for the differentiation of culture isolates but also for the rapid detection and identification of pathogenic mycobacteria in primary clinical specimens.
The genus Mycobacterium comprises more than 70 species, some of which are pathogenic or potentially pathogenic to humans and animals, and some of which are saprobes. Human infections are mainly caused by slowly growing mycobacteria such as Mycobacterium tuberculosis, M. avium complex (MAC), and M. kansasii. Recently, reports of infections due to mycobacteria other than M. tuberculosis complex (MOTT) have been increasing. Because of their clinical importance, in terms of strategies for treatment and the epidemiological implications, the rapid differentiation of causative mycobacteria is important. However, isolation and identification procedures usually require several weeks.
Various PCR-mediated methods had been applied for the rapid detection and identification (or differentiation) of mycobacterial species. Among these different methods PCR-restriction analysis (PRA) is preferred, as a simple and cost-effective method that does not involve radioisotopes. It has been applied to several genes, such as 16S ribosomal DNA (rDNA) (6, 9, 10, 33), hsp65 (2, 4, 8, 18, 19, 22, 28, 30, 31), and dnaJ (29), for the rapid differentiation of closely related clinical isolates, such as Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis (8), and subspecies of M. kansasii (18). Previously we reported that comparative sequence analysis of the RNA polymerase gene (rpoB) DNA (342 bp), encompassing the region related to rifampin resistance (Rifr) in M. tuberculosis, is useful for the identification of mycobacterial species (12). The present study demonstrates that the same region can be directly used for the differential identification of mycobacteria in sputa and culture isolates. PRA of amplified rpoB DNA can differentiate the two naturally divided mycobacterial groups (rapidly growing and slowly growing mycobacteria) and can distinguish M. tuberculosis from MOTT. According to the identification scheme established in this study, most of the human pathogens of MOTT can be differentiated.
MATERIALS AND METHODS
Mycobacteria and sputa.
Forty-nine reference strains and 240 clinical isolates of mycobacteria, as well as 50 sputum specimens and 3 specimens from bronchoalveolar lavage (BAL) from patients suspected of having mycobacterial infections, were provided by the Korean Institute of Tuberculosis (KIT), Department of Clinical Pathology, and the Department of Internal Medicine, Seoul National University Hospital. Five heat-killed reference strains of M. kansasii (subspecies I to V) and two strains of subspecies VI were kindly provided by Veronique Vincent (TB and Mycobacteria Lab, Institut Pasteur, Paris, France) and Elvira Richter (Forschungszentrum Borstel, National Reference Center for Mycobacteria, Borstel, Germany) respectively. Clinical isolates were identified by growth characteristics, conventional biochemical tests, and molecular biological methods (Table 1).
TABLE 1.
Methods used for the identification of clinical isolates
| Method |
|---|
| Growth characteristics and conventional biochemical tests |
| Pigmentation on subculture |
| Growth temperature |
| Semiquantitative catalase |
| Iron uptake |
| Growth on p-nitrobenzoic acid |
| Tween 80 hydrolysis test |
| Tellurite reduction test |
| 5% NaCl tolerance test |
| Growth rate |
| 3- and 14-day arylsulfatase |
| Heat-stable catalase (pH 7, 68°C) |
| Niacin production |
| Nitrate reduction test |
| Urease test |
| Growth on ethambutol and MacConkey agar |
| Pyrazinamidase test |
| Molecular biological methods |
| IS6110 PCR |
| hsp65 PRA (BstEII and AccII) |
| rpoB sequencing |
| DT1–DT6 PCR |
| 16S rDNA sequencing |
All sputa which had been requested for the culture of M. tuberculosis or MOTT were processed by 1% NaOH liquefaction, decontamination, and sedimentation (at 12,000 × g for 15 min) (17). Sediments were resuspended in 1.5 ml of phosphate buffer (pH 6.8). DNAs were isolated from 1.0 ml of the sediments, and 0.5 ml of the residual sediments were inoculated onto Löwenstein-Jensen media. Culture isolates of M. tuberculosis were simultaneously identified by the methods listed in Table 1 by KIT. Results obtained from cultures were compared to the results of rpoB PRA (HindII) and IS6110 PCR by blind testing.
Preparation of DNA.
Mycobacterial DNAs from the cultures and sediments of sputa were prepared by the method previously described (12). A loopful of culture of each mycobacterium or sediment was suspended with 200 μl of TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl [pH 8.0]), placed in a 2.0-ml screw-cap microcentrifuge tube filled with 100 μl (packed volume) of glass beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.) and 100 μl of phenol-chloroform-isopropyl alcohol (50:49:1). To disrupt the mycobacteria, the tube was oscillated on a Mini-Bead beater (Biospec Products) for 1 min, and to separate phases, the tube was centrifuged (at 12,000 × g for 5 min). After the aqueous phase was transferred into another clean tube, 10 μl of 3 M sodium acetate and 250 μl of ice-cold ethanol were added; to enable the DNA to precipitate, the mixture was kept at −20°C for 10 min. The DNA pellet was then washed with 70% ethanol, dissolved in 60 μl of TE buffer (10 mM Tris-HCl–1 mM EDTA [pH 8.0]) and used as a template for PCR. DNAs from sputa were prepared as above and dissolved in 60 μl of TE buffer (pH 8.0).
Amplification of rpoB and IS6110 DNA.
A set of mycobacterium-specific primers (MF and MR) which had been used for the sequence analysis of rpoB DNA (342 bp) (12) was also used in this study. Template DNA (50 ng for the culture and 5 to 10 μl for the sputa) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Choongbuk, Korea) which contained 1 U of Taq DNA polymerase, 250 μM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and gel loading dye, and the volume was adjusted to 20 μl with distilled water. The reaction mixture was subjected to 30 cycles of amplification (30 s at 95°C, 30 s at 60°C, and 45 s at 72°C) followed by a 5-min extension at 72°C (Thermocycler model 9600; Perkin-Elmer Cetus). M. tuberculosis IS6110 DNAs (536 bp) (32) were separately amplified using an IS6110 kit (catalog no. N5811; Bioneer).
Restriction analysis.
Enzyme restriction sites for the sizing of DNA fragments on each of the rpoB sequences of mycobacteria (GenBank accession no. AF057449 to AF057493) were generated by MapDraw, version 3.14 (DNASTAR, Madison, Wis.). Restriction enzymes which produced the most discernible fragments between clusters or strains were selected. Ten microliters of the amplified PCR products was transferred to a fresh microcentrifuge tube and digested with restriction enzymes, according to the supplier's instructions. Following digestion, the mixtures were electrophoresed on a 3% agarose gel (at 100 V for 25 min). DNA bands were visualized by ethidium bromide staining and photographed. The results obtained from the analysis of MAC and M. kansasii isolates were compared with those of DT1–DT6 PCR (5) and PRA (BstEII and HaeIII) of the hsp65 gene (2, 4), respectively.
Nucleotide sequencing.
The rpoB DNA sequences for the five reference strains of M. kansasii (subspecies II to VI) and clinical isolates were directly determined as previously described (12). A 373A automatic sequencer and a BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom) were used. For the sequencing reaction, 60 ng of PCR-amplified DNA, 3.2 pmol of either the forward or the reverse primer, and 8 μl of BigDye Terminator RR mix (Perkin-Elmer Applied Biosystems; catalog no. 4303153) were mixed, and made up to 20 μl with distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 10 s at 50°C, and 4 min at 60°C. The sequences determined were aligned by using the multiple alignment algorithm in the MegAlign package (Windows, version 3.12e; DNASTAR).
Nucleotide sequence accession numbers.
The rpoB sequences of five M. kansasii subspecies (II to VI) were deposited in GenBank (accession no. AF173084 to AF173088).
RESULTS
Restriction enzyme sites.
The nucleotide sequences of 49 rpoB DNAs were compared by computer-aided analysis. Four useful restriction enzyme sites (HaeIII, HindII, MvaI, and AccII), which produced the most discernible fragments among the tested mycobacterial strains, were selected.
Differentiation of rapidly growing mycobacteria from slowly growing mycobacteria.
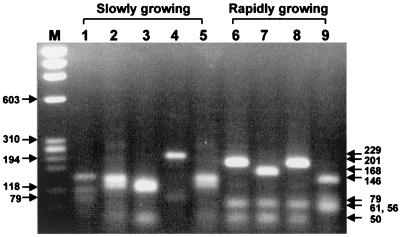
Mycobacteria belonging to the natural division of rapidly and slowly growing groups were differentiated by the PRA patterns of HaeIII. All of the rapidly growing mycobacteria had G467L468 encoded by GGCCT(G/C) (12). HaeIII specifically cleaves the rpoB DNAs of rapidly growing mycobacteria, yielding a 61-bp DNA fragment, but does not cleave those of slowly growing mycobacteria. In addition to the 61-bp fragment, another useful marker DNA fragment for rapidly growing mycobacteria upon HaeIII digestion was the 201-bp (or 202-bp) fragment. Most rapidly growing mycobacteria produced the 201/202 bp fragment except M. abscessus and M. fallax (168 bp) and M. chitae (146 bp). Thus, M. abscessus, formerly Mycobacterium chelonae subsp. abscessus, was easily distinguished from M. chelonae and M. fortuitum upon HaeIII digestion (Fig. 1). Nineteen clinical isolates (11 strains of M. fortuitum and 8 strains of M. chelonae) were confirmed as rapidly growing mycobacteria by PRA of the rpoB DNA. Interestingly, eight strains of M. chelonae complex which had been identified by culture were identified as M. abscessus by PRA of rpoB (HaeIII and AccII), yielding characteristic bands (Table 2). Furthermore, HaeIII was also useful for distinguishing closely related species such as M. avium-M. paratuberculosis-M. intracellulare and M. celatum type 1–M. celatum type 2. Five subspecies of M. kansasii, except subspecies V, were differentiated from M. gastri.
FIG. 1.
Differentiation of rapidly and slowly growing mycobacteria by PRA of rpoB (HaeIII). Amplified rpoB DNAs (342 bp) of reference strains were digested with HaeIII and electrophoresed on a 3% agarose gel. DNA fragments of 61 or 201 bp are observed in the lanes of rapidly growing species (lanes 6 to 9). Lanes: M; φX174/RF DNA/HaeIII digest; 1, M. tuberculosis; 2, M. avium; 3, M. intracellulare; 4, M. kansasii; 5, M. szulgai; 6, M. fortuitum; 7, M. abscessus; 8, M. chelonae; 9, M. chitae.
TABLE 2.
Restriction fragments of rpoB DNAs (342 bp)
| Reference strain | Restriction fragment sizes (bp)a
|
|||
|---|---|---|---|---|
| HaeIII | AccII | MvaI | HindII | |
| Rapidly growing mycobacteria | ||||
| M. abscessus (CAP97E-03) | 168, 61, 50, 33, 30 | 207, 79, 38, 18 | 342 | 342 |
| M. aurum (ATCC 23366) | 202, 61, 50, 29 | 218, 55, 38, 18, 13 | 237, 105 | 342 |
| M. chelonae (ATCC 35749) | 202, 61, 50, 29 | 207, 68, 38, 18, 11 | 342 | 342 |
| M. chitae (ATCC 19627) | 146, 79, 61, 56 | 198, 79, 56, 9 | 237, 105 | 342 |
| M. fortuitum (ATCC 6841) | 201, 61, 50, 30 | 207, 55, 38, 24, 18 | 237, 105 | 342 |
| M. fortuitum 49403 (ATCC 49403) | 201, 61, 50, 30 | 207, 79, 38, 18 | 237, 105 | 342 |
| M. fallax (ATCC 35219) | 168, 61, 50, 34, 29 | 137, 79, 38, 31, 18, 9 | 189, 105, 48 | 342 |
| M. flavescens (ATCC 14474) | 201, 61, 50, 30 | 207, 68, 56, 11 | 237, 105 | 342 |
| M. neoaurum (ATCC 25795) | 202, 79, 61 | 234, 68, 20 | 237, 105 | 342 |
| M. peregrinum (ATCC 14467) | 201, 61, 50, 30 | 218, 68, 38, 18 | 237, 105 | 342 |
| M. phlei (ATCC 11758) | 201, 61, 50, 30 | 198, 68, 38, 18, 11, 9 | 237, 105 | 342 |
| M. senegalense (ATCC 35796) | 201, 61, 50, 30 | 225, 79, 38 | 237, 105 | 342 |
| M. smegmatis (ATCC 19420) | 201, 61, 50, 30 | 231, 55, 38, 18 | 237, 105 | 342 |
| M. thermoresistibile (ATCC 19527) | 201, 61, 50, 30 | 198, 79, 38, 18, 9 | 237, 105 | 342 |
| M. vaccae (ATCC 15483) | 201, 61, 50, 30 | 198, 55, 38, 20, 18, 13 | 237, 105 | 342 |
| Slowly growing mycobacteria | ||||
| M. africanum (ATCC 25420) | 149, 114, 79 | 142, 94, 68, 38 | 237, 105 | 232, 110 |
| M. asiaticum (ATCC 25276) | 149, 113, 50, 30 | 174, 79, 74, 9 | 294, 48 | 342 |
| M. avium (ATCC 25291) | 146, 117, 54, 25 | 216, 79, 38, 9 | 237, 105 | 342 |
| M. bovis (ATCC 19210) | 149, 114, 79 | 142, 94, 68, 38 | 237, 105 | 232, 110 |
| M. celatum type 1 (ATCC 51131) | 262, 50, 30 | 216, 88, 38 | 342 | 342 |
| M. celatum type 2 (ATCC 51130) | 162, 100, 50, 26, 4 | 167, 88, 49, 38 | 189, 105, 48 | 342 |
| M. gastri (ATCC 15754) | 117, 112, 80, 30 | 225, 79, 38 | 274, 68 | 342 |
| M. genavense (ATCC 51233) | 146, 117, 50, 29 | 236, 68, 38 | 342 | 342 |
| M. gordonae (ATCC 14470) | 117, 114, 79, 32 | 180, 79, 74, 9 | 342 | 342 |
| M. haemophilum (ATCC 29548) | 149, 79, 70, 34, 10 | 116, 94, 68, 38, 26 | 342 | 342 |
| M. interjectum (ATCC 51457) | 117, 113, 80, 32 | 116, 79, 74, 64, 9 | 342 | 342 |
| M. intermedium (ATCC 51848) | 149, 80, 74, 50, 25, 4 | 118, 83, 79, 38, 24 | 237, 68, 37 | 342 |
| M. intracellulare (ATCC 13950) | 117, 114, 50, 32, 29 | 124, 74, 68, 56, 11, 9 | 294, 48 | 342 |
| M. kansasii subsp. I (ATCC 12478) | 229, 79, 34 | 342 | 342 | 342 |
| M. kansasii subsp. IIb | 229, 50, 33, 26, 4 | 261, 81 | 294, 48 | 342 |
| M. kansasii subsp. IIIb | 229, 50, 34, 29 | 342 | 294, 48 | 342 |
| M. kansasii subsp. IVb | 229, 50, 33, 30 | 225, 81, 36 | 294, 48 | 342 |
| M. kansasii subsp. Vb | 117, 112, 50, 34, 29 | 257, 85 | 294, 48 | 342 |
| M. kansasii subsp. VIc | 229, 50, 33, 30 | 124, 74, 70, 36, 20, 18 | 294, 48 | 342 |
| M. leprae (Thai 53 strain) | 229, 80, 33 | 210, 68, 64 | 237, 105 | 342 |
| M. malmoense (ATCC 29571) | 149, 114, 79 | 116, 83, 64, 55, 13, 11 | 237, 105 | 342 |
| M. marinum (ATCC 927) | 263, 79 | 218, 68, 56 | 237, 105 | 342 |
| M. microti (ATCC 19422) | 149, 114, 79 | 142, 94, 68, 38 | 237, 105 | 232, 110 |
| M. nonchromogenicum (ATCC 19530) | 145, 117, 50, 30 | 190, 79, 56, 9, 8 | 189, 105, 48 | 342 |
| M. paratuberculosis (ATCC 19698) | 146, 117, 79 | 216, 79, 38, 9 | 237, 105 | 342 |
| M. scrofulaceum (ATCC 19981) | 149, 70, 54, 39, 26, 10 | 118, 88, 74, 62 | 342 | 342 |
| M. shimoidei (ATCC 27962) | 263, 79 | 180, 94, 68 | 237, 105 | 342 |
| M. simiae (ATCC 25275) | 146, 117, 50, 29 | 236, 55, 38, 13 | 342 | 342 |
| M. szulgai (ATCC 35799) | 149, 114, 54, 24 | 180, 79, 74, 9 | 237, 105 | 342 |
| M. terrae (ATCC 15755) | 145, 117, 80 | 180, 88, 74 | 237, 105 | 342 |
| M. triviale (ATCC 23292) | 263, 54, 25 | 142, 88, 74, 38 | 237, 105 | 342 |
| M. tuberculosis (ATCC 27294) | 149, 114, 79 | 142, 94, 68, 38 | 237, 105 | 232, 110 |
| M. ulcerans (ATCC 19423) | 263, 79 | 124, 94, 68, 56 | 237, 105 | 342 |
| M. xenopi (ATCC 19250) | 159, 129, 54 | 116, 88, 74, 64 | 237, 105 | 342 |
Exact restriction fragment sizes were determined from the sequences.
From V. Vincent.
From E. Richter.
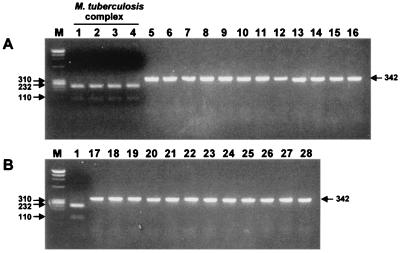
Identification of M. tuberculosis complex.
Identification of M. tuberculosis complex was simply performed with HindII. This restriction enzyme was very useful for differentiating between M. tuberculosis and MOTT. The rpoB DNA sequences of M. tuberculosis complex had only one HindII restriction site (GGG523TT↓G524ACC525), which was not found in MOTT and other bacteria (12). Thus, two fragments (232 and 110 bp) were generated from the DNAs of M. tuberculosis complex by HindII, while the MOTT type strains were not cleaved (342 bp) (Fig. 2). When this procedure was applied to 134 strains of M. tuberculosis isolates (104 rifampin-susceptible and 30 rifampin-resistant strains), they were easily identified (Table 3), showing characteristic PRA patterns (data not shown).
FIG. 2.
Differentiation of M. tuberculosis complex from MOTT by the PRA of rpoB (HindII). Amplified rpoB DNAs (342 bp) of 28 reference mycobacteria were digested with HindII and electrophoresed on a 3% agarose gel. Only DNAs from the members of M. tuberculosis complex (A; lanes 1 to 4) were digested (232 and 110 bp), while those of MOTT were not. Lanes: M, φX174/RF DNA/HaeIII digest; 1, M. tuberculosis H37Rv; 2, M. bovis; 3, M. bovis BCG; 4, M. africanum; 5, M. avium; 6, M. paratuberculosis; 7, M. intracellulare; 8, M. scrofulaceum; 9, M. celatum; 10, M. xenopi; 11, M. kansasii; 12, M. gastri; 13, M. nonchromogenicum; 14, M. terrae; 15, M. triviale; 16, M. fortuitum; 17, M. chelonae; 18, M. gordonae; 19, M. szulgai; 20, M. ulcerans; 21, M. marinum; 22, M. simiae; 23, M. haemophilum; 24, M. malmoense; 25, M. smegmatis; 26, M. phlei; 27, M. vaccae; 28, M. genavense.
TABLE 3.
Results of identification for the culture isolates
| Result for:
| |||
|---|---|---|---|
| Conventional biochemical test
|
rpoB PRA
|
||
| Species | No. of isolates | Species | No. of isolates |
| M. tuberculosisa | 134 | M. tuberculosis | 134 |
| M. avium complex | 40 | M. avium | 8 |
| M. intracellulare | 32 | ||
| M. kansasii | 47 | M. kansasii subsp. I | 45 |
| M. kansasii subsp. II | 2 | ||
| M. fortuitum | 11 | M. fortuitum | 11 |
| M. chelonae | 8 | M. abscessus | 8 |
Comprising 104 rifampin-susceptible and 30 rifampin-resistant strains.
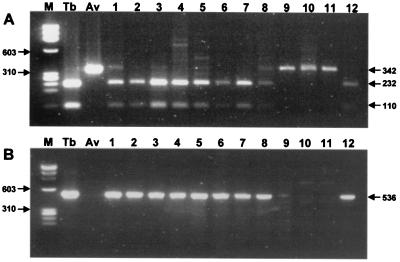
PRA of rpoB DNA (HindII) and IS6110 PCR were directly applied to 50 sputa and 3 BAL samples, and these results were compared to the results of culture (Table 4). Nothing was amplified from the culture-negative sputa, which were also smear negative for acid-fast bacilli (AFB). However, rpoB DNAs (342 bp) were amplified from the three smear-positive sputa identified as MOTT by culture. rpoB PRA was compared with the IS6110 PCR for the ability to detect the most common pathogen, M. tuberculosis. The sensitivity of rpoB PRA for detection and identification of M. tuberculosis (93.2%) was slightly lower than that of IS6110 PCR (97.7%). This discrepancy possibly originates from the different copy numbers of rpoB and IS6110. However, it should be noted that the existence of MOTT in the three sputum samples was revealed by rpoB PRA (HindII) (Fig. 3A, lanes 9, 10, and 11), while it was not detected by IS6110 PCR (Fig. 3B, lanes 9, 10, and 11). Both methods specifically detected M. tuberculosis. M. tuberculosis was also detected and cultivated from the three BAL samples.
TABLE 4.
Comparison of the results obtained by two different methods for detecting and identifying M. tuberculosis from 50 sputa and 3 BAL specimens
| Test and result | No. of isolates, by culture
|
Sensitivity (%) | Specificity (%) | PPV (%)a | NPV (%)a | |
|---|---|---|---|---|---|---|
| M. tuberculosis(n = 44) | MOTT/no mycobacterial growth (n = 3/6) | |||||
| rpoB PRA (HindII) | ||||||
| M. tuberculosis | 41 | 0/0 | 93.2 | 100 | 100 | 19.2 |
| MOTT | 0 | 3/0b | ||||
| Not detected | 3 | 0/6c | ||||
| IS6110 PCR | ||||||
| M. tuberculosis | 43 | 0 | 97.7 | 100 | 100 | 18.4 |
| Not detected | 1 | 9 | ||||
PPV and NPV, positive and negative predictive values, respectively.
Sputum samples were AFB positive, rpoB DNAs were amplified.
Sputum samples were AFB negative.
FIG. 3.
Simultaneous detection and identification of M. tuberculosis in sputa by rpoB PRA (HindII). (A) rpoB DNAs (342 bp) amplified from sputa were digested with HindII. Nine specimens (lanes 1 to 8 and lane 12) showed two fragments (232 and 110 bp), while the others (lanes 9 to 11) remained intact, showing that the former originated from M. tuberculosis and the latter from MOTT. (B) For comparison with the rpoB PRA results, IS6110 DNA (536 bp) was amplified from the same specimens. Nothing was observed in three lanes (lanes 9 to 11). Lane M, φX174/RF DNA/HaeIII digest; lane Tb, M. tuberculosis H37Rv; lane Av, M. avium.
Differentiation of MOTT.
Many of the species pathogenic for humans and animals belong to the slowly growing mycobacteria. These could be differentiated by the simultaneous digestion of rpoB DNA with MvaI, HaeIII, and AccII. For example M. avium, M. paratuberculosis, and M. intracellulare are closely related and are not easy to differentiate, but they were easily differentiated by digestion with MvaI. Two DNA fragments of different sizes were produced from the reference strains of M. avium (237 and 105 bp) and M. intracellulare (294 and 48 bp). rpoB DNA from M. scrofulaceum was not digested (342 bp) (Table 2). Only HaeIII could distinguish M. avium (146, 117, 54, and 25 bp) from M. paratuberculosis (146, 117, and 79 bp). Forty clinical isolates of MAC showed one of the PRA (MvaI) patterns (data not shown) and thus were identified as either M. avium (8 strains) or M. intracellulare (32 strains) (Table 3). The results were concordant with those obtained with DT1–DT6 PCR.
M. terrae, M. nonchromogenicum, and M. triviale, grouped as the Terrae complex, members of which are rarely associated with human disease, were also easily distinguished (data not shown). Other closely related pairs of clades, such as types 1 and 2 of M. celatum, and M. marinum and M. ulcerans, could be differentiated by MvaI and AccII.
Differentiation of M. kansasii subspecies and M. gastri.
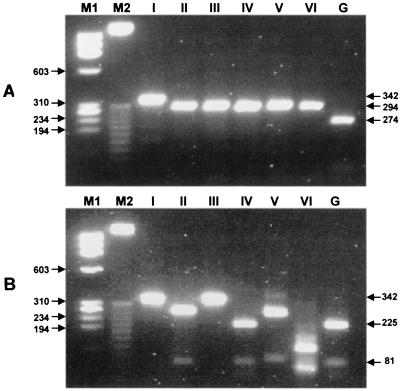
The rpoB DNA sequences of five subspecies of M. kansasii (subspecies II to VI) were determined in this study and deposited in GenBank (accession no. AF173084 to AF173088). There were marked variations (90.5 to 98.4% similarity) in the rpoB sequences of six type strains. Subspecies VI of M. kansasii showed low levels of similarity with the other five subspecies (90.5 to 93.1%). However, it was clustered with five other subspecies and M. gastri in the phylogenetic tree constructed by either the neighborhood joining method or the UPGMA method (data not shown). M. gastri, with a 16S rDNA sequence identical with subspecies I, IV, and V, was most similar to subspecies IV with respect to its rpoB sequence (95.8% similarity).
PRA was applied on the basis of these rpoB sequences from six type strains of subspecies. Although subspecies I of M. kansasii was simply differentiated from M. gastri by HaeIII digestion, MvaI and AccII could distinguish the six subspecies of M. kansasii and M. gastri (Table 2 and Fig. 4). This procedure was also applied to 47 clinical isolates of M. kansasii. All strains showed identically one of the fragment polymorphisms of subspecies I (45 strains) or II (2 strains) (Table 3). This result was concordant with those of the sequence analysis and PRA of hsp65 (data not shown). The rpoB sequences of two subspecies II strains were identical, and sequence variations among the wild-type strains were less than 0.6%.
FIG. 4.
Differentiation of M. kansasii subspecies and M. gastri by PRA of rpoB DNA. Amplified DNAs from six subspecies of M. kansasii (lanes I to VI) and M. gastri (lane G) were digested with MvaI (A) and AccII (B) and electrophoresed on a 3% agarose gel. Lanes M1, φX174/RF DNA/HaeIII digest; lanes M2, 25-bp DNA ladder.
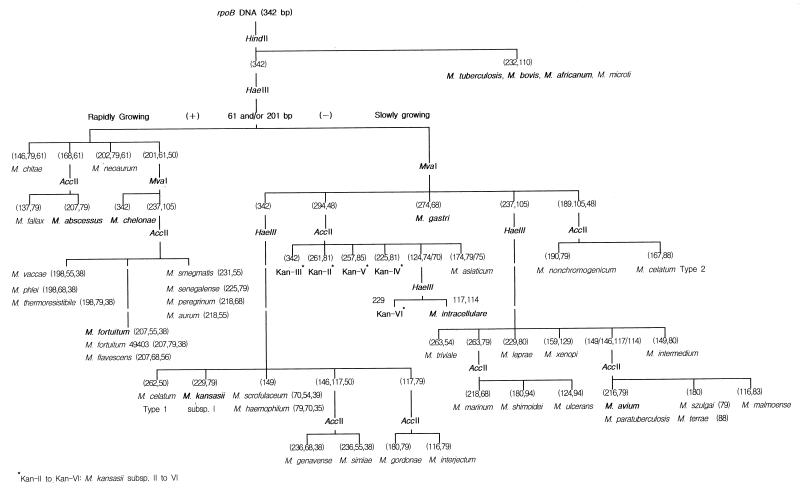
Based on the above results, we constructed a scheme for the differential identification of mycobacteria, including almost all of the major human pathogenic species, by rpoB PRA (Fig. 5).
FIG. 5.
Scheme for the differential identification of mycobacteria by PRA of rpoB DNA (342 bp) using four restriction enzymes (HaeIII, HindII, MvaI, and AccII).
DISCUSSION
The most common mycobacterial infection in the world is caused by M. tuberculosis. There are also increasing reports of nontuberculous mycobacterial infection not only in the immunocompromised but also in immunocompetent hosts (25, 27). Among those species that are currently recognized, slowly growing nontuberculous mycobacteria such as MAC and M. kansasii and rapidly growing species such as M. fortuitum, M. chelonae, and M. abscessus are frequently encountered and are clinically important. To identify these mycobacteria, rapid, simple, and sensitive methods based on molecular biology have been introduced. Methods employing PCR are feasible for the specific detection and identification or differentiation of mycobacteria. PRA is one of these methods and is simple and convenient. A scheme that was derived from the PRA of hsp65 has been widely used. Many reports have shown that this method is useful for species identification from the cultures of type strains, clinical isolates (2, 4, 8, 19, 22, 28), and isolates growing in BACTEC cultures (30, 31). It also allowed the elucidation of genetic heterogeneity of many M. kansasii strains by grouping them into five subspecies (4, 18, 31). However, although the use of the BACTEC system reduced the culturing period, all reports published to date have characterized only the cultured mycobacteria.
In this study, we developed a PRA method using rpoB DNA (342 bp), which comprises the region related to rifampin resistance (Rifr) in M. tuberculosis. This DNA was previously used for the identification of mycobacteria and the determination of the rifampin susceptibility of M. tuberculosis by sequence analysis (12). Compared to the other methods PRA of rpoB has unique characteristics, which are practical enough for routine use in the clinical laboratory. First of all, it can not only detect mycobacteria but also determine their natures early in the laboratory process, and it proved to be very useful for identifying rapidly growing mycobacteria and M. tuberculosis. The growth of mycobacteria could be predicted by the PRA of rpoB as by the sequence analysis (12). According to the deduced amino acid sequences, only the M. terrae complex had the marker amino acid (underlined) for rapidly growing mycobacteria, i.e., G467L468 [GGG(T/C)TG] and not G467M468 [GGC(T/A)TG], which was found only in slowly growing mycobacteria (12). So HaeIII (GG↓CCTG) could not cleave the site in the M. terrae complex as it did in the rapidly growing mycobacteria. Because other bacteria also have L468, it is reasonable to use the unique DNA fragment (61 and/or 201 bp) to identify the rapidly growing mycobacteria by HaeIII digestion. Some of the rapidly growing mycobacteria were cultivated from skin biopsy specimens, but unfortunately the biopsy materials were not available. However, considering that only a few members of the rapidly growing mycobacteria cause human infections, PRA of rpoB could be applied directly to biopsy materials as with sputa and BAL specimens.
Second, the differentiation of M. tuberculosis and MOTT could be performed on sputa as well as on primary cultures. Thus, the rapid detection and identification of M. tuberculosis or MOTT should provide a prompt differential diagnosis and settle proper management issues depending on the diagnosis. By testing sputa and BAL specimens, we were able to demonstrate the existence of mycobacteria and to identify the mycobacteria detected. Identification of M. tuberculosis in sputa proved to be very useful. For the specific detection of M. tuberculosis, several genes had been used. Of these, IS6110, a multicopy gene in M. tuberculosis, has been most widely used (20). Although several cases of false-positive results due to amplification from MOTT (11, 15, 16) and false-negative results due to the absence of IS6110 in certain strains of M. tuberculosis had been reported (35), M. tuberculosis is efficiently detected by the amplification of IS6110 (32). The sensitivity of IS6110 PCR for the detection of M. tuberculosis in sputa was slightly higher than that of rpoB PRA in this study. This was possibly due to the different copy numbers of rpoB and IS6110. However, it is noteworthy that the existence of MOTT in sputa was revealed not by IS6110 PCR but by PRA of rpoB (HindII). Because IS6110 PCR can detect only M. tuberculosis, MOTT was not detected in sputa (Fig. 3). Considering the increasing incidence of MOTT infection, the detection of MOTT and its differentiation from M. tuberculosis should be of value. So far, of the bacteria which reside in the respiratory tract, only M. tuberculosis is known to have a HindII restriction site in the 342-bp region.
While this report was being prepared, a method using rpoB DNA was reported (14). However, it was not based on the rpoB sequences of MOTT and nonmycobacteria. Thus, the investigators had to use only the culture isolates and could not apply PRA to sputa or other primary clinical specimens. The target rpoB DNA differs from ours and does not comprise the Rifr region. The target region we chose has very important information on the rifampin resistance of M. tuberculosis and other naturally rifampin resistant bacteria. Thus, the region can be used for many purposes. Because HindII specifically cleaves the PCR products from M. tuberculosis in sputa, the cleaved fragments of rpoB DNA could be directly sequenced to detect the mutations related to rifampin resistance (data not shown). On occasion, the undigested DNA could be used for MOTT identification. Therefore, we suggest that it would be very useful to apply PRA of rpoB directly to the primary specimens early in the diagnostic process. This would save the several weeks that are required for primary culture. Amplification of rpoB DNA with mycobacterium-specific primers (MF-MR) and the sequence information of MOTT and rifampin-resistant M. tuberculosis would facilitate the procedure.
The results of this study also demonstrated that PRA of rpoB DNA usefully distinguished many closely related mycobacterial species. Besides differentiating among M. abscessus-M. chelonae-M. fortuitum, M. avium-M. paratuberculosis-M. intracellulare, M. marinum-M. ulcerans, and M. celatum types 1 and 2, we found that rpoB PRA allowed the differentiation of six subspecies of M. kansasii. Although it may be isolated from the environment (7, 13, 21), M. kansasii has been increasingly reported to cause infection among patients infected with human immunodeficiency virus (3). Molecular biology methods are also feasible for the rapid identification of M. kansasii. However, such methods are not simple because of the genetic heterogeneity of the species (1, 4, 34) and the identical 16S rDNA sequences of M. kansasii and M. gastri (24). The existence of a genetic subspecies of M. kansasii was suggested by information on 16S rDNA (26), the hsp65 gene (4, 18), and the spacer region between the 16S–23S rRNA genes (2). In addition, several strains of M. kansasii have recently been proposed to constitute M. kansasii subspecies VI (23). This report also showed that subspecies I, IV, and V have the same 16S rDNA sequences as M. gastri; for this reason the authors recommended sequencing the hsp65 gene or spacer or performing classical tests to distinguish these species. However, all reference strains and clinical isolates were simply identified by PRA of rpoB. Not only do the results of rpoB PRA correspond with those for the hsp65 gene, but they are also concordant with the fact that M. kansasii subspecies I is the most common human isolate (2). Unfortunately, isolates of the other subspecies were not available.
Results reaffirming the analytical advantages of rpoB DNA have been demonstrated. Not only can rpoB PRA be applied to culture isolates, but it is also applicable directly to sputa. Its use on sputa will save time and expense. Thus, this technique is suggested as a simple and useful method for the rapid detection and simultaneous identification of pathogenic mycobacteria in primary clinical specimens or for the identification of culture isolates.
ACKNOWLEDGMENTS
This work was supported by grant 98-N1-02-01-A-08 from the National Project for Medical Research, funded by the Ministry of Science and Technology (MOST) of Korea, by grant 1999 from Seoul National University College of Medicine and the Hospital Research Fund, and in part by project BK21 for Medicine, Dentistry and Pharmacy.
We thank V. Vincent and E. Richter for providing the type strains of M. kansasii.
REFERENCES
- 1.Abed Y, Bollet C, de Micco P. Demonstration of Mycobacterium kansasii species heterogeneity by the amplification of the 16S–23S spacer region. J Med Microbiol. 1995;43:156–158. doi: 10.1099/00222615-43-2-156. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide F, Richter I, Bernasconi C, Springer B, Hagenau C, Schulze-Robbecke R, Tortoli E, Martin R, Böttger E C, Telenti A. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J Clin Microbiol. 1997;35:1959–1964. doi: 10.1128/jcm.35.8.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloch K C, Zwerling L, Pletcher M J, Hahn J A, Gerberding J L, Ostroff S M, Vugia D J, Reingold A L. Incidence and clinical implications of isolation of Mycobacterium kansasii: results of a 5-year, population-based study. Ann Intern Med. 1998;129:698–704. doi: 10.7326/0003-4819-129-9-199811010-00004. [DOI] [PubMed] [Google Scholar]
- 4.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devallois A, Picardeau M, Paramasivan C N, Vincent V, Rastogi N. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1–DT6 PCR. J Clin Microbiol. 1997;35:2767–2772. doi: 10.1128/jcm.35.11.2767-2772.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domenech P, Menendez M C, Garcia M J. Restriction fragment length polymorphisms of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol Lett. 1994;116:19–24. doi: 10.1111/j.1574-6968.1994.tb06669.x. [DOI] [PubMed] [Google Scholar]
- 7.Engel H W, Berwald L G, Havelaar A H. The occurrence of Mycobacterium kansasii in tapwater. Tubercle. 1980;61:21–26. doi: 10.1016/0041-3879(80)90055-0. [DOI] [PubMed] [Google Scholar]
- 8.Eriks I S, Munck K T, Besser T E, Cantor G H, Kapur V. Rapid differentiation of Mycobacterium avium and M. paratuberculosis by PCR and restriction enzyme analysis. J Clin Microbiol. 1996;34:734–737. doi: 10.1128/jcm.34.3.734-737.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes M S, Skuce R A, Beck L A, Neill S D. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol. 1993;31:3216–3222. doi: 10.1128/jcm.31.12.3216-3222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanaujia G V, Katoch V M, Shivannavar C T, Sharma V D, Patil M A. Rapid characterization of Mycobacterium fortuitum-chelonei complex by restriction fragment length polymorphism of ribosomal RNA genes. FEMS Microbiol Lett. 1991;61:205–208. doi: 10.1016/0378-1097(91)90552-l. [DOI] [PubMed] [Google Scholar]
- 11.Kent L, McHugh T D, Billington O, Dale J W, Gillespie S H. Demonstration of homology between IS6110 of Mycobacterium tuberculosis and DNAs of other Mycobacterium spp.? J Clin Microbiol. 1995;33:2290–2293. doi: 10.1128/jcm.33.9.2290-2293.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B J, Lee S H, Lyu M A, Kim S J, Bai G H, Chae G T, Kim E C, Cha C Y, Kook Y H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubalek I, Mysak J. The prevalence of environmental mycobacteria in drinking water supply systems in a demarcated region in Czech Republic, in the period 1984–1989. Eur J Epidemiol. 1996;12:471–474. doi: 10.1007/BF00143998. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Park H J, Cho S N, Bai G H, Kim S J. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000;38:2966–2971. doi: 10.1128/jcm.38.8.2966-2971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebana E, Aranaz A, Francis B, Cousins D. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:933–938. doi: 10.1128/jcm.34.4.933-938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHugh T D, Newport L E, Gillespie S H. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J Clin Microbiol. 1997;35:1769–1771. doi: 10.1128/jcm.35.7.1769-1771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte F S, Metchock B. Mycobacterium. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: American Society for Microbiology; 1995. pp. 400–437. [Google Scholar]
- 18.Picardeau M, Prod'Hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plikaytis B B, Plikaytis B D, Yakrus M A, Butler W R, Woodley C L, Silcox V A, Shinnick T M. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J Clin Microbiol. 1992;30:1815–1822. doi: 10.1128/jcm.30.7.1815-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plikaytis B B, Crawford J T, Woodley C L, Butler W R, Eisenach K D, Cave M D, Shinnick T M. Rapid, amplification-based fingerprinting of Mycobacterium tuberculosis. J Gen Microbiol. 1993;139:1537–1542. doi: 10.1099/00221287-139-7-1537. [DOI] [PubMed] [Google Scholar]
- 21.Powell B L, Jr, Steadham J E. Improved technique for isolation of Mycobacterium kansasii from water. J Clin Microbiol. 1981;13:969–975. doi: 10.1128/jcm.13.5.969-975.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi N, Goh K S, Berchel M. Species-specific identification of Mycobacterium leprae by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J Clin Microbiol. 1999;37:2016–2019. doi: 10.1128/jcm.37.6.2016-2019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter E, Niemann S, Rusch-Gerdes S, Hoffner S. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J Clin Microbiol. 1999;37:964–970. doi: 10.1128/jcm.37.4.964-970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogall T, Wolters J, Flohr T, Böttger E C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 25.Rosenzweig D Y. Nontuberculous mycobacterial disease in the immunocompetent adult. Semin Respir Infect. 1996;11:252–261. [PubMed] [Google Scholar]
- 26.Ross B C, Jackson K, Yang M, Sievers A, Dwyer B. Identification of a genetically distinct subspecies of Mycobacterium kansasii. J Clin Microbiol. 1992;30:2930–2933. doi: 10.1128/jcm.30.11.2930-2933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saubolle M A, Kiehn T E, White M H, Rudinsky M F, Armstrong D. Mycobacterium haemophilum: microbiology and expanding clinical and geographic spectra of disease in humans. Clin Microbiol Rev. 1996;9:435–447. doi: 10.1128/cmr.9.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steingrube V A, Gibson J L, Brown B A, Zhang Y, Wilson R W, Rajagopalan M, Wallace R J., Jr PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J Clin Microbiol. 1995;33:149–153. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takewaki S, Okuzumi K, Manabe I, Tanimura M, Miyamura K, Nakahara K, Yazaki Y, Ohkubo A, Nagai R. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol. 1994;44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 30.Taylor T B, Patterson C, Hale Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telenti A, Marchesi F, Balz M, Bally F, Bottger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaneechoutte M, De Beenhouwer H, Claeys G, Verschraegen G, De Rouck A, Paepe N, Elaichouni A, Portaels F. Identification of Mycobacterium species by using amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1993;31:2061–2065. doi: 10.1128/jcm.31.8.2061-2065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Ross B C, Dwyer B. Identification of an insertion. sequence-like element in a subspecies of Mycobacterium kansasii. J Clin Microbiol. 1993;31:2074–2079. doi: 10.1128/jcm.31.8.2074-2079.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen L K, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]