Abstract
Objective
Although immunoglobulin A (IgA) is abundantly expressed in the gut and known to be an important component of mucosal barriers against luminal pathogens, its precise function remains unclear. Therefore, we tried to elucidate the effect of IgA on gut homeostasis maintenance and its mechanism.
Design
We generated various IgA mutant mouse lines using the CRISPR/Cas9 genome editing system. Then, we evaluated the effect on the small intestinal homeostasis, pathology, intestinal microbiota, cytokine production, and immune cell activation using intravital imaging.
Results
We obtained two lines, with one that contained a <50 base pair deletion in the cytoplasmic region of the IgA allele (IgA tail-mutant; IgAtm/tm) and the other that lacked the most constant region of the IgH α chain, which resulted in the deficiency of IgA production (IgA−/−). IgA−/− exhibited spontaneous inflammation in the ileum but not the other parts of the gastrointestinal tract. Associated with this, there were significantly increased lamina propria CD4+T cells, elevated productions of IFN-γ and IL-17, increased ileal segmented filamentous bacteria and skewed intestinal microflora composition. Intravital imaging using Ca2+ biosensor showed that IgA−/− had elevated Ca2+ signalling in Peyer’s patch B cells. On the other hand, IgAtm/tm seemed to be normal, suggesting that the IgA cytoplasmic tail is dispensable for the prevention of the intestinal disorder.
Conclusion
IgA plays an important role in the mucosal homeostasis associated with the regulation of intestinal microbiota and protection against mucosal inflammation especially in the ileum.
INTRODUCTION
It is suggested that immunoglobulin A (IgA), the most abundantly expressed antibody (Ab) in both humans and mice, is known to be important for mucosal defence.1-5 In humans, IgA deficiency is associated with various diseases such as inflammatory bowel diseases (IBD), allergies, autoimmune diseases and recurrent infections.6 Further understanding of the role of IgA would benefit from animal model(s) that mimic human pathophysiology. Although a previous study of the IgA-deficient mice did not show a remarkable phenotype,7 a more recent study found that IgA is crucial for gut homeostasis, especially in neonatal mice, where IgA deficiency increased susceptibility to colon injury.8 Other studies had suggested the importance of secreted Ig in the lumen of the gut. A study of activation-induced cytidine deaminase (AID)–deficient mice, which exhibit impaired Ig class switching and hyper-somatic mutation, suggested that IgA is required to maintain the gut microbiota.9 Other studies with AIDG23S mutant mice, which also exhibit impaired hyper-somatic mutation, confirmed that high-affinity Abs are crucial for gut microbiota maintenance.10 A lack of programmed cell death protein 1 (PD-1), which mediates a T cell–B cell interaction that affects Ig production, can lead to impaired high-affinity Ab production and dysbiosis.11 In summary, high-affinity Abs appear to be crucial for gut flora maintenance.
The membrane-bound forms of class-switched IgG, IgE and IgA contain longer cytoplasmic tails than that of IgM/IgD. In IgG and IgE, these tails are composed of 28 amino acids (aa) and found to be crucial for Ab production.12 13 In contrast, IgA has a relatively short cytoplasmic tail of 14 aa,14 and its role in IgA responses has not been defined.
Our research group has recently established a conditional expression of Ca2+ biosensor, named Yellow Cameleon 3.60 (YC3.60), in a transgenic mouse line that facilitates the analyses of both dynamics and signal transduction in living animals.15 We previously used these mice to demonstrate abnormal Ca2+ signalling as an indicator of predisposition to autoimmune disease at the very early phase of disease development. Furthermore, we also established an intravital imaging system to evaluate Peyer’s patches (PP) and epithelial layer in intestine.15-17
Several novel genome editing technologies have recently been developed. Among these, the CRISPR/Cas9 system comprises a powerful strategy for the generation of genetically engineered mice.18 One advantage of this method is the ability to introduce multiple allelic mutations during a one-step experiment. In this study, we aimed to generate various IgA mutations in mice, including deletion of either the entire Ig heavy chain (H) α or the cytoplasmic domain of IgA, using the CRISPR/Cas9 system in order to clarify the role of IgA. Moreover, we analysed CRISPR/Cas9-mediated IgA mutant mice using conventional and novel experimental techniques, including our newly established intravital imaging system.15-17 Here, we show spontaneous pathology in the intestines of mice that are IgA deficient.
MATERIALS AND METHODS
Mice
Various IgA mutant mouse lines were obtained using the CRISPR/Cas9 genome editing system. Details were described in online supplemental materials and methods. The CD19-Cre/YC3.60flox mouse line was described previously.15
Antibiotic (ABx) treatment of the mice was performed as follows. Mice were administrated ABx via drinking water containing neomycin (1.0 mg/mL), ampicillin (1.0 mg/mL), metronidazole (1.0 mg/mL) and vancomycin (0.5 mg/mL) for 3 weeks. Alternatively, mice were administered vancomycin (0.5 mg/mL) for 1 week.
All mice were maintained in the animal facility of Tokyo Medical and Dental University (TMDU) under specific pathogen-free conditions in accordance with the animal care guidelines of TMDU.
Flow cytometry
Cells were analysed on a CyAn ADP (Beckman Coulter, Brea, CA),19 MACSQuant (Miltenyi Biotec, Bergish Gladbach, Germany) or FACSCanto II (BD Biosciences, San Jose, CA) flow cytometer. The specific antibodies (Abs) used are described in online supplemental materials and methods.
Measurement of immunoglobulin levels
Serum Ig levels were measured as described previously,15 using enzyme-linked immunosorbent assays (ELISAs). Alternatively, 100 mg of ileum and faeces were suspended in 1 mL of phosphate-buffered saline (PBS) and their supernatants were subjected to ELISAs. The results were subjected to an unpaired Tukey-Kramer's test for statistical analysis. P value <0.05 was considered statistically significant.
Histopathological assessment
Male and female mice were euthanised at 6–10 weeks of age, and organs and tissues were collected for histopathological assessment. All specimens were fixed with 4% paraformaldehyde (Wako, Tokyo, Japan) for 24 hours and subsequently embedded in paraffin. Later, samples were cut into 4 μm thick sections and stained with hematoxylin and eosin (H&E). The stained sections were analysed without prior knowledge of genotype. The severity of inflammation in small and large intestines was defined using modified versions of previously described scoring systems.17 20 21 Details are described in online supplemental materials and methods.
Scanning electron microscopy
Scanning electron microscopy was performed using the modified version of a previously described protocol22 and described in online supplemental materials and methods.
Cell isolation
Single cells were isolated from spleens (SPL), mesenteric lymph nodes (MLN) and PP using 40 μm cell strainers (BD Biosciences) and ACK solution for red blood cell depletion. Lamina propria lymphocytes (LPL) from either small or large intestine were also isolated as previously described.17 23
Cytokine assay
To measure cytokine production, 5×105 lymphocytes from SPL, MLN or intestinal LPL were cultured in 100 μL of complete RPMI 1640 (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 500 U/mL penicillin, 100 μg/mL streptomycin (Sigma-Aldrich), 10 mM HEPES, 1% nonessential aa and 50 μM β-mercaptoethanol (Life Technologies, Carlsbad, CA) in the presence of 5 μg/mL plate-bound anti-CD3ε (145-2 C11) and 2 μg/mL soluble anti-CD28 (37.51) Abs on flat-bottomed 96-well plates at 37°C in 5% CO2 for 48 hours; LPL were additionally treated with 10 μg/mL gentamycin (Nacalai Tesque, Kyoto, Japan). Culture supernatants were harvested and analysed to assess the production of cytokines using ELISAs specific for interferon (IFN)-γ, interleukin (IL)–4, IL-10 (BD Biosciences) and IL-17A (R&D Systems, Minneapolis, MN) according to the manufacturers' instructions. Data are expressed as means±standard error of the mean (SEM), and statistical significance was determined using Tukey-Kramer’s test. A p value <0.05 was considered significant.
DNA extraction, preparation of 16S rRNA gene amplicon sequence library and metagenomic analysis
Details are described in online supplemental materials and methods.
Intravital microscopy
Intravital imaging of PP was performed using A1 laser scanning confocal microscope with a ×20 objective and NIS-Elements AR software (Nikon, Tokyo, Japan) as described previously.15 The yellow fluorescent protein (YFP):cyan fluorescent protein (CFP) ratio was obtained at an excitation wavelength of 458 nm. Data are expressed as means±standard deviation (SD), and statistical significance was determined using Student’s t-test. P values < 0.05 were considered significant.
RESULTS
Generation of mice lacking either the cytoplasmic domain or the entire heavy chain in IgA genes
To generate IgA-deficient and tail-mutant mouse lines, we designed guide RNAs for the IgE and IgA tails (online supplemental figure S1) and injected these and Cas9 mRNA into C57BL/6 zygotes. Fourteen pups were born from 261 embryos that had been transferred into foster mothers. At the end, we were able to obtain eight surviving mice, which produced offspring. They were analysed by sequencing PCR products of the corresponding region between IgE and IgA tails (online supplemental figure S2A,B). All of these mice harboured at least one mutant allele, indicating that the CRISPR/Cas9-mediated system had worked efficiently. Most of these mice harboured multiple mutant alleles, and two (lines 4 and 8) possessed all four mutant alleles. Although most mutations were short deletions or insertions, lines 5 and 12 harboured long deletions. Line 5 lacked a sequence of > 5 kilobase pairs (Kb) between the 3′ region of IgHε and IgHα, resulting in an entire deletion of the IgHα constant region (online supplemental figure 3A) with a slight mutation in the IgE carboxy (C) terminus. Line 12 lacked the cytoplasmic tail of IgA and consequently possessed additional aa residues (online supplemental figure 3B). We generated the homozygous IgHα deletion (IgA−/−) and IgA tail-mutant (IgAtm/tm) mice derived from lines 5 and 12, respectively.
Characterisation of IgA mutant mouse lines
The cytoplasmic tails of IgHγ and IgHε were previously shown to exert important functions.12 13 Therefore, in addition to the mice with deficiency of the entire IgHα (IgA−/−), we also analysed another mice lacking the cytoplasmic region (IgAtm/tm). We initially examined serum levels of different Ig classes and found that IgA−/− had similar or slightly higher levels of IgM, IgG and IgE when compared with the wild-type (WT) mice (figure 1A), and as expected, it did not harbour detectable levels of IgA. In contrast, IgAtm/tm and WT mice had similar serum IgA levels (figure 1B), which suggests that the tail sequence is dispensable for serum IgA production.
Figure 1.
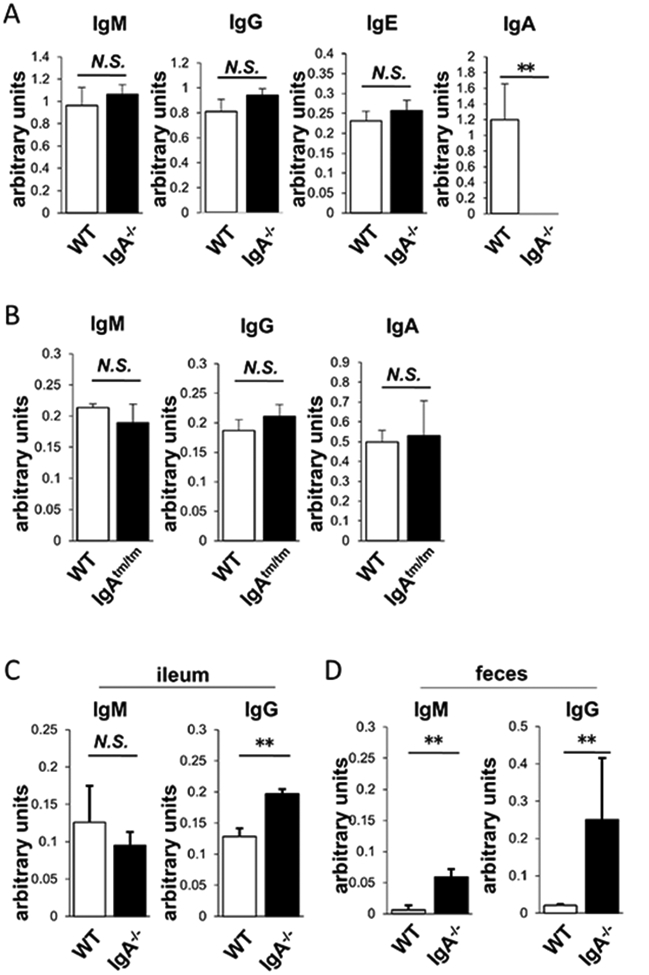
Ig levels in IgA−/− and IgAtm/tm. (A) Serum IgM, IgG, IgE and IgA levels in WT (open) and IgA−/− (closed). (B) Serum IgM, IgG and IgA levels in WT (open) and IgAtm/tm (closed). (C) Ileal IgM and IgG levels in WT (open) and IgA−/− (closed). (D) Fecal IgM and IgG levels in WT (open) and IgA−/− (closed) (n>3). Values are presented as means±SD. N.S., not significant. **p<0.01.
We also analysed ileal and faecal IgM and IgG in IgA−/− mice whether they compensate IgA deficiency in gut lumen. Ileal IgG was significantly increased in IgA−/− mice, although ileal IgM was not altered (figure 1C). Furthermore, both of faecal IgM and IgG were increased in IgA−/− mice (figure 1D). These results indicate that other classes of Igs compensate IgA deficiency in the gut.
We further analysed T cells and B cells in lymphoid tissues of IgA−/− mice (figure 2). B cell populations were not altered in the SPL and PP, but increased in the MLN of IgA−/− (figure 2A-C). CD21+CD23− marginal zone B cells of IgA−/− were also increased (figure 2D). Furthermore, B220+GL7+ germinal centre B cells were increased in the MLN of IgA−/−, but B220−CD138+ plasma cells and B220+CD86+ B cells were not altered (figure 2E-G). On the other hand, CD4+ T cells and CD8+ T cells were not altered in IgA−/−. CD4+CD69+ T cells were also not altered (figure 2H). CD4+CD44+CD62L− effecter memory T cells were slightly increased while CD4+CD44−CD62L+ naive T cells were not altered in PP (figure 2I,J). Furthermore, follicular helper T (Tfh) cells were not altered (figure 2K,L). Therefore, some B and T populations were slightly altered in some lymphoid tissues as a result of IgA deficiency. On the other hand, in IgAtm/tm mice, B populations were substantially the same as those of WT mice (online supplemental figure S4).
Figure 2.
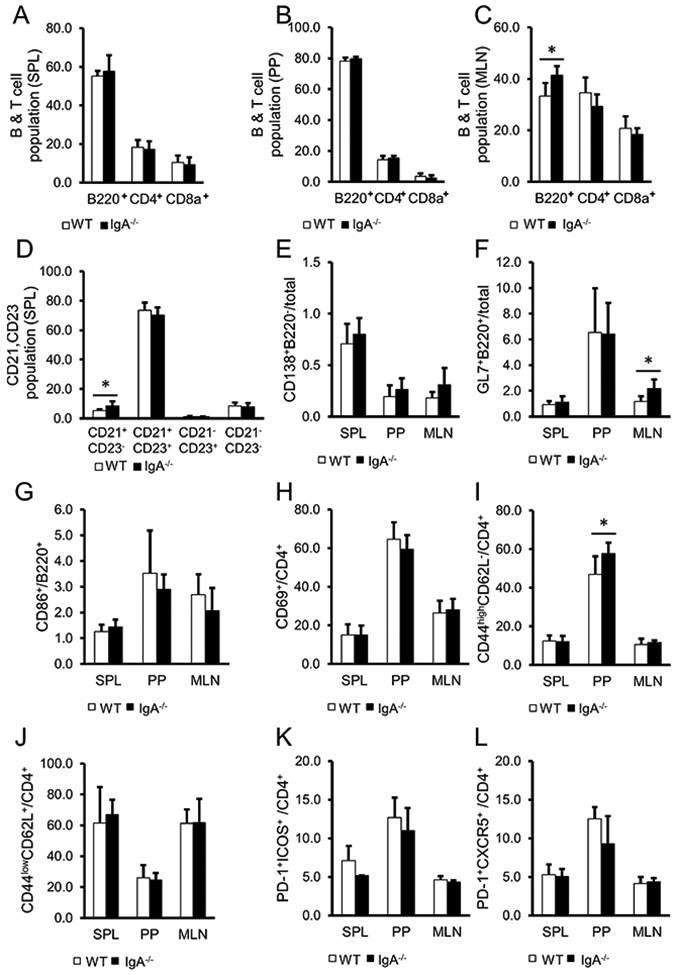
Analysis of lymphoid tissue cell populations in the spleen (SPL), Peyer’s patches (PP) and mesenteric lymph nodes (MLN) in WT (open) and IgA−/− (closed). (A-C) Percentage of either B220+, CD4+ or CD8a+ cells in SPL (A), PP (B) and MLN (C). (D) Percentage of CD21+CD23− marginal zone B cells and CD21+CD23+ follicular B cells in CD19+ population at SPL. (E-L) Percentage of B220−CD138+ plasma cells (E), and B220+GL7+ germinal centre B cells (F) in total cells, CD86+ B cells in B220+ population (G), CD69+T cells (H), CD44highCD62L− effecter memory T cells (I), CD44lowCD62L+ naive T cells (J), PD-1+ICOS+ Tfh cells (K), PD-1+CXCR5+ Tfh cells (L) in CD4+ population . Data are expressed as means±SD. WT: n=7, IgA−/−: n=8. *p<0.05 (t-test).
IgA−/− mice exhibited inflammatory disorder in the small intestine
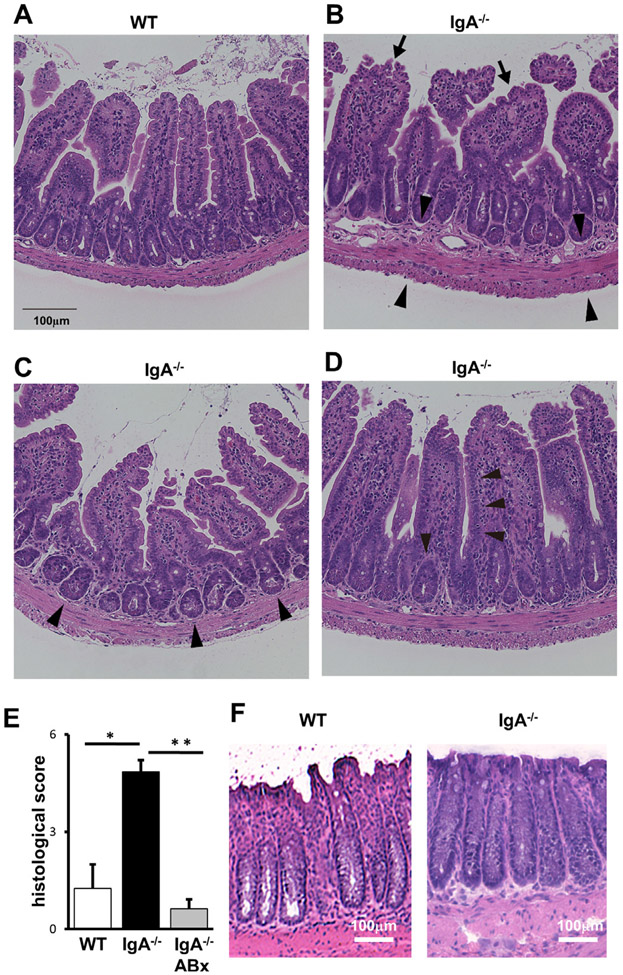
We next performed a histological assessment of the intestines in IgA−/− and IgAtm/tm mice. Compared with WT (figure 3A), histopathological analysis of IgA−/− revealed several inflammatory lesions including mild lymphocyte infiltrations with slightly thickened wall (figure 3B), cryptitis and/or crypt abscesses (figure 3C), and reduced goblet cells (figure 3D) specifically in the ileum. These pathological findings were significant based on histopathological quantification of these changes defined by the scoring system in the online supplemental materials and methods section. IgA−/− treated with antibiotics (ABx) ameliorated these histological scores (figure 3E), suggesting that microbiota may mediate these abnormalities. Interestingly, these pathological abnormalities were not observed in the other parts of the gastrointestinal tract including the jejunum (data not shown) and the colon (figure 3F) of IgA−/−. We also analysed younger IgA−/− mice and found the pathological abnormalities in 6-week-old mice (online supplemental figure S5). The pathological abnormalities appear to be in an age-dependent manner. In contrast, these pathological abnormalities were not observed in IgAtm/tm mice (data not shown).
Figure 3.
Pathological changes in the ileum tissues of IgA−/−. Tissues of WT (A) and IgA−/− (B–D) were fixed and stained with H&E. Normal appearance at ileum of WT (A), mild infiltration with mononuclear cells in the lamina propria (arrows in B), thickened submucosal, muscular and serosal layers (arrowheads in B), crypt abscesses (arrowheads in C), reduced goblet cells (arrowheads in D) at ileum of IgA−/− are indicated. Representative results are shown (n>7). (E) Pathological evaluation of the ileum. WT (left), IgA−/− (middle) and antibiotic (ABx)-treated IgA−/− (right) mice were subjected for the analysis. Scores were determined as described in online supplemental materials and methods. Values are presented as means±SEM. *p<0.05. **p<0.01. (F) Histological evaluation of the colons from WT (left) and IgA−/− (right). Representative results are shown (n>7).
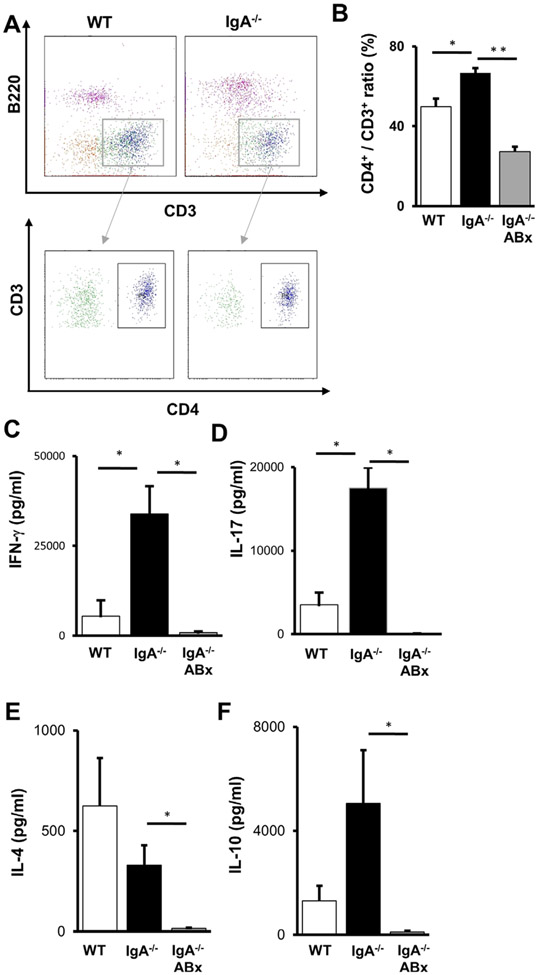
CD4+T cells were increased in the ileal lamina propria of IgA−/− mice
We next analysed the properties of lymphocytes isolated from the ileal tissues. CD4+ T-cell populations were increased in the ileal lamina propria of IgA−/− (figure 4A,B). Next, we evaluated the ability of these T cells to produce cytokines including IFN-γ, IL-4, IL-10 and IL-17. Production of IFN-γ and IL-17 were significantly increased in the ileal lamina propria CD4+ T cells from IgA−/− (figure 4C,D). A tendency towards decreased IL-4 production was also observed in IgA−/− (figure 4E). IgA−/− and WT mice exhibited similar levels of IL-10 production (figure 4F). ABx treatment lowered the cytokine production in IgA−/−, suggesting that microbiota were responsible for the cytokine induction. In summary, IgA deficiency also affected cytokine production by the CD4+ T cells located at the ileum.
Figure 4.
Analysis of ileal lamina propria CD4+ T cells of IgA−/−. (A) Flow cytometric analysis of the ileal lamina propria lymphocytes of WT (left) and IgA−/− (right). B220−CD3+ cells were gated and then opened by CD3 and CD4. Representative data from each group are shown. (B) The ratios of stained CD3+ and CD4+ T cells from the lamina propria of WT (left), IgA−/− (middle) and ABx-treated IgA−/− (right). Data are expressed as means±SEM. (n>3). *p<0.05. **p<0.01. (C–F) Cytokine production from the ileal lamina propria CD4+ T cells of WT (left), IgA−/− (middle) and ABx-treated IgA−/− (right) after stimulation with anti-CD3e and anti-CD28 Abs. The concentrations of IFN-γ (C), IL-17 (D), IL-4 (E) and IL-10 (F) in culture supernatants were determined (n>3). Values are presented as means±SEM (n>3). N.S., not significant. *p<0.05.
Microbial composition in the gut was skewed by IgA deficiency
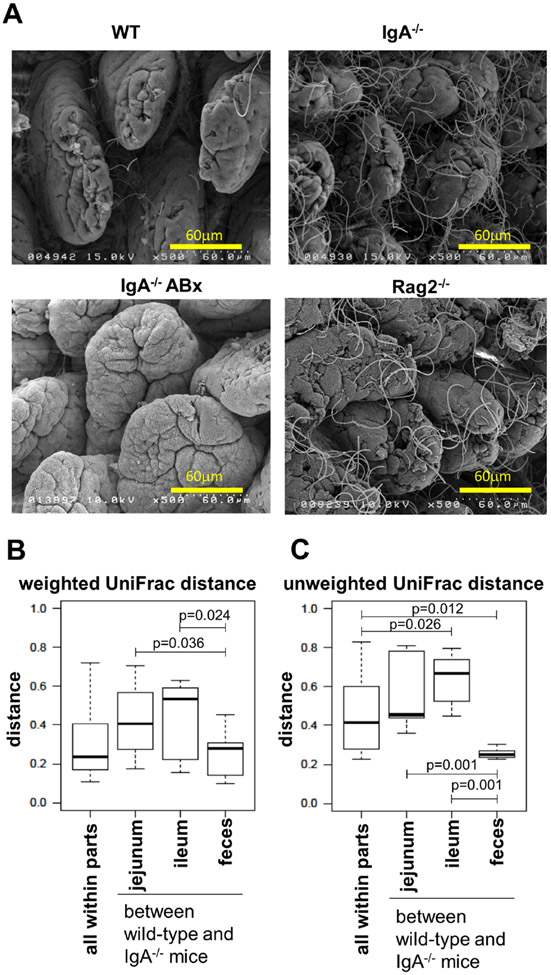
To assess the influence of small intestinal inflammation on gut microbiota composition in IgA−/−, we initially evaluated segmented filamentous bacteria (SFB), which has been reported to be associated with helper T (Th) 17 inflammation.24 Scanning electron microscopic analysis revealed profoundly increased SFB in the ileum of IgA−/− similar to those of recombination activating gene 2 (Rag2)–deficient mice as a positive control, whereas these microbes were rarely detected in WT (figure 5A). However, almost no SFB were observed in the IgA−/− when treated with ABx. In addition, SFB was not increased in the ileum of IgAtm/tm mice (online supplemental figure S6). These studies suggest SFB may be one of factors associated with the inflammation observed in the ileum of IgA−/−.
Figure 5.
Analysis of microbiota in the gut. (A) Analysis of SFB on the ileal mucosa. The ileal samples from WT (top left), IgA−/− (top right), ABx-treated IgA−/− (bottom left) and Rag2−/− (bottom right) mice were subjected to scanning electromicroscopic analysis. Representative results from each group are shown (n>3). (B and C) Weighted (B) and unweighted (C) UniFrac distances between WT and IgA−/− mice. The extracted samples from either the mixture of small intestine and colon, the isolated jejunum, ileum or faeces were subjected to metagenomic analysis as described in online supplemental materials and methods.
We further analysed the bacterial composition in the small intestines and faeces using 16S rRNA gene amplicon sequencing (online supplemental figures S7, S8 and table 1). In the small intestine, WT and IgA−/− differed significantly with respect to three taxa in the jejunum (table 1 and online supplemental figure S8) and six taxa in the ileum. In faeces, Desulfovibrio, Sutterella and Adlercreutzia species were more frequently detected in IgA−/−, whereas Faecalibacterium and Lachnobacterium species and Oscillospira guilliermondii were more frequently observed in WT. The differences observed with Adlercreutzia and Sutterella were significant in all of the jejunum, ileum and the faeces between WT and IgA−/−. However, Sturella and Desulfovibrio, and Adlercureutzia belonging to Proteobacteria and Actinobacteria phyla, respectiviely, may not be coated by IgA significantly.25 26
Table 1.
Taxonomic groups of significant differences at species level
| Phylum | Class | Order | Family | Genus | Species | WT (%) | IgA−/− (%) | Stamp p value | maaslin2 p value |
maaslin2 q value |
Gram |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Jejunum | |||||||||||
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.2 erell | 1.2 erell | 0.017 | 0.009716 | 0.466376 | (−) | |
| Bacteroidetes | Bacteroidia | Bacteroidales | S24-7 | 18.4 roidal | 50.6 roida | 0.022 | 0.141442 | 0.741488 | (−) | ||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Adlercreutzia | 0.1 rcreu | 0.4 rcreu | 0.029 | 0.021543 | 0.513532 | (+) | |
| Ileum | |||||||||||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Adlercreutzia | 0.0 rcreu | 0.3 rcreu | 0.018 | 0.032124 | 0.600348 | (+) | |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | C21_c20 | 0.1 c20vi | 0.7 c20vi | 0.034 | 0.128786 | 0.600348 | (−) |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.0 erell | 0.7 erell | 0.037 | 0.052296 | 0.600348 | (−) | |
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | 0.0 obact | 0.2 obact | 0.043 | 0.151946 | 0.600348 | (+) | ||
| Faeces | |||||||||||
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | ND | 0.0 lfovi | 0.004 | 0.868091 | 0.931884 | (−) | |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.2 erell | 0.5 erell | 0.004 | 0.029101 | 0.641393 | (−) | |
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Adlercreutzia | 0.1 rcreu | 0.3 rcreu | 0.009 | 0.072939 | 0.641393 | (+) | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | guilliermondii | 0.6 lierm | 0.3 lierm | 0.031 | 0.839028 | 0.929958 | (+) |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Faecalibacterium | 0.1 aliba | 0.0 aliba | 0.043 | 0.223489 | 0.641393 | (+) | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnobacterium | 0.1 nobac | 0.0 nobac | 0.046 | 0.446701 | 0.694329 | (+) | |
| Bacteroidetes | Bacteroidia | Bacteroidales | Paraprevotellaceae | (Prevotella) | 0.2 otell | 0.6 otell | 0.049 | 0.173527 | 0.641393 | (−) | |
ND, not detected.
Next, UniFrac distance was calculated to estimate the magnitude of the effect observed in the context of IgA deficiency. A weighted UniFrac analysis showed that the distance between WT and IgA−/− was slightly greater in the jejunum and ileum compared with that in the faeces (p = 0.036 and p = 0.024, respectively; figure 5B and online supplemental figure S9A). An unweighted UniFrac analysis also observed a smaller inter-group distance for faeces compared with that for the small intestine (p = 0.001; figure 5C and online supplemental figure S9B). Furthermore, the absolute number of bacteria appear to increase in the IgA−/− ileum (online supplemental figure S10). These results indicate that IgA deficiency causes distinctive changes in the bacterial communities associated with the small intestine, especially in the ileum.
Furthermore, administration of vancomycin which kills Gram-positive bacteria significantly decreased the pathological abnormalities in IgA−/− mice (online supplemental figure Sll). However, some extent of the pathological abnormalities remained, suggesting that both Gram-positive and Gram-negative bacteria contributed to its pathology.
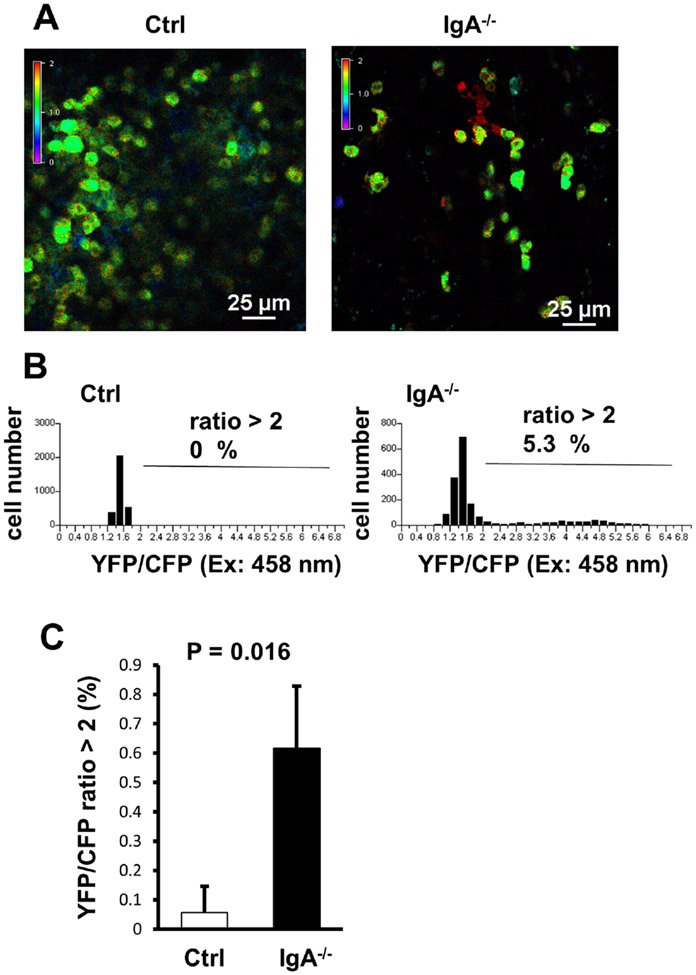
Intravital imaging of PP B cells in IgA−/− mice revealed abnormal Ca2+ signalling
We also investigated the impact on B-cell function in the context of IgA deficiency using a conditional Ca2+ biosensor YC3.60 transgenic model, which we have previously generated. This transgenic animal model has been shown by us to be able to detect subtle changes in Ca2+ signalling in vivo with high sensitivity before the onset of a clinical disorder of systemic autoimmune and IBD models.15 17 To evaluate the effects of IgA deficiency on Ca2+ signalling in B cells, we crossed IgA−/− with CD19-Cre/YC3.60flox mice and analysed Ca2+ signalling in PP B cells using intravital imaging. In the IgA−/−CD19-Cre/YC3.60flox mice, some of PP B cells exhibited constitutively increased intracellular Ca2+ concentrations (figure 6A,B) despite the lack of salient differences in the size of B-cell population and in the numbers of T cells and B cells (figure 2). Furthermore, Ca2+ signalling was significantly elevated in randomly selected fields in PP of IgA−/−YCD19-Cre/YC3.60flox compared with that of the control (Ctrl) CD19-Cre/YC3.60flox mice (figure 6C). In contrast, IgAtm/tmCD19-Cre/YC3.60flox mice, Ca2+ signalling was comparable with that of CD19-Cre/YC3.60flox mice (data not shown). In summary, the PP B cells in the context of IgA deficiency demonstrated significant activation.
Figure 6.
Intravital imaging of Ca2+ signalling in Peyer’s patch (PP) B cells. (A) Representative images of Ca2+ signalling in the PP of the control (Ctrl, left) and IgA−/− (right) CD19-Cre/YC3.60flox mice. Intravital imaging analysis of PP was performed under a confocal microscopy. Time-lapse images were obtained at every 2 s. Representative images reflecting the Ca2+ concentration based on the YFP:CFP ratio (excitation: 458 nm) are shown (n=3). (B) Distribution of the time-integrated intracellular Ca2+ concentrations of the PP B cells from Ctrl (left) and IgA−/− (right) CD19-Cre/YC3.60flox mice (n=20, frame=147 for Ctrl; n=20, frame=89 for IgA−/−). Percentages of cells with YFP/CFP ratios >2 are indicated. (C) YFP/CFP ratiometric intensities (excitation: 458 nm) of the PP B cells from Ctrl (left) and IgA−/− (right) CD19-Cre/YC3.60flox mice. Randomly selected fields from three mice were analysed. p<0.05 (t-test).
DISCUSSION
In this study, we generated IgA−/− and IgAtm/tm mice through a single-step CRISPR/Cas9 system experiment. Interestingly, IgA−/− showed significantly increased SFB in the ileum and skewed microflora composition in the jejunum, ileum and faeces. Furthermore, spontaneous ileitis was also observed in IgA−/− in association with significantly increased IFN-γ-producing and IL-17-producing CD4+ T cells in the lamina propria. These abnormalities were restored by ABx treatment. Intravital imaging of PP in IgA−/− ileum revealed an increase in B-cell activation based on the evidence for constitutively elevated Ca2+ concentrations.15 17 In contrast, IgAtm/tm mice exhibited a comparable phenotype with that of WT. Together, these results indicate that IgA is crucial to homeostatic maintenance in the gut, especially in the ileum, although the cytoplasmic tail sequence does not appear to be crucial.
Although most animal models of IBD manifest as colitis, there are some notable exceptions. The IgA−/− mouse model we report here is reminiscent of that observed in Xbp1−/− mice, which also develop spontaneous ileitis.27 The XBP1 gene has been identified as a genetic risk factor for both types of IBD, Crohn’s disease (CD) and ulcerative colitis.27 CD is one form of IBD characterised by immune-related disorder involving the entire alimentary tract especially the small intestine, although its aetiology is uncertain. More than half of the patients with CD exhibit pathological condition involving the terminal ileum. The Xpb1−/− model is also IgA dependent in that loss of IgA results in proximal extension of the inflammation into the duodenum.28 In addition, the SAMP1/Yit mice have been defined as the model for spontaneous CD enteritis.27 29 Excess IFN-γ production in the pathological site was also observed in SAMP1/Yit mice.29 Similar to Xbp1−/− mice and SAMP1/Yit mice, IgA−/− mice in our study showed spontaneous inflammation specifically in the small intestine and most notably in the ileum, which was associated with increased pro-inflammatory cytokine production, including IFN-γ and IL-17, from the infiltrating lamina propria CD4+ T cells in the ileum. Thus, the IgA−/− mice appear to be a potential model for CD. In fact, several clinical studies have reported that selective IgA deficiency in humans is correlated with increased risk for IBD, especially CD.30 31 Interestingly, coeliac disease can also involve inflammation in the small intestine and previous reports have suggested a correlation between selective IgA deficiency and coeliac disease.32 33 However, a recent review of several prior studies on selective IgA deficiency in humans suggested a weaker association with coeliac disease than that of CD.34
A previous report demonstrated that IgA deficiency led to an increase in the serum levels of other Ig classes, such as IgM and IgG.7 Our findings are consistent with this and also provide evidence that is associated with spontaneous ileitis occurred. Previous associations between IgA deficiency and human IBD have not focused on specific anatomical sites. It is notable that the pathological disorder in the small intestine of IgA−/− observed in our study closely correlated with the areas of greatest IgA expression in the whole GI tract.25 Specifically, there are estimated to be 10-fold to 15-fold more IgA+ plasma cells in the small intestinal lamina propria relative to that observed in the colonic lamina propria.25 Consistent with previous studies,7 8 we did not observe any pathological changes in the alimentary tract, including colon, of IgA−/− mice, despite previous predictions.8 In this study, we analysed the small intestine using recently established several techniques including direct microinjection of C57BL/6 zygotes, CRISPR/Cas9 genome editing system, metagenomic analysis with 16S rRNA sequencing and intravital microscopy system with a recently established biosensor involving fluorescence resonance energy transfer technology. Importantly, we were able to obtain the mouse lines with a pure strain background directly without need for back-crossing into C57BL/6. It should be noted that the IgA-deficient mice in previous reports were established using conventional procedures at that time, and thus, such mice may have slightly mixed background with different strains such as 129/Sv.7 Therefore, this may reflect the differences in the pathologic phenotypes between ours and previous reports.7 8
Our analyses revealed remarkably increased SFB in the terminal ileum of IgA−/− compared with that of WT. A previous study with AID-deficient mice also implied an aberrant expansion of SFB,9 even though there was no direct evidence showing that this increase of SFB would be caused by specific lack of IgA, because AID deficiency may also result in decreased production of all Ig classes, except IgM and IgD, in all organs including the intestines. However, the increased SFB and skewed microflora composition in the small intestine of IgA−/− in our study are consistent with the hypothesis of this previous report, and thus, this strongly suggests that IgA secretion in the gut suppresses these bacterial populations. On the other hand, spontaneous ileitis was not described in the same previous report9 as another previous study with conventional IgA-deficient mice,7 and therefore our current study is the first to show such pathological phenotype caused by IgA specific deficiency. A recent study demonstrated an abundance of IgA in the small intestine but not in the colon.25 It also showed production of IgA specifically against SFB in a T cell–dependent manner. Therefore, IgA deficiency is suggested to lead to an expansion of SFB specifically in the small intestine. Furthermore, SFB has been reported to induce Th17 cells in the ileal lamina propria.24 Indeed, our study also showed an increased IL-17 production from the ileal lamina propria CD4+ T cells of IgA−/− mice. In addition, CD4+ T-cell production of another pro-inflammatory cytokine, INF-γ, which is not induced by SFB efficiently24 was also significantly increased in the IgA−/− mice. Furthermore, IgA−/− mice showed slight histological abnormalities in the SFB-free condition by vancomycin treatment. Taken together, these findings suggest that IgA deficiency leads to increased SFB and other bacteria in association with CD4+ T-cell activation, even though how these changes culminate in ileal inflammation is still unknown.
We also observed more Gram-negative bacterial taxa (22 taxa) in IgA−/− compared with that of WT (8 taxa). Presumably, IgA deficiency may have provided a symbiotic milieu for Gram-negative bacteria, and this is consistent with a previous report.8 Notably among these microbes, Sutterella species, which are increased in IgA−/− mice, were also previously found to correlate with human autism.35 And interestingly, IgA has also been suggested to correlate with human autism.36 37 Furthermore, another microbial species, Proteobacteria S24-7, a commensal bacteria that is coated with IgA in a T cell–dependent and T cell–independent manner was increased in IgA−/− mice.25 It seems that IgA deficiency may therefore cause an expansion of these bacteria in the gut.
In this study, we show that the cytoplasmic tail of IgA is dispensable for IgA Ab production. The cytoplasmic tail of IgA (14 aa) is shorter than those of IgG and IgE (28 aa) but longer than those of IgM and IgD (3 aa).14 Although the cytoplasmic tails of IgG and IgE are known to be crucial for Ab production,12 13 IgA production is not dependent on the presence of the tail, as shown in our current study with IgAtm/tm mice, suggesting that this element may differ substantially in function (as well as in length and sequence) from those of IgG and IgE. Therefore, our study here identifies this as an important distinguishing characteristic of IgA relative to IgG and IgE.
We also observed B cells with constitutively increased intracellular Ca2+ levels in the PP of IgA−/− mice, and this is reminiscent of our findings in the B cells associated with autoimmune-prone lpr/lpr mice and oxazolone-induced colitis model15 17 We previously demonstrated a significant increase in B cells exhibiting constitutively elevated intracellular Ca2+ concentrations in lpr/lpr mice despite an absence of any clinical findings. In IgA−/− mice, abnormally augmented Ca2+ signalling was observed in B cells from the PP, which coincides with ileal inflammation. The appearance of constitutively activated B cells is consistent with the inflammation observed.
In summary, our study highlights the role of IgA in the gut. Our findings indicate that IgA deficiency elicits the expansion of some commensal bacterial strains and promotes inflammation in the small intestine. This suggests that IgA is an important regulator of commensal bacteria, especially in the small intestine. Further analysis of commensal bacterial expansion is needed to precisely define the role of IgA in gut homeostasis.
Supplementary Material
Significance of this study.
What is already known on this subject?
Immunoglobulin A (IgA) is the most abundantly expressed antibody (Ab) in both humans and mice. IgA deficiency has been reported to increase susceptibility to colon injury in mice, and is associated with various diseases in humans such as inflammatory bowel diseases (IBD), allergies, autoimmune diseases and recurrent infections.
What are the new findings?
IgA deficiency in mice exhibited spontaneous inflammation in the ileum but not in the colon.
Gut microbiota were affected significantly in the ileum by IgA deficiency.
Ca2+ signalling in B cells in Peyer’s patches was augmented by IgA deficiency.
IgA deficiency impacts the ileum rather than the colon.
How might it impact on clinical practice in the foreseeable future?
Our findings underlie a novel target for the diagnosis and treatment of patients with Crohn’s disease and/or IgA deficiency through the mechanism by which ileitis may be induced.
Acknowledgements
We are grateful to Dr K Rajewsky (Max Delbrück Center for Molecular Medicine) for the CD19-Cre mice, Dr A Miyawaki (RIKEN) for the YC3.60 gene, Dr Y Tanaka (TMDU) for critical reading, Dr N M Tsuji for discussion (AIST) and A Onimaru, Y Kojima, J Takahashi, S Watanabe and T Shirasaki (TMDU) for technical assistance.
Funding
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Area (MW, 22117508), Challenging Exploratory Research (TN, 15K15288), Scientific Research-S (MW, 26221307), Scientific Research-B (TN, 16H05286 and 20H03658) and Scientific Research-C (TA, 15K08526 and TW, 18K07997) from the Japanese Ministry of Education, Culture, Sports, Science and Technology, Practical Research Project for Rare/Intractable Disease (MW, 16eK0109047h0003) from Japan Agency for Medical Research and Development, RO1 Grants from the National Institutes of Health (RSB, DK088199, DK44319 and DK51362), Yamada Foundation (TA), Memorial Fund of Nihon University Medical Alumni Association (TN), Abbott Japan Allergy Research Award (TN), Foundation for Advancement of International Science (TN), Takeda Science Foundation (TN), Joint Usage/Research Programme of TMDU Medical Research Institute (SY, TN and TA), Japan Foundation for Applied Enzymology (TW), Nipponham Foundation for the Future of Food (TA), Tojuro Iijima Foundation for Food Science and Technology (TA), Mishima Kaiun Memorial Foundation (TA), and Naoki Tsuchida Memorial Research Grant (SY, TN and TA).
Footnotes
Competing interests None declared.
Ethics approval All animal experiments were approved by the institutional committee for animal care and use of TMDU.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as online supplemental information. DNA sequences were deposited in the DDBJ Sequence Read Archive under accession number DRA005447.
REFERENCES
- 1.Fagarasan S, Honjo T. Regulation of IgA synthesis at mucosal surfaces. Curr Opin Immunol 2004;16:277–83. [DOI] [PubMed] [Google Scholar]
- 2.Fagarasan S, Kawamoto S, Kanagawa O, et al. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol 2010;28:243–73. [DOI] [PubMed] [Google Scholar]
- 3.Kurashima Y, Kiyono H. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev Immunol 2017;35:119–47. [DOI] [PubMed] [Google Scholar]
- 4.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol 2012;12:821–32. [DOI] [PubMed] [Google Scholar]
- 5.Okai S, Usui F, Yokota S, et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol 2016;1:16103. [DOI] [PubMed] [Google Scholar]
- 6.Yel L. Selective IgA deficiency. J Clin Immunol 2010;30:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harriman GR, Bogue M, Rogers P, et al. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J Immunol 1999;162:2521–9. [PubMed] [Google Scholar]
- 8.Mirpuri J, Raetz M, Sturge CR, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014;5:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K, Meek B, Doi Y, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A 2004;101:1981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei M, Shinkura R, Doi Y, et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol 2011;12:264–70. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto S, Tran TH, Maruya M, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 2012;336:485–9. [DOI] [PubMed] [Google Scholar]
- 12.Achatz G, Nitschke L, Lamers MC. Effect of transmembrane and cytoplasmic domains of IgE on the IgE response. Science 1997;276:409–11. [DOI] [PubMed] [Google Scholar]
- 13.Kaisho T, Schwenk F, Rajewsky K. The roles of gamma 1 heavy chain membrane expression and cytoplasmic tail in IgG1 responses. Science 1997;276:412–5. [DOI] [PubMed] [Google Scholar]
- 14.Sato M, Adachi T, Tsubata T. Augmentation of signaling through BCR containing IgE but not that containing IgA due to lack of CD22-mediated signal regulation. J Immunol 2007;178:2901–7. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa S, Usami T, Kikuta J, et al. Intravital imaging of Ca(2+) signals in lymphocytes of Ca(2+) biosensor transgenic mice: indication of autoimmune diseases before the pathological onset. Sci Rep 2016;6:18738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi T, Kakuta S, Aihara Y, et al. Visualization of probiotic-mediated Ca2+ signaling in intestinal epithelial cells in vivo. Front Immunol 2016;7:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watabe T, Nagaishi T, Tsugawa N, et al. B cell activation in the cecal patches during the development of an experimental colitis model. Biochem Biophys Res Commun 2018;496:367–73. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013;153:910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adachi T, Tsubata T. Fret-based Ca2+ measurement in B lymphocyte by flow cytometry and confocal microscopy. Biochem Biophys Res Commun 2008;367:377–82. [DOI] [PubMed] [Google Scholar]
- 20.Onizawa M, Nagaishi T, Kanai T, et al. Signaling pathway via TNF-alpha/NF-kappaB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am J Physiol Gastrointest Liver Physiol 2009;296:G850–9. [DOI] [PubMed] [Google Scholar]
- 21.Yamaji O, Nagaishi T, Totsuka T, et al. The development of colitogenic CD4(+) T cells is regulated by IL-7 in collaboration with NK cell function in a murine model of colitis. J Immunol 2012;188:2524–36. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M, Nagaishi T, Yamazaki M, et al. Myosin light chain kinase expression induced via tumor necrosis factor receptor 2 signaling in the epithelial cells regulates the development of colitis-associated carcinogenesis. PLoS One 2014;9:e88369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaishi T, Pao L, Lin S-H, et al. SHP1 phosphatase-dependent T cell inhibition by CEACAM1 adhesion molecule isoforms. Immunity 2006;25:769–81. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunker JJ, Flynn TM, Koval JC, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015;43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaser A, Lee A-H, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008;134:743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grootjans J, Krupka N, Hosomi S, et al. Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science 2019;363:993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pizarro TT, Pastorelli L, Bamias G, et al. SAMP1/YitFc mouse strain: a spontaneous model of Crohn’s disease-like ileitis. Inflamm Bowel Dis 2011;17:2566–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manfredi R, Coronado OV, Marinacci G, et al. Chron’s disease, rare association with selective IgA immunodeficiency, and development of life-threatening bacterial infections. Scand J Infect Dis 2004;36:523–4. [DOI] [PubMed] [Google Scholar]
- 31.Iizuka M, Itou H, Sato M, et al. Crohn’s disease associated with selective immunoglobulin A deficiency. J Gastroenterol Hepatol 2001;16:951–2. [PubMed] [Google Scholar]
- 32.Mulder SJ, Mulder-Bos GC. Most probable origin of coeliac disease is low immune globulin A in the intestine caused by malfunction of Peyer’s patches. Med Hypotheses 2006;66:757–62. [DOI] [PubMed] [Google Scholar]
- 33.Ludvigsson JF, Neovius M, Hammarström L. Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol 2014;34:444–51. [DOI] [PubMed] [Google Scholar]
- 34.Odineal DD, Gershwin ME. The epidemiology and clinical manifestations of autoimmunity in selective IgA deficiency. Clin Rev Allergy Immunol 2020;58:107–33. [DOI] [PubMed] [Google Scholar]
- 35.Williams BL, Hornig M, Parekh T, et al. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012;3. doi: 10.1128/mBio.00261-11. [Epub ahead of print: 10 01 2012]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren RP, Odell JD, Warren WL, et al. Brief report: immunoglobulin A deficiency in a subset of autistic subjects. J Autism Dev Disord 1997;27:187–92. [DOI] [PubMed] [Google Scholar]
- 37.Wasilewska J, Kaczmarski M, Stasiak-Barmuta A, et al. Low serum IgA and increased expression of CD23 on B lymphocytes in peripheral blood in children with regressive autism aged 3–6 years old. Arch Med Sci 2012;8:324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as online supplemental information. DNA sequences were deposited in the DDBJ Sequence Read Archive under accession number DRA005447.